Apoptotic epithelial cells signal to neighboring cells to induce dying cell extrusion by releasing sphingosine-1-phosphate.
Abstract
To maintain an intact barrier, epithelia eliminate dying cells by extrusion. During extrusion, a cell destined for apoptosis signals its neighboring cells to form and contract a ring of actin and myosin, which squeezes the dying cell out of the epithelium. Here, we demonstrate that the signal produced by dying cells to initiate this process is sphingosine-1-phosphate (S1P). Decreasing S1P synthesis by inhibiting sphingosine kinase activity or by blocking extracellular S1P access to its receptor prevented apoptotic cell extrusion. Extracellular S1P activates extrusion by binding the S1P2 receptor in the cells neighboring a dying cell, as S1P2 knockdown in these cells or its loss in a zebrafish mutant disrupted cell extrusion. Because live cells can also be extruded, we predict that this S1P pathway may also be important for driving delamination of stem cells during differentiation or invasion of cancer cells.
Introduction
Epithelia comprised of one or two cell layers cover and protect the organs that they encase. The cells making up epithelia are constantly turning over by cell division and apoptosis, yet cell death could compromise the barrier function of the epithelium. We previously found that epithelia use a process termed “apoptotic cell extrusion” to remove apoptotic cells from a layer, while preserving their barrier function (Rosenblatt et al., 2001). Specifically, an early apoptotic epithelial cell triggers formation of an actin and myosin ring in the live neighboring cells surrounding it. Contraction of this ring then squeezes the dying cell out of the epithelium. Apoptotic cell extrusion is conserved in all in vivo epithelia we have examined ranging from Drosophila to human.
We previously showed that extrusion depends on a chemical signal from the apoptotic cells, which activates the Rho pathway in the neighboring cells (Rosenblatt et al., 2001; Slattum et al., 2009). Specifically, addition of early apoptotic cells onto an epithelial monolayer induces actin assembly in the live contacted cells. Furthermore, inhibition of Rho in the cells surrounding an apoptotic cell blocks extrusion (Rosenblatt et al., 2001). We recently determined that Rho activation during extrusion requires p115 RhoGEF (Slattum et al., 2009), a protein activated downstream of the G12/13 G protein–coupled receptor (Holinstat et al., 2003). Thus, a signal on the surface of the dying cell triggers p115 RhoGEF to activate Rho-mediated actin–myosin assembly and contraction in the live surrounding cells to remove the dying cell. However, we did not know the identity of the signal produced in early apoptotic cells that activates apoptotic cell extrusion.
Here, we report that the signal produced by dying cells is the bioactive lipid sphingosine-1-phosphate (S1P), which activates actomyosin contraction in surrounding cells via the S1P2 receptor. Inhibition of S1P synthesis or extracellular S1P signaling blocks apoptotic cell extrusion. The cells surrounding the dying cell require S1P2 to bind S1P and activate formation and contraction of the actomyosin-extruding ring in both tissue culture and zebrafish epithelia. Together, our data reveal the signaling pathway that drives a cell to extrude from an epithelial monolayer.
Results and discussion
Blocking S1P signaling inhibits extrusion of apoptotic cells
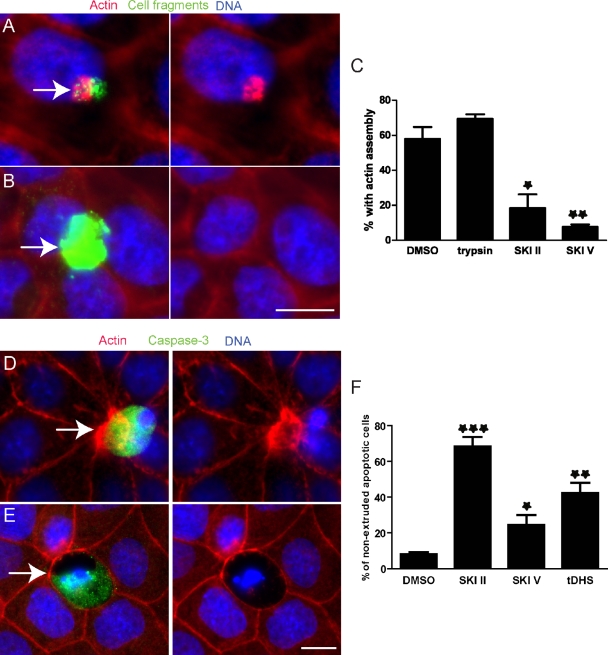
To characterize the extracellular apoptotic signal that triggers formation of the actin–myosin extruding ring, we used a modified version of our previous “cell addition assay.” In that assay, addition of early apoptotic cells, but not late apoptotic cells or live cells, to an intact Madin Darby canine kidney (MDCK) epithelial monolayer induced actin assembly in the contacted living cells. Necrotic cells likely use the same signal that apoptotic cells use to produce an extruding ring in epithelia, as laser-ablated or mechanically wounded necrotic cells are extruded identically to apoptotic cells (Tamada et al., 2007). Therefore, we added necrotic cell fragments, prepared by scraping and needle shearing cells and found that they induced accumulation of actin in the contacted monolayer and with the same kinetics (Fig. 1). When added to a cell monolayer, ∼60% of added control cell fragments (green) resulted in actin (red) accumulation in the contacted cells (Fig. 1, A and C). Pre-digestion of the dead cell fragments with trypsin did not significantly alter actin accumulation (Fig. 1 C), suggesting that the signal triggering the response is not a protein. We then postulated that the signal is a bioactive lipid.
Figure 1.
Inhibitors of SphKs block actin assembly and apoptotic cell extrusion. (A and B) Alexa Fluor 488–labeled cell fragments (green) prepared from MDCK cells pretreated with DMSO (A) or SKI II (B) were added to an intact MDCK monolayer. Arrows point to added cell fragments. (C) The percentage of cell fragments causing actin assembly from three independent experiments; n = 100 cell fragments per experiment and error bars are standard deviations (SDs). *, P < 0.05; **, P < 0.01. (D and E) Extrusion in an MDCK monolayer in the presence of DMSO (D) or SKI II (E). Arrows point to active caspase-3–positive dying cells in each case. (F) Quantification of nonextruded active caspase-3–positive apoptotic cells with DMSO or SphK inhibitor treatment from three independent experiments; n = 100, error bars = SDs. ***, P < 0.001. Bars, 10 µm.
Based on the fact that a bioactive lipid within this cell fragment should activate Rho-dependent actomyosin assembly and contraction during extrusion, we investigated several candidate lipids and found that addition of S1P caused actin accumulation when added to monolayers, as in our cell addition assay (Fig. S1). Conversely, a total lipid extraction from Escherichia coli that did not contain S1P did not elicit the same reaction. Based on this experiment, the fact that a precursor to S1P, ceramide, is pro-apoptotic (Kolesnick and Hannun, 1999; Hannun and Obeid, 2002), and recent findings that S1P is produced in apoptotic cells (Kolesnick and Hannun, 1999; Hannun and Obeid, 2002; Gude et al., 2008; Weigert et al., 2010), we investigated whether this lipid is required for extrusion. To directly test if S1P is required for the actin reaction in the cell addition assay, we used SKI II, a specific, potent noncompetitive inhibitor of SphK that blocks conversion of sphingosine to S1P (French et al., 2003). Pre-treatment of cells with SKI II before making cell fragments significantly inhibited the formation of actin cables when added to monolayers (Fig. 1, B and C). SKI V, another SphK inhibitor, also reduced the percentage of cell fragments that induce actin accumulations with even stronger efficiency than SKI II (Fig. 1 C and Fig. S2, A and B). Therefore, we conclude that SphK is required in dying cells to trigger actin accumulation in the live cells they contact.
To test if SphK is required for extrusion of apoptotic cells from an MDCK monolayer, we pretreated the monolayer with SKI II and induced apoptosis with short-wave UV light. Extrusion was evaluated by immunostaining the resulting monolayers for active caspase-3 Ab to identify apoptotic cells, Alexa Fluor 568–phalloidin to analyze actin-based extruding rings, and Hoechst for DNA. Fig. 1 D shows a control extruding apoptotic cell, focusing only on the dying cell for the caspase-3 and DNA (which is fragmented) and in another plane on the actin ring, which is around and beneath the dying cell. Treatment with SKI II inhibited formation of the actin extrusion ring and resulted in holes in the monolayer wherever there were apoptotic cells (Fig. 1 E). Note that when a cell extrudes, DNA and caspase-3 do not exist in the same plane as its neighboring cells, whereas all nuclei are in focus when extrusion is blocked. SKI II, SKI V, and d,l-threo-dihydrosphingosine (tDHS), a competitive inhibitor of SphKs, all dramatically increased the percentage of unextruded apoptotic cells compared with control treatment (Fig. 1 F). Inhibition of apoptotic cell extrusion by SKIs was dose dependent (Fig. S2, C–E). These results show that extrusion of apoptotic cells requires SphK activity, presumably because it catalyzes S1P synthesis.
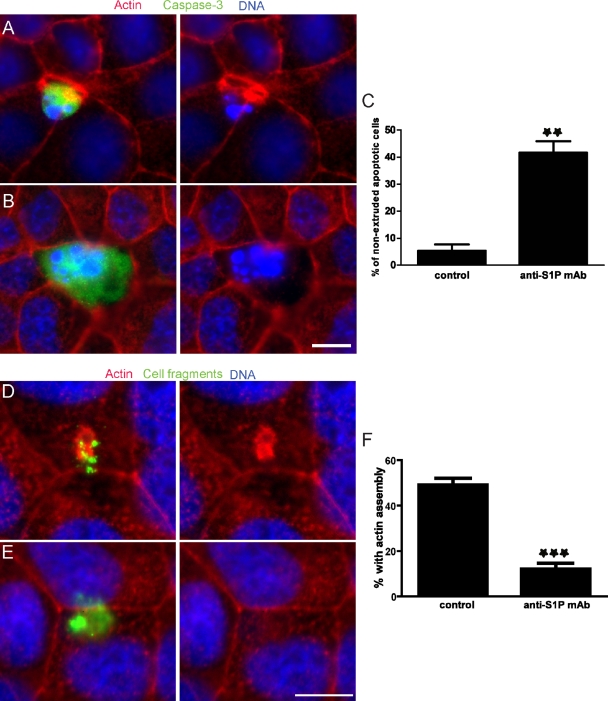
To regulate many cellular responses, S1P is exported out of cells and binds to its receptors on the same or neighboring cells. To test whether extracellular S1P is necessary for apoptotic cell extrusion, the MDCK monolayer was treated with UV in the presence of an S1P-neutralizing mAb (Visentin et al., 2006; O’Brien et al., 2009). Formation of the extrusion ring was inhibited by the anti-S1P mAb, and consequently the apoptotic cell failed to extrude and was instead retained in the monolayer (Fig. 2, B and C). Additionally, the anti-S1P mAb also blocks actin accumulation induced by cell fragments (Fig. 2, E and F). These results suggest that extracellular S1P is critical for extrusion.
Figure 2.
An inhibitory anti-S1P mAb blocks apoptotic cell extrusion. (A and B) Extrusion in an MDCK monolayer treated with short-wave UV to induce apoptosis in the presence of a mouse IgG isotype control (A) or 10 µg/ml anti-S1P mAb (B). (C) Quantification of nonextruded apoptotic cells from three independent experiments; n = 100 active caspase-3–positive cells where error bars are SDs; **, P < 0.01. (D and E) Alexa Fluor 488–labeled cell fragments (green) prepared from MDCK cells were added to an intact MDCK monolayer in the presence of a mouse IgG isotype control (D) or 10 µg/ml anti-S1P mAb (E). (F) The percentage of cell fragments causing actin assembly from three independent experiments; n = 100 cell fragments per experiment and error bars = SDs. ***, P < 0.001. Bars, 10 µm.
Extrusion requires signaling through the S1P2 receptor
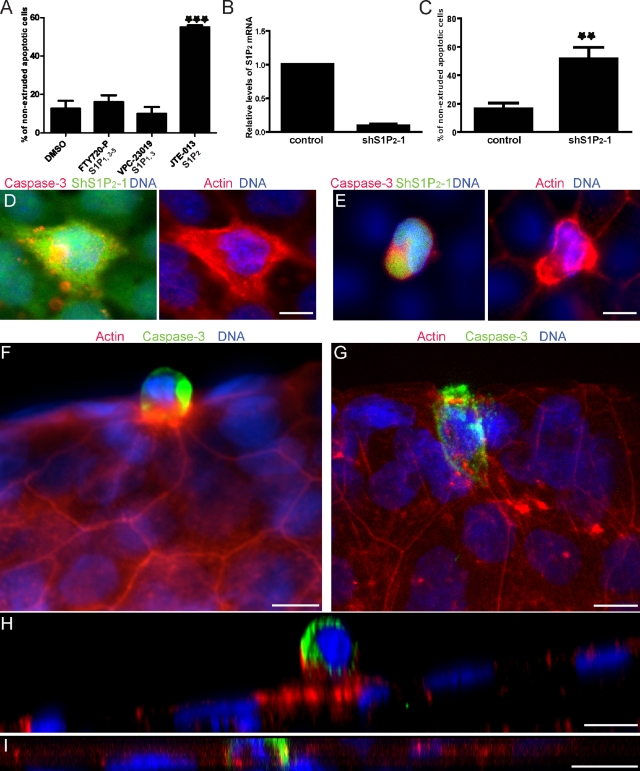
We next investigated which of the five known high-affinity cell surface S1P receptors (S1P1–5) might mediate extrusion signaling. S1P1–3 are ubiquitously expressed, whereas S1P4 and S1P5 are expressed mainly in lymphoid cells and neuronal cells, respectively. By RT-PCR we determined that mRNAs from S1P1,2,3&5 are expressed in MDCK cells and another epithelial cell line, human bronchial epithelial (HBE) cells (unpublished data). To investigate the involvement of S1P receptors in extrusion, we treated an HBE monolayer with UV to induce apoptosis in conjunction with several antagonists that are specific for different S1P receptors, and evaluated extrusion of apoptotic cells. Neither FTY720-P (up to 10 µM), a functional antagonist of all S1P receptors except S1P2 (Brinkmann et al., 2002; Mandala et al., 2002), nor VPC-23019 (up to 10 µM), an antagonist specific for S1P1 and S1P3 (Davis et al., 2005), notably affected extrusion of apoptotic cells (Fig. 3 A). In contrast, an S1P2-specific antagonist, JTE-013 (Osada et al., 2002), increased the proportion of un-extruded apoptotic cells by approximately threefold (Fig. 3 A). We obtained similar results with these S1P inhibitors in MDCK monolayers (not depicted). These results suggested that S1P signaling through S1P2, but not the other S1P receptors, is necessary for cell extrusion.
Figure 3.
Apoptotic cell extrusion requires the S1P2 receptor. (A) HBE cells induced to undergo apoptosis with UV in the presence of DMSO or the indicated S1P receptor antagonists. (B) qRT-PCR confirms shRNA-mediated knockdown of S1P2 in HBE cells. (C) Quantification of nonextruded apoptotic cells in HBE monolayers expressing control or S1P2-specific shRNA after UV treatment. (D and E) A dying HBE cell is not extruded by S1P2-silenced cells (D, green), but is extruded successfully by normal surrounding cells (E). When S1P2 shRNA is only in the dying cell (E), it extrudes and is in a higher plane than the actin ring below it, but is in the same plane when the surrounding cells are knocked down for S1P2 (D). Projections of extruding and nonextruding apoptotic cells from WT (F) or mil (G) zebrafish larvae, respectively. (H and I) Cross sections (XZs) of an apoptotic extruding cell (H) and a nonextruding cell (I) from WT (H) and mil zebrafish larvae (I), respectively. For all bar graphs, each bar represents the average percentage of nonextruded apoptotic cells to total apoptotic cells with each treatment from three independent experiments; n = 100 dying cells per experiment, error bars = SDs. **, P < 0.01; ***, P < 0.001. Bars, 10 µm.
To confirm the involvement of S1P2 in extrusion and to determine which cells require this receptor, we used shRNA to silence S1P2 expression in HBE cells. GFP-positive cells were FAC-sorted and knockdown of S1P2 was confirmed by quantitative RT-PCR (Fig. 3 B). Knockdown of S1P2 dramatically increased the proportion of nonextruded apoptotic cells, compared with control shRNA knockdown cells (Fig. 3, C and D). To control for off-target effects of hairpins, we used two additional shRNA sequences to knock down S1P2 and obtained similar results (Fig. S3, A and B). To test which cells require S1P2, we examined extrusion in monolayers that were mosaically knocked down for S1P2. A dying cell with S1P2 knockdown surrounded by cells with wild-type (WT) levels of S1P2 extrudes successfully (Fig. 3 E, representing nine cases in three independent experiments). By contrast, complete S1P2 knockdown of all cells within the monolayer blocked extrusion of dying cells (Fig. 3 D). These results support the conclusion that extrusion requires S1P2 in the cells surrounding a dying cell, but not in the apoptotic cell itself.
Our previous work showed that the zebrafish larval epidermis provides an excellent in vivo model system to study extrusion (Slattum et al., 2009). To test if S1P2-mediated signaling is also required for apoptotic cell extrusion in vivo, we tested if zebrafish that have a mutation in S1P2 could extrude epidermal cells. Treating WT 3 d post-fertilization (dpf) zebrafish with apoptotic stimuli resulted in extrusion of dying cells (Fig. 3, F and H), whereas induction of apoptosis in miles apart (mil) zebrafish, which carry a loss-of-function mutation in S1P2 (Kupperman et al., 2000), resulted in apoptotic cells that do not extrude (Fig. 3, G and I, and Videos 1 and 2). Instead, brightly staining active capase-3–positive cells remain in the plane of the epidermis with no obvious actin ring around or below them. Of 60 apoptotic cells from WT zebrafish, 59 extruded and one did not, whereas of 88 apoptotic cells from mil zebrafish, none extruded. Addition of JTE-013 to WT zebrafish also blocked extrusion of apoptotic epidermal cells (Fig. S3 C). Of 20 apoptotic cells from WT zebrafish treated with JTE-013, one extruded and the rest did not. These data indicate that S1P signaling through the S1P2 is required for extrusion in a number of vertebrate epithelia both in vivo and in culture.
S1P localizes to the extruding apoptotic cell and its surrounding epithelial cells
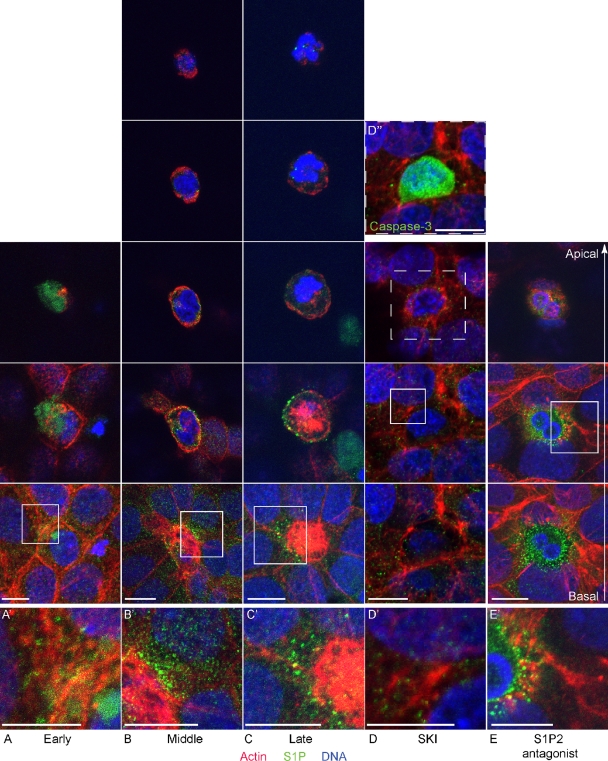
To visualize S1P formation during apoptotic cell extrusion, we stained UV-irradiated HBE monolayers with anti-S1P mAb and imaged them by confocal microscopy. Fig. 4 shows representative 3D projections of images of early, middle, and late stages of extrusion, where apoptotic cells are marked by condensed and increasingly fragmented nuclei over time while the actin ring that surrounds the dying cell gradually constricts inward over time. Apoptotic cells from early to late stages of extrusion contained prominent intracellular pools of S1P that existed in granular puncta. No staining was observed in secondary Ab–only treated cells (not depicted). From early to middle stages of extrusion, S1P was distributed along the plasma membrane predominantly near the actin ring at the basolateral surface (Fig. 4, A and B and insets; and Videos 3 and 4). Notably, S1P puncta could be seen in the surrounding cells, especially during the middle and late stages of extrusion (Fig. 4 C and Video 5). Interestingly, ∼10% of cells extrude basally instead of apically, which produce much less S1P than apically extruding cells (not depicted). As expected, addition of SKI blocked S1P accumulation in and around dying cells, in addition to blocking extrusion (Fig. 4 D and Video 6). We also note that when signaling via the S1P2 was blocked with the antagonist JTE-013, high levels of S1P accumulated in the dying, unextruded cells but not in surrounding cells (Fig. 4 E and Video 7). Thus, early apoptotic cells appear to produce and transmit S1P, which is taken up in neighboring cells via the S1P2 to drive extrusion.
Figure 4.
Apoptotic cells produce and transmit S1P during extrusion. (A–C) Confocal fluorescence images during early (A), middle (B), and late (C) stages of extrusion of apoptotic cells from an HBE monolayer. (D and E) Confocal fluorescence images of blocked apoptotic cell extrusion by SKI II (D) or the S1P2 antagonist JTE-013 (E). Each experimental sample was visualized with five (B and C) or three (A, D, and E) consecutive 3D projections (comprising 2-µm thickness each), as necessary to span the full distance from the most basal to the most apical section (second-to-bottom and top images, respectively). Note that total cell height under the different conditions varies: during early extrusion (A) and when extrusion is blocked with SKI II and JTE-013 (D and E), the dying cell is not squeezed out of the epithelium and therefore does not inhabit as great an apical-to-basal distance as when the dying cell is extruding (B and C). A, B, C, and E were obtained using a confocal microscope (TCS SP5; Leica), whereas D was taken using an inverted microscope (Eclipse TE300; Nikon) converted for spinning disc confocal microscopy. A′–E′ represent zoomed-in region (square) from each montage. (D′′) Inset denoting that the unextruded cell in D is apoptotic. Bars, 10 µm.
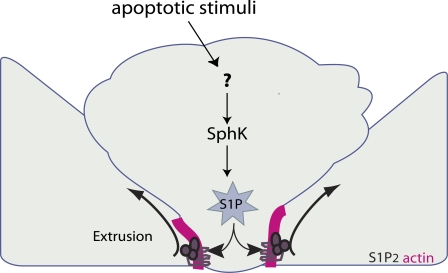
Taken together, our results suggest that S1P produced by apoptotic cells binds and activates S1P2 in neighboring cells to trigger contraction of an intercellular actin–myosin ring, which then squeezes the dying cell out of the monolayer (Fig. 5, see schematic). Pharmacological inhibition, mutation, or shRNA silencing of the S1P2 receptor significantly increases the frequency of nonextruded apoptotic cells, suggesting that S1P signals via S1P2 to drive apoptotic cell extrusion.
Figure 5.
Model for how S1P triggers extrusion. S1P is produced in the dying cell and exported to neighboring cells. S1P binds to S1P2 and triggers formation and contraction of an actin-and-myosin II extruding ring to squeeze the dying cell out.
Based on our data, we propose a model showing how S1P triggers apoptotic cell extrusion (Fig. 5). S1P produced within the apoptotic cells is transported to the extracellular surface, where it binds S1P2 on the surface of the surrounding cells and activates Rho, presumably through p115 RhoGEF (Rosenblatt et al., 2001; Slattum et al., 2009), to trigger assembly and contraction of the actin–myosin extrusion ring at the live–dying cell interface. Contraction of the extrusion ring drives the dying cell out of the epithelium, thereby detaching it from the epithelium and underlying matrix. As S1P is known to promote survival (Kolesnick and Hannun, 1999; Hannun and Obeid, 2002; Weigert et al., 2010), loss of S1P in the extruded cell may help promote its death, whereas the increase of S1P in the surrounding cells could promote their survival, ensuring that they can extrude the dying cell. How S1P is produced and exported outside the cell will be important future goals.
Extrusion is critical for maintaining proper epithelial barrier function; therefore, we expect defects in the S1P2 signaling pathway may result in diseases associated with poor mucosal barriers. Aberrant levels of S1P have been associated with asthma, which many believe may initiate from poor barrier function in airway epithelia (Gitter et al., 2001; Proksch et al., 2006; Sartor, 2006; Kim et al., 2009; Swindle et al., 2009; Voelkel and Spiegel, 2009). Further, aberrant S1P signaling is known to be involved in a variety of pathways that could further aggravate asthma, including immune cell migration (Matloubian et al., 2004; Gude et al., 2008; Weigert et al., 2009), epithelial proliferation (Shida et al., 2008), and vasoconstriction (Watterson et al., 2005). S1P2 knockout in mice also results in defective epithelial barrier function in the cochlea (Kono et al., 2007). Therefore, the identification of the S1P2 pathway in controlling extrusion will allow us to test if defective extrusion could bring about a variety of epithelial barrier diseases in animal models.
On the other hand, extrusion could be activated normally during development or inappropriately during cancer progression. Several reports have shown that cells can extrude without dying (Gibson and Perrimon 2005; Shen and Dahmann 2005; Monks et al., 2008). Further, neuroblasts delaminate from a neuroepithelium in Drosophila embryos by a process that appears similar to basal extrusion (Hartenstein et al., 1994). In advanced tumors where cell death is blocked or survival signaling is up-regulated (LoPiccolo et al., 2008; Liu et al., 2009), extrusion could enable exit of tumor cells to initiate metastasis. Given that SphK1 is considerably increased in multiple types of cancers (French et al., 2003; Johnson et al., 2005; Van Brocklyn et al., 2005) and that S1P is associated with proliferation, survival, migration, and invasion of cancer cells (Maceyka et al., 2002; Spiegel and Milstien, 2003; Radeff-Huang et al., 2004), increased S1P signaling could enable tumors to invade by misregulating extrusion. Therefore, our work defining the signaling that drives extrusion of an epithelial cell from a layer may also be important for driving other delamination events crucial to developmental differentiation or invasion of tumor cells.
Materials and methods
Cell culture
MDCK II cells were cultured in Dulbecco’s minimum essential medium (DMEM) high glucose with 5% FBS and 100 µg/ml penicillin/streptomycin (all from Invitrogen) at 5% CO2, 37°C. HBE cells were cultured in MEM supplemented with 10% FBS and l-glutamine in a flask coated with human fibronectin type I (BD), bovine collagen I (Purecol; Inamed biomaterials), and BSA (Invitrogen).
Drug and UV treatment
Cells were treated with 30 µM SKI II (EMD), 4 µM SKI V (Sigma-Aldrich), 40 µM tDHS (Avanti Polar Lipids, Inc.), 10 µM JTE-013 (Tocris Bioscience), 10 µM VPC-23019 (Avanti Polar Lipids, Inc.), 10 µM FTY720-P (Echelon Biosciences), or 10 µg/ml murine anti-S1P mAb (LPath) for 10 min before UV treatment. To induce apoptosis, cultured monolayers were exposed to 1,200 µJ/cm2 UV254 irradiation in a UV series II (Spectroline) and incubated for 2 h before fixation.
Cell staining
Cells were fixed with 4% formaldehyde in PBS at 37°C for 20 min, permeabilized for 10 min with 0.5% Triton in PBS, blocked with AbDil (PBS with 0.1% Triton X-100 and 2% BSA) for 10 min, and incubated with primary antibody for 1 h. Antibody concentrations used for immunostaining were: 1:200 rabbit anti-active caspase-3 (BD) and 50 µg/ml anti-S1P mAb (LPath Inc.). Alexa Fluor 488 goat anti–rabbit IgG and Alexa Fluor 488 goat anti–mouse IgG were used as secondary antibodies to detect active caspase-3 and S1P, respectively. Actin was detected with Alexa Fluor 568–phalloidin (Invitrogen). DNA was detected with 1 µg/ml Hoechst 33342 (Sigma-Aldrich) or 5 µM DRAQ5 (Axxora).
Zebrafish staining
To induce apoptosis, we treated 3-d-old zebrafish larvae with 1% DMSO on ice for 30 min, followed by recovery at 28°C for 10 min. Embryos were then fixed in 4% formaldehyde, 0.15% glutaraldehyde, 1 mM MgCl2, 0.2% Triton X-100, and 25 mM Pipes, pH 6.9, for 2 h, blocked in 0.25% casein overnight. Cells undergoing apoptosis were identified using an anti-activated caspase-3 rabbit polyclonal Ab (BD) followed by incubation with Alexa Fluor 488 anti–rabbit IgG Ab (Invitrogen). Actin was visualized using 0.1 µg/ml Alexa Fluor 568–phalloidin (Invitrogen). DNA was visualized using 1 µg/ml DAPI (Sigma-Aldrich) or 5 µM DRAQ5 (Enzo Life Sciences).
Addition of dead cell fragments or S1P to monolayers
To make dead cell fragments, MDCK cells were scraped from the culture dish and sheared with a 27G1/2 needle seven times. Cell fragments were transferred to a microfuge tube and centrifuged for 1 min at 8,000 rpm in a microfuge (Eppendorf). Cells were fluorescently labeled by resuspending them in 50 µl DMEM containing 40 µg/ml FITC snail Lectin (Invitrogen) for 5 min and washed three times with 1 ml of DMEM. The labeled cell fragments were then added to confluent MDCK monolayers cultured on glass coverslips, and incubated for 90 min. The coverslips were fixed and stained with Alexa Fluor 568–phalloidin and Hoechst dye. For drug inhibition in the dead cell fragment addition, cells were pretreated for 10 min with SKI at concentrations listed above before scraping and needle shearing. For S1P mAb addition, cell fragments were added to MDCK monolayers in the presence of 10 µg/ml S1P mAb. For the lipid addition experiment, 20 µg/ml S1P (Avanti Polar Lipids, Inc.) or total E. coli lipid extraction (Avanti Polar Lipids, Inc.) resuspended in DMEM was added to confluent MDCK monolayers, incubated for 1 h, and fixed and stained with phalloidin and Hoechst dye, as above.
Quantification of cell extrusion
To quantify the ratio of nonextruded apoptotic cells, we counted 100 active caspase-3–positive cells that were associated with cultured monolayers. Apoptotic cells that possessed clearly shrunken nuclei but were not surrounded by a distinguishable actin ring were defined as nonextruded apoptotic cells. Apoptotic cells that came out of the plane of the monolayer with strong actin staining around and/or underneath the cells were defined as extruded cells. Old apoptotic cells with strong caspase-3 staining floating above the monolayer that died before we treated monolayers were excluded.
Microscopy
Fluorescence micrographs of fixed, cultured cells were obtained using a microscope (DM 6000B; Leica) with an HCX PL Fluotar 63x/1.25 oil lens (Leica) and captured using a Micromax charge-coupled device camera (Roper Scientific). IP Laboratory software was used to control the camera and to process images. Fluorescence micrographs of zebrafish larvae were obtained using a microscope (model 90i; Nikon) with a 40x PHI lens (Nikon) and captured using a charge-coupled device camera (Retiga 2000R; Q Imaging). Confocal micrographs were obtained using a microscope (TCS SP5; Leica) with a 63x oil lens or an inverted microscope (Eclipse TE300; Nikon) converted for spinning disc confocal microscopy (Andor Technologies) using a 60x Plan Fluor 0.95 oil lens with an electron-multiplied cooled CCD camera 1,000 x 1,000, 8 x 8 mm2 driven by the IQ software (Andor Technologies). We used ImageJ to stack confocal sections into Z series that were then color combined and reconstructed into 3D image using MetaMsorph (GE Healthcare). For HBE cells stained with anti-S1P, we displayed five consecutive projections of 2-µm thickness each using the “montage” function on MetaMorph software. All images were processed further using ImageJ, Photoshop (Adobe), Illustrator (Adobe), and Quicktime Pro (Quicktime) software.
RT-PCR
Total RNA was isolated from cultured cells using the RNeasy kit (QIAGEN) and reverse-transcribed using the SuperScript III first strand synthesis kit (Invitrogen) with random hexamers according to the manufacturer’s guidelines. PCR detection of S1P1–5 was conducted as described previously (Estrada et al., 2008). In brief, PCR amplification of the targeted fragments was performed with 30 cycles of denaturation at 95°C (30 s), annealing at 58°C (30 s), and extension at 72°C (30 s). PCR primer pairs used were: S1P1: sense, 5′-GCACCAACCCCATCATTTAC-3′, antisense, 5′-TTGTCCCCTTCGTCTTTCTG-3′; S1P2: sense, 5′-CAAGTTCCACTCGGCAATGT-3′, antisense, 5′-CAGGAGGCTGAAGACAGAGG-3′; S1P3: sense, 5′-TCAGGGAGGGCAGTATGTTC-3′, antisense, 5′-GAGTAGAGGGGCAGGATGGT-3′; S1P4: sense, 5′-AGCCTTCTGCCCCTCTACTC-3′, antisense, 5′-ATCAGCACCGTCTTCAGCA-3′; and S1P5: sense, 5′-ACAACTACACCGGCAAGCTC-3′, antisense, 5′-GCCCCGACAGTAGGATGTT-3′. Quantitative real-time PCR was performed using a LightCycler480 (Roche) and the SYBR Green PCR master mix (SABiosciences). Relative mRNA expression was quantified using the comparative threshold method with the content of actin mRNA as internal control.
shRNA-mediated gene silencing of S1P2
We designed sense and antisense hairpin oligonucleotides specific for S1P2 (shS1P2-1) according to a published work (Estrada et al., 2008). Two additional pairs of hairpin oligonucleotides were used to knockdown the S1P2 receptor. The oligonucleotide sequences were: shS1P2-2: sense, 5′-GCGCCATTGTGGTGGAAAA-3′, antisense, 5′-TTTTCCACCACAATGGCGC-3′; shS1P2-3: sense, 5′-GCAAGTTCCACTCGGCAAT-3′, antisense: 5′-ATTGCCGAGTGGAACTTGC-3′. The sense and antisense oligonucleotides were annealed and cloned into a pLentilox 5.0–based lentiviral vector. Transducing lentiviral particles carrying the shRNA oligonucleotides were produced by packaging in 293T cells. Lentiviral particles carrying nonspecific shRNA oligonucleotides were used as a control. HBE cells were transduced with lentiviral particles for 6 h in the presence of 2 µg/ml polybrene, washed, and replaced with fresh medium. Stably transduced cells were FAC-sorted based on GFP fluorescence.
Statistics
The statistical analysis was performed using an unpaired t test. Values of P < 0.05 were considered significant.
Online supplemental material
Fig. S1 shows that S1P directly induces actin assembly at the apex of an MDCK monolayer. Fig. S2 shows that inhibition of actin assembly and apoptotic cell extrusion by inhibitors of SphKs is dose dependent. Fig. S3 shows that apoptotic cell extrusion requires the signaling mediated by S1P2. Video 1 shows a tilting 3D projection movie of two apoptotic cells that do not extrude from the epidermis of a mil zebrafish embryo. Video 2 shows the Z planes of an apoptotic cell that extrudes normally from the epidermis of a WT zebrafish embryo and an apoptotic cell that fails to extrude from the epidermis of a mil zebrafish embryo. Videos 3–5 show the localization of S1P during early, middle, and late stages of apoptotic cell extrusion. Video 6 shows that an apoptotic cell does not produce S1P or extrude in the presence of SKI II. Video 7 shows that an apoptotic HBE cell that generates high levels of S1P fails to extrude in the presence of JTE-013. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201010075/DC1.
Acknowledgments
We thank Dr. Jean Marie Delalande and Amy Carr for help on zebrafish experiments and to Mark Metzstein, Katie Ullman, and Thomas Marshall for helpful comments on our manuscript. We also thank Dr. Diana Stafforini for helpful advice regarding lipids, Dr. Carl Thummel for use of his LSM confocal microscope, and Dr. James Bear for providing the lentiviral constructs used for shRNA experiments.
This work was supported by a National Institutes of Health Innovator Award no. DP2 OD002056-01 to J. Rosenblatt and P30 CA042014 awarded to The Huntsman Cancer Institute for core facilities. R. Sabbadini has stock options in Lpath, Inc.
Footnotes
Abbreviations used in this paper:
- HBE
- human bronchial epithelial
- mil
- miles apart
- S1P
- sphingosine-1-phosphate
- SKI
- sphingosine kinase inhibitor
- WT
- wild type
References
- Brinkmann V., Davis M.D., Heise C.E., Albert R., Cottens S., Hof R., Bruns C., Prieschl E., Baumruker T., Hiestand P., et al. 2002. The immune modulator FTY720 targets sphingosine 1-phosphate receptors. J. Biol. Chem. 277:21453–21457 10.1074/jbc.C200176200 [DOI] [PubMed] [Google Scholar]
- Davis M.D., Clemens J.J., Macdonald T.L., Lynch K.R. 2005. Sphingosine 1-phosphate analogs as receptor antagonists. J. Biol. Chem. 280:9833–9841 10.1074/jbc.M412356200 [DOI] [PubMed] [Google Scholar]
- Estrada R., Zeng Q., Lu H., Sarojini H., Lee J.F., Mathis S.P., Sanchez T., Wang E., Kontos C.D., Lin C.Y., et al. 2008. Up-regulating sphingosine 1-phosphate receptor-2 signaling impairs chemotactic, wound-healing, and morphogenetic responses in senescent endothelial cells. J. Biol. Chem. 283:30363–30375 10.1074/jbc.M804392200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French K.J., Schrecengost R.S., Lee B.D., Zhuang Y., Smith S.N., Eberly J.L., Yun J.K., Smith C.D. 2003. Discovery and evaluation of inhibitors of human sphingosine kinase. Cancer Res. 63:5962–5969 [PubMed] [Google Scholar]
- Gibson M.C., Perrimon N. 2005. Extrusion and death of DPP/BMP-compromised epithelial cells in the developing Drosophila wing. Science. 307:1785–1789 10.1126/science.1104751 [DOI] [PubMed] [Google Scholar]
- Gitter A.H., Wullstein F., Fromm M., Schulzke J.D. 2001. Epithelial barrier defects in ulcerative colitis: characterization and quantification by electrophysiological imaging. Gastroenterology. 121:1320–1328 10.1053/gast.2001.29694 [DOI] [PubMed] [Google Scholar]
- Gude D.R., Alvarez S.E., Paugh S.W., Mitra P., Yu J., Griffiths R., Barbour S.E., Milstien S., Spiegel S. 2008. Apoptosis induces expression of sphingosine kinase 1 to release sphingosine-1-phosphate as a “come-and-get-me” signal. FASEB J. 22:2629–2638 10.1096/fj.08-107169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannun Y.A., Obeid L.M. 2002. The Ceramide-centric universe of lipid-mediated cell regulation: stress encounters of the lipid kind. J. Biol. Chem. 277:25847–25850 10.1074/jbc.R200008200 [DOI] [PubMed] [Google Scholar]
- Hartenstein V., Younossi-Hartenstein A., Lekven A. 1994. Delamination and division in the Drosophila neurectoderm: spatiotemporal pattern, cytoskeletal dynamics, and common control by neurogenic and segment polarity genes. Dev. Biol. 165:480–499 10.1006/dbio.1994.1269 [DOI] [PubMed] [Google Scholar]
- Holinstat M., Mehta D., Kozasa T., Minshall R.D., Malik A.B. 2003. Protein kinase Calpha-induced p115RhoGEF phosphorylation signals endothelial cytoskeletal rearrangement. J. Biol. Chem. 278:28793–28798 10.1074/jbc.M303900200 [DOI] [PubMed] [Google Scholar]
- Johnson K.R., Johnson K.Y., Crellin H.G., Ogretmen B., Boylan A.M., Harley R.A., Obeid L.M. 2005. Immunohistochemical distribution of sphingosine kinase 1 in normal and tumor lung tissue. J. Histochem. Cytochem. 53:1159–1166 10.1369/jhc.4A6606.2005 [DOI] [PubMed] [Google Scholar]
- Kim R.H., Takabe K., Milstien S., Spiegel S. 2009. Export and functions of sphingosine-1-phosphate. Biochim. Biophys. Acta. 1791:692–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolesnick R., Hannun Y.A. 1999. Ceramide and apoptosis. Trends Biochem. Sci. 24:224–225, author reply :227 10.1016/S0968-0004(99)01408-5 [DOI] [PubMed] [Google Scholar]
- Kono M., Belyantseva I.A., Skoura A., Frolenkov G.I., Starost M.F., Dreier J.L., Lidington D., Bolz S.S., Friedman T.B., Hla T., Proia R.L. 2007. Deafness and stria vascularis defects in S1P2 receptor-null mice. J. Biol. Chem. 282:10690–10696 10.1074/jbc.M700370200 [DOI] [PubMed] [Google Scholar]
- Kupperman E., An S., Osborne N., Waldron S., Stainier D.Y. 2000. A sphingosine-1-phosphate receptor regulates cell migration during vertebrate heart development. Nature. 406:192–195 10.1038/35018092 [DOI] [PubMed] [Google Scholar]
- Liu P., Cheng H., Roberts T.M., Zhao J.J. 2009. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 8:627–644 10.1038/nrd2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoPiccolo J., Blumenthal G.M., Bernstein W.B., Dennis P.A. 2008. Targeting the PI3K/Akt/mTOR pathway: effective combinations and clinical considerations. Drug Resist. Updat. 11:32–50 10.1016/j.drup.2007.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maceyka M., Payne S.G., Milstien S., Spiegel S. 2002. Sphingosine kinase, sphingosine-1-phosphate, and apoptosis. Biochim. Biophys. Acta. 1585:193–201 [DOI] [PubMed] [Google Scholar]
- Mandala S., Hajdu R., Bergstrom J., Quackenbush E., Xie J., Milligan J., Thornton R., Shei G.J., Card D., Keohane C., et al. 2002. Alteration of lymphocyte trafficking by sphingosine-1-phosphate receptor agonists. Science. 296:346–349 10.1126/science.1070238 [DOI] [PubMed] [Google Scholar]
- Matloubian M., Lo C.G., Cinamon G., Lesneski M.J., Xu Y., Brinkmann V., Allende M.L., Proia R.L., Cyster J.G. 2004. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature. 427:355–360 10.1038/nature02284 [DOI] [PubMed] [Google Scholar]
- Monks J., Smith-Steinhart C., Kruk E.R., Fadok V.A., Henson P.M. 2008. Epithelial cells remove apoptotic epithelial cells during post-lactation involution of the mouse mammary gland. Biol. Reprod. 78:586–594 10.1095/biolreprod.107.065045 [DOI] [PubMed] [Google Scholar]
- O’Brien N., Jones S.T., Williams D.G., Cunningham H.B., Moreno K., Visentin B., Gentile A., Vekich J., Shestowsky W., Hiraiwa M., et al. 2009. Production and characterization of monoclonal anti-sphingosine-1-phosphate antibodies. J. Lipid Res. 50:2245–2257 10.1194/jlr.M900048-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada M., Yatomi Y., Ohmori T., Ikeda H., Ozaki Y. 2002. Enhancement of sphingosine 1-phosphate-induced migration of vascular endothelial cells and smooth muscle cells by an EDG-5 antagonist. Biochem. Biophys. Res. Commun. 299:483–487 10.1016/S0006-291X(02)02671-2 [DOI] [PubMed] [Google Scholar]
- Proksch E., Fölster-Holst R., Jensen J.M. 2006. Skin barrier function, epidermal proliferation and differentiation in eczema. J. Dermatol. Sci. 43:159–169 10.1016/j.jdermsci.2006.06.003 [DOI] [PubMed] [Google Scholar]
- Radeff-Huang J., Seasholtz T.M., Matteo R.G., Brown J.H. 2004. G protein mediated signaling pathways in lysophospholipid induced cell proliferation and survival. J. Cell. Biochem. 92:949–966 10.1002/jcb.20094 [DOI] [PubMed] [Google Scholar]
- Rosenblatt J., Raff M.C., Cramer L.P. 2001. An epithelial cell destined for apoptosis signals its neighbors to extrude it by an actin- and myosin-dependent mechanism. Curr. Biol. 11:1847–1857 10.1016/S0960-9822(01)00587-5 [DOI] [PubMed] [Google Scholar]
- Sartor R.B. 2006. Mechanisms of disease: pathogenesis of Crohn’s disease and ulcerative colitis. Nat. Clin. Pract. Gastroenterol. Hepatol. 3:390–407 10.1038/ncpgasthep0528 [DOI] [PubMed] [Google Scholar]
- Shen J., Dahmann C. 2005. Extrusion of cells with inappropriate Dpp signaling from Drosophila wing disc epithelia. Science. 307:1789–1790 10.1126/science.1104784 [DOI] [PubMed] [Google Scholar]
- Shida D., Takabe K., Kapitonov D., Milstien S., Spiegel S. 2008. Targeting SphK1 as a new strategy against cancer. Curr. Drug Targets. 9:662–673 10.2174/138945008785132402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattum G., McGee K.M., Rosenblatt J. 2009. P115 RhoGEF and microtubules decide the direction apoptotic cells extrude from an epithelium. J. Cell Biol. 186:693–702 10.1083/jcb.200903079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel S., Milstien S. 2003. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 4:397–407 10.1038/nrm1103 [DOI] [PubMed] [Google Scholar]
- Swindle E.J., Collins J.E., Davies D.E. 2009. Breakdown in epithelial barrier function in patients with asthma: identification of novel therapeutic approaches. J. Allergy Clin. Immunol. 124:23–34, quiz :35–36 10.1016/j.jaci.2009.05.037 [DOI] [PubMed] [Google Scholar]
- Tamada M., Perez T.D., Nelson W.J., Sheetz M.P. 2007. Two distinct modes of myosin assembly and dynamics during epithelial wound closure. J. Cell Biol. 176:27–33 10.1083/jcb.200609116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Brocklyn J.R., Jackson C.A., Pearl D.K., Kotur M.S., Snyder P.J., Prior T.W. 2005. Sphingosine kinase-1 expression correlates with poor survival of patients with glioblastoma multiforme: roles of sphingosine kinase isoforms in growth of glioblastoma cell lines. J. Neuropathol. Exp. Neurol. 64:695–705 10.1097/01.jnen.0000175329.59092.2c [DOI] [PubMed] [Google Scholar]
- Visentin B., Vekich J.A., Sibbald B.J., Cavalli A.L., Moreno K.M., Matteo R.G., Garland W.A., Lu Y., Yu S., Hall H.S., et al. 2006. Validation of an anti-sphingosine-1-phosphate antibody as a potential therapeutic in reducing growth, invasion, and angiogenesis in multiple tumor lineages. Cancer Cell. 9:225–238 10.1016/j.ccr.2006.02.023 [DOI] [PubMed] [Google Scholar]
- Voelkel N.F., Spiegel S. 2009. Why is effective treatment of asthma so difficult? An integrated systems biology hypothesis of asthma. Immunol. Cell Biol. 87:601–605 10.1038/icb.2009.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson K.R., Ratz P.H., Spiegel S. 2005. The role of sphingosine-1-phosphate in smooth muscle contraction. Cell. Signal. 17:289–298 10.1016/j.cellsig.2004.09.013 [DOI] [PubMed] [Google Scholar]
- Weigert A., Weis N., Brüne B. 2009. Regulation of macrophage function by sphingosine-1-phosphate. Immunobiology. 214:748–760 10.1016/j.imbio.2009.06.003 [DOI] [PubMed] [Google Scholar]
- Weigert A., Cremer S., Schmidt M.V., von Knethen A., Angioni C., Geisslinger G., Brüne B. 2010. Cleavage of sphingosine kinase 2 by caspase-1 provokes its release from apoptotic cells. Blood. 115:3531–3540 10.1182/blood-2009-10-243444 [DOI] [PubMed] [Google Scholar]