HIV-1 infection of host cells leads to significant reorganization and redistribution of components of the nuclear envelope and nuclear pore complex.
Abstract
Human immunodeficiency virus type 1 (HIV-1) commandeers host cell proteins and machineries for its replication. Our earlier work showed that HIV-1 induced the cytoplasmic retention of nucleocytoplasmic shuttling and ribonucleic acid (RNA)–binding proteins. This retention is dependent on nuclear export of the viral genomic RNA and on changes in the localization and expression level of the nucleoporin (Nup) p62 (Nup62). To further characterize the extent of perturbation induced by HIV-1, we performed proteomics analyses of nuclear envelopes (NEs) isolated from infected T cells. Infection induced extensive changes in the composition of the NE and its associated proteins, including a remarkable decrease in the abundance of Nups. Immunogold electron microscopy revealed the translocation of Nups into the cytoplasm. Nup62 was identified as a component of purified virus, and small interfering RNA depletion studies revealed an important role for this Nup in virus gene expression and infectivity. This detailed analysis highlights the profound effects on NE composition induced by HIV-1 infection, providing further evidence of the magnitude of viral control over the cell biology of its host.
Introduction
Human immunodeficiency virus type 1 (HIV-1) infection induces changes in the host cell transcriptome (Giri et al., 2006; Li et al., 2009) and proteome (Chan et al., 2007; Ringrose et al., 2008; Rasheed et al., 2009). Both biochemical studies and genome-wide short hairpin/siRNA screens have identified nucleoporins (Nups) as HIV-1 dependency factors that assist nuclear import of the preintegration complex (PIC; Ebina et al., 2004; Goff, 2008; König et al., 2008; Kok et al., 2009; Woodward et al., 2009; Yeung et al., 2009). Nups are also required for the nuclear export of viral factors during the late stages of HIV-1 replication (Zolotukhin and Felber, 1999; Hofmann et al., 2001; Le Rouzic et al., 2002; Kiss et al., 2003; Hutten and Kehlenbach, 2006; Hutten et al., 2009). Approximately 30 different Nups present in multiple copies and organized in an eightfold radial symmetry compose nuclear pore complexes (NPCs), which stud and span nuclear envelopes (NEs) and act as selective barriers for the nucleocytoplasmic shuttling of macromolecules (Wente and Rout, 2010). Hepatitis B virus, herpes simplex virus, influenza virus, and adenovirus also use Nups to access nuclei (Trotman et al., 2001; Copeland et al., 2009; König et al., 2010; Schmitz et al., 2010), whereas poliovirus and cardiovirus induce their rearrangement or degradation (Gustin and Sarnow, 2001, 2002; Bardina et al., 2009; Porter and Palmenberg, 2009).
Studies using organellar proteomics can reveal proteins that are otherwise masked during whole cell analyses and provide information on protein localization and function, an especially useful tool for studying a virus that usurps many cellular machineries (Brunet et al., 2003; Gilchrist et al., 2006). This work extends our previous finding that HIV-1 replication imposes a blockade to the nuclear import of heterogeneous nuclear RNP A1 (hnRNP A1) and its transport receptor Transportin-1 via alterations in the localization and abundance of Nup p62 (Nup62; Monette et al., 2009). The persistence of Nup62 at the NPC core, where it authenticates passing cargo, depends on the surrounding scaffolding and anchoring Nups (Wente and Rout, 2010). To determine whether the block in nuclear shuttling imposed by HIV-1 was limited to Nup62 expression or to the malfunctioning of other Nups, we undertook a proteomic study to compare the composition of purified NEs from mock- and HIV-1–infected Jurkat T cells. This has enabled the identification of 413 NE-associated host proteins, with 68% showing significant changes in abundance, among which many were those associated with NPCs. Immunogold EM revealed that at least one Nup is dislodged from NPCs and is redirected to budding virions. Immunofluorescence (IF) experiments suggest that Nup62 is essential for viral genomic RNA (vRNA) export and may take leave of NPCs as part of the growing HIV-1 vRNA–RNP complex, where it may ensure its replicative success during viral egress, gene expression, and assembly.
Results and discussion
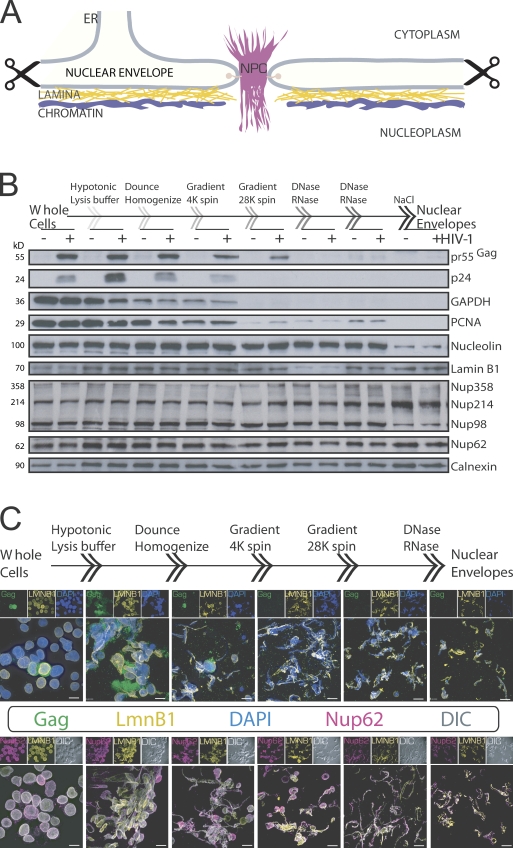
Isolation of NEs from HIV-1–infected T cells
To define changes to NPC composition in HIV-1–infected T cells, we used a recently published method to isolate NEs and accompanying NPCs, associated nuclear lamina, and contiguous ER from T cells for a comparative liquid chromatography (LC)/tandem mass spectrometry (MS/MS) study (Fig. 1 A; Korfali et al., 2009). Because much of the starting material is lost from the purification procedure, we first tested the method for its ability to enrich NEs by collecting cellular products isolated at each step of the procedure from mock- or lowly infected cells (day 3 after transfection of proviral DNA). These were normalized for protein content and loaded onto SDS-PAGE gels for Western analysis, which validated the purification procedure of NEs from cells by highlighting the gradual loss of plasma membrane–associated viral (e.g., p24 Capsid [CA] and pr55Gag), cytoplasmic (e.g., pr55Gag and glyceraldehyde 3-phosphate dehydrogenase), and chromatin-binding and nuclear proteins (e.g., proliferating cell nuclear antigen and Nucleolin) and the accompanying enrichment of NE, NPC, and ER proteins Lamin B1, Nup, and Calnexin, respectively (Fig. 1 B). To further validate the method, again, at low infection stages (day 3) mirrored by only two cells labeled by the anti-Gag antibody (Fig. 1 C, top row, first panel), products from each step of the procedure were analyzed by IF. The staining for NE-associated Lamin B1 and the decrease in DAPI staining demonstrate the enrichment in NE proteins and the removal of the majority of chromosomal DNA contaminants, respectively (Fig. 1 C, top row). In addition, differential interference contrast (DIC) confirmed that all visible endpoint structures from NE preparations were immunopositive for Nup62 and Lamin B1 (Fig. 1 C, bottom row).
Figure 1.
Procedure taken for isolating and verifying NEs from HIV-1–infected cells. (A) A diagram depicting the architecture of NEs with associated ER, nuclear lamina, and chromatin that were purified during the isolation procedure (indicated by scissors). (B) Western analysis of proteins isolated at each step of the NE purification method using mock (−)- or minimally HIV-1 (+)–infected cells to show the elimination of plasma membrane (pr55Gag and p24), cytoplasmic (pr55Gag and glyceraldehyde 3-phosphate dehydrogenase [GAPDH]), nuclear (chromatin-binding proliferating cell nuclear antigen [PCNA]), and nucleolar (Nucleolin) proteins and the enrichment of NE (Lamin B1), NPC (Nup62 and phenylalanine-glycine–Nups), and ER (Calnexin) proteins. (C) Stepwise IF analysis of HIV-1 Gag, Lamin B1 (LMNB1), and Nup62 with DAPI staining (top row) or DIC (bottom row) of isolated products. DIC shows all visible endpoint structures that stain positively for LMNB1 and Nup62. 3D renderings of z stacks are shown. Bars, 10 µm.
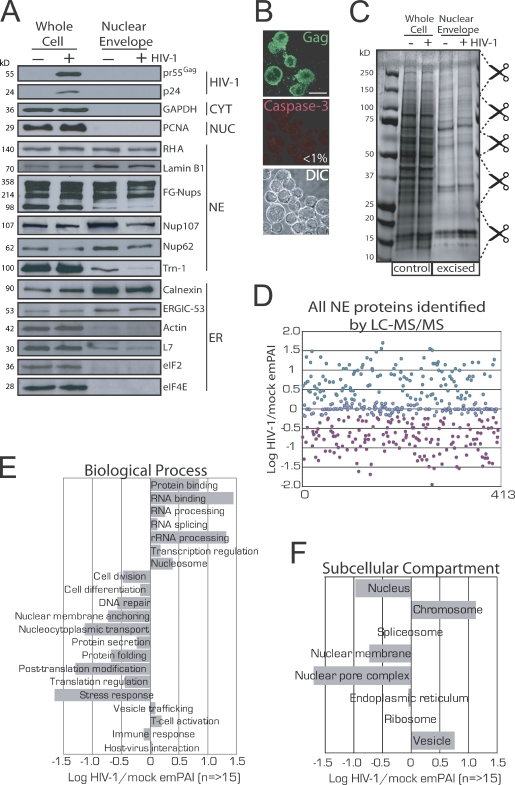
Satisfied with the successful isolation of NEs, we next repeated the procedure using mock- or peak-infected T cells. Western analysis identified viral proteins from peak-infected whole cell lysates but not from NEs derived from peak-infected cells deficient in plasma membrane, cytoplasmic, and nuclear proteins and enriched with NE and NPC proteins (Fig. 2 A). Importantly, and as previously observed in our earlier work, Nup62 levels were reduced in peak-infected whole cells (Fig. 2 A, left; Monette et al., 2009), and this reduction was also apparent in NEs isolated from these cells (Fig. 2 A, right). As an additional control to verify peak-infected cell populations, coimmunostaining for Gag and cleaved Caspase-3 revealed that <1% (n = 5,000) of cells was apoptotic at peak infection (Fig. 2 B). Protein gels were stained, and gel slices were excised for LC-MS/MS analysis (Fig. 2 C). Mascot server–generated exponentially modified protein abundance index (emPAI) values for individual proteins were used to calculate how HIV-1 infection affects their abundance at NEs (Table S3, last column). Globally, HIV-1 infection dramatically impacted NE protein composition by increasing or decreasing total protein abundance by 38 and 30%, respectively (Fig. 2 D). The classification of protein hits into Gene Ontology (GO)–term biological processes, and the means of their cumulative emPAI values provided evidence that proteins involved in cell division, differentiation, and DNA repair were less prevalent in NEs from infected cells (Fig. 2 E). Other down-regulated processes included protein secretion, posttranslational modification, translation regulation, stress response, immune response, and nucleocytoplasmic shuttling, all of which might otherwise elicit an antiviral state. Conversely, proteins participating in T cell activation and vesicular trafficking were up-regulated, as were nucleosomal, transcription, RNA-binding, processing, splicing, and ribosomal proteins, suggesting that HIV-1 has far-reaching effects on nuclear events that may contribute to its replicative success. In support of this, when protein hits were organized according to subcellular compartments, those known to be important for HIV-1 replication, such as chromosomal and vesicular proteins, were found to be up-regulated (Fig. 2 F). Importantly, proteins involved in nucleocytoplasmic transport were most underrepresented, and this was reflected by a lowered abundance of nuclear and nuclear membrane–associated proteins. Our observation that HIV-1 infection affects the expression of nucleocytoplasmic shuttling proteins is reflected by earlier studies using CD4+ and primary cells (Chan et al., 2007, 2009). Although Chan et al. (2007, 2009) used whole cells for their analysis, >46% of the common hits are modified in the same direction by HIV-1 infection (Table I). In all, we find that 54, 100, 100, and 50% of our identified Nups, importins, Ran GTP–GDP cycle, and hnRNP proteins, respectively, are modified in the same direction by HIV-1 infection, lending additional support to our virological methods and use of emPAI values to report our findings (Table I). More recently, several Nups and importins were found to be decreased by human respiratory syncycial virus infection (Munday et al., 2010), and it has been speculated that the blocking of nuclear import may be a common feature of infections by RNA viruses (Gustin, 2003).
Figure 2.
Verification of the purity of isolated NEs, the apoptosis levels of peak-infected T cells, and GO analysis of LC-MS/MS–identified NE-associated proteins. (A) Western analysis of whole cell lysates and purified NEs in mock- and peak-infected cells. (B) IF for Gag and cleaved Caspase-3 (apoptosis marker) and DIC in peak-infected cells. Less than 1% of cells was apoptotic. 3D renderings of z stacks are shown. Bar, 10 µm. (C) Coomassie blue–stained SDS-PAGE of whole cell and purified NEs for LC-MS/MS. Scissors demarcate excised gel plugs. (D) A scatterplot of all NE-associated proteins identified by LC-MS/MS. Increases and decreases are shown in green and red. (E and F) NE-associated proteins are identified and organized according to GO UniProt/SwissProt biological processes (E) or subcellular compartments (F).
Table I.
Comparison of NPC and shuttling proteins identified from LC-MS/MS studies
| Type | Down-regulated by HIV-1 | Up-regulated by HIV-1 | ||
| This study (purified NEs) | Chan et al.a (whole cells) | This study (purified NEs) | Chan et al.a (whole cells) | |
| Nups (54%)b | KTN1, NUP43, NUP58, NUP45, NUP54, GP210, POM121, NUP107, NUP133, NUP155, NUP160, NUP37, NUP85, NUP93 | KTN1, NUP43, NUP58/NUP45, NUP54, NUP50, NUP62 | NUP35, NUP98, TPR | NUP35, NUP98, TPR, NUP153, NUP214, NUP93, POM121 |
| Importins (100%) | KNPB1, KPNA2 | KPNB1, KPNA2, KPNA3, KPNA4, KPNA6, IPO5, IPO7, IPO9, TNPO1, TNPO2, TNPO3 | N/D | KPNA1 |
| Ran GTP–GDP cycle (100%) | RANGAP1, RCC1 | RANBP2(NUP358), RANGAP1, RCC1, NUTF2, RANBP1, RANBP3, RAP1GDS1, SUMO1 | N/D | RAN, RAE1 |
| Exportins | RIP | N/D | NXF1 | RIP, XPO1, XPO2, XPO5, XPO7 |
| hnRNP proteins (50%) | HNRNPG, HNRNPK, HNRNPL, HNRNPR | HNRNPG, HNRNPF | HNRNPA1, HNRNPA2, HNRNPC, HNRNPM, HNRNPU | HNRNPA1, HNRNPA2, HNRNPC, HNRNPM, HNRNPU, HNRNPH, HNRNPK, HNRNPL, HNRNPQ, HNRNPR |
Boldface text indicates common proteins between studies that are modulated in a similar direction by HIV-1. Italicized proteins were found to be regulated in an opposite manner. N/D, not detected.
Mass spectrometry hits found in Chan et al. (2007) and/or Chan et al. (2009).
Quantitative correspondence between this study and those of Chan et al. (2007, 2009).
HIV-1 disperses Nups into the cytoplasm and into progeny virus
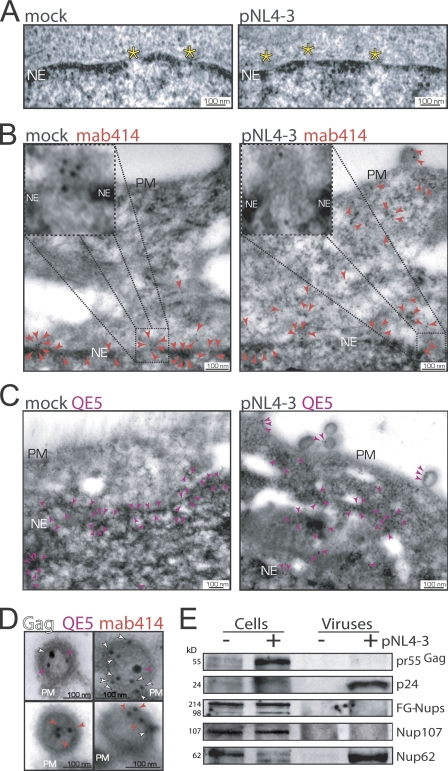
The most salient effect induced by HIV-1 infection was on the abundance of NPC proteins with observed decreases of 18 of the 30 main Nups. Therefore, thin-section transmission EM was performed to evaluate the integrity of NEs and NPCs from mock- or pNL4-3–transfected HeLa cells, but no detectable differences in these nanostructures between samples could be seen (Fig. 3 A). However, when immunogold EM was performed using two different phenylalanine-glycine–rich Nup-specific antibodies (i.e., mab414 and QE5), whereas Nup labeling was specifically and predominantly associated with the NPCs of mock-transfected cells (Fig. 3, B and C, left), analysis of the distribution of Nups in HIV-1–expressing cells (identified by a budding virus at the plasma membranes) showed them to be dispersed throughout the cytoplasm and in budding viruses (Fig. 3, B and C, right). Lamin B2 labeling, used as a control, remained associated with NEs (unpublished data). Quantitative software-assisted measurements of the distance between mab414/QE5 immunogold labeling and the inner nuclear membrane confirmed a fivefold increase in HIV-1–infected cells (n = 459, mock; n = 498, HIV-1 expressing). Double immunogold labeling with anti–CA p24 and QE5 or mab414 antibodies colabeled budding viruses (Fig. 3 D). Though mab414 most readily recognizes Nup62 as well as other Nups, Western blotting analysis revealed that Nup62 was selectively incorporated in the virus (Fig. 3 E).
Figure 3.
HIV-1 disperses Nups into the cytoplasm and progeny virions. (A) Thin-section electron micrographs of NE cross sections from mock- or pNL4-3–transfected HeLa cells. (B) Immunogold labeling of Nups using mab414 antibody (10-nm gold labels; red arrowheads) in mock-transfected and in pNL4-3–expressing HeLa cells. (C) Immunogold labeling of Nups using the QE5 antibody (purple arrowheads). Nups associated with NEs in mock-transfected (91 ± 15.7%, SD) versus pNL4-3–transfected (32 ± 5.9%) cells; Nups within the cytoplasm in mock-transfected (8.9 ± 4.3%) versus pNL4-3–transfected (68 ± 8.5%) cells. (D) Immunogold labeling of budding virus for Gag (10- or 15-nm gold labels; white arrowheads), QE5 (10- or 15-nm gold labels; purple arrowheads), and mab414 (10-nm gold labels; red arrowheads). Yellow asterisks, NPC; PM, plasma membrane; NE, nuclear envelope. (E) Western analysis for Gag and Nups in cell lysates and purified viruses collected from mock (−)- and pNL4-3 (+)–transfected HeLa cells.
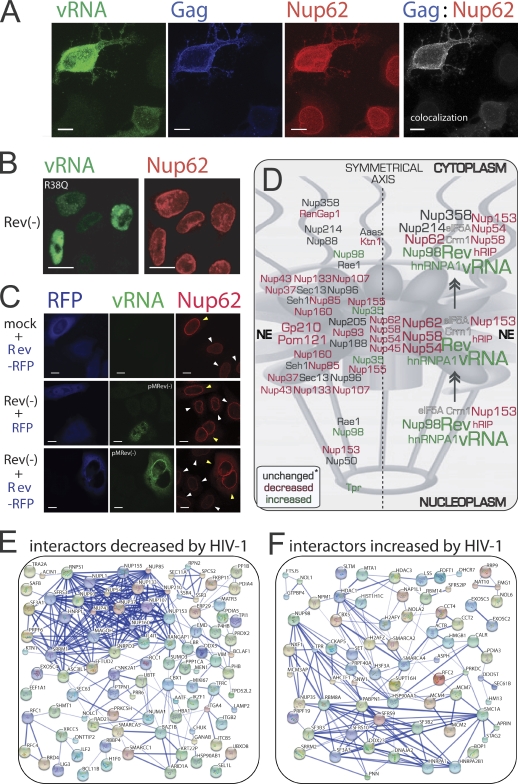
In untransfected cells, Nup62 localizes exclusively to NEs. However, HIV-1 induces the translocation of Nup62 to the cytoplasm and plasma membrane where it colocalizes with Gag, indicating that Nup62 may be a resident of the vRNA–Gag complex as it traffics intracellularly (Fig. 4 A). Our earlier work suggested a tight correlation between Rev gene expression and the cytoplasmic retention of hnRNP A1 (Monette et al., 2009). Therefore, to identify the viral gene responsible for altered Nup62 distribution, we tested a battery of pNL4-3 mutants, and only Rev-deficient mutants failed to translocate Nup62 to the cytoplasm (Fig. 4 B and not depicted). We next examined whether Rev alone could recapitulate these findings. The overexpression (OE) of Rev-RFP in mock-transfected cells was able to cause the thickened Nup62 staining at the NE but not its cytoplasmic localization (Fig. 4 C, top row). The OE of RFP empty vector control in Rev(−)-expressing cells did not affect Nup62 staining, nor did it induce vRNA nuclear export (Fig. 4 C, middle panel). However, the OE of Rev-RFP with Rev(−) rescued vRNA nuclear export and elicited both the thickened NE staining and cytoplasmic translocation of Nup62 (Fig. 4 C, bottom panel). These results suggest that Nup62 becomes engaged with the vRNA before nuclear export. Colocalization analyses revealed that Nup62 localizes with Rev, the vRNA, and Gag (Fig. 4 A and not depicted), suggesting that they may be part of the same cytoplasmic RNP and, thus, supporting a cytoplasmic role for Nup62 (Hubert et al., 2009).
Figure 4.
HIV-1 Rev–vRNA RNP nuclear export leads to the dispersal of Nup62. (A) Wild-type HIV-1 DNA–transfected HeLa cells were stained for vRNA, Gag, and Nup62. Nup62-Gag colocalization is shown in gray. (B) HeLa cells transfected with Rev(−) (R38Q) DNA and stained for vRNA and Nup62. 3D renditions of z stacks with deconvolution (A and B) are shown. (C) HeLa cells were transfected as indicated in the figure: RFP and Rev-RFP (blue), vRNA (green), and Nup62 (red). Yellow arrowheads identify cells expressing Rev-RFP (top and bottom panels) or RFP alone (middle panel), and white arrowheads identify untransfected cells. (D) Color-coded effects on Nups by HIV-1 infection, as determined by LC-MS/MS of the NE. A predictive model is shown on how the Rev–vRNA RNP may displace Nups during nuclear export. Asterisk, unchanged or not detected. (E and F) Protein interaction maps for LC-MS/MS hits that are decreased (E) or increased (F) by HIV-1 infection. Bars, 10 µm.
The observations that many Nups are decreased at NEs by HIV-1 infection and the reported interactions between Nups and the Rev–vRNA RNP (Hofmann et al., 2001) raise the possibility that this RNP may, during nuclear export, dislodge Nups essential for NPC integrity. This could lead to, among other things, the encapsidation of Nups in progeny virus (Fig. 4 D). Such a dismantling of the NPC could be possible because of the finding that two of the three anchoring Nups that we detected (i.e., Gp210 and Pom121) are decreased at the NE by HIV-1 infection. Gp210 was validated by Western blotting in this study (unpublished data) and has also been identified as an HIV-1 dependency factor (Kok et al., 2009).
The results presented here using both HIV-1–infected T cells and pNL4-3–transfected CD4− HeLa cells, along with the biochemical and physical evidence for virion-associated Nup62, indicate that NPC remodeling is induced by HIV-1 during the late replication stages. Further support for this comes from the observation that Nup358 is also decreased by HIV-1 (Fig. 4 D and Table I; Chan et al., 2007). Nup358 is required for the nuclear import of Rev, so it would be counterproductive to modulate NE levels early, resulting in a block in vRNA export (Hutten et al., 2009). Nup98, in contrast, is required for both Rev–vRNA RNP export and PIC import, and its increased representation at NEs by HIV-1 suggests a safeguard to guarantee its persistence at the NPC. Nup98 has also been shown to be up-regulated in expression by interferons during the course of vesicular stomatitis virus infection to release the block on nuclear mRNA export (Enninga et al., 2002).
Further support pointing to the late replication stages comes from the finding that newly synthesized Rev contributes to the resistance to superinfection by binding to and exporting Integrase (Levin et al., 2010), introducing the possibility that if Nup-interacting proteins are exported as a single RNP, they may together negatively influence NPC integrity.
Based on the prediction that the export of the HIV-1 RNP leads to the loss of Nups from the NPC, we investigated the possibility that other NE-associated proteins may also be affected as a result of extensive protein–protein interaction networks. The STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database v8.3–generated protein interaction maps show that 76 and 69% of hits up- or down-regulated by HIV-1 interact with others within these categories (Fig. 4, E and F). This further suggests that proteins from micro or macro RNPs may move together toward or away from NEs and that this movement may be greatly enhanced by infection. We performed literature searches to show those of our hits that interact with viral proteins or with the vRNA in case transient interactions with viral proteins have effects on the abundance at the NEs (Table S1). HIV-1–interacting proteins most affected by infection are CBX1, CSNK2A1, KKBP11, NUMA1, PPIB, RCC1, PP1CA, SNRPD3, and XRCC5, and others that interact with the Rev–vRNA RNP are DHX9, HNRNPs, HRIP1, ILF3, MATR3, NPM1, and SR proteins. We then compared our hits with proteins known to be encapsidated in case some have their RNP-joining origins at NEs (Table S2). We found that a small set of our hits were encapsidated and that those most decreased at NEs were EEF1A1, HIST1H4, HSP90AB1, HSPA1L, KPNB1, and VIM. Hoping to accelerate the discovery of central participants of HIV-1 replication, we further refined our findings by performing literature searches to define those of our hits that had known functions during HIV-1 replication and that inhibit HIV-1 or have modified expression levels as a result of infection (Table S2). One potential caveat to our approach has been the necessity to use the Jurkat T cell line instead of primary cells because of the large amount of starting material required for the isolation of NEs. In addition, though the majority of our protein hits were those that could be associated in some way with NEs or with the ER, others are known to have entirely different cellular roles and, therefore, may be contaminants arising from the isolation procedure. Therefore, the development of better strategies for the isolation of subcellular compartments for minimal loss of specimens and preservation of intact organelles will help advance the important field of organellar proteomics (Gauthier and Lazure, 2008).
Nup62 is essential for HIV-1 vRNA export
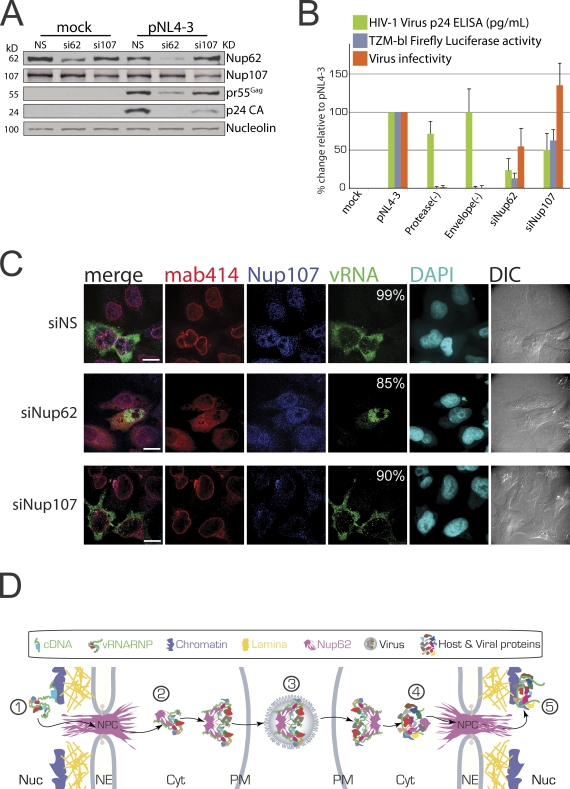
Our results provide evidence that the Rev–vRNA RNP disrupts NPCs, leading to the cytoplasmic translocation of Nup62 and its encapsidation into virions. To determine whether Nup62 is essential for HIV-1 gene expression and infectivity, we used siRNAs to perform siRNA-mediated knockdown (KD) on Nup62 in HeLa (CD4−) cells that were then transfected with mock or proviral DNA, and purified viruses produced by these cells were used to infect native HeLa–TZM-bl (CD4+) cells. Western analysis revealed that a >82% Nup62 KD led to a substantial decrease in pr55Gag and p24 CA, whereas the 60% Nup107 KD only modestly reduced p24 CA levels (Fig. 5 A). The depletion of either Nup decreased virus production (Nup62 KD, 23.7 ± 18.6%; Nup107 KD, 43.0 ± 25.6%; Fig. 5 B). Nup62 KD also decreased virus infectivity to 54.8 ± 25% of wild-type levels, whereas the Nup107 KD modestly enhanced infectivity to 134.3 ± 29%. It is important to add that, as shown in Figs. 2 A and 3 E, although HIV-1 infection lowers the abundance of both Nups at NEs, KD lowers total cellular levels. Additional cellular loss of Nup62 by HIV-1 may be a result of its encapsidation into viruses. The observation that Nup62 KD halves virus infectivity suggests that it has important roles in HIV-1 replication. Nup107 KD may increase virus infectivity by causing more Nup62 to be available or by limiting the number of NPCs, thereby causing a temporal shift that blocks further nuclear import and that accelerates viral expression and packaging.
Figure 5.
Nup62 is essential for vRNA nuclear export and virus infectivity. Mock- and HIV-1–transfected HeLa cells were depleted of Nup62 and Nup107 using siRNA. (A) Western analysis for Nup62, Nup107, pr55Gag, and Nucleolin (loading control). (B) Effects of Nup62 and Nup107 KDs on virus production, TZM-bl FLuc activity, and infectivity. Protease(−) and Envelope(−) virus were used as negative infectivity controls. Error bars represent SD. (C) IF/FISH analysis of Nup62, Nup107, vRNA, DAPI, and DIC. Percentages in the vRNA column represent the frequency observed for each condition, and n = 500 vRNA-positive cells in three independent experiments. Bars, 10 µm. (D) A predictive model on how viruses may exploit encapsidated Nup62. Integrated proviral DNA (green; 1) is transcribed into vRNA (squiggly green) that is then exported with Nup62 (purple) to the cytoplasm as an RNP (2). vRNA dimerizes and is encapsidated into budding virus (3) that can now infect new cells, in which Nup62 may promote efficient nuclear import of the PIC (4) and integration into the host DNA (5). Cyt, cytoplasm; NE, nuclear envelope; Nuc, nucleus; PM, plasma membrane.
We next examined the effects of Nup62 KD on vRNA localization using IF/FISH (Fig. 5 C). In the control, nonsilencing condition, we observed thickened staining for Nup107 at NEs as we do for Nup62 in HIV-1–expressing cells. Because HIV-1 does not cause a decrease in whole cell levels of Nup107, this staining likely represents Nup mislocalization at NPCs (Fig. 5 C, top row). The effects of the Nup62 KD are remarkable (Fig. 5 C, middle row). For one, the KD does not completely diminish Nup62 staining, which is suggested to be a result of preferential replenishment of an essential Nup62 pool (Fig. 5 C, middle row; Hubert et al., 2009). Even more striking is the fact that Nup62 KD causes the nuclear retention of the vRNA similar to what is observed in Rev(−) conditions (Fig. 4 C), providing an explanation for the dramatic reduction in Gag expression and viral production. Nup107 KD has no effect on vRNA localization but does cause the improper positioning of Nup62, which accumulates at distinct perinuclear regions (Fig. 5 C, bottom row). In addition, under Nup107 KD, HIV-1 does not cause the same Nup62 staining pattern as in nonsilencing conditions, indicating that only properly positioned Nup62 at NPCs may become cytoplasmically diffused as a result of HIV-1 infection. Altogether, these results indicate that Nup62 is essential for vRNA export and virus infectivity and suggest that its role during HIV-1 replication begins at the nucleus.
Though our data suggest that Nups take leave of the NPC by virtue of their affinity to the Rev–vRNA RNP, other alternatives are possible. For instance, NPC destabilization may result from the association of Nups and integrated proviral cDNA or nuclear vRNA during transcription (Capelson et al., 2010; Hou and Corces, 2010; Strambio-De-Castillia et al., 2010). HIV-1 may disrupt NPCs to evade antiviral responses, as seen in other viruses (Gustin and Sarnow, 2002; Fontoura et al., 2005; Bardina et al., 2009; Porter and Palmenberg, 2009; Atasheva et al., 2010; Ren et al., 2010). Another possibility is that encapsidated Nup62 may be required for efficient delivery of the PIC into the nuclei of newly infected cells (Fig. 5 D). Consistently, Transportin-SR2 (TRN-SR2/TNPO3), the importin found to be responsible for the nuclear import of the HIV-1 PIC, was found to bind exclusively to Nup62 (Lai et al., 2001; Christ et al., 2008). Lai et al. (2001) suggested that the import route used by TRN-SR2 is distinct from that taken by other importins and that Nup62 may function as the docking site for TRN-SR2–cargo complexes at NPCs.
In this study, we find that several Nups are decreased at NEs by HIV-1 infection and that at least one of them is essential for HIV-1 replication and virus infectivity. Because HIV-1 requires Nups for the nuclear entry of the PIC, the nuclear export of the vRNA RNP, and for virus infectivity, the prospect of targeting the interacting protein domains of Nups and viral components against HIV-1 propagation may be even more effective than is presently considered, and targeting the right Nup may bring the HIV-1 replication cycle to a halt.
Materials and methods
Cell culture and transfections
HeLa and HeLa–TZM-bl cells grown in DME supplemented with FBS (Invitrogen) or CD4+/CXCR4+ Jurkat CE6.1 T cells (American Type Culture Collection) were grown in RPMI 1640 media supplemented with 10% FBS (Invitrogen). Cells were transfected with 1 µg/ml plasmid DNA and/or 100 nM siRNA using Lipofectamine 2000 (Invitrogen). HeLa cells were collected 24 h later unless otherwise indicated. T cells were counted and reseeded at 300,000/ml every 3 d until peak infection at day 12 after transfection. For imaging studies, HeLa cells were seeded onto sterile coverslips, and, for T cells, sterile poly-l-lysine–coated (Sigma-Aldrich) coverslips were dropped into wells, and cells were allowed to settle onto these for 1 h before fixing with 4% PFA.
NE isolation
A recently published technique was used for NE isolation (Korfali et al., 2009). In brief, equal numbers of mock- and peak-infected T cells were collected, washed with PBS, incubated in hypotonic lysis buffer (10 mM Hepes, pH 7.4, 1.5 mM MgCl2, and 10 mM KCl) for 10 min, Dounce homogenized (40-ml capacity Wheaton type; VWR), and resuspended in 2.2 M SHKM (2.2 M sucrose, 50 mM Hepes, pH 7.4, 25 mM KCl, and 5 mM MgCl2). The lot was transferred to 38.5-ml ultracentrifuge tubes (Beckman Coulter), underlayed with 30% SHKM (0.9 M sucrose, 50 mM Hepes, pH 7.4, 25 mM KCl, and 5 mM MgCl2), and centrifuged at 4,000 rpm for 20 min at 4°C (SW28 Ti Rotor; Beckman Coulter). Pellets were resuspended in 1.8 M SHKM and were underlayed with 2.2 M SHKM for centrifugation at 25,000 rpm for 2.5 h at 4°C. Pellets were resuspended in 0.25 M SHKM (50 mM Hepes, pH 7.4, 25 mM KCl, and 5 mM MgCl2), treated with RNase A and DNase I (Invitrogen) for 15 min, and resuspended in 10% SHM (0.3 M sucrose, 10 mM Hepes, pH 7.4, and 2 mM MgCl2) for centrifugation at 5,000 rpm for 10 min at 4°C. Pellets were resuspended in 10% SHM, again treated with RNase A and DNase I for 15 min, underlayed with 30% SHM, and centrifuged at 5,000 rpm for 30 min at 4°C. Pellets were resuspended in 1 ml of 10% SHM, aliquoted, and centrifuged at 8,000 rpm for 10 min at 4°C, after which the supernatant was aspirated, and pellets were stored at −80°C until further processing. All solutions had 2 mM of fresh DTT and protease inhibitor (Roche) added to them. For salt extractions yielding higher numbers of Nups in mass spectrometry, pellets were resuspended in 0.25 M SHKM containing 1 M NaCl (Dreger et al., 2001; Schirmer and Gerace, 2005), incubated on a rotating platform for 30 min at 4°C, and centrifuged at 8,000 rpm for 10 min at 4°C, after which the supernatants were aspirated. For IF analyses of products from the NE isolations, 20 µl of material obtained at each step was dropped at the center of poly-d-lysine–treated coverslips, sandwiched by another nontreated coverslip, and incubated for 1 h at 4°C before fixation with 4% PFA. NE isolations were performed seven times, and IF experiments were performed four times.
Antibodies, plasmids, and siRNA sequences
The following antibodies were used in this study: α–p24 CA (a gift from M. Phelan, National Institutes of Health AIDS Reference and Reagent Program, Germantown, MD); cleaved α–Caspase-3 (a gift from J. Galipeau, McGill University, Montréal, Quebec, Canada); α–Lamin B1 (a gift from V. Blank, McGill University, Montréal, Quebec, Canada); α-RHA (a gift from J. Ortin, Universidad de Madrid, Madrid, Spain); α-Gp210 (a gift from D. Görlich and K. Paulsen, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany); α-GAPDH (Techni-Science, Inc.); α–ERGIC-53 and α-Nup62 (Sigma-Aldrich); α-Actin, α-eIF4E, α-eIF2, α–Transportin-1, α-Nup107, QE5, and mab414 (Abcam); α–CA p24 (ABT-Trinity Biotechnology); α-Nucleolin and α-PCNA (Santa Cruz Biotechnology, Inc.); α-Calnexin (Stressgen); α-L7 (Novus Biologicals); HRP conjugates (Rockland Immunochemicals); Alexa Fluor fluorophore conjugates and DAPI (Invitrogen); and gold conjugates (Ted Pella, Inc.). The plasmids used in this study were pcDNA3.1 (Invitrogen); pNL4-3 and pMRev(−) (National Institutes of Health AIDS Reference and Reagent Program); pNL4-3 Rev(−) (R38Q; a gift from D. Purcell, University of Melbourne, Melbourne, Australia); pNL4-3vif(−), nef(−), vpu(−), and vpr(−) (a gift from K. Strebel, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD); pro(−) (a gift from M. Laughrea, McGill University, Montréal, Quebec, Canada); env(–) (Yao et al., 1998); pNL4-XX (a gift from D. Ott, National Cancer Institute, National Institutes of Health, Frederick, MD); and RFP and Rev-RFP (provided by R. Brack-Werner, Helmholtz Zentrum München, Neuherberg, Germany). Nonsilencing control and siRNAs targeting Nup62 (5′-GCAACTGCTCCAACCTCAT-3′) and siNup107 (5′-CAAATACTTCTTTTGCAGC-3′) were purchased from QIAGEN.
Western, IF, FISH, and imaging analyses
For Western analysis, cells were collected at the indicated times after transfection, washed with PBS (Invitrogen), and lysed in NTEN buffer (100 mm NaCl, 10 mm Tris, pH 7.5, 1 mm EDTA, 0.5% Nonidet P-40 Protease inhibitor cocktail [Roche], and RNaseOUT [Invitrogen]). Cell lysates were quantified for protein content by the micro Bradford assay (Bio-Rad Laboratories), and equal quantities of protein were loaded into SDS-polyacrylamide gels. Proteins were then transferred onto nitrocellulose membranes (Pall), and blocked membranes were first incubated with primary antibodies of interest and then with secondary antibodies conjugated to HRP (Rockland Immunochemicals). The Western Lightning Chemiluminescence Reagent Plus kit (PerkinElmer) was used for protein detection via films (BioMax; Kodak).
IF and FISH analyses have been described previously (Bériault et al., 2004; Lévesque et al., 2006; Lehmann et al., 2009). In brief, cells on coverslips were fixed in 4% PFA in PBS for 20 min followed by permeabilization with 0.2% Triton X-100 for 10 min at the indicated times after transfection. All reagents used were treated with diethylpyrocarbonate (Sigma-Aldrich). For FISH analyses of the HIV-1 vRNA, coverslips were treated with DNase I (Invitrogen) for 30 min and washed. A digoxigenin-labeled RNA probe was synthesized via in vitro transcription from the plasmid pKS-Pol236nt in the presence of digoxigenin-labeled UTP from the RNA Labeling Mix (Roche) and was annealed to permeabilized cells overnight at 42°C. After hybridization, the vRNA was visualized by staining with a mouse antidigoxigenin antibody (Sigma-Aldrich) followed by staining with an Alexa Fluor fluorochrome-conjugated donkey IgG anti–mouse antibody (Invitrogen). For IF, coverslips were blocked with blocking reagent (Roche) in PBS before being incubated with primary antibodies against antigens of interest and then with secondary fluorophore-conjugated donkey IgG Alexa Fluor antibodies. Coverslips were stained with DAPI (Sigma-Aldrich), dried, and mounted onto slides using a mounting medium (Immu-mount; Thermo Fisher Scientific).
Images appearing in Figs. 1, 2, and 5 were acquired at RT using a microscope (DM16000B; Leica) equipped with a spinning disk confocal head (WaveFX; Quorum), a 63× (1.4 numerical aperture oil immersion) plan apochromat objective lens, and an EM charge-coupled device camera (ImageEM; Hamamatsu Photonics). Volocity Imaging software (v4.3.2; PerkinElmer) was the acquisition software used to acquire images, in which Alexa Fluor 633, 594, and 488 antibody fluorochromes and DAPI were scanned using excitation wavelengths of 646, 561, 491, and 405 nm, and emission spectra were filtered with 665–715-, 570–620-, 500–550-, and 435–485-nm bandpass filters, respectively and sequentially. Images appearing in Fig. 4 were acquired at RT using a laser-scanning confocal microscope (LSM 5 PASCAL; Carl Zeiss) equipped with a 63× (1.4 numerical aperture oil immersion) plan apochromat objective lens and an Ar/Kr laser. The acquisition software used was LMS Image Browser (Carl Zeiss), in which Alexa Fluor 633, 594, and 488 antibody fluorochromes were scanned using excitation wavelengths of 633, 543, and 488 nm, and emission spectra were filtered with 650-, 560–615-, and 505–530-nm long- and bandpass filters, respectively and sequentially. Images were recorded at a thickness of 1 µm (z stacks were taken at 0.2-µm intervals) and were digitized at a resolution of 1,024 × 1,024 pixels. AutoDeblur software (Autoquant; Media Cybernetics) was used for deconvolution of Fig. 4 (A and B). The LSM (Carl Zeiss) and Liff (Volocity; PerkinElmer) files were imported into Imaris software (v7; Bitplane) for pseudocoloring of channels, 3D reconstitutions of z stacks, colocalization analysis using Mander’s coefficient, and image export for figure montage in Illustrator (CS3; Adobe).
LC-MS/MS analysis
Coomassie blue–stained SDS-PAGE LC-MS/MS analysis was performed as previously described (Zulak et al., 2009). In brief, gel plugs were rinsed with HPLC-grade water with 25 mM ammonium bicarbonate in 50% (vol/vol) acetonitrile followed by dehydration using acetonitrile before lyophilization. These were rehydrated in 25 mM ammonium bicarbonate, pH 8.0, containing 12.5 ng/µl trypsin and incubated overnight at 37°C in 25 mM ammonium bicarbonate. Peptides were extracted by two rounds of incubation with 1% formic acid in 50% acetonitrile for 15 min. Pooled extracts were dried and reconstituted in mobile phase buffer A for LC. Digested peptides were analyzed using an integrated 1100 LC Ion Trap XCT Ultra system (Agilent Technologies) fitted with an integrated fluidic cartridge for peptide capture, separation, and nanospray (HPLC Chip; Agilent Technologies). Injected samples were trapped and desalted on a precolumn channel (40 nl Zorbax 300SB-C18) for 5 min with 3% acetonitrile in 0.2% formic acid delivered by an auxiliary pump at 4 µl/min. The peptides were then reverse eluted from the trapping column and separated on the analytical column (150 mm Zorbax 300SB-C18) at 0.3 µl/min. Peptides were eluted using a 5–70% (vol/vol) acetonitrile gradient in 0.2% (vol/vol) formic acid for >45 min. MS/MS spectra were collected by data-dependent acquisition, with parent ion scans of 8,100 Th/s over m/z 300–2,000 and MS/MS scans at the same rate over m/z 100–2,200. Peak list data were extracted from these files by DataAnalysis software for the 6300 series ion trap (v3.4, build 175; Agilent Technologies).
Data analysis and bioinformatics
Mascot v2.1 (Matrix Science) was used to probe the MS/MS data using 1.6-D precursor ion mass tolerance, 0.8-D fragment ion mass tolerance, one potential missed cleavage, and oxidized methionine as a variable modification as parameters. Major primary peptide sequences generated by Mascot were verified for protein identification using the National Center for Biotechnology Information basic local alignment search tool protein database, and protein hits were verified to match the expected molecular weights of proteins derived from excised bands of Coomassie blue–stained gels. The numerical emPAI values generated by the Mascot algorithm were used to normalize all datasets from several experiments for their pooling. The emPAI is generated from the number of the observable peptides to the number of experimentally observed peptide ions for a given protein (Shinoda et al., 2010). As the log of the emPAI provides linear relationships to protein amounts, providing a direct comparison of those highly and lowly expressed, we took the log of the HIV-1–infected emPAI divided by the mock-infected emPAI to generate numerical values designating how our individual protein hits were being affected by HIV-1 infection. Peptides generated for several proteins in both conditions were manually counted to ensure that final numerical values reflected the original raw data. The cut-off value for significant changes in protein abundance was set at an emPAI score of ±0.2, corresponding to a 1.5-fold change. UniProt accession numbers were gathered for the organization of hits according to GO terms, biological processes, and subcellular compartments with the help of the Software Tool for Rapid Annotation of Proteins (STRAP; Bhatia et al., 2009). Tables S1 and S2 were generated using PubMed, the National Center for Biotechnology Information, the HIV-1 Human Protein Interaction Database, and Google. Protein–protein interaction networks were generated by using the STRING v8.3 database.
Transmission EM
For thin-section transmission EM, HeLa cells were fixed with 2% glutaraldehyde for 1 h, fixed with 1% OsO4 in PBS for 1 h before progressive dehydration through ethanol series (30, 50, 70, 85, 95, and 100% for 20 min each) followed by 100% acetone for 20 min, and then embedded in Epon 812 (Fluka). Thin 70–90-nm sections were cut on an Ultramicrotome (Leica) using a diamond knife, mounted on Parlodion-and carbon-coated cooper grids, and stained with 2% uranyl acetate and 2% lead citrate.
For immunogold labeling, cells were fixed with 2% PFA followed by progressive dehydration through ethanol series (50, 70, and 100% for 20 min each), infiltration, and embedding in LR White (Ted Pella, Inc.), which was then polymerized by 365 nm UV light for 2.5 d. Thin sections, obtained as indicated in this section, were immunogold labeled at RT in humid chambers (Au et al., 2010). Grids containing EM sections were blocked (2% BSA, three times for 10 min) and labeled with primary antibody (diluted in 2% BSA in PBS and incubated for 1 h) and secondary antibody conjugated to gold particles (Ted Pella, Inc.). Finally, samples were fixed with 1% glutaraldehyde in PBS for 5 min and stained as indicated in this section.
Samples were examined using a transmission electron microscope (H7600; Hitachi) operated at 100 kV. Images were acquired digitally using a 1-megapixel AMT Advantage CCD camera (ORCA; Hamamatsu Photonics). Cells exhibiting virus particles at their plasma membranes were those used for assessment of NE structure and gold labeling from HIV-1 expression. Imaris software was used to measure particle distances from the inner nuclear membrane.
Virus purification
Supernatants were collected from mock- or pNL4-3–transfected HeLa and T cells and were clarified by centrifugation at 2,000 g for 10 min at 4°C followed by filtration (0.2 µm). These were then underlayed with a 20% wt/vol sucrose and centrifuged at 125,000 g for 1 h at 4°C. Virus pellets were resuspended in TNE buffer (10 mM Tris-HCl, pH 8.0, 100 mM NaCl, and 2 mM EDTA) and separated using SDS-PAGE.
Virus infectivity assay
CD4− HeLa cells were transfected with 100 nM siRNAs for 24 h and were then cotransfected with siRNAs and mock or pNL4-3, Pro(−), or Env(−) proviral DNAs. 48 h later, cells were collected for Western analysis, and supernatants were processed as described in the previous section before being added to growing TZM-bl CD4+ cells containing Tat-inducible Fluc reporter. At 24 h after infection (Kim et al., 1989), TZM-bl cells were lysed in passive lysis buffer for Fluc activity according to the manufacturer (Promega). Virus from supernatants was quantified using the HIV-1 p24 Capture ELISA kit (Advanced BioScience Laboratories, Inc.). Infectivity was determined by dividing Fluc activity by virus concentration.
Online supplemental material
Table S1 shows identified NE-associated proteins that interact with HIV-1 proteins or the vRNA, and Table S2 shows identified NE-associated proteins with known functions during HIV-1 replication. Table S3 shows all proteins identified by LC-MS/MS with associated emPAI values in the last column. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201008064/DC1.
Acknowledgments
We thank Ruth Brack-Werner, Dirk Görlich, Katharina Paulsen, Juan Ortin, Klaus Strebel, Damian Purcell, and Volker Blank for reagents and Mark Wainberg, David Ott, Morgan Khan, David Schreimer, Sue Germinario, and Valerie Doye for technical contributions and advice. We also thank Iain Mattaj, Daniel Legault-Coutu, and members of the lab for helpful discussions.
A. Monette is a recipient of an HIV-1/AIDS Doctoral Research Award from the Canadian Institutes of Health Research (CIHR). N. Panté is a Michael Smith Foundation for Health Research Senior Scholar, and A.J. Mouland is a Fraser, Monat, and McPherson McGill University Scholar. This work was supported by a Natural Sciences and Engineering Research Council of Canada grant to N. Panté and by a CIHR grant to A.J. Mouland.
Footnotes
Abbreviations used in this paper:
- DIC
- differential interference contrast
- emPAI
- exponentially modified protein abundance index
- hnRNP
- heterogeneous nuclear RNP
- IF
- immunofluorescence
- KD
- knockdown
- LC
- liquid chromatography
- MS/MS
- tandem mass spectrometry
- NE
- nuclear envelope
- NPC
- nuclear pore complex
- Nup
- nucleoporin
- OE
- overexpression
- PIC
- preintegration complex
- vRNA
- viral genomic RNA
References
- Atasheva S., Fish A., Fornerod M., Frolova E.I. 2010. Venezuelan equine Encephalitis virus capsid protein forms a tetrameric complex with CRM1 and importin alpha/beta that obstructs nuclear pore complex function. J. Virol. 84:4158–4171 10.1128/JVI.02554-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au S., Cohen S., Panté N. 2010. Microinjection of Xenopus laevis oocytes as a system for studying nuclear transport of viruses. Methods. 51:114–120 10.1016/j.ymeth.2010.02.001 [DOI] [PubMed] [Google Scholar]
- Bardina M.V., Lidsky P.V., Sheval E.V., Fominykh K.V., van Kuppeveld F.J., Polyakov V.Y., Agol V.I. 2009. Mengovirus-induced rearrangement of the nuclear pore complex: hijacking cellular phosphorylation machinery. J. Virol. 83:3150–3161 10.1128/JVI.01456-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bériault V., Clément J.F., Lévesque K., Lebel C., Yong X., Chabot B., Cohen E.A., Cochrane A.W., Rigby W.F., Mouland A.J. 2004. A late role for the association of hnRNP A2 with the HIV-1 hnRNP A2 response elements in genomic RNA, Gag, and Vpr localization. J. Biol. Chem. 279:44141–44153 10.1074/jbc.M404691200 [DOI] [PubMed] [Google Scholar]
- Bhatia V.N., Perlman D.H., Costello C.E., McComb M.E. 2009. Software tool for researching annotations of proteins: open-source protein annotation software with data visualization. Anal. Chem. 81:9819–9823 10.1021/ac901335x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S., Thibault P., Gagnon E., Kearney P., Bergeron J.J., Desjardins M. 2003. Organelle proteomics: looking at less to see more. Trends Cell Biol. 13:629–638 10.1016/j.tcb.2003.10.006 [DOI] [PubMed] [Google Scholar]
- Capelson M., Liang Y., Schulte R., Mair W., Wagner U., Hetzer M.W. 2010. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 140:372–383 10.1016/j.cell.2009.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.Y., Qian W.J., Diamond D.L., Liu T., Gritsenko M.A., Monroe M.E., Camp D.G., II, Smith R.D., Katze M.G. 2007. Quantitative analysis of human immunodeficiency virus type 1-infected CD4+ cell proteome: dysregulated cell cycle progression and nuclear transport coincide with robust virus production. J. Virol. 81:7571–7583 10.1128/JVI.00288-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan E.Y., Sutton J.N., Jacobs J.M., Bondarenko A., Smith R.D., Katze M.G. 2009. Dynamic host energetics and cytoskeletal proteomes in human immunodeficiency virus type 1-infected human primary CD4 cells: analysis by multiplexed label-free mass spectrometry. J. Virol. 83:9283–9295 10.1128/JVI.00814-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christ F., Thys W., De Rijck J., Gijsbers R., Albanese A., Arosio D., Emiliani S., Rain J.C., Benarous R., Cereseto A., Debyser Z. 2008. Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 18:1192–1202 10.1016/j.cub.2008.07.079 [DOI] [PubMed] [Google Scholar]
- Copeland A.M., Newcomb W.W., Brown J.C. 2009. Herpes simplex virus replication: roles of viral proteins and nucleoporins in capsid-nucleus attachment. J. Virol. 83:1660–1668 10.1128/JVI.01139-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger M., Bengtsson L., Schöneberg T., Otto H., Hucho F. 2001. Nuclear envelope proteomics: novel integral membrane proteins of the inner nuclear membrane. Proc. Natl. Acad. Sci. USA. 98:11943–11948 10.1073/pnas.211201898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebina H., Aoki J., Hatta S., Yoshida T., Koyanagi Y. 2004. Role of Nup98 in nuclear entry of human immunodeficiency virus type 1 cDNA. Microbes Infect. 6:715–724 10.1016/j.micinf.2004.04.002 [DOI] [PubMed] [Google Scholar]
- Enninga J., Levy D.E., Blobel G., Fontoura B.M. 2002. Role of nucleoporin induction in releasing an mRNA nuclear export block. Science. 295:1523–1525 10.1126/science.1067861 [DOI] [PubMed] [Google Scholar]
- Fontoura B.M., Faria P.A., Nussenzveig D.R. 2005. Viral interactions with the nuclear transport machinery: discovering and disrupting pathways. IUBMB Life. 57:65–72 10.1080/15216540500078608 [DOI] [PubMed] [Google Scholar]
- Gauthier D.J., Lazure C. 2008. Complementary methods to assist subcellular fractionation in organellar proteomics. Expert Rev. Proteomics. 5:603–617 10.1586/14789450.5.4.603 [DOI] [PubMed] [Google Scholar]
- Gilchrist A., Au C.E., Hiding J., Bell A.W., Fernandez-Rodriguez J., Lesimple S., Nagaya H., Roy L., Gosline S.J., Hallett M., et al. 2006. Quantitative proteomics analysis of the secretory pathway. Cell. 127:1265–1281 10.1016/j.cell.2006.10.036 [DOI] [PubMed] [Google Scholar]
- Giri M.S., Nebozhyn M., Showe L., Montaner L.J. 2006. Microarray data on gene modulation by HIV-1 in immune cells: 2000-2006. J. Leukoc. Biol. 80:1031–1043 10.1189/jlb.0306157 [DOI] [PubMed] [Google Scholar]
- Goff S.P. 2008. Knockdown screens to knockout HIV-1. Cell. 135:417–420 10.1016/j.cell.2008.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K.E. 2003. Inhibition of nucleo-cytoplasmic trafficking by RNA viruses: targeting the nuclear pore complex. Virus Res. 95:35–44 10.1016/S0168-1702(03)00165-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K.E., Sarnow P. 2001. Effects of poliovirus infection on nucleo-cytoplasmic trafficking and nuclear pore complex composition. EMBO J. 20:240–249 10.1093/emboj/20.1.240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin K.E., Sarnow P. 2002. Inhibition of nuclear import and alteration of nuclear pore complex composition by rhinovirus. J. Virol. 76:8787–8796 10.1128/JVI.76.17.8787-8796.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann W., Reichart B., Ewald A., Müller E., Schmitt I., Stauber R.H., Lottspeich F., Jockusch B.M., Scheer U., Hauber J., Dabauvalle M.C. 2001. Cofactor requirements for nuclear export of Rev response element (RRE)- and constitutive transport element (CTE)-containing retroviral RNAs. An unexpected role for actin. J. Cell Biol. 152:895–910 10.1083/jcb.152.5.895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou C., Corces V.G. 2010. Nups take leave of the nuclear envelope to regulate transcription. Cell. 140:306–308 10.1016/j.cell.2010.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert T., Vandekerckhove J., Gettemans J. 2009. Exo70-mediated recruitment of nucleoporin Nup62 at the leading edge of migrating cells is required for cell migration. Traffic. 10:1257–1271 10.1111/j.1600-0854.2009.00940.x [DOI] [PubMed] [Google Scholar]
- Hutten S., Kehlenbach R.H. 2006. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol. Cell. Biol. 26:6772–6785 10.1128/MCB.00342-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutten S., Wälde S., Spillner C., Hauber J., Kehlenbach R.H. 2009. The nuclear pore component Nup358 promotes transportin-dependent nuclear import. J. Cell Sci. 122:1100–1110 10.1242/jcs.040154 [DOI] [PubMed] [Google Scholar]
- Kim S.Y., Byrn R., Groopman J., Baltimore D. 1989. Temporal aspects of DNA and RNA synthesis during human immunodeficiency virus infection: evidence for differential gene expression. J. Virol. 63:3708–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss A., Li L., Gettemeier T., Venkatesh L.K. 2003. Functional analysis of the interaction of the human immunodeficiency virus type 1 Rev nuclear export signal with its cofactors. Virology. 314:591–600 10.1016/S0042-6822(03)00531-2 [DOI] [PubMed] [Google Scholar]
- Kok K.H., Lei T., Jin D.Y. 2009. siRNA and shRNA screens advance key understanding of host factors required for HIV-1 replication. Retrovirology. 6:78 10.1186/1742-4690-6-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R., Zhou Y., Elleder D., Diamond T.L., Bonamy G.M., Irelan J.T., Chiang C.Y., Tu B.P., De Jesus P.D., Lilley C.E., et al. 2008. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 135:49–60 10.1016/j.cell.2008.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- König R., Stertz S., Zhou Y., Inoue A., Hoffmann H.H., Bhattacharyya S., Alamares J.G., Tscherne D.M., Ortigoza M.B., Liang Y., et al. 2010. Human host factors required for influenza virus replication. Nature. 463:813–817 10.1038/nature08699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfali N., Fairley E.A., Swanson S.K., Florens L., Schirmer E.C. 2009. Use of sequential chemical extractions to purify nuclear membrane proteins for proteomics identification. Methods Mol. Biol. 528:201–225 10.1007/978-1-60327-310-7_15 [DOI] [PubMed] [Google Scholar]
- Lai M.C., Lin R.I., Tarn W.Y. 2001. Transportin-SR2 mediates nuclear import of phosphorylated SR proteins. Proc. Natl. Acad. Sci. USA. 98:10154–10159 10.1073/pnas.181354098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M., Milev M.P., Abrahamyan L., Yao X.J., Pante N., Mouland A.J. 2009. Intracellular transport of human immunodeficiency virus type 1 genomic RNA and viral production are dependent on dynein motor function and late endosome positioning. J. Biol. Chem. 284:14572–14585 10.1074/jbc.M808531200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Rouzic E., Mousnier A., Rustum C., Stutz F., Hallberg E., Dargemont C., Benichou S. 2002. Docking of HIV-1 Vpr to the nuclear envelope is mediated by the interaction with the nucleoporin hCG1. J. Biol. Chem. 277:45091–45098 10.1074/jbc.M207439200 [DOI] [PubMed] [Google Scholar]
- Lévesque K., Halvorsen M., Abrahamyan L., Chatel-Chaix L., Poupon V., Gordon H., DesGroseillers L., Gatignol A., Mouland A.J. 2006. Trafficking of HIV-1 RNA is mediated by heterogeneous nuclear ribonucleoprotein A2 expression and impacts on viral assembly. Traffic. 7:1177–1193 10.1111/j.1600-0854.2006.00461.x [DOI] [PubMed] [Google Scholar]
- Levin A., Hayouka Z., Friedler A., Brack-Werner R., Volsky D.J., Loyter A. 2010. A novel role for the viral Rev protein in promoting resistance to superinfection by human immunodeficiency virus type 1. J. Gen. Virol. 91:1503–1513 10.1099/vir.0.019760-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Smith A.J., Schacker T.W., Carlis J.V., Duan L., Reilly C.S., Haase A.T. 2009. Microarray analysis of lymphatic tissue reveals stage-specific, gene expression signatures in HIV-1 infection. J. Immunol. 183:1975–1982 10.4049/jimmunol.0803222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monette A., Ajamian L., López-Lastra M., Mouland A.J. 2009. Human immunodeficiency virus type 1 (HIV-1) induces the cytoplasmic retention of heterogeneous nuclear ribonucleoprotein A1 by disrupting nuclear import: implications for HIV-1 gene expression. J. Biol. Chem. 284:31350–31362 10.1074/jbc.M109.048736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday D., Emmott E., Surtees R., Lardeau C.H., Wu W., Duprex W.P., Dove B.K., Barr J.N., Hiscox J.A. 2010. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol. Cell. Proteomics. 9:2438–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter F.W., Palmenberg A.C. 2009. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. J. Virol. 83:1941–1951 10.1128/JVI.01752-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed S., Yan J.S., Hussain A., Lai B. 2009. Proteomic characterization of HIV-modulated membrane receptors, kinases and signaling proteins involved in novel angiogenic pathways. J. Transl. Med. 7:75 10.1186/1479-5876-7-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y., Seo H.S., Blobel G., Hoelz A. 2010. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc. Natl. Acad. Sci. USA. 107:10406–10411 10.1073/pnas.1005389107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose J.H., Jeeninga R.E., Berkhout B., Speijer D. 2008. Proteomic studies reveal coordinated changes in T-cell expression patterns upon infection with human immunodeficiency virus type 1. J. Virol. 82:4320–4330 10.1128/JVI.01819-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer E.C., Gerace L. 2005. The nuclear membrane proteome: extending the envelope. Trends Biochem. Sci. 30:551–558 10.1016/j.tibs.2005.08.003 [DOI] [PubMed] [Google Scholar]
- Schmitz A., Schwarz A., Foss M., Zhou L., Rabe B., Hoellenriegel J., Stoeber M., Panté N., Kann M. 2010. Nucleoporin 153 arrests the nuclear import of hepatitis B virus capsids in the nuclear basket. PLoS Pathog. 6:e1000741 10.1371/journal.ppat.1000741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda K., Tomita M., Ishihama Y. 2010. emPAI Calc—for the estimation of protein abundance from large-scale identification data by liquid chromatography-tandem mass spectrometry. Bioinformatics. 26:576–577 10.1093/bioinformatics/btp700 [DOI] [PubMed] [Google Scholar]
- Strambio-De-Castillia C., Niepel M., Rout M.P. 2010. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat. Rev. Mol. Cell Biol. 11:490–501 10.1038/nrm2928 [DOI] [PubMed] [Google Scholar]
- Trotman L.C., Mosberger N., Fornerod M., Stidwill R.P., Greber U.F. 2001. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 3:1092–1100 10.1038/ncb1201-1092 [DOI] [PubMed] [Google Scholar]
- Wente S.R., Rout M.P. 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb. Perspect. Biol. 2:a000562 10.1101/cshperspect.a000562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward C.L., Prakobwanakit S., Mosessian S., Chow S.A. 2009. Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. J. Virol. 83:6522–6533 10.1128/JVI.02061-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.J., Mouland A.J., Subbramanian R.A., Forget J., Rougeau N., Bergeron D., Cohen E.A. 1998. Vpr stimulates viral expression and induces cell killing in human immunodeficiency virus type 1-infected dividing Jurkat T cells. J. Virol. 72:4686–4693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolotukhin A.S., Felber B.K. 1999. Nucleoporins nup98 and nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J. Virol. 73:120–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zulak K.G., Khan M.F., Alcantara J., Schriemer D.C., Facchini P.J. 2009. Plant defense responses in opium poppy cell cultures revealed by liquid chromatography-tandem mass spectrometry proteomics. Mol. Cell. Proteomics. 8:86–98 10.1074/mcp.M800211-MCP200 [DOI] [PubMed] [Google Scholar]
- Yeung M.L., Houzet L., Yedavalli V.S., Jeang K.T. 2009. A genome-wide short hairpin RNA screening of jurkat T-cells for human proteins contributing to productive HIV-1 replication. J. Biol. Chem. 284:19463–19473 10.1074/jbc.M109.010033 [DOI] [PMC free article] [PubMed] [Google Scholar]