Plk1-dependent modification of centrioles early in mitosis is necessary for accurate centriole duplication and segregation.
Abstract
Centrioles are self-reproducing organelles that form the core structure of centrosomes or microtubule-organizing centers (MTOCs). However, whether duplication and MTOC organization reflect innate activities of centrioles or activities acquired conditionally is unclear. In this paper, we show that newly formed full-length centrioles had no inherent capacity to duplicate or to organize pericentriolar material (PCM) but acquired both after mitosis through a Plk1-dependent modification that occurred in early mitosis. Modified centrioles initiated PCM recruitment in G1 and segregated equally in mitosis through association with spindle poles. Conversely, unmodified centrioles segregated randomly unless passively tethered to modified centrioles. Strikingly, duplication occurred only in centrioles that were both modified and disengaged, whereas unmodified centrioles, engaged or not, were “infertile,” indicating that engagement specifically blocks modified centrioles from reduplication. These two requirements, centriole modification and disengagement, fully exclude unlimited duplication in one cell cycle. We thus uncovered a Plk1-dependent mechanism whereby duplication and segregation are coupled to maintain centriole homeostasis.
Introduction
The centrosome, which is comprised of one or two centrioles and the surrounding pericentriolar material (PCM), is the major microtubule-organizing center (MTOC) and is essential for the assembly of cilia in animal cells. The number of centrosomes or centrioles is stably maintained in cycling cells, in part through strict regulation of centriole biogenesis. Centriole formation requires a group of assembly factors (Nigg and Raff, 2009), which are able to drive centriole formation either independent of preexisting centrioles, through the so-called de novo assembly pathway (Khodjakov et al., 2002; Peel et al., 2007; Rodrigues-Martins et al., 2007), or dependent on preexisting centrioles, to promote localized assembly or duplication (Cizmecioglu et al., 2010; Dzhindzhev et al., 2010; Hatch et al., 2010). Centriole-dependent centriole duplication promotes the assembly of new centrioles exactly once per cell cycle and serves as the dominant pathway in proliferating cells. De novo assembly, which often generates highly variable numbers of centrioles, is normally suppressed in cycling cells (Khodjakov et al., 2002; La Terra et al., 2005; Tsou and Stearns, 2006a).
The centriole duplication cycle in animal cells follows a stereotypical program. Cells begin G1 phase with two centrioles that were mother and daughter centrioles in the previous cell cycle. Each of these two preexisting centrioles duplicates in the following S phase. During duplication, a new daughter centriole grows from the lateral surface of each mother centriole, reaches full length in early mitosis (Vorobjev and Chentsov, 1982), and remains engaged to its mother, exhibiting an orthogonal configuration, until disengagement occurs in late mitosis (Kuriyama and Borisy, 1981a). A combination of centriole engagement-dependent block and low concentrations of centriole assembly factors ensures that a mother centriole supports the formation of only one daughter centriole during interphase (Wong and Stearns, 2003; Tsou and Stearns, 2006b; Strnad et al., 2007; Loncarek et al., 2008; Cunha-Ferreira et al., 2009; Rogers et al., 2009; Tsou et al., 2009). However, it is not fully understood how a daughter centriole is prevented from producing its own daughter centriole (granddaughter) in the same cell cycle. For example, overexpression of centriole assembly factors can overcome the centriole engagement block, resulting in the formation of extra daughter centrioles, but this does not lead to the formation of granddaughter centrioles (Kleylein-Sohn et al., 2007). This suggests that a mechanism aside from centriole engagement prevents the duplication of the daughter. In the Drosophila melanogaster wing disc, cells that have lost Cdk1 activity produce centrioles with abnormal configurations, including the formation of granddaughters, but the underlining cause is not clear (Vidwans et al., 2003). After centriole duplication, cells enter mitosis with two centrosomes, each of which contains two centrioles (a mother and a daughter). The physiological importance, if any, of having two centrioles per mitotic centrosome is unclear.
To maintain a constant number of centrioles in proliferating cells, not only centriole biogenesis during interphase but also centriole segregation in mitosis must be precisely regulated. In wild-type cells, these two processes occur perfectly; centrioles duplicate exactly once in S phase and segregate equally through their association with spindle poles during mitosis. Centrioles are able to recruit PCM that nucleates microtubules from the beginning of the cell cycle in G1 (Piel et al., 2000). Such activity increases dramatically around G2/M phase in preparation for organizing mitotic spindles, a process called centrosome maturation (Snyder and McIntosh, 1975; Telzer and Rosenbaum, 1979; Kuriyama and Borisy, 1981b; Palazzo et al., 2000). Interestingly, evidences have clearly shown that centrosomes are not absolutely required for spindle assembly and cell division (Debec and Abbadie, 1989; Heald et al., 1996; Khodjakov et al., 2000; La Terra et al., 2005; Basto et al., 2006). This is consistent with the idea, proposed more than 30 yr ago, that centrosomes associate with spindle poles to facilitate segregation of centrioles during cell division (Pickett-Heaps, 1975). Following on this idea, it seems that PCM recruitment can be thought as an accessory activity acquired by centrioles in dividing cells to ensure correct segregation. Analyses of centriole cycles by EM in vertebrate cells have demonstrated that in mitosis, mother and daughter centrioles behave very differently in their ability to associate with PCM (Rieder and Borisy, 1982; Vorobjev and Chentsov, 1982). Most of mother centrioles are fully embedded within PCM (covered by an electron-dense halo), whereas full-length daughter centrioles leave a large portion of their distal ends uncovered and have only their proximal ends embedded in PCM (Rieder and Borisy, 1982; Vorobjev and Chentsov, 1982). This suggests that either the ability of centrioles in recruiting PCM is regulated in cell cycle–dependent manners (Rieder and Borisy, 1982; Vorobjev and Chentsov, 1982), or the PCM recruitment at daughter centrioles is somehow reduced by centriole engagement. In any case, the exact timing of the centriole to centrosome transition, its molecular requirement, and the impact of this transition on centriole biogenesis and maintenance remain unclear.
We demonstrate here that full-length centrioles formed in interphase can neither duplicate nor organize PCM regardless of their age, configuration, or how they are formed. Instead, these centrioles are modified early in mitosis through a Plk1-dependent activity and, thereby, acquire competence to recruit PCM at the end of mitosis. We show that only modified centrioles, which are competent to organize MTOC, can duplicate in S phase and segregate in the following mitosis through association with spindle poles. In contrast, unmodified centrioles can do neither, regardless of whether they are in an engaged or disengaged state, and thus, must associate with MTOC-competent (modified) centrioles for correct segregation. Our results thus reveal a novel Plk1-dependent mechanism whereby only centrioles that can segregate themselves are allowed to duplicate. This regulation leads to a coupling between centriole duplication and segregation, the two determining factors for centriole homeostasis in cycling cells.
Results
Engaged daughter centrioles have no contribution to PCM recruitment
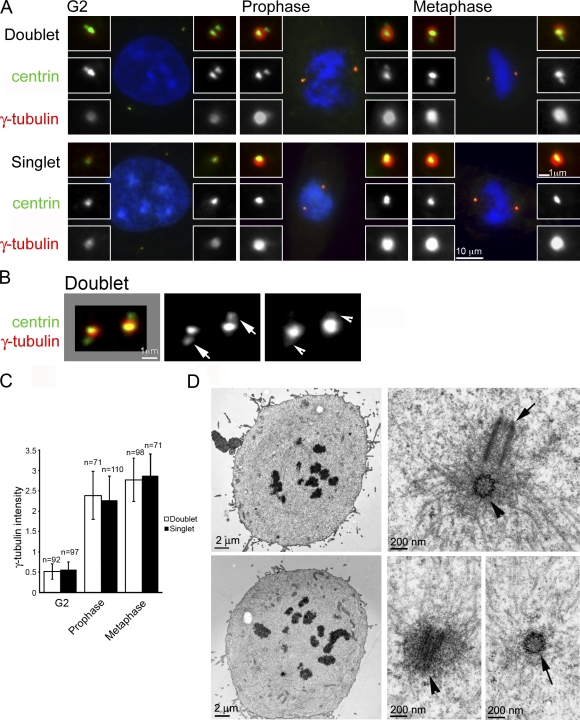
Previous EM analysis showed that daughter centrioles in mitotic centrosomes are not fully embedded within PCM, unlike their mother centrioles to which they are engaged (Rieder and Borisy, 1982; Vorobjev and Chentsov, 1982). To determine whether daughter centrioles play a role in PCM recruitment, the centriole duplication factor hSas-6 was first depleted using RNAi (Dammermann et al., 2004; Leidel et al., 2005). Under this condition, cells in late G2 or mitosis contain centrosomes with a single mother centriole (Fig. 1 A). PCM size in these centrosomes was measured by quantifying γ-tubulin immunoreactivity and compared with that of wild-type centrosomes at the same cell cycle stages. Strikingly, from G2 to early mitosis (metaphase), there was no difference in the amount of γ-tubulin in these two types of centrosomes (Fig. 1, A and C). No difference was also observed for pericentrin, another major PCM component (Fig. S1 A). Overexpression of Plk4, on the other hand, promotes the formation of supernumerary (five to seven) daughter centrioles that still engage to a mother centriole (Bettencourt-Dias et al., 2005; Habedanck et al., 2005), described hereafter as “centriole rosettes” (Fig. S1 B). Similarly, the amount of PCM recruited to these centriole rosettes was similar to that acquired by wild-type centriole doublets (Fig. S1 B), confirming that engaged daughter centrioles do not contribute to PCM recruitment at these stages of the cell cycle.
Figure 1.
Daughter centrioles do not contribute to PCM recruitment. (A) Untreated (doublets) or hSas-6–depleted (singlets) RPE1 cells stably expressing centrin::GFP at indicated cell cycle stages were stained with antibodies against γ-tubulin. G2 cells were obtained by treatment with the Cdk1 inhibitor RO-3306, which arrested cells at G2/M. Mitotic cells were enriched by RO-3306 washout and identified by DAPI staining. Insets show a higher magnification of centrosomes. (B) Higher magnification of a pair of centrosomes from a G2/M cell extracted with Pipes buffer before fixation and stained for centrin and γ-tubulin. Arrowheads indicate weak centriolar γ-tubulin associated with daughter centrioles (arrows). (C) Quantification of γ-tubulin signals associated with centrosomes in different cell cycle stages. Numbers of centrosomes are indicated. Error bars indicate standard deviations. (D) Electron micrographs of mitotic cells. We obtained random sections of >20 mitotic centrosomes. Two representatives are shown here, one pair of centrioles from each cell. Mother (arrowheads) and daughter (arrows) centrioles are shown in both cross and longitudinal sections. Note that most of the microtubules and electron-dense material associate with mother centrioles.
To investigate the relationship between PCM and daughter centrioles, centrosomes were examined with immunofluorescence localization of γ-tubulin. Daughter centrioles were mostly located outside of the major γ-tubulin focus (Fig. 1 B, arrows), though a weak signal associated with them was detected after removal of cytoplasmic γ-tubulin to reduce the background (see Materials and Methods; Fig. 1 B, arrowheads). Because γ-tubulin is also present in the core of centrioles (Fuller et al., 1995), these weak signals likely reflect centriolar γ-tubulin (see the following results for Fig. S4). Furthermore, reexamination of mitotic centrosomes with serial sectioning and EM confirmed previous observations (Rieder and Borisy, 1982; Vorobjev and Chentsov, 1982), in which the majority of microtubules and PCM (electron-dense material) associated with mother centrioles (Fig. 1 D and Fig. S2, arrowheads). These results together indicate that full-length daughter centrioles, when engage to their mothers, serve little or no function in constructing mitotic centrosomes.
Disengaged daughter centrioles lack PCM-organizing activity
Daughter centrioles are engaged to mother centrioles during centrosome maturation (Kuriyama and Borisy, 1981a; Tsou and Stearns, 2006b), with their proximal ends covered by PCM. To test the possibility that the physical association of daughters with their mothers inhibits their ability to recruit PCM, freestanding or de novo–formed centrioles were induced in S phase by conditionally expressing a more stable form of Plk4 (Plk4ΔSCF; Cunha-Ferreira et al., 2009; Rogers et al., 2009; Holland et al., 2010; Sillibourne et al., 2010) and allowed to proceed to G2/M (Fig. 2, A and B, arrows). Such de novo centrioles contained centriolar markers (Fig. 2 A) and were full length and freestanding as judged by correlative light EM (Fig. S3). Plk4ΔSCF expression also generates centriole rosettes (Fig. 2, A and B, arrowheads; and Fig. S3), each of which contains a mother centriole. Strikingly, whereas mother centrioles were associated with large amounts of PCM-associated γ-tubulin (Fig. 2 A, arrowheads), de novo centrioles contained only a minimal signal detectable after removal of cytoplasmic γ-tubulin (Fig. 2 A, arrows), similar to that of engaged daughter centrioles (see Materials and methods; Fig. 1 B). This minimal γ-tubulin signal detected in de novo centrioles was insensitive to cell cycle changes (Fig. S4), in contrast to the centrosomal γ-tubulin whose amount fluctuated during cell cycle (Fig. S4). This is consistent with the notion that such a minimal signal reflects centriolar γ-tubulin (Fuller et al., 1995). More importantly, microtubule regrowth assays revealed that de novo centrioles were unable to nucleate microtubules (Fig. 2 B). The inability of de novo centrioles to organize microtubule arrays in interphase has been reported previously (La Terra et al., 2005). We conclude that full-length daughter centrioles are unable to recruit PCM or act as MTOCs regardless of whether they are engaged to a mother centriole or are freestanding.
Figure 2.
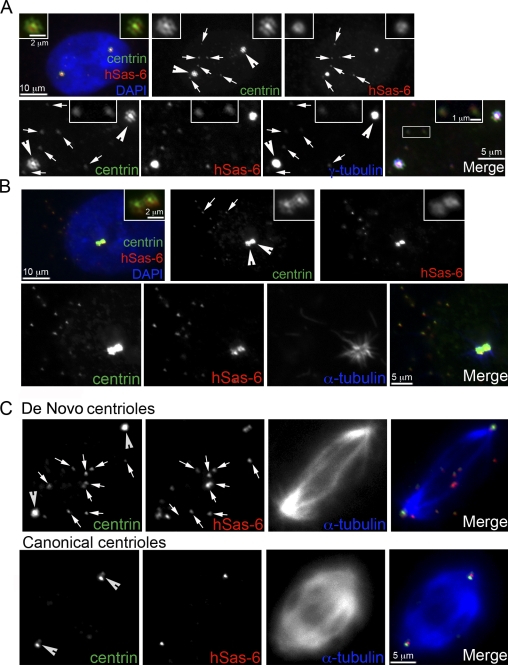
Equal segregation of daughter centrioles depends on mother centrioles. (A) Freestanding de novo centrioles, like engaged daughter centrioles, do not actively recruit PCM. Centriole rosettes (arrowheads) and de novo centrioles (arrows) were induced in RPE1 cells transiently expressing a more stable form of Plk4 (Plk4ΔSCF; the recognition motif of the SCF ubiquitin ligase is abolished; Cunha-Ferreira et al., 2009; Rogers et al., 2009; Holland et al., 2010; Sillibourne et al., 2010) during S phase and released into G2 (see Materials and methods). Before fixation, cells were extracted with Pipes buffer, and centrioles were visualized with centrin::GFP and antibodies against hSas-6 and γ-tubulin. Note that two major γ-tubulin foci formed around the mother centrioles located at the center of each centriole rosette, whereas de novo centrioles had only faint centriolar γ-tubulin labeling (arrows and top insets). Two de novo centrioles were magnified for better visualization (bottom insets). (B) Centriole rosettes (arrowheads) and de novo centrioles (arrows) in RPE1 cells were analyzed by a microtubule regrowth assay. Cells were incubated in 0°C medium for 30 min, transferred to 37°C medium for 1 min, and then fixed immediately. Centrioles were visualized with centrin::GFP and antibodies against hSas-6. Note that microtubule asters labeled with antibodies against α-tubulin formed only at centriole rosettes in which mother centrioles recruited large amounts of γ-tubulin. Insets show a higher magnification of centrosomes. (C) Canonical or de novo centrioles induced in S phase were allowed to enter mitosis. Centrioles were marked with GFP::centrin and antibodies against hSas-6, and spindles were labeled with antibodies against α-tubulin. Note that de novo centrioles (arrows) are scattered around the spindle, whereas engaged daughter centrioles and their mothers (arrowheads) occupy the spindle poles to allow proper segregation.
Equal segregation of daughter centrioles depends on mother centrioles
Given that newly formed centrioles do not contribute to PCM recruitment, it is unclear whether they play any role in mitosis. To explore this, the fate of freestanding de novo centrioles was examined in mitosis. De novo centrioles remained unable to nucleate microtubules (Fig. 2 C) and scattered around the spindle (Fig. 2 C), resulting in random segregation during cell division. In contrast, engaged daughter centrioles, although not able to organize PCM themselves (Fig. 1 D), were carried passively by mother centrioles (Fig. 2 C) and segregated equally. Random segregation of centrioles that resulted from a lack of PCM has also been observed in vivo (Basto et al., 2008). These results indicate that correct segregation of centrioles relies on the ability of centrioles to recruit PCM and localize to spindle poles (Pickett-Heaps, 1975). Furthermore, it suggests that the attachment between daughter and mother centrioles in mitosis is not to enhance centrosome function as an MTOC but rather to ensure the correct segregation of daughter centrioles themselves during cell division.
Localization of hSas-6 and C-Nap1 differentiates MTOC-competent from MTOC-noncompetent centrioles
Cycling cells begin in G1 with two centrioles that, despite their differences in age and ability to anchor interphase microtubules, are both capable of recruiting PCM and nucleating microtubules (Piel et al., 2000). Indeed, wild-type G1 cells exiting from mitosis inherit two centrioles (a mother and a daughter) that recruit similar amounts of γ-tubulin (Fig. 3 A, left), indicating that an unknown modification enables active PCM recruitment at the end of mitosis around previously inactive daughter centrioles to establish a functional centrosome. We hereafter use “modified” or “unmodified” to describe centrioles that have or have not passed mitosis and, therefore, are MTOC competent or MTOC noncompetent, respectively, in their ability to recruit PCM and nucleate microtubules.
Figure 3.
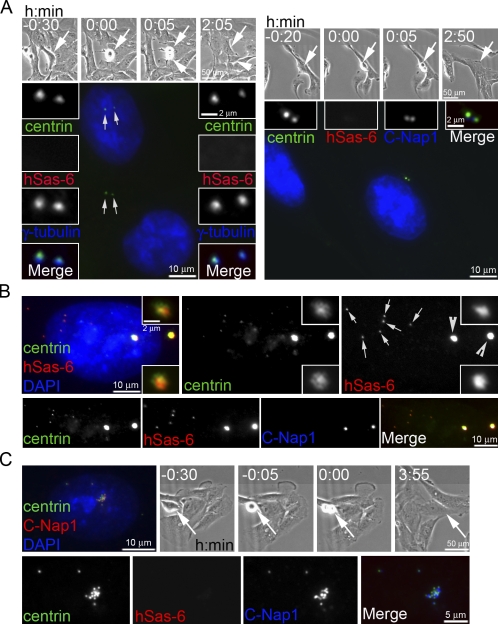
Localization of hSas-6 and C-Nap1 differentiates MTOC-competent from noncompetent centrioles. (A) RPE1 cells going through cell division and exiting mitosis were recorded using time-lapse microscopy. The daughter cells (marked by arrows and arrowheads in phase images) were located and analyzed for γ-tubulin recruitment at the two inherited centrioles (arrows) that were previously mother and daughter centrioles. Note that all centrioles recruited similar amounts of γ-tubulin (left), indicating that daughter centrioles had converted to motherlike centrioles that were active in recruiting PCM. Centrioles in these early G1 cells were also examined for hSas-6 and C-Nap1 localization (right), which negatively and positively correlate, respectively, with modified centrioles that recruit PCM. (B) De novo centrioles (arrows) and centriole rosettes (arrowheads) induced in RPE1 cells as described in Fig. 2 were analyzed for hSas-6 and C-Nap1 localization. All unmodified centrioles, freestanding or engaged, were labeled with hSas-6 but lacked C-Nap1, a reverse pattern to that of modified centrioles shown (top). (A and B) Insets show a higher magnification of centrosomes. (C) RPE1 cells induced to form de novo centrioles and centriole rosettes during interphase were traced by time-lapse microscopy and allowed to pass through mitosis. After division, centrioles in one of the daughter cells (arrows) were analyzed for hSas-6 and C-Nap1 localization. Note that all centrioles displayed a pattern for modified centrioles (strong C-Nap1 and no hSas-6).
To further investigate the centriole to MTOC conversion, we examined known centrosomal proteins for their ability to differentiate modified from unmodified centrioles. Interestingly, the localizations of two proteins, hSas-6 and C-Nap1, correlate tightly with the ability of centrioles to recruit PCM (Fig. 3). All daughter centrioles, either freestanding (de novo) or engaged, were marked with the daughter centriole–specific protein hSas-6 (Fig. 3 B) but not with C-Nap1 (Fig. 3 B, bottom), a protein that tethers two centrosomes in somatic cells (Mayor et al., 2000). Strikingly, this localization pattern was reversed (hSas-6 negative and C-Nap1 positive) once centrioles passed through mitosis and become MTOC competent (Fig. 3 C). Centriole doublets exiting mitosis display the same pattern (Fig. 3 A). Because of this all or none localization pattern, C-Nap1 and hSas-6 in addition to γ-tubulin recruitment were used to differentiate MTOC-competent from MTOC-noncompetent centrioles.
Plk1 is required for centriole modification leading to MTOC conversion
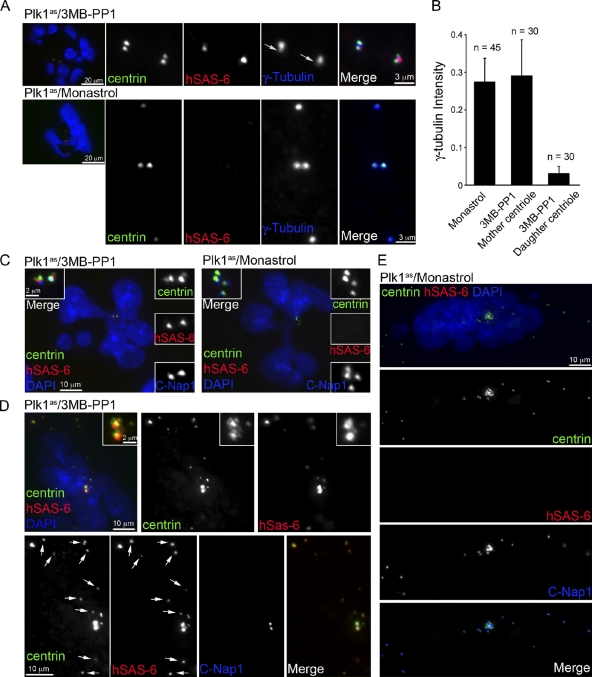
The temporal correlation between centriole disengagement (Tsou and Stearns, 2006b; Tsou et al., 2009) and centriole to MTOC conversion in late mitosis prompted us to examine whether the two processes are molecularly coupled. We previously showed that early mitotic Plk1 is essential for centriole disengagement, whereas, during anaphase, separase plays a supporting but nonessential role in human cells in promoting timely disengagement during mitotic exit (Tsou et al., 2009). To examine whether Plk1 activity is also required for daughter centriole modification leading to PCM recruitment, Plk1 (or Eg5 as the control) was rapidly inactivated in G2/M phase, and the cells were released from mitotic arrest and entered G1 phase by Cdk1 inhibition as described previously (Tsou et al., 2009). Similar results were obtained through inhibition of the aurora kinase with ZM447439 (unpublished data), which triggers mitotic exit by inactivating the spindle assembly checkpoint (Ditchfield et al., 2003). Cells that undergo this process fail centriole disengagement (Fig. 4 A; Tsou et al., 2009) and display donut-shaped, multilobed, or multiple small nuclei (Fig. 4 A; Tsou et al., 2009), differentiating them from unaffected cells. Strikingly, in Plk1-inhibited cells in which centrioles remained engaged, the γ-tubulin signal of mother centrioles was no different from that of centrioles in control (Eg5 inhibited) cells (Fig. 4, A and B) but was seven- to eightfold brighter than that of their engaged daughters (Fig. 4, A and B). Furthermore, all engaged daughter centrioles were labeled with hSas-6 and not with C-Nap1 (Fig. 4 C), a pattern associated with unmodified centrioles. In contrast, engaged mother centrioles in Plk1-inhibited cells and all disengaged centrioles in control cells displayed the pattern of modified centrioles (C-Nap1 positive and hSas-6 negative; Fig. 4 C). These results indicate that early mitotic Plk1 activity is essential for the modification of daughter centrioles, which promotes MTOC conversion later in mitosis. Conversely, inhibition of Plk1 has no effect on PCM recruitment on mother centrioles that have already been modified.
Figure 4.
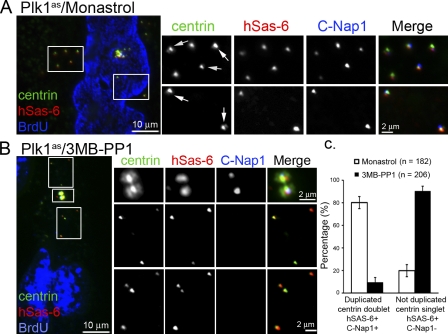
Plk1 is required for centriole to MTOC conversion. RPE1 cells in which the endogenous Plk1 gene has been replaced with an analogue-sensitive allele (Plk1as) that can be inhibited by bulky purine analogues (Burkard et al., 2007) were used in these experiments. (A) Plk1as cells were treated with the purine analogue 3MB-PP1 (10 µM) or the Eg5 inhibitor monastrol (50 µM) as a control during late G2. Note that Plk1 or Eg5 inactivation in late G2 or prophase activates the spindle assembly checkpoint and arrests cells in prometaphase (Burkard et al., 2007; Tsou et al., 2009). To allow analysis of centriole to MTOC transition in G1, cells were induced to exit mitosis using the Cdk1-selective inhibitor RO-3306 for 3 h as shown previously (Vassilev, 2006; Tsou et al., 2009). Under these conditions, cells displayed multilobed nuclei, and each cell inherits four centrioles. Cells with multilobed nuclei were examined for the centrosomal proteins indicated. Although monastrol-treated cells had four disengaged and modified centrioles that recruited similar amounts of γ-tubulin, Plk1-inhibited cells received two pairs of engaged centrioles within which daughter centrioles remained unmodified and only had minimal centriolar γ-tubulin labeling (arrows). (B) Quantification of γ-tubulin signals associated with centrioles, including centrioles in control cells (monastrol), mother centrioles in Plk1-inhibited cells (3MB-PP1 mother), and daughter centrioles in Plk1-inhibited cells (3MB-PP1 daughter). Numbers of centrioles are indicated. Error bars indicate standard deviations. (C) Centrioles in cells treated as in A were analyzed for hSas-6 and C-Nap1 localization. C-Nap1 labeled only modified centrioles, including mother centrioles of 3MB-PP1–treated cells and all centrioles of monastrol-treated cells. hSas-6 only labeled unmodified centrioles, the daughter centrioles of 3MB-PP1–treated cells. Insets show a higher magnification of centrosomes. (D and E) De novo–formed centrioles induced in Plk1as cells transiently expressing Plk4ΔSCF were allowed to pass through mitosis under Plk1 (3MB-PP1) or Eg5 (monastrol) inhibition and analyzed for hSas-6 and C-Nap1 localization. In 3MB-PP1–treated cells, centriole disengagement failed as the two centriole rosettes remained (insets). Only the two mother centrioles at the center of each rosette had C-Nap1 labeling (modified), and other centrioles (arrows) were labeled with hSas-6 (unmodified). In control cells (monastrol), all centrioles were labeled with C-Nap1 but lacked hSas-6 labeling, a pattern of modified centrioles. Therefore, de novo–formed freestanding centrioles behave identically to engaged daughter centrioles.
To examine whether centriole disengagement is required for centriole to MTOC conversion, cells containing de novo centrioles and centriole rosettes (Fig. 2) were allowed to enter mitosis either with Plk1 inhibition or with Eg5 inactivation as a control (Tsou et al., 2009). Every centriole in control cells became modified after exiting from mitosis, showing strong C-Nap1, but no hSas-6, labeling (Fig. 4 E). Most importantly, all these modified centrioles were equally active and acquired similar amounts of γ-tubulin regardless of their age and how they were formed (Fig. S5 A, arrows indicate old mother centrioles marked by centriolin). In contrast, when Plk1 was inhibited, all freestanding centrioles exiting from mitosis remained as unmodified singlets (C-Nap1 negative and hSas-6 positive; Fig. 4 D, arrows) and could not organize PCM or nucleate microtubules, even when these centrioles had progressed into late interphase for >16 h (Fig. S5, B and C). These results indicate that the failure of centriole to MTOC conversion upon Plk1 inhibition is not a result of the defect in centriole disengagement; rather, centriole to MTOC conversion is upstream or in parallel to centriole disengagement.
MTOC-noncompetent centrioles are unable to duplicate
Centriole duplication normally occurs only to disengaged centrioles (Tsou and Stearns, 2006b; Loncarek et al., 2008; Tsou et al., 2009). Because centriole disengagement and centriole to MTOC conversion are coupled through Plk1, it is not clear whether lack of MTOC conversion alone affects centriole duplication. To address this question, G1 cells containing modified or unmodified de novo centrioles that were generated by allowing cells to pass through mitosis with or without Plk1 inhibition, respectively (Fig. 4, D and E), were allowed to enter late S phase as judged by BrdU labeling (Fig. 5; Tsou et al., 2009). Centriole duplication in BrdU-labeled cells was determined by immunolocalization of centrin, C-Nap1, and hSas-6. As expected, modified freestanding centrioles fully duplicated (Fig. 5, A and C), as seen by centrin-marked centriole doublets, C-Nap1 labeled the mother centriole, and hSas-6 staining identified the newly formed daughter centrioles (Fig. 5 A). In contrast, unmodified de novo centrioles remained as free singlets (Fig. 5, B and C), indicating that centrioles lacking Plk1-dependent modification are unable to duplicate even when they are disengaged. The requirement of such a modification for duplication explains how a daughter centriole is prevented from producing its own daughter centriole (granddaughter) in the same cell cycle.
Figure 5.
MTOC-noncompetent centrioles are unable to support duplication. (A and B) Plk1as cells induced to form de novo centrioles were treated with 3MB-PP1 or monastrol in late G2 for 3 h, treated with RO-3306 for 2 h to cause mitotic exit, incubated for 10 h to allow S-phase entry, and pulse labeled with BrdU for 1 h followed by a 4-h chase. BrdU-positive cells containing multilobed nuclei were identified, and their freestanding centrioles were analyzed for duplication using the centriolar markers centrin (centrin::GFP), hSas-6, and C-Nap1. A duplicated centriole pair is defined as a centrin doublet that is hSas-6 positive (marking newly formed daughter centrioles) and C-Nap1 positive (marking mother centrioles). Note that although centrin labels all centrioles, at some viewing angles, immature daughter centrioles containing small amounts of centrin may be blocked by mother centrioles and, therefore, not visible. In control cells (monastrol treated), most centriole pairs were labeled with hSas-6 and C-Nap1 (arrows), although a few of them were viewed as single centrin foci. Nevertheless, because all these centrioles lost hSas-6 labeling in early G1 (Fig. 4 E), the regaining of hSas-6 signal in S phase indicates that they had initiated duplication. Conversely, in 3MB-PP1–treated cells, almost all of the freestanding centrioles are centrin singlets. These centriole singlets have hSas-6 labeling and lack C-Nap1 labeling, indicating that unmodified centrioles are unable to support duplication. Centriole rosettes in 3MB-PP1–treated cells containing C-Nap1–labeled mother centrioles are shown in the insets. (C) Quantification of centriole duplication after down-regulation of Plk1 or Eg5. Numbers of centrioles are indicated. Error bars indicate standard deviations.
MTOC-competent centrioles, when disengaged, can duplicate normally without Plk1
An alternative interpretation of the aforementioned result is that inhibition of mitotic Plk1 disrupts centriole duplication, regardless of whether centrioles are MTOC competent or noncompetent. To address this, freestanding mother centrioles (MTOC competent) generated in hSas6-depleted cells (as shown in Fig. 1 A) were allowed to pass through mitosis with Plk1 inhibition (Fig. 6). These mother centrioles not only recruited PCM normally as seen in Fig. 4 B (not depicted) but, most importantly, supported the assembly of new centrioles in the following S phase when an RNAi-resistant form of hSas-6 was conditionally expressed (Fig. 6). We conclude that MTOC-competent centrioles, when disengaged, can support duplication irrespective of whether Plk1 is present or not. Conversely, MTOC-noncompetent centrioles cannot do so, even when disengaged, unless modified by Plk1 (Fig. 5). Together, centriole biogenesis in cycling cells is under a two-step control: a centriole is modified first in mitosis to activate its self-reproduction, after which centriole engagement blocks further self-reproduction (Tsou et al., 2009), producing exactly one new (unmodified) centriole that is itself not competent to duplicate (Fig. 5).
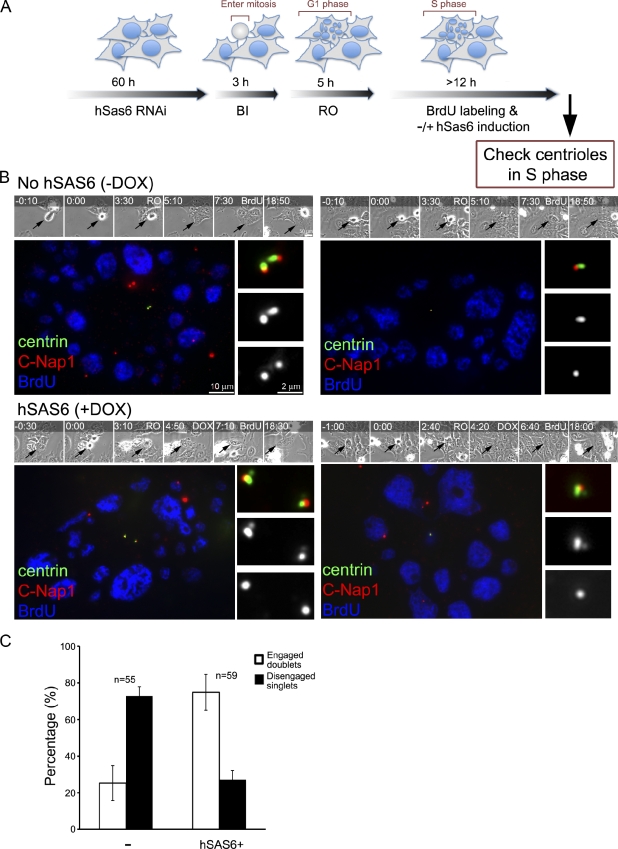
Figure 6.
MTOC-competent centrioles, when disengaged, can duplicate normally without Plk1. (A) Experimental scheme. To generate freestanding mother centrioles, asynchronously proliferating cells stably transfected with constructs that direct the expression of an RNAi-resistant form of hSas-6 (hSas-6R) from a tetracycline-inducible promoter were depleted of endogenous hSas-6 by RNAi. The hSas-6–depleted cells were filmed by time-lapse microscopy and then treated with a Plk1 inhibitor (BI-2536; BI) during mitotic entry followed by a Cdk1 inhibitor (RO-3306; RO), which induce mitotic exit. 5 h after mitotic exit, cells were allowed to progress to S/G2 phase (marked by BrdU labeling) for another 12 h, during which cells were either maintained as hSas-6 depleted or treated with doxycycline (DOX) to induce hSas-6R expression. (B) Representing images of cells in S/G2 phase had gone through the experimental scheme described in Fig. 3 A and been treated with (+) or without (–) doxycycline. Manipulated cells (arrows) were filmed (times are given at the top in hours and minutes) and identified by time-lapse microscopy. Cells were stained with anti-GFP (centrin::GFP) and anti–C-Nap1 antibodies. Freestanding mother centrioles (or singlets) exhibit a 1:1 ratio of centrin and C-Nap1 foci, whereas duplicated centriole pairs (or doublets) display a 2:1 ratio. Our knockdown experiments blocked centriole duplication in >70% of cells that had gone through mitosis during the recording period. In these cells, either one or two mother centriole singlets were left (singlets; see C), depending on when the RNAi had worked in each cell, either one or two cell cycles before. The remaining ∼25% of cells were unaffected and still had two pairs of engaged centrioles (doublets; not depicted; see C). Note that when the expression of hSas-6R was turned on (DOX+), most of centriole singlets were able to duplicate in S phase (BrdU) and became doublets. (C) Quantification of centriole configuration and duplication for these S/G2 cells. Error bars indicate standard deviations from three independent experiments. The minus sign indicates lack of doxycycline treatment (no hSAS6).
Discussion
Our results reveal that a Plk1-dependent modification, which occurs in early mitosis, is required to convert centrioles to centrosomes/MTOCs at late mitosis (Fig. 7). Without such modifications, centrioles cannot organize PCM regardless of their age, configuration, or how they are formed (canonical or de novo assembly). Importantly, only modified centrioles, which are competent to organize MTOC, can duplicate in S phase and segregate in the following mitosis through association with spindle poles. Newly formed daughter centrioles (unmodified) have no ability to duplicate from S to M phase, which excludes the assembly of granddaughter centrioles in the same interphase (Fig. 7) and explains why, in noncycling cells, centrioles are produced primarily through de novo assembly rather than rounds of duplication (Dirksen, 1991; Hagiwara et al., 2004). Importantly, unmodified centrioles must associate with MTOC-competent centrioles if they are to be segregated properly during mitosis (Fig. 7). This regulation leads to a tight coupling between centriole duplication and segregation, i.e., only centrioles that can segregate themselves are allowed to duplicate, thus maintaining centriole homeostasis in cycling cells.
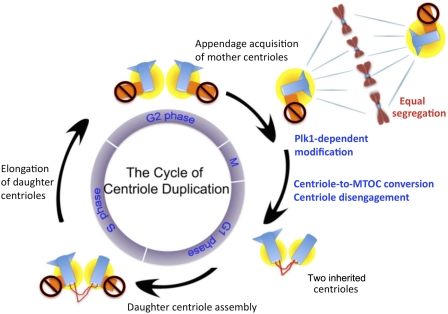
Figure 7.
Centriole duplication and segregation cycle. A Plk1-dependent modification during G2/M is required to produce MTOC-competent centrioles during late mitosis and early G1 (modified centrioles [blue] and surrounded by PCM [yellow]). Only modified centrioles can duplicate in the following S phase, in which the capacity of the duplication is limited by centriole engagement to form one daughter centriole per mother centriole. Newly formed daughter centrioles can neither duplicate nor recruit PCM (orange and marked by prohibited signs), which prevents the assembly of their own daughter. These unmodified centrioles segregate equally during cell division by tethering to MTOC-competent centrioles that are capable of associating with spindle poles.
We show that centriole duplication in cycling cells generates hybrid centriole pairs consisting of one modified and one unmodified centriole. This inherent asymmetry may be linked to the acquisition of appendages during G2/M, which selectively occurs at mother centrioles (Dawe et al., 2007), and could possibly provide a mechanism by which duplicated centrioles (hybrids) are differentiated from unduplicated centrioles (no asymmetry). One speculation is that unmodified centrioles may carry unknown inhibitory activities that locally suppress duplication. This short-range feedback inhibition model would explain how centriole engagement, through which daughter centrioles bring such inhibitory activities close to mother centrioles, blocks the ability of the mother centriole to duplicate. This is consistent with our observations that unmodified (daughter) centrioles not only cannot duplicate (Fig. 5) but also suppress the duplication potential of otherwise active mother centrioles (Fig. 6) with which they associate tightly via centriole engagement (Tsou et al., 2009). In this sense, Plk1-dependent modification would be expected to remove the inhibitory activity from daughter centrioles, which then allows mother centrioles to duplicate again. This is consistent with the fact that ectopic expression of active Plk1 in S/G2-arrested cells induces centriole reduplication (Loncarek et al., 2010).
The centriole to MTOC conversion described here involves Plk1-dependent activities in early mitosis that convert full-length centrioles to MTOCs at late mitosis. A recent discovery described that ectopic Plk1 activity is involved in some forms of procentriole maturation that accompany procentriole elongation during a prolonged S-phase arrest in transformed cells in which centriole reduplication occurs (Loncarek et al., 2010). In these S/G2 phase–arrested systems, the ectopic Plk1 activity is required for centriole reduplication (Loncarek et al., 2010), which is consistent with the essential role of Plk1 in centriole disengagement and duplication licensing (Tsou et al., 2009). The nature of procentriole maturation in these arrested systems has not been clearly defined (Loncarek et al., 2010). We noted that in the Xenopus laevis egg extracts, neither centriole disengagement nor MTOC formation seems to be dependent on procentriole elongation (Tsou and Stearns, 2006b), as procentrioles purified from S phase–arrested HeLa cells, despite being small in size, could efficiently disengage from their mothers and organize active MTOCs within 20 min of anaphase entry (Tsou and Stearns, 2006b). Similarly, in hPoc5-depleted cells in which centrioles fail to reach full length, both MTOC formation and centriole duplication can occur, suggesting that the length of centrioles does not play a critical role in either of the two processes. In light of our current experiments, we suggest that procentriole maturation previously described in these arrested cells (Loncarek et al., 2010) may constitute, in part, the conversion of centrioles to MTOC, which normally occurs on full-length centrioles in wild-type cells but can also occur prematurely and leads to centriole reduplication in systems where Plk1 is ectopically activated.
The mitotic requirement for centriole to centrosome conversion suggests that centrosomes are normally made only in dividing cells, in which centriole segregation relies on mitotic spindles. For noncycling cells or organisms that do not use spindle poles to segregate centrioles, the centriole to MTOC conversion would be unnecessary or might not even exist. It will be interesting to explore this prediction in unicellular eukaryotes, such as paramecia and trypanosome, in which centrioles are transmitted through cortical inheritance or cytotaxis during cell division (Sonneborn, 1964; Beisson and Sonneborn, 1965; Ng and Frankel, 1977; Moreira-Leite et al., 2001; Feldman et al., 2007; Beisson, 2008; González-Robles et al., 2009), i.e., in these organisms, the machinery that converts centrioles to centrosomes would be absent (Moreira-Leite et al., 2001).
The cell cycle timing of centriole to MTOC conversion at late mitosis is critical. Premature conversion before mitosis may trigger the assembly of granddaughter centrioles within the same cell cycle (Balczon et al., 1995). This restriction on the ability of daughter centrioles to organize PCM between their formation during S phase and segregation in mitosis can provide an explanation for why cells need to begin the cell cycle with not one but two MTOC-competent centrioles, namely to avoid monopolar spindle formation during mitosis. In addition, in vertebrates, the strict separation of the two processes, centriole assembly in interphase and conversion in mitosis, could allow noncycling cells, such as the ciliated epithelium, to produce centrioles in interphase that function solely as basal bodies for cilia formation (Marshall, 2008). The requirement for such a separation may underlie the cell cycle timing of centriole to MTOC conversion.
Materials and methods
Cell culture, drug treatments, and time-lapse microscopy
Human telomerase-immortalized retinal pigment epithelial cells were cultured in DME/F-12 (1:1) medium supplemented with 10% FBS and 1% penicillin-streptomycin. For drug treatments, the following compounds and concentrations were used: 10 µM 3MB-PP1, 200 nM BI-2536, 50 µM monastrol, and 10 µM RO-3306. The Plk1as cell (RPE1, retinal pigment epithelial human cells) was obtained from P. Jallepalli (Memorial Sloan-Kettering Cancer Center, New York, NY; Burkard et al., 2007). For correlative time-lapse experiments, cells were grown on gridded coverslips and imaged on a microscope (Axiovert; Carl Zeiss) configured with a 10× phase objective, motorized temperature-controlled stage, environmental chamber, and CO2 enrichment system (Carl Zeiss). Image acquisition and processing were performed using Axiovision software (Carl Zeiss). 60 fields of cells were filmed with 2 × 2 binning during each experiment.
RNAi and expression of RNAi-resistant hSas-6
A lentivirus-based small hairpin RNA set of five clones that target hSas-6 was obtained from Thermo Fisher Scientific (RHS4533). Viruses derived from three of these clones were mixed and used to infect cells. The RNAi-resistant construct (hSas-6R) was made by introducing nucleotides changes in the targeted regions without changing the corresponding amino acids using site-directed mutagenesis (QuickChange; Agilent Technologies). The following three pairs of mutagenic primers were used: (1) 5′-CAGAGAGATGGAACATTGGGGGCATTACATAC-3′ and 5′-GTATGTAATGCCCCCAATGTTCCATCTCTCTG-3′, (2) 5′-GAAAATCAGCTAGTAAGGAAACAGGATGTATTGGGCTAC-3′ and 5′-GTAGCCCAATACATCCTGTTTCCTTACTAGCTGATTTTC-3′, and (3) 5′-CTAGATGATGCTACAAAACAGCTTGACTTTACACGAAAG-3′ and 5′-CTTTCGTGTAAAGTCAAGCTGTTTTGTAGCATCATCTAG-3′. To generate an inducible expression system, stable clones of RPE1 and HeLa cells expressing hSas-6R from the tetracycline-inducible promoter were obtained through in vivo gene delivery using the lentiviral vector pLVX-Tight-Puro (Takara Bio Inc.).
The induction of de novo centrioles
The full-length cDNA of human Plk4 was obtained from OriGene, Inc. The Plk4 mutant resistant to the SCF ubiquitin ligase (Plk4ΔSCF) was made by replacing S285 and T289 with alanine using site-directed mutagenesis (QuickChange). To generate an inducible expression system, stable clones of wild-type RPE1 cells and Plk1as cells expressing either Plk4ΔSCF or wild-type Plk4 from the tetracycline-inducible promoter were obtained through in vivo gene delivery using the lentiviral vector pLVX-Tight-Puro. De novo centrioles were induced in S phase by treating cells with 2 mM thymidine or 2 µg/ml aphidicolin and 1 µg/ml doxycycline for 12–16 h. Cells were then released to G2 or mitosis by removing both drugs.
Antibodies
A rabbit polyclonal antibody against human C-Nap1 was produced as previously described (Mayor et al., 2000) and used at a 1:500 dilution. Other antibodies used in this study include mouse anti–α-tubulin (1:1,000; Sigma-Aldrich), rat anti-BrdU (1:500; Novus Biologicals), rabbit anticentriolin (1:1,000; a gift from G. Fang, Genentech, South San Francisco, CA), rabbit anti–human pericentrin (Lüders et al., 2005), mouse anti–γ-tubulin (1:1,000; Santa Cruz Biotechnology, Inc.), and mouse anti–hSas-6 (1:500; Santa Cruz Biotechnology, Inc.).
Immunofluorescence microscopy
Cells were fixed with methanol at −20°C and blocked with 3% bovine serum albumin (wt/vol) and 0.1% Triton X-100 in PBS for 30 min. DNA was visualized using DAPI (Invitrogen). For centrosomal staining, cells were treated with Pipes buffer, pH 6.8, containing 0.1% Triton X-100 for 1 min before the methanol fixation. For visualizing replicated DNA, 20 µM BrdU was added to the cells as a 1-h pulse. After staining for centrosomal antigens, cells were fixed again with −20°C methanol for 10 min and then treated with 2 N HCl for 30 min at room temperature followed by BrdU staining with anti-BrdU antibodies. Fluorescent images were acquired on an upright microscope (Axio imager; Carl Zeiss) equipped with 100× oil objectives, NA of 1.45, a camera (ORCA ER; Hamamatsu Photonics), and a computer loaded with image-processing software (Axiovision). Individual images were cropped and assembled into figures using Photoshop (CS2; Adobe). For γ-tubulin quantification, all cells were treated the same during the process of immunocytochemistry and image acquisition. The images were analyzed using ImageJ software (National Institutes of Health). The minimum pixel value displayed was increased until only the centrosome was labeled, thus defining the PCM-associated γ-tubulin; the same setting was applied to all images. The selection tool was then used to mark the γ-tubulin foci, and the total pixel value of the marked region was measured.
EM
Mitotic cells grown on coverslips made of Aclar film (Electron Microscopy Sciences) were fixed in 2.5% glutaraldehyde and 0.5% tannic acid in 0.1 M sodium cacodylate buffer overnight, postfixed in 1% OsO4 in sodium cacodylate buffer for 1 h, dehydrated in graded series of ethanol, infiltrated with EMbed 812 resin (Electron Microcopy Sciences), and embedded in the resin. Serial sections (∼90-nm thickness) were cut on a microtome (Ultracut UC6; Leica) and stained with 2% uranyl acetate as well as 1% lead citrate. Samples were examined on a microscope (Tecnai Spirit G2; FEI), and electron micrographs were captured with the digital imaging system (UltraScan 4000; Gatan, Inc.) and the associated software (Digital Micrograph 3.9.0.; Gatan, Inc.). For correlative light EM, cells were traced by phase-contrast microscopy (10× phase on an Axiovert) on gridded coverslips made of Aclar film, permeabilized in Pipes buffer, pH 6.8, containing 0.1% Triton X-100, stained with anticentrin and hSas-6 antibodies as described in the previous paragraph, and then fixed in modified Karnovsky’s fixative (Murphy et al., 2000) consisting of 4% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer. After the acquisition of fluorescent images of centrosomes, cells were maintained on coverslips and further processed for EM as described in the previous paragraph.
Online supplemental material
Fig. S1 shows that daughter centrioles do not contribute to PCM recruitment. Fig. S2 shows serial section transmission EM of mitotic centrosomes. Fig. S3 shows analyses of de novo–formed centrioles and centriole rosettes by the correlative light and transmission EM. Fig. S4 shows that de novo–formed daughter centrioles contain minimal γ-tubulin signals that are insensitive to cell cycle changes. Fig. S5 shows that Plk1 is required for the conversion of de novo–formed daughter centrioles to MTOCs. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201101109/DC1.
Acknowledgments
We thank P. Jallepalli for sharing the Plk1as cell line.
This work was supported by the National Institutes of Health grant GM088253 to M.-F. B. Tsou.
Footnotes
Abbreviations used in this paper:
- MTOC
- microtubule-organizing center
- PCM
- pericentriolar material
References
- Balczon R., Bao L., Zimmer W.E., Brown K., Zinkowski R.P., Brinkley B.R. 1995. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol. 130:105–115 10.1083/jcb.130.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W. 2006. Flies without centrioles. Cell. 125:1375–1386 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J.W. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell. 133:1032–1042 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson J. 2008. Preformed cell structure and cell heredity. Prion. 2:1–8 10.4161/pri.2.1.5063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisson J., Sonneborn T.M. 1965. Cytoplasmic inheritance of the organization of the cell cortex in Paramecium aurelia. Proc. Natl. Acad. Sci. USA. 53:275–282 10.1073/pnas.53.2.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M., Rodrigues-Martins A., Carpenter L., Riparbelli M., Lehmann L., Gatt M.K., Carmo N., Balloux F., Callaini G., Glover D.M. 2005. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 15:2199–2207 10.1016/j.cub.2005.11.042 [DOI] [PubMed] [Google Scholar]
- Burkard M.E., Randall C.L., Larochelle S., Zhang C., Shokat K.M., Fisher R.P., Jallepalli P.V. 2007. Chemical genetics reveals the requirement for Polo-like kinase 1 activity in positioning RhoA and triggering cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 104:4383–4388 10.1073/pnas.0701140104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cizmecioglu O., Arnold M., Bahtz R., Settele F., Ehret L., Haselmann-Weiss U., Antony C., Hoffmann I. 2010. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 191:731–739 10.1083/jcb.201007107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha-Ferreira I., Rodrigues-Martins A., Bento I., Riparbelli M., Zhang W., Laue E., Callaini G., Glover D.M., Bettencourt-Dias M. 2009. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 19:43–49 10.1016/j.cub.2008.11.037 [DOI] [PubMed] [Google Scholar]
- Dammermann A., Müller-Reichert T., Pelletier L., Habermann B., Desai A., Oegema K. 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 7:815–829 10.1016/j.devcel.2004.10.015 [DOI] [PubMed] [Google Scholar]
- Dawe H.R., Farr H., Gull K. 2007. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J. Cell Sci. 120:7–15 10.1242/jcs.03305 [DOI] [PubMed] [Google Scholar]
- Debec A., Abbadie C. 1989. The acentriolar state of the Drosophila cell lines 1182. Biol. Cell. 67:307–311 10.1016/0248-4900(89)90068-3 [DOI] [PubMed] [Google Scholar]
- Dirksen E.R. 1991. Centriole and basal body formation during ciliogenesis revisited. Biol. Cell. 72:31–38 10.1016/0248-4900(91)90075-X [DOI] [PubMed] [Google Scholar]
- Ditchfield C., Johnson V.L., Tighe A., Ellston R., Haworth C., Johnson T., Mortlock A., Keen N., Taylor S.S. 2003. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J. Cell Biol. 161:267–280 10.1083/jcb.200208091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzhindzhev N.S., Yu Q.D., Weiskopf K., Tzolovsky G., Cunha-Ferreira I., Riparbelli M., Rodrigues-Martins A., Bettencourt-Dias M., Callaini G., Glover D.M. 2010. Asterless is a scaffold for the onset of centriole assembly. Nature. 467:714–718 10.1038/nature09445 [DOI] [PubMed] [Google Scholar]
- Feldman J.L., Geimer S., Marshall W.F. 2007. The mother centriole plays an instructive role in defining cell geometry. PLoS Biol. 5:e149 10.1371/journal.pbio.0050149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller S.D., Gowen B.E., Reinsch S., Sawyer A., Buendia B., Wepf R., Karsenti E. 1995. The core of the mammalian centriole contains gamma-tubulin. Curr. Biol. 5:1384–1393 10.1016/S0960-9822(95)00276-4 [DOI] [PubMed] [Google Scholar]
- González-Robles A., Cristóbal-Ramos A.R., González-Lázaro M., Omaña-Molina M., Martínez-Palomo A. 2009. Naegleria fowleri: light and electron microscopy study of mitosis. Exp. Parasitol. 122:212–217 10.1016/j.exppara.2009.03.016 [DOI] [PubMed] [Google Scholar]
- Gromley A., Jurczyk A., Sillibourne J., Halilovic E., Mogensen M., Groisman I., Blomberg M., Doxsey S. 2003. A novel human protein of the maternal centriole is required for the final stages of cytokinesis and entry into S phase. J. Cell Biol. 161:535–545 10.1083/jcb.200301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R., Stierhof Y.D., Wilkinson C.J., Nigg E.A. 2005. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 7:1140–1146 10.1038/ncb1320 [DOI] [PubMed] [Google Scholar]
- Hagiwara H., Ohwada N., Takata K. 2004. Cell biology of normal and abnormal ciliogenesis in the ciliated epithelium. Int. Rev. Cytol. 234:101–141 10.1016/S0074-7696(04)34003-9 [DOI] [PubMed] [Google Scholar]
- Hatch E.M., Kulukian A., Holland A.J., Cleveland D.W., Stearns T. 2010. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 191:721–729 10.1083/jcb.201006049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., Karsenti E. 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 382:420–425 10.1038/382420a0 [DOI] [PubMed] [Google Scholar]
- Holland A.J., Lan W., Niessen S., Hoover H., Cleveland D.W. 2010. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 188:191–198 10.1083/jcb.200911102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Cole R.W., Oakley B.R., Rieder C.L. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10:59–67 10.1016/S0960-9822(99)00276-6 [DOI] [PubMed] [Google Scholar]
- Khodjakov A., Rieder C.L., Sluder G., Cassels G., Sibon O., Wang C.L. 2002. De novo formation of centrosomes in vertebrate cells arrested during S phase. J. Cell Biol. 158:1171–1181 10.1083/jcb.200205102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.D., Nigg E.A. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Kuriyama R., Borisy G.G. 1981a. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J. Cell Biol. 91:814–821 10.1083/jcb.91.3.814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R., Borisy G.G. 1981b. Microtubule-nucleating activity of centrosomes in Chinese hamster ovary cells is independent of the centriole cycle but coupled to the mitotic cycle. J. Cell Biol. 91:822–826 10.1083/jcb.91.3.822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Terra S., English C.N., Hergert P., McEwen B.F., Sluder G., Khodjakov A. 2005. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J. Cell Biol. 168:713–722 10.1083/jcb.200411126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gönczy P. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7:115–125 10.1038/ncb1220 [DOI] [PubMed] [Google Scholar]
- Loncarek J., Hergert P., Magidson V., Khodjakov A. 2008. Control of daughter centriole formation by the pericentriolar material. Nat. Cell Biol. 10:322–328 10.1038/ncb1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J., Hergert P., Khodjakov A. 2010. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr. Biol. 20:1277–1282 10.1016/j.cub.2010.05.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüders J., Patel U.K., Stearns T. 2005. GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8:137–147 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- Marshall W.F. 2008. Basal bodies platforms for building cilia. Curr. Top. Dev. Biol. 85:1–22 10.1016/S0070-2153(08)00801-6 [DOI] [PubMed] [Google Scholar]
- Mayor T., Stierhof Y.D., Tanaka K., Fry A.M., Nigg E.A. 2000. The centrosomal protein C-Nap1 is required for cell cycle–regulated centrosome cohesion. J. Cell Biol. 151:837–846 10.1083/jcb.151.4.837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira-Leite F.F., Sherwin T., Kohl L., Gull K. 2001. A trypanosome structure involved in transmitting cytoplasmic information during cell division. Science. 294:610–612 10.1126/science.1063775 [DOI] [PubMed] [Google Scholar]
- Murphy K.P., Carter R.J., Lione L.A., Mangiarini L., Mahal A., Bates G.P., Dunnett S.B., Morton A.J. 2000. Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington’s disease mutation. J. Neurosci. 20:5115–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng S.F., Frankel J. 1977. 180 degrees rotation of ciliary rows and its morphogenetic implications in Tetrahymena pyriformis. Proc. Natl. Acad. Sci. USA. 74:1115–1119 10.1073/pnas.74.3.1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., Raff J.W. 2009. Centrioles, centrosomes, and cilia in health and disease. Cell. 139:663–678 10.1016/j.cell.2009.10.036 [DOI] [PubMed] [Google Scholar]
- Palazzo R.E., Vogel J.M., Schnackenberg B.J., Hull D.R., Wu X. 2000. Centrosome maturation. Curr. Top. Dev. Biol. 49:449–470 10.1016/S0070-2153(99)49021-0 [DOI] [PubMed] [Google Scholar]
- Peel N., Stevens N.R., Basto R., Raff J.W. 2007. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 17:834–843 10.1016/j.cub.2007.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps J.D. 1975. Aspects of spindle evolution. Ann. NY Acad. Sci. 253:352–361 10.1111/j.1749-6632.1975.tb19213.x [DOI] [PubMed] [Google Scholar]
- Piel M., Meyer P., Khodjakov A., Rieder C.L., Bornens M. 2000. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149:317–330 10.1083/jcb.149.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C., Borisy G.G. 1982. The centrosome cycle in PtK2 cells: asymmetric distribution and structural changes in the pericentriolar material. Biol. Cell. 44:117–132 [Google Scholar]
- Rodrigues-Martins A., Riparbelli M., Callaini G., Glover D.M., Bettencourt-Dias M. 2007. Revisiting the role of the mother centriole in centriole biogenesis. Science. 316:1046–1050 10.1126/science.1142950 [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rusan N.M., Roberts D.M., Peifer M., Rogers S.L. 2009. The SCFSlimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 184:225–239 10.1083/jcb.200808049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillibourne J.E., Tack F., Vloemans N., Boeckx A., Thambirajah S., Bonnet P., Ramaekers F.C., Bornens M., Grand-Perret T. 2010. Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol. Biol. Cell. 21:547–561 10.1091/mbc.E09-06-0505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder J.A., McIntosh J.R. 1975. Initiation and growth of microtubules from mitotic centers in lysed mammalian cells. J. Cell Biol. 67:744–760 10.1083/jcb.67.3.744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn T.M. 1964. The determinants and evolution of life. The differentiation of cells. Proc. Natl. Acad. Sci. USA. 51:915–929 10.1073/pnas.51.5.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., Gönczy P. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 13:203–213 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telzer B.R., Rosenbaum J.L. 1979. Cell cycle-dependent, in vitro assembly of microtubules onto pericentriolar material of HeLa cells. J. Cell Biol. 81:484–497 10.1083/jcb.81.3.484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou M.F., Stearns T. 2006a. Controlling centrosome number: licenses and blocks. Curr. Opin. Cell Biol. 18:74–78 10.1016/j.ceb.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., Stearns T. 2006b. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 442:947–951 10.1038/nature04985 [DOI] [PubMed] [Google Scholar]
- Tsou M.F., Wang W.J., George K.A., Uryu K., Stearns T., Jallepalli P.V. 2009. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell. 17:344–354 10.1016/j.devcel.2009.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L.T. 2006. Cell cycle synchronization at the G2/M phase border by reversible inhibition of CDK1. Cell Cycle. 5:2555–2556 10.4161/cc.5.22.3463 [DOI] [PubMed] [Google Scholar]
- Vidwans S.J., Wong M.L., O’Farrell P.H. 2003. Anomalous centriole configurations are detected in Drosophila wing disc cells upon Cdk1 inactivation. J. Cell Sci. 116:137–143 10.1242/jcs.00204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev I.A., Chentsov Y.S. 1982. Centrioles in the cell cycle. I. Epithelial cells. J. Cell Biol. 93:938–949 10.1083/jcb.93.3.938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C., Stearns T. 2003. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat. Cell Biol. 5:539–544 10.1038/ncb993 [DOI] [PubMed] [Google Scholar]