Abstract
In flowering plants, the developing embryo consists of growing populations of cells whose fates are determined in a position-dependent manner to form the adult organism. Mutations in the FACKEL (FK) gene affect body organization of the Arabidopsis seedling. We report that FK is required for cell division and expansion and is involved in proper organization of the embryo. We isolated FK by positional cloning. Expression analysis in embryos revealed that FK mRNA becomes localized to meristematic zones. FK encodes a predicted integral membrane protein related to the vertebrate lamin B receptor and sterol reductases across species, including yeast sterol C-14 reductase ERG24. We provide functional evidence that FK encodes a sterol C-14 reductase by complementation of erg24. GC/MS analysis confirmed that fk mutations lead to accumulation of intermediates in the biosynthetic pathway preceding the C-14 reductase step. Although fk represents a sterol biosynthetic mutant, the phenotype was not rescued by feeding with brassinosteroids (BRs), the only plant sterol signaling molecules known so far. We propose that synthesis of sterol signals in addition to BRs is important in mediating regulated cell growth and organization during embryonic development. Our results indicate a novel role for sterols in the embryogenesis of plants.
Keywords: FACKEL (FK), embryogenesis, sterol reductase, brassinosteroids, GC/MS, erg24
The basic body plan of multicellular organisms is established during embryogenesis. In the flowering plant Arabidopsis, stereotyped cell divisions are coordinated with cell expansions as cells adopt specific fates within the developing embryo (Jürgens and Mayer 1994). This results in an apical–basal pattern of distinct elements, superimposed by a perpendicular radial pattern of concentric tissue layers in the seedling. The apical–basal axis of the seedling is subdivided into major components: shoot meristem, cotyledons, hypocotyl, root, and root meristem. These components are established through partitioning of the axis in successive steps. During this process, cell fates are singled out in a position-dependent manner that involves communication between neighboring cells (Laux and Jürgens 1997). Although cell–cell communication is thought to play an important role in patterning of the plant embryo, the underlying molecular mechanisms, as well as the nature of the positional information are not understood but may be presumed to involve signaling across cell membranes.
Sterols are the major components modulating the fluidity and permeability of eukaryotic cell membranes and in addition serve as precursors for steroid-signaling molecules that are known to elicit a variety of physiological processes. In vertebrates the major sterol is cholesterol, and derived steroid hormones are crucial for embryonic development and homeostasis of the adult organism (Goldstein and Brown 1990; Farese and Herz 1998). In plants, several types of structurally related phytosterols have been characterized. The principal sterol is sitosterol and campesterol is the second most abundant (Patterson et al. 1993). Functional roles for the campesterol-derived brassinosteroids (BRs) are established for integrating light signals in the regulation of postembryonic plant growth (Clouse and Sasse 1998). Specific biological roles for sitosterol and other less abundant sterols have not been reported.
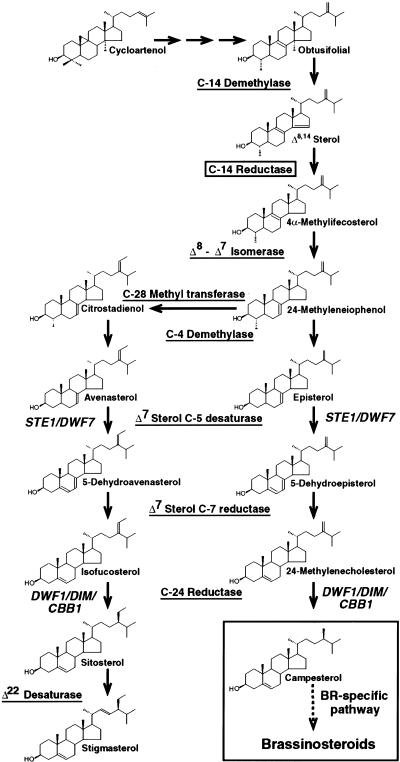
The biosynthesis of plant sterols and BRs has been studied extensively and genes for many of the enzymatic steps have been identified through biochemical and genetic approaches (Benveniste 1986; Ourisson 1994; Bach and Benveniste 1997; Sakurai and Fujioka 1997). The biosynthetic pathway from cycloartenol to sterols and BRs diverges at 24-methyleneIophenol, after which parallel branches produce sitosterol and campesterol (Fig. 1). From campesterol a specific pathway leads to BR biosynthesis. Mutations blocking this BR-specific pathway result in dwarf plants (Altmann 1999). However, two genes corresponding to dwarf mutants, STE1/DWF7 (Choe et al. 1999b; Husselstein et al. 1999) and DWF1/DIM/CBB1 (Klahre et al. 1998; Choe et al. 1999a), act upstream of the BR-specific pathway and thus are involved in the synthesis of both sitosterol and campesterol (Fig. 1). The enzyme sterol C-14 reductase acts in the sterol biosynthetic pathway upstream of STE1/DWF7 (Fig. 1). The C-14 reductase step is thought to be essential for sterol biosynthesis (Lees et al. 1999) and is conserved among eukaryotes (Benveniste 1986; Taton et al. 1989). Although C-14 reductase activity has been reported in an enzymatic assay using plant microsomal fractions (Taton et al. 1989), a gene encoding a plant C-14 reductase has not been identified.
Figure 1.
Sterol biosynthesis in Arabidopsis. The pathway to sterols and BRs is shown from the first cyclic precursor, cycloartenol. Sterol molecules are labeled with their common names. Enzymes involved are underlined and genes identified by mutants are shown in italics. The sterol C-14 reductase step is boxed. The BR-specific pathway from campesterol to BRs is shown in a large box.
A systematic screen for mutations affecting the body organization of the Arabidopsis seedling led to the identification of the FK gene (Mayer et al. 1991). Mutant fk seedlings display a severely shortened hypocotyl and show defects in patterning of apical and basal structures. To determine the cellular basis of the defect, we examined fk embryos that showed marked disturbances in cell division and cell expansion, as well as defects in the organization of the embryonic pattern. We cloned and identified the FK gene product as a functional sterol C-14 reductase. Our data provide evidence that FK is involved in the production of sterols with important functions in the embryonic development of Arabidopsis.
Results
The fk mutant seedling phenotype
Wild-type seedlings display an organized body pattern with apical (cotyledons and shoot meristem), central (hypocotyl), and basal (root and root meristem) regions (Fig. 2a). In fk mutant seedlings the hypocotyl is severely shortened and the cotyledons appear to be directly attached to a short root (Fig. 2b). Seven fk alleles induced in the Ler ecotype showed similar seedling phenotypes, although one EMS-induced allele, fk-T329, displayed a slightly weaker phenotype. fk-T329 seedlings developed longer roots (Fig. 2c) and could be propagated on rich medium to form shoots and flowers (data not shown). Two fk alleles induced in the Wassilewskija (WS) ecotype also displayed a weaker phenotype, as measured by their ability to grow postembryonically in vitro. None of the fk mutants could be grown to flowering stage on soil. Necrotic tissues were not observed in fk seedlings, suggesting that their aborted development cannot be attributed to cell death.
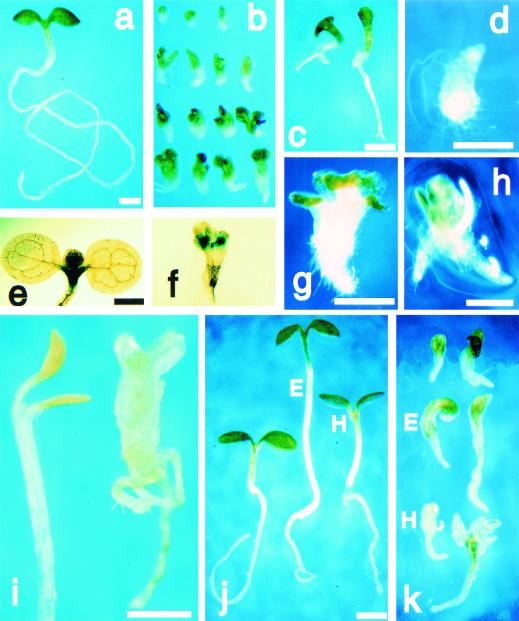
Figure 2.
fk seedling phenotype. (a–i) Seedlings were germinated on 1.7% agar/water for 11 days. (a,e) Wild type. (b–d,f–h) fk. (b) Phenotypic variability: The strong phenotypes (top) are severely stunted with a short root, malformed cotyledons, and appear to lack the hypocotyl. The weaker phenotypes (bottom) are larger; (c) weak allele fk-T329 shows longer root; (d) rooty seedling. KNAT2–GUS staining: (e) Wild type shows staining in shoot apical meristem; (f) fk shows multiple meristems; (g) seedling with multiple apices; (h) seedling with multiple roots; (i) etiolation response to germination in the dark: wild type (left) shows apical hook and elongation of the hypocotyl (one-third of the hypocotyl length is shown); fk (right) lacks apical hook and shows slightly elongated hypocotyl region; (j) wild-type control (left); treated with BRs 24-epibrassinolide (E, middle) or 22S,23S-homobrassinolide (H, right); (k) fk control (top); treatment with 24-epibrassinolide (E, center) or 22S,23S-homobrassinolide (H, bottom). Bars, 0.5 mm (a = b, e = f, g = h, j = k).
Phenotypic variability was observed within each of the nine mutant alleles (Fig. 2b). Cotyledons were malformed: Instead of two symmetrical cotyledons (Fig. 2a), fk apices had either one cotyledon, multiple cotyledons, and/or pin-shaped cotyledons. Roots were short and wider than in wild type. Rare “rooty” fk mutants lacking apical structures were also observed (Fig. 2d). Staining for KNAT2 (Dockx et al. 1995) promoter GUS-fusion expression showed multiple shoot meristems in some fk seedlings (Fig. 2f). We often observed twin or multiple structures such as multiple apices or roots (Fig. 2g,h).
As a test for the presence of hypocotyl tissue in fk mutants, we examined fk germination in the dark. Wild-type seedlings exhibited a characteristic etiolation response: apical hook formation and elongation of the hypocotyl (Fig. 2i). fk seedlings displayed a defective etiolation response: They exhibited reduced elongation of presumptive hypocotyl tissue in comparison to wild-type and callus-like tissue (Fig. 2i) suggesting that fk mutants, whereas able to respond to dark growth, are unable to organize elongation in a coordinated manner.
The fk mutant phenotype derives from defective cell divisions in embryogenesis
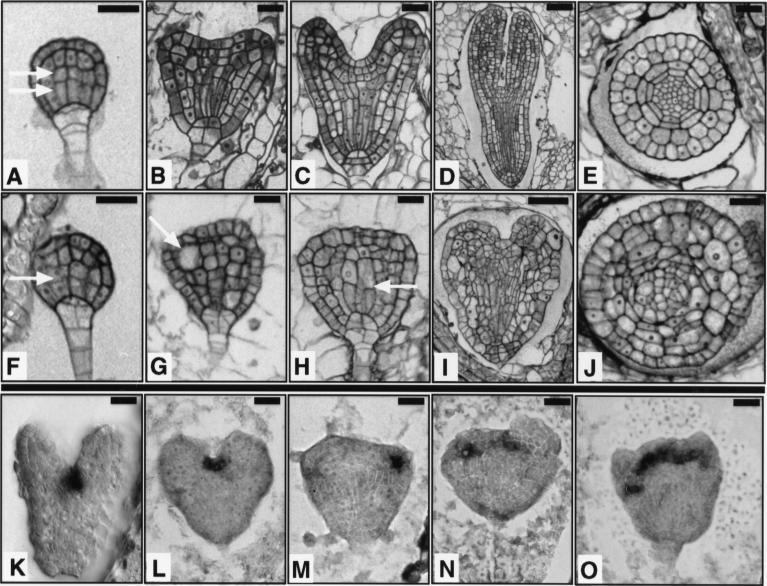
Siliques from heterozygous FK/fk plants containing embryos at various stages of development were analyzed to determine the stage at which we could detect the first deviation in fk mutants from their wild-type siblings. We traced the fk mutant phenotype back to the globular stage of embryogenesis (Fig. 3A,F). At this stage, wild-type cell elongations are followed by asymmetric cell divisions in the central cells, giving small apical and elongated basal cells. In fk embryos the innermost cells failed to elongate and produced daughter cells of similar sizes. fk embryos appeared to be delayed in shape changes associated with subsequent stages of development. fk embryos did not exhibit the characteristic heart shape (Fig. 3B,C) and appeared rounded (Fig. 3G,H). Elongations of cell files in the central region failed to occur or were defective in fk at the heart and later stages, and cell morphologies were abnormal. Mutant embryos appeared smaller and wider than wild-type embryos. Unlike the torpedo shape observed for their wild-type siblings (Fig. 3D), fk embryos displayed a heart-like shape (Fig. 3I). The characteristic wild-type bent-cotyledon or mature embryo stages were not observed for mutant embryos. The radial organization of the vasculature was also abnormal in fk. In transverse sections of fk embryos we observed cells with aberrant morphology and disorganized tissue layers (Fig. 3E,J).
Figure 3.
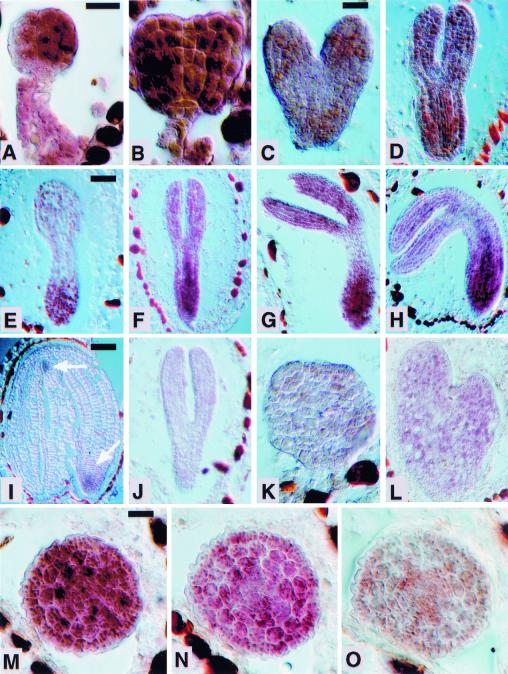
Embryogenesis is abnormal in fk mutants. (A–E,K) Wild type. (F–J,L–O) fk. (A,F) Globular stage embryo. In wild type, the central vascular precursor cells divide asymmetrically to produce small apical and elongated basal cells (two arrows). In fk, the central cells divide to give cells of similar sizes (single arrow). (B,G) Early heart stage embryo. In wild type, the cotyledon primordia become visible and the central cells are elongated. In fk, cotyledon primordia are not visible and cells in the center of the embryo fail to elongate. Some cells are grossly enlarged (arrow); (C,H) Heart stage embryo. In wild type, the cotyledon primordia mark the bilateral symmetry of the embryo. fk embryos exhibit abnormal cell morphologies and incomplete cell walls (arrow); (D,I) Torpedo stage embryo. Wild-type embryo shows a characteristic torpedo shape. The fk embryo displays an abnormal heart shape and cell elongations leading to longitudinal outgrowth are defective. (E,J) Transverse sections of torpedo stage embryos. Wild-type embryo shows radial organization of tissue layers. The fk embryo lacks an organized radial pattern. Cells are misshapen and either larger or smaller than cells of the corresponding layer in wild type. (K–O) Heart stage embryos showing expression of STM mRNA, a shoot apical meristem marker. Wild-type expression is in the center, between the cotyledon primordia. fk embryos often show aberrant placement, multiple, or widened meristems. Bars, 20 μm (A–C,E–H,J); 40 μm (D,I).
At all stages we observed what appeared to be cytokinesis defects: enlarged cells, random orientations of cell division, and incomplete cell walls (Fig. 3F–J). However, fk cells could be propagated in callus-inducing medium (data not shown), unlike cytokinesis mutants (Assaad et al. 1996). This finding suggested that fk mutants are not defective in cell division per se, but seem to be specifically defective in organized cell divisions within a developmental context.
In situ hybridization with a STM RNA probe that marks the shoot apical meristem (Long et al. 1996; Fig. 3K) was used to examine the presence of this organ in mutant embryos. In some fk embryos we observed the wild-type staining pattern (Fig. 3L). More often we detected abnormal positioning of the presumptive shoot apical meristem in fk embryos (Fig. 3M). Multiple or widened staining regions were also detected (Fig. 3N,O), consistent with our observations of multiple shoot meristems in fk seedlings (Fig. 2f).
Identification of the FK gene
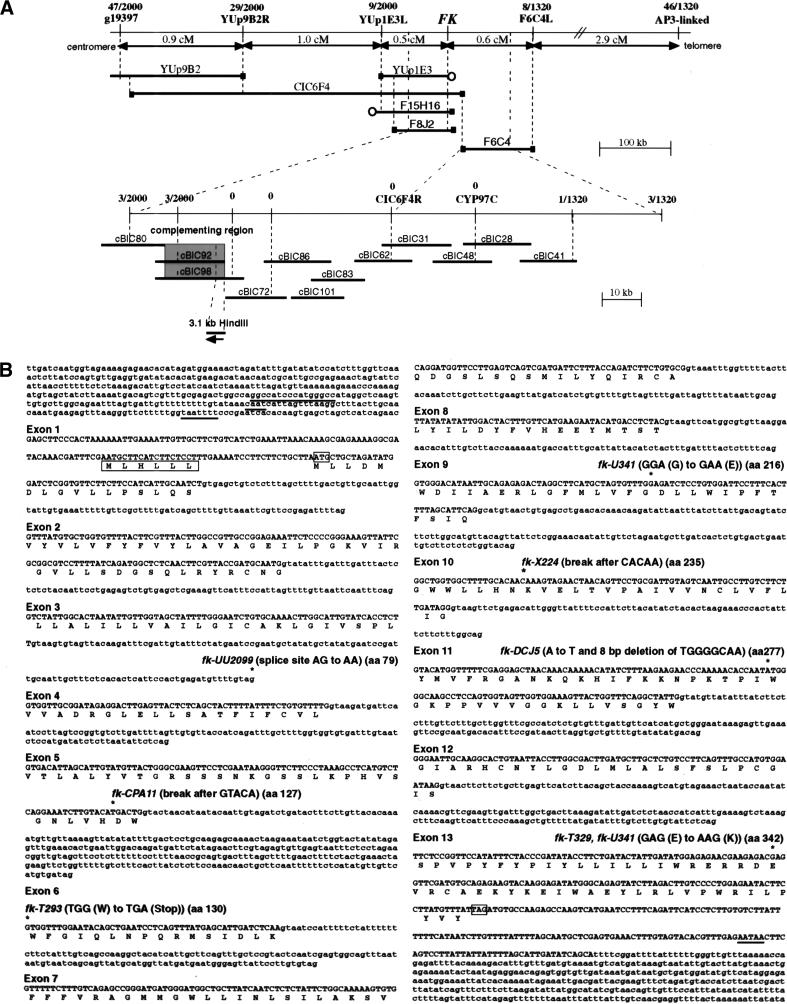
A positional cloning strategy was applied to isolate the FK gene (Fig. 4A). The cosmid contig established during this work completes the physical map between BACs F8J2 and F6C4 on chromosome 3 (cBIC62 in Figure 4A; http://www.arabidopsis.org/cgi-bin/maps/Pmap?contig=5S_rDNA-F28P10-Sp6&clone=F8J2). Overlapping cosmids from this contig were used to identify FK by complementation. A 3.1-kb HindIII fragment from the complementing region showed allele-specific RFLPs in fk-X224 and fk-CPA11 (data not shown), and contained a single gene, FK (Fig. 4A,B). Figure 4B shows the sites of the corresponding mutations within the FK sequence from seven alleles. All EMS-induced mutations represent single base exchanges predicted to result in splice site ablation (fk-UU2099), a stop codon (fk-T293), or amino acid substitutions (fk-T329, fk-U341). An 8-bp deletion was found in fk-DCJ5, an allele from a T-DNA mutagenesis. Genomic rearrangements were found for the T-DNA- and X-ray-induced alleles fk-CPA11 and fk-X224, with break points predicted to remove the last two-thirds and one third of the gene, respectively. In fk-X224, the rearrangement is an inversion because the flanking DNA corresponds to the 3′ end of EST FAFJ61 (GenBank accession no. Z33977), about 3-kb downstream from the FK gene (data not shown).
Figure 4.
Molecular identification of the FK gene. (A) Positional cloning of the FK gene. RFLP and CAPS markers used for determining recombination break points and corresponding numbers of recombinant chromosomes are indicated. Genetic distances between markers are indicated in cm. The physical map represented by YAC, BAC and cosmid contigs is diagrammed. (█) Mapped YAC or BAC ends, (○) unmapped ends. The two complementing cosmids (shaded) and the 3.1-kb HindIII fragment corresponding to the FK gene are shown (bottom). (B) Nucleotide sequence of the FK gene and deduced amino acid sequence of the encoded protein. Nucleotides in exons are shown in uppercase letters with single-letter amino acid codes below each codon, nucleotides of introns and 5′ and 3′ untranscribed and spliced regions in lowercase letters. Putative GC rich, CAAT and TATA (TAATTTT) boxes in the promoter of FK are underlined. A putative upstream open reading frame (uORF) of six amino acids is boxed. Exons are numbered at their 5′ ends. For exon 1, the inferred 5′ end is the start of the longest 5′ RACE product sequenced. The translation initiation ATG and termination TAG codons are boxed. A putative poly(A) signal (AATAA) is underlined. Mutations identified in the genomic DNAs of fk alleles are marked (*) followed by the allele designation above the DNA sequence.
Transcription of the FK gene in embryos
The mutant phenotype suggested a role for the FK gene in cell elongation and cell division during embryogenesis. We used in situ hybridization of embryos to examine the expression of the FK gene. FK mRNA was detected in early stages of embryogenesis but was absent from the cellularizing endosperm (Fig. 5). Globular and early heart stage embryos showed nearly uniform distribution of the hybridization signal (Fig. 5A,B). At the mid-heart stage, the signal became more intense towards the top and bottom of the embryo (Fig. 5C,D). We observed clearly localized expression at later stages (Fig. 5E–H). Transcripts accumulated in the basal end of torpedo and bent-cotyledon stage embryos and to a lesser extent in the cotyledons. The basal expression was confirmed in a series of transverse sections that show a transition zone within the torpedo-stage embryo (Fig. 5M–O). In the mature embryo expression was diminished and confined to the presumptive shoot meristem and root tip (Fig. 5I). A control with a sense probe did not give staining at the torpedo stage (Fig. 5J) and all other stages examined (data not shown). We hybridized the antisense probe to fk-X224 mutant embryos at various stages and did not detect staining (Fig. 5K,L), indicating that the gene is not expressed in this allele.
Figure 5.
Expression of FK mRNA in Arabidopsis embryo development. Sections were hybridized with RNA probes containing the entire FK coding sequence. Wild-type embryos hybridized to antisense probe: (A) early globular stage; (B) late globular stage; (C) heart stage; (D) early torpedo stage; (E) torpedo parasagittal section; (F,G) torpedo stages; (H) bent-cotyledon stage; (I) mature embryo [arrows indicate weak hybridization signals in the shoot meristem (top arrow) and root tip (bottom arrow)]. (J) Wild-type torpedo stage embryo hybridized to FK sense probe as a negative control; (K,L) fk-X224 mutant embryos hybridized to antisense probe. Expression is absent; (K) fk heart stage; (L) fk torpedo stage; (M–O) wild-type torpedo stage embryo transverse serial sections of a transition zone in the hypocotyl region hybridized to the antisense probe. FK expression is stronger in the first (M), more basal section and becomes weaker in the following sections (N,O). Scale bars, 20 μm (A = B, C = D = K, M = N = O); 40 μm (E = F = G = H, I = J).
The FK gene encodes a membrane protein with homology to sterol reductases across species
The FK gene encodes a conceptual protein of 369 amino acids with a predicted molecular weight of 41–42 kD. The FK protein is highly leucine-rich (16.5%) and almost entirely hydrophobic. Hydropathy plot analysis suggested that FK encodes a membrane protein with eight to nine transmembrane segments and a carboxy-terminal cytosolic tail of about 43 amino acids. Computer analysis predicted an uncleavable amino-terminal signal sequence and protein localization to the endoplasmic reticulum (ER) membrane or plasma membrane.
Sequence comparison revealed homology to members of the lamin B receptor/sterol reductase multigene family, with the most significant homology to sterol C-14 reductases across species. A phylogenetic tree that summarizes our findings is shown in Figure 6A. FK showed significant identities (38%–34%) and similarities (56%–53%) to the 400 amino acid carboxy-terminal sterol reductase domain of lamin B receptors from human, rat, Xenopus, and chicken. The FK protein also showed significant identities (39%–28%) and similarities (54%–43%) to the human putative sterol reductase SR-1 and sterol C-14 reductases from Ascobolus, Schizosaccharomyces pombe, Saccharomyces cerevisiae, Nectria haematococca, Neurospora, and Septoria lycopersici. In addition, FK showed similarities, from amino acids 77–369, to Δ7 C-7 reductases from mouse, humans, rat, and to a putative Δ7 C-7 reductase from Caenorhabditis elegans. Arabidopsis ST7R, encoding a sterol Δ7 C-7 reductase, showed similarity along the length of FK from amino acid 33. The FK protein showed weaker similarities to C-24(28) reductases from S. cerevisiae and S. pombe.
Figure 6.
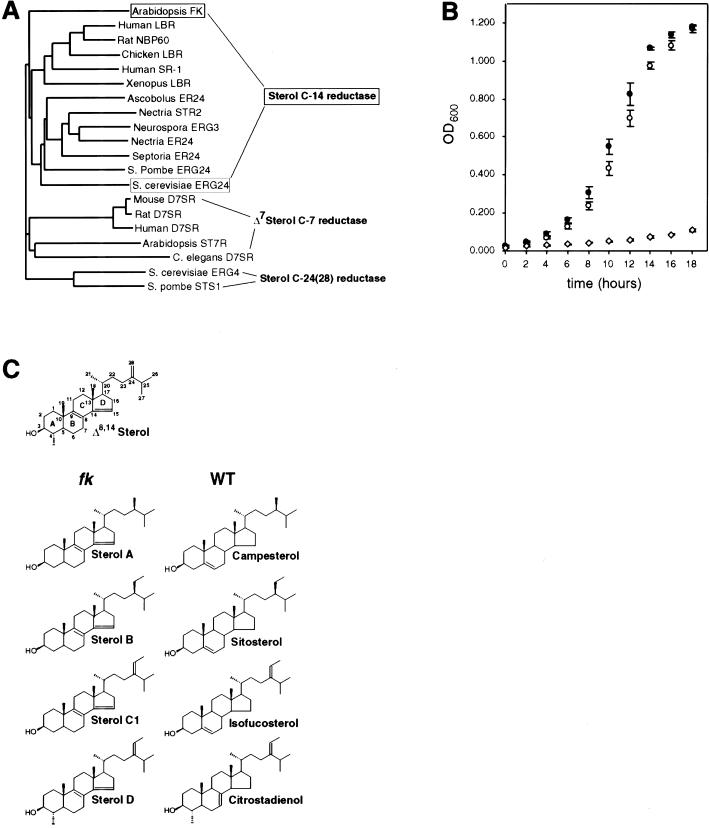
FK encodes a sterol C-14 reductase: phylogenetic relationship, functional assay in yeast, and accumulation of biosynthetic intermediates in planta. (A) Phylogenetic tree of sterol reductases with FK and S. cerevisiae ERG24 proteins boxed. The sterol reductase family is grouped into sterol C-14 reductases, Δ7 sterol C-7 reductases, and yeast sterol C-24(28) reductases. FK is most closely related to the C-14 reductase branch. Accession nos.: Human LBR, AAA59495; Rat NBP60, BAA20471; Chicken LBR, P23913; Human SR-1, AAD09765; Xenopus LBR, CAB44317; Ascobolus ER24, P78575; Nectria STR2, CAA66943; Neurospora ERG3, P38670; Nectria ER24, Q01447; Septoria ER24, O13597; S. pombe ERG24, JC4057; S. cerevisiae ERG24, 6324049; Mouse D7SR, AAC40164; Rat D7SR, BAA34306; Human D7SR, AAD02816; Arabidopsis ST7R, U49398; S. cerevisiae ERG4, 6321426; S. pombe STS1, A43765; C. elegans D7SR, CAB03797. (B) The FK cDNA complements the growth defect of a S. cerevisiae erg24 strain. An erg24 strain was transformed with expression vector plasmids containing the FK cDNA (●), ERG24 (○), or empty vector control (⋄). Growth under restrictive conditions was measured as a function of optical density (OD600) with time. Data points represent averages for four independent transformants in two trials. Error bars indicate s.d. (C) Δ8,14 sterols in fk mutants. A reference Δ8,14 sterol with numbered carbons and lettered rings is diagrammed (top). Molecular formulas for the Δ8,14 sterols (A,B,C1,D; Table 1) that accumulate in fk mutants in comparison with sterols from wild type (WT) are shown. Modifications of their side chains indicate that these Δ8,14 sterols are substrates for enzymes that act downstream of the C-14 reductase step (Fig. 1). For sterols A, B, and C1, the side chain at C-24 is equivalent to that of campesterol, sitosterol, and isofucosterol, respectively. Sterols C2 and C3, whose putative structures are not shown because the position of the third double bond is uncertain (see Results), have a side chain at C-24 which is equivalent to that of sitosterol. For sterol D, modifications at C-4 and C-24 are equivalent to those of citrostadienol, a sterol intermediate that was not detected in our analysis.
The FK cDNA complements the growth defect of yeast erg24 cells
We examined the function of the FK cDNA in an S. cerevisiae erg24 null mutant that is defective for sterol C-14 reductase. erg24 cells are unable to grow on calcium-depleted medium but may be propagated conditionally in medium containing CaCl2 (Crowley et al. 1998).To examine whether expression of the FK gene product can alleviate the growth defect and calcium dependency of erg24 cells, we cloned the FK cDNA and ERG24 sequence into a yeast expression vector. Yeast transformants were grown under permissive conditions and shifted to a calcium-poor medium (Fig. 6B). erg24 cells containing a control vector showed residual growth after 18 hr. In contrast, erg24 cells transformed with either FK cDNA or ERG24 exhibited growth to stationary phase within the same time course. Colony formation and growth kinetics under permissive conditions were also improved for the erg24 transformants harboring the FK cDNA or ERG24 relative to the control vector (data not shown). The three plasmids showed no differences in colony formation or in growth kinetics when expressed in an erg4 strain that is defective in C-24(28) reductase (Lai et al. 1994), suggesting that the growth enhancement by the FK cDNA is specific to erg24.
Sterol analysis of fk mutants indicates an accumulation of intermediates in sterol biosynthesis
The complementation of the erg24 phenotype by FK suggested that it can function as a sterol C-14 reductase in a heterologous system. To determine whether fk mutations block sterol biosynthesis, we analyzed sterol extracts from wild type and fk by GC/MS (Table 1). We examined both seedling and callus cell extracts, and these showed different sterol profiles. For both fk seedlings and callus cell extracts, the mass spectral data indicated an abnormal accumulation of Δ8,14 sterols with two double bonds at carbon positions 8 and 14 (Figs. 1 and 6C), a structural feature of biosynthetic intermediates that accumulate in plant cell suspensions treated with an inhibitor of C-14 reductase, 15-azasterol (Schmitt et al. 1980; Schaller et al. 1994). The molecules detected in fk mutants but not in wild type are listed as sterols A, B, C1, C2, C3, and D in Table 1 and Figure 6C. The EI mass spectra of abnormal sterols A, B, C1, and D were in agreement with those reported by Schmitt et al. (1980). Sterols C2 and C3 represent novel C-29 sterols with three double bonds in the sterol nucleus as indicated by the M+ ion at m/z 452 and key ions at m/z 311 and 251 in the EIMS of the acetate. These mass spectra do not reveal the exact position of the third double bond in C2 and C3.
Table 1.
Mass spectral analysis of sterols from fk mutants shows accumulation of Δ8,14 sterols
| Sterol
|
M+
|
Seedlings
|
Callus Cells
|
|||
|---|---|---|---|---|---|---|
| WT
|
fk-X224
|
WT
|
fk-X224
|
fk-UU2099
|
||
| Cholesterol | 368* | 8.6 | 16.1 | 0.5 | 2.5 | 1.1 |
| 24-Methylcholesta-5,22-dien-3β-ol | 380* | 2.0 | − | − | − | − |
| 24-Methylenecholesterol | 380* | (+) | − | − | − | − |
| Campesterol | 382* | 13.1 | (+) | 16.2 | (+) | − |
| Campestanol | 444 | (+) | − | 0.5 | − | − |
| Stigmasterol | 394* | 8.9 | 10.5 | 0.5 | 0.8 | 0.8 |
| Sitosterol | 396* | 67.4 | 51.9 | 59.8 | (+) | 4.2 |
| Sitostanol/Isofucosterol | 458/456 | (+) | − | 1.3 | − | − |
| Cycloartenol | 468 | − | − | 2.3 | 9.1 | 6.6 |
| 24-Methylenecycloartanol | 482 | − | − | 18.6 | 30.4 | 11.6 |
| Sterol A | 440 | − | (+) | − | 3.6 | 6.4 |
| Sterol B | 454 | − | 6.8 | − | 39.8 | 59.0 |
| Sterol C1 | 452 | − | (+) | − | 0.6 | 0.5 |
| Sterol C2 | 452 | − | 8.5 | − | 5.3 | 4.8 |
| Sterol C3 | 452 | − | 6.1 | − | 3.9 | 1.7 |
| Sterol D | 466 | − | − | − | 4.0 | 3.3 |
| Total sterols A-D | − | 21.4 | − | 57.2 | 75.7 | |
Percentages of the relative sterol composition are shown. (*) (M-HOAc)+; (−) not present; (+) traces. (WT) Wild type; fk-X224 and fk-UU2099 alleles are indicated; Sterols are listed according to mass (M); Sterols A, B, C1, C2, C3, and D are Δ8,14 sterols that were found in fk. A = 5α-Ergosta-8,14-dien-3β-ol; B = 5α-Stigmasta-8,14-dien-3β-ol; C1 = 5α-Stigmasta-8,14,Z-24(28)-trien-3β-ol; C2 = unknown Stigmastatrien-3β-ol; C3 = unknown Stigmastatrien-3β-ol; D = 4α-Methyl-5α-stigmasta-8,14,Z-24(28)-trien-3β-ol.
The accumulation of abnormal Δ8,14 sterols was enhanced in fk callus cells: Sterol B was at a level of 39.8% (fk-X224) and 59.0% (fk-UU2099), and the total percentage of Δ8,14 sterols was 57.2% (fk-X224) and 75.7% (fk-UU2099). In fk seedlings, the accumulation of Δ8,14 sterols was less dramatic (21.4%), and sterol D was not detected. Sterol B was detected at 6.8%, a sixfold lower level as compared to callus cells.
Relative to the wild-type profiles, fk seedlings and callus cells showed a reduction in the BR precursors campesterol and campestanol and in the sitosterol precursors sitostanol and isofucosterol (Table 1), consistent with an upstream block at the C-14 reductase step (Fig. 1). Both also showed a reduction in sitosterol, although this was enhanced in callus cells, in which the amount of sitosterol was reduced from a wild-type level of about 60% to traces (fk-X224) and 4.2% (fk-UU2099). In addition, an increase in the sterol precursor cycloartenol was detected in callus cells from both fk alleles (Table 1), consistent with a downstream block at the C-14 reductase step (Fig. 1).
fk mutant seedlings respond to BRs but are not rescued
BR biosynthesis mutants can be rescued by feeding steroid compounds postembryonically (Kauschmann et al. 1996). We examined whether we could rescue the fk mutant phenotype by feeding sterol compounds including two BRs that have been shown to rescue dwarf mutants. Seeds from heterozygous FK/fk plants were sown on media containing one of five sterol compounds: campesterol, cholesterol, sitosterol, stigmastanol, stigmasterol, and the two BRs, 24-epibrassinolide and 22S,23S-homobrassinolide. We also examined seedlings germinated on high concentrations of campesterol and sitosterol, major sterols that are depleted in fk. On plates containing the non-BR sterols we saw no effect on wild-type seedlings nor a change from the characteristic fk phenotype. Treatment with the two BRs resulted in hypocotyl elongation and shortened roots in wild-type seedlings (Fig. 2j; Clouse et al. 1993). In fk seedlings we did not observe a rescue of the mutant phenotype (Fig. 2k). However, the mutant seedlings were elongated or larger, callus-like, and often curled. Therefore, fk mutant seedlings are responsive to these BR compounds, but appear unable to coordinate their growth in a regulated manner.
Discussion
A screen for mutants affecting body organization of the Arabidopsis seedling resulted in the isolation of nine alleles of a single genetic locus, FK. We cloned the FK gene and found that it encodes a functional sterol C-14 reductase. fk mutations affecting the C-14 reductase step represent the earliest genetic block in sterol biosynthesis in a multicellular organism. We address our molecular data supporting the role of FK as a sterol C-14 reductase and discuss how a primary defect in sterol biosynthesis could result in the fk phenotypes observed.
FK is a member of the sterol C-14 reductase family
The predicted FK protein shows homology with sterol reductases across species with strongest similarity to sterol C-14 reductases. Sequence analysis of the almost entirely hydrophobic FK protein predicts an integral membrane protein localized to the ER or plasma membrane. ER localization has been reported for enzymes involved in plant sterol biosynthesis (Hartmann and Benveniste 1987), and for the human SR-1 putative C-14 reductase when expressed transiently in COS-7 cells (Holmer et al. 1998).
To date there have been no reports of sequenced point mutations in other C-14 reductases nor studies addressing structure–function analysis of a related sterol reductase in any system. We identified mutations throughout the FK sequence after the first 78 amino acids of the gene. Of the alleles characterized molecularly, five lack portions of the FK gene or predicted protein and give essentially the same phenotype. FK mRNA is not expressed in fk-X224 mutant embryos, indicating that it is a null allele. Missense mutations representing changes in conserved amino acids were found in two alleles, demonstrating functional roles for these residues. The weak allele (fk-T329) has a change from a negatively charged glutamic acid to a positively charged lysine within the predicted carboxy-terminal tail. This residue is either glutamic or aspartic acid in all sterol reductases reported, suggesting that a negatively charged residue is critical at this site.
The role of FK in sterol biosynthesis
FK shares 33% identity with S. cerevisiae ERG24, a functional sterol C-14 reductase (Lorenz and Parks 1992; Marcireau et al. 1992). The FK cDNA complemented the growth defect of S. cerevisiae erg24, strongly suggesting that the encoded protein can function as a C-14 reductase. The human lamin B receptor, which shares 39% identity with ERG24, rescues erg24 (Silve et al. 1998) and Neurospora erg3 (Prakash et al. 1999) mutants. Thus the catalytic domain of C-14 reductase appears to be conserved among eukaryotes.
A sterol C-14 reductase activity for FK in Arabidopsis is further indicated by the accumulation of unreduced biosynthetic intermediates, Δ8,14 sterols, in samples from two different fk tissues. Schmitt et al. (1980) reported a comparable level of Δ8,14 sterols and reduction in end products of sterol biosynthesis after treatment of plant cell cultures with an inhibitor of C-14 reductase, 15-azasterol. Therefore, lack of C-14 reductase activity because of fk mutations has the same effect as application of a specific inhibitor of the enzyme.
Absence of the FK gene product does not eliminate the entire downstream pathway. Thus, sterol biosynthesis may occur through a C-14 reductase independent pathway or a second C-14 reductase can provide partial function. In BLAST searches we find an Arabidopsis EST (accession no. AA395510), which shares similarity with FK on both nucleotide and amino acid levels. There is precedence for two sterol C-14 reductase genes in the phytopathogenic fungus Nectria haematococca (Fig. 6A). The identification of a second C-14 reductase in Arabidopsis will enable testing whether these genes are functionally redundant or display specialized roles in sterol biosynthesis.
The role of FK in development
fk embryos exhibit a range of cellular defects illustrating that FK is needed for organized cell division and expansion. However, FK is not required for basic cell division since fk mutations did not affect the ability of callus cells to proliferate in culture. Thus, FK appears to play a role in cell growth in a developmental context. All components of the apical–basal axis are represented in fk mutant seedlings, suggesting that FK is not strictly required for the establishment of cell fates. However, partitioning of groups of cells with specific fates, such as the shoot meristem, is dramatically disturbed in fk embryos. This suggests that in addition to a role in cell division and expansion, FK is involved in generating positional cues for the organization of pattern within the embryo.
fk phenotypes are due to a defect in sterol biosynthesis
A lesion in sterol C-14 reductase due to fk mutations results in a reduction of downstream sterols and an accumulation of biosynthetic intermediates, Δ8,14 sterols. These intermediates, although they are substrates for downstream biosynthetic modifications (Fig. 6C), probably do not functionally substitute for normal sterols in vivo. Callus cells can tolerate high levels of Δ8,14 sterols (Table 1), suggesting that Δ8,14 sterols are unlikely to be toxic to dividing plant cells. Furthermore, Δ8,14 sterols have been reported to occur naturally as constituents of plants such as Senita cactus (Campbell and Kircher 1980). However, our data do not exclude the possibility that Δ8,14 sterols interfere with normal cell membrane function in the developing embryo (see below).
We propose that the defects in fk embryos are primarily caused by a reduction in downstream sterols, which have either cell structural or signaling roles in embryogenesis. The sterol biosynthetic pathway yields high levels of sitosterol, which may play a structural role in stabilizing plant cell membranes (Bloch 1983; Schuler et al. 1991). Aberrant cell expansion in fk embryos might be explained if sitosterol is limiting in expanding cells. In addition, abnormal membranes in fk mutants could hinder the function of important signaling molecules that mediate either cell expansion or embryonic partitioning. Below we discuss the possibility that fk mutations affect signaling more directly, by disrupting the synthesis of a sterol with a signaling role.
The only sterol biosynthetic products with known physiological effects on plant growth are BRs, which act as signals for postembryonic cell expansion (Clouse and Sasse 1998). BR feeding elicited a response in fk seedlings but did not rescue their phenotype. This is in contrast to the rescue, by BR treatment of dim/dwf1/cbb1 and ste1/dwf7 dwarf mutant seedlings, which, like fk mutants, show reduced levels of both sitosterol and campesterol (Fig. 1; Gachotte et al. 1995; Klahre et al. 1998). Mutations affecting the BR-specific biosynthetic pathway or putative BR receptor (Li and Chory 1997) cause a generic dwarf plant phenotype but do not cause defects in the embryo (Altmann 1999). Therefore it is unlikely that the fk embryonic phenotypes are caused solely by a shortage of, or a defective response to BRs.
A novel sterol signal required for embryogenesis?
The patterning defects, as exemplified by multiple shoot meristems and apices in fk mutants, may reflect the absence of a novel sterol signal needed for embryonic cell–cell communication. Partitioning of the apex and formation of a meristematic zone in the embryo depends on positional cues from surrounding tissues (Laux and Jürgens 1997). The shoot meristem may receive positional information from the hypocotyl (Moussian et al. 1998). Could the partitioning defects in fk be attributed to a lesion in signaling between the hypocotyl and meristem? The embryonic root is also thought to receive patterning information from the hypocotyl (Hamann et al. 1999). fk seedlings with multiple roots probably arise from incorrectly partitioned basal regions, suggesting a defect in cell–cell interactions between the hypocotyl and developing root. Notably, the earliest visible phenotype in fk embryos is irregular cell expansion in globular stage central cells, the hypocotyl primordium. Thus, the fk patterning alterations could result from defective sterol signaling between the hypocotyl primordium and opposing meristematic zones.
In addition, the cellular defects in fk embryos may be explained by the lack of a novel sterol signal that regulates cell expansion. It is striking that cell expansion defects in fk embryos do not affect the polarity of the zygote and are not obvious until the globular stage, when about 60 cells have divided successfully. This observation argues against the possibility that fk affects only the synthesis of structural sterols.
FK mRNA expression is regulated during embryogenesis; not what one might expect of an enzymatic function that is simply coupled with membrane biosynthesis. Differential expression of the FK mRNA is first visible at the heart stage and is localized to apical and basal domains of the embryo as bilateral symmetry of the developing embryo becomes pronounced. In the torpedo-stage embryo, expression is strongest in the basal region and spreads as a diffuse signal into the hypocotyl primordium. However, in the mature embryo, when cell divisions have ceased, FK expression is conspicuously present in the shoot apical meristem and root tip. Thus, the FK expression pattern is consistent with the localized biosynthesis of a regulatory sterol in postglobular stages of embryogenesis.
In summary, fk represents a novel mutant phenotype associated with loss of sterol C-14 reductase activity in a multicellular organism. We propose that the embryonic defects observed in fk are due to lack of end-pathway sterol supply that needs to be present from the globular stage on. Mechanisms underlying embryonic development in animals include the use of sterols as signals for cell fates and growth of defined cell populations. However, analogous sterol molecules have not been identified to play a role in embryogenesis of plants. We provide the first genetic evidence that sterol production, which may include the synthesis of a sterol signal, is crucial for embryogenesis in Arabidopsis. Future studies will address whether FK is required solely for sterol biosynthesis or plays a more intimate role in a putative sterol signaling pathway.
Materials and methods
Plant strains and growth conditions
The Arabidopsis thaliana ecotypes Landsberg erecta (Ler) and WS were used as wild-type controls; Niederzenz (Nd) was used for genetic mapping. The alleles fk-X224 and fk-G214, fk-G251, fk-T293, fk-T329, and fk-U341 were induced in Ler by X-ray and EMS mutagenesis, respectively (Mayer et al. 1991). Allele fk-UU2099 was from a separate EMS mutagenesis (Mayer et al. 1999.) A T-DNA mutagenesis in the WS ecotype gave the untagged alleles fk-DCJ5 and fk-CPA11 (C. Bellini, unpubl.). fk alleles were maintained as FK/fk heterozygotes. Plants were grown on soil under continuous illumination at 18°C or 25°C as described previously (Mayer et al. 1993).
Phenotypic analyses
Seedling phenotypes were assayed on 1.7% agar, 100 mg/ml ampicillin, or on rich medium: 0.8% agar, MS salts (Murashige and Skoog 1962), 1% sucrose, 100 μg/ml ampicillin. The KNAT2–GUS marker was crossed to FK/fk plants. GUS staining was as in Sundaresan et al. (1995). For callus induction, seeds were surface sterilized (Hamann et al. 1999), germinated on rich medium, and transferred to liquid-rich medium containing 1 mg/l 2,4-D and 0.25 mg/l kinetin. Callus formation was evaluated after three weeks. Whole-mount preparations, histological analysis, and microscopy of ovules were as described previously (Mayer et al. 1993; Hamann et al. 1999). Images were processed with Photoshop 3.0 (Adobe, Mountain View, CA) and Aldus Freehand 7.0 software.
Molecular cloning of FK
FK was mapped to 77 cm on chromosome 3 by linkage to morphological markers gl1 to the left and tt5 to the right. FK/fk F2 plants from a cross of FK/fk-X224 (Ler) to wild type (Nd) were examined with CAPS markers g19397 and AP3-linked for (Ler/Nd) heterozygosity at both loci. Plants with this criterium were selected as parents of the recombinant population. An F3 population of 1000 plants derived from six parents was screened for recombination in the g19397–AP3 interval. One of these parents was homozygous (Nd) for the AP3 locus, reducing the mapping population to the right of FK to 660 plants. yUP (Ecker 1990) and CIC (Creusot et al. 1995) YAC libraries were used. The Resource Center/Primary Database of the German Human Genome Project, Berlin (Mozo et al. 1998) was the source of the F-BAC DNA library filter and clones. pBIC20 cosmid libraries containing Ler and Rld genomic inserts, respectively, were used (Meyer et al. 1994). YAC, BAC, and cosmid clones were isolated and aligned to a contig by hybridization experiments with end and internal fragments. YAC and BAC end probes were generated by TAIL–PCR (Liu and Whittier 1995). Two cDNA clones confirming the 3′ end of the mRNA were isolated from flower (Weigel et al. 1992) and silique-specific (Grebe et al. 2000) libraries, respectively. The silique-specific cDNA library was used to amplify and sequence a corresponding 1.1-kb cDNA fragment. Gene-specific primers used to amplify and sequence fk alleles from mutant seedlings are available upon request. Vectorette PCR (Devon et al. 1995; G. Martin and G. Jürgens, unpubl.) was used to isolate mutational break points in fk-X224 and fk-CPA11.
CAPS markers
The g19397 marker was derived from cosmid g19397 (Olszewski et al. 1988), by subcloning a 1.1-kb HindIII fragment into pBluescript KS+ (Stratagene). Primers 5′-CCGACAGTGGAATGCAGAGTTC-3′ and 5′-AGATGTAAGCAAGGCAAGCACC-3′ (annealing T = 60°) amplifed a 0.9-kb fragment. BsuRI digestion resulted in three fragments in Ler (0.55-, 0.28-, 0.07-kb) and two fragments in Nd (0.55, 0.28). The AP3-linked marker was derived from pD714, a 7.5-kb genomic BglII fragment (5′ of AP3) cloned into the BamHI site of pGEM3z+. It contains an 800-bp Sau3A1 fragment that is polymorphic between Ler (800 bp) and Nd, Col, WS (650 bp) (T. Jack, pers. comm.), which was cloned into the BamHI site of pBluescript KS+. Primers 5′-GCGTAAAGCGATAAGTGC-3′ and 5′-AGTGACGAACCACGATTCG-3′ (annealing T = 50°) amplified a Sau3A1 polymorphic fragment. The CYP97C marker is closely linked and 3′ of FK. Primers 5′-CTTCACTCTTTTCTCCATCTTCC-3′ and 5′-CCAAGTGAGGACAGATCCAG-3′ (Annealing T = 52°) amplified a 1.4-kb fragment from genomic DNA. Digestion with TaqI resulted in three fragments in Ler (0.6-, 0.2-, 0.1-kb) and four fragments in Nd (0.34, 0.26, 0.2, 0.1), and with DpnII resulted in two fragments in Ler (0.8, 0.3) and three fragments in Nd (0.6, 0.3, 0.2).
Complementation of fk-X224
The progeny of heterozygous FK/fk (Nd/Ler) plants were transformed by Agrobacterium GV3101 (pMP90) (Koncz and Schell 1986) mediated vacuum infiltration (Bechtold and Pelletier 1998) with 18 overlapping cosmids from the FK region. Transformants were selected on 0.8% agar, MS salts (Murashige and Skoog 1962), 1% sucrose, and 50 μg/ml kanamycin, and assayed for their genotype at the FK locus using the CAPS marker CYP97C. Progeny of six transformed plants showed complete correlation of the wild-type seedling phenotype with kanamycin resistance and GUS expression. Plants exhibiting complementation contained cosmids cBIC92 (one heterozygote) and cBIC98 (four heterozygotes and one fk homozygote).
5′ RACE PCR
The predicted transcription start of the FK gene was determined by 5′-RACE PCR using the 5′ RACE System for Rapid Amplification of cDNA Ends (Version 2.0 from Life Technologies). Stop codons upstream of the initiating methionine indicated that the cDNA contains the complete ORF. RNA from two different tissues, stems, and flowers, gave the same result for reverse transcription of the 5′ FK sequence. Gene-specific primers used for RT and PCR amplification of FK were 5′-TACAAGAATTCCTGAGAC-3′ and 5′-AACTTCGCCCAGTAACATACAATGC-3′.
In situ hybridization
For in situ hybridization (Mayer et al. 1998), 1266 bp of the FK cDNA including the entire coding sequence was amplified with the primers 5′-ACGATTTCGAATTCTTCATCTTCTCCTTTG-3′ and 5′-GTACTACAAAGTTTCACTCGAGCATTGCTA-3′, digested with XhoI/EcoRI and cloned into Bluescript pKS+ and pSK+ vectors (Stratagene) to generate pKS-FK1266 and pSK-FK1266. An antisense probe was synthesized by T7 RNA polymerase in vitro transcription using pSK-FK1266 as a template; a sense probe was synthesized using pKS-FK1266 as a template.
Heterologous expression of FK in S. cerevisiae
A total of 1266 bp containing the FK cDNA was amplified with the same primers as above, digested with XhoI/EcoRI and cloned into pYX212 (Ingenius MBV-028-10), which has a TPI promoter, 2 μ origin and URA3 selectable marker. 1480 bp containing the ERG24 gene was amplified from yeast genomic DNA with the primers 5′-ACGAAAAGGGGAGCTCTGCATAGATAGACC-3′ and 5′-CATAGGAGTAAAGACTTCGGTGTAGAG-3′, digested with EcoRI/SacI and cloned into pYX212. Yeast strains were: erg24::LEU2 his3Δ200 leu2Δ1 trp1Δ63 ade2-101 lys2-801 ura3-52 and erg4-1 his7-2 ura3-52. Yeast transformation was performed as in Gietz et al. (1992), except that 10 μg of plasmid DNA was used for transformation of erg24. Media were according to Sherman (1991). Permissive growth conditions were in −URA + 20 mm CaCl2 synthetic medium. To assay growth under restrictive conditions, cells were grown overnight in permissive medium and inoculated into fresh calcium poor medium (YPD) at 2.5 × 106 cells/ml.
Sterol extraction and GC/MS analysis
The plant material (100–500 mg) was extracted in acetone for a period of 72 hr. After evaporation in vacuo, the residue was partitioned between CHCl3 and water. The CHCl3-soluble fraction was evaporated in vacuo, resuspended in n-hexane, purified with a silica gel (60) (0.040–0.063 mm) cartridge column, and eluted with 7/3 n-hexane/ethyl acetate. The eluate was evaporated in vacuo, resuspended in 98/2 MeOH/water, purified with a RP18 (Merck) cartridge and eluted with 98/2 MeOH/water. Extracts were assayed by thin-layer chromatography (running buffer 95/5 CHCl3/MeOH). Acetylation of 0.2–1.0 mg of the purified fraction was in 1 ml of 1/1 pyridine/acetic acid anhydride overnight at room temperature, followed by evaporation in vacuo. GC/MS analysis of the residue was under the following conditions: MD-800, Fisons Instruments (70 eV EI, source temperature 200°C), column DB-5MS (J&W, 15 m × 0.32 mm, 0.25 μm film thickness), injection temperature 250°C, interface temperature 300°C, carrier gas He, flow rate 1 ml/min, splitless injection, column temperature program (170°C for 1 min, then raised to 270°C at a rate of 25°C min−1 and then to 290°C at a rate of 2°C min−1).
Treatment with sterols and BRs
Seeds from heterozygous FK/fk plants were germinated on sterol-containing half-concentrated MS (Murashige and Skoog 1962) medium with 0.7% agarose. Sterols (from Sigma, except stigmasterol, from Roth) were dissolved in absolute ethanol, filter-sterilized and added to the medium at 25 μm for campesterol and sitosterol, and 1 μm for cholesterol, stigmastanol, stigmasterol, sitosterol, 24-epibrassinolide, and 22S,23S-homobrassinolide.
Computer analyses
Sequence data were analyzed with MacVector (Kodak Scientific Imaging Systems, New Haven, CT). PSORT version 6.4 (www) was used for prediction of protein sorting. BLAST searches were done using the databases at the NCBI (www). Protein topology predictions of the FK protein were generated with TMpred (ISREC). Protein alignments and phylogenetic trees were generated with Multiple Sequence Alignment v.5.0.3 (InforMax Biosuite).
Accession numbers
The following accession numbers have been deposited with GenBank: AF256536 (FK genomic region); AF256535 (FK cDNA).
Acknowledgments
We thank Michel Delseny, David Bouchez, and Marcel Salanoubat for providing YAC contig data for chromosome 3 prior to publication; Tom Jack for providing plasmid pD714; Erwin Grill for cosmid libraries; Martin Bard for S. cerevisiae strains; Karen Deuschle, Achim Haecker, Rita Gross-Hardt, Uli Hiller, Christian Leischner, Titus Neumann, and Marion Zobel for technical assistance; and Niko Geldner, Markus Grebe, Maren Heese, Viktor Kirik, Thomas Laux, Michael Lenhard, Hubert Schaller, and ArpSchnittger for critical reading of the manuscript. This work was supported by a National Science Foundation Postdoctoral Research Fellowship in Plant Biology to K.S., a Beningsen Research Prize to J.L.D., and a Leibniz Award to G.J.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kathrin.schrick@uni-tuebingen.de; FAX 49 7071 29 5797.
References
- Altmann T. Molecular physiology of brassinosteroids revealed by the analysis of mutants. Planta. 1999;208:1–11. doi: 10.1007/s004250050528. [DOI] [PubMed] [Google Scholar]
- Assaad FF, Mayer U, Wanner G, Jürgens G. The KEULE gene is involved in cytokinesis in Arabidopsis. Mol Gen Genet. 1996;253:267–277. doi: 10.1007/pl00008594. [DOI] [PubMed] [Google Scholar]
- Bach TJ, Benveniste P. Cloning of cDNAs or genes encoding enzymes of sterol biosynthesis from plants and other eukaryotes: Heterologous expression and complementation analysis of mutations for functional characterization. Prog Lipid Res. 1997;36:197–226. doi: 10.1016/s0163-7827(97)00009-x. [DOI] [PubMed] [Google Scholar]
- Bechtold N, Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol Biol. 1998;82:259–266. doi: 10.1385/0-89603-391-0:259. [DOI] [PubMed] [Google Scholar]
- Benveniste P. Sterol biosynthesis. Annu Rev Plant Physiol. 1986;37:275–308. [Google Scholar]
- Bloch KE. Sterol structure and membrane function. Crit Rev Biochem. 1983;14:47–91. doi: 10.3109/10409238309102790. [DOI] [PubMed] [Google Scholar]
- Campbell CE, Kircher HW. Senita cactus: A plant with interrupted sterol biosynthetic pathways. Phytochemistry. 1980;19:2777–2779. [Google Scholar]
- Choe S, Dilkes BP, Gregory BD, Ross AS, Yuan H, Noguchi T, Fujioka S, Takatsuto S, Tanaka A, Yoshida S, et al. Arabidopsis dwarf1 is defective in the conversion of 24-methylenecholesterol to campesterol in brassinosteroid biosynthesis. Plant Physiol. 1999a;119:897–907. doi: 10.1104/pp.119.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Noguchi T, Fujioka S, Takatsuto S, Tissier CP, Gregory BD, Ross AS, Tanaka A, Yoshida S, Tax FE, et al. The Arabidopsis dwf7/ste1 mutant is defective in the Δ7 sterol C-5 desaturation step leading to brassinosteroid biosynthesis. Plant Cell. 1999b;11:207–221. [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. Brassinosteroids: Essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- Clouse SD, Hall AF, Langford M, McMorris TC, Baker MEJ. Physiological and molecular effects of brassinosterioids on Arabidopsis thaliana. J Plant Growth Regul. 1993;12:61–66. [Google Scholar]
- Creusot F, Fouilloux E, Dron M, Lafleuriel J, Picard G, Billault A, Lepaslier D, Cohen D, Chaboute ME, Durr A, et al. The CIC library—a large insert YAC library for genome mapping in Arabidopsis thaliana. Plant J. 1995;8:763–770. doi: 10.1046/j.1365-313x.1995.08050763.x. [DOI] [PubMed] [Google Scholar]
- Crowley JH, Tove S, Parks LW. A calcium-dependent ergosterol mutant of Saccharomyces cerevisiae. Curr Genet. 1998;34:93–99. doi: 10.1007/s002940050371. [DOI] [PubMed] [Google Scholar]
- Devon RS, Porteous DJ, Brookes AJ. Splinkerettes—improved vectorettes for greater efficiency in PCR walking. Nucleic Acids Res. 1995;23:1644–1645. doi: 10.1093/nar/23.9.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dockx J, Quaedvlieg N, Keultjes G, Kock P, Weisbeek P, Smeekens S. The homeobox gene ATK1 of Arabidopsis thaliana is expressed in the shoot apex of the seedlings and in flowers and inflorescence stems of mature plants. Plant Mol Biol. 1995;28:723–737. doi: 10.1007/BF00021196. [DOI] [PubMed] [Google Scholar]
- Ecker JR. PFGE and YAC analysis of the Arabidopsis genome. Methods Enzymol. 1990;1:186–194. [Google Scholar]
- Farese RV, Jr, Herz J. Cholesterol metabolism and embryogenesis. Trends Genet. 1998;3:115–120. doi: 10.1016/s0168-9525(97)01377-2. [DOI] [PubMed] [Google Scholar]
- Gachotte D, Meens R, Benveniste P. An Arabidopsis mutant deficient in sterol biosynthesis: Heterologous complementation by ERG3 encoding a Δ7-sterol-C-5-desaturase from yeast. Plant J. 1995;8:407–416. doi: 10.1046/j.1365-313x.1995.08030407.x. [DOI] [PubMed] [Google Scholar]
- Gietz D, St. Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;15:1543–1558. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Grebe M, Gadea J, Steinmann T, Kientz M, Rahfeld J-U, Salchert K, Koncz C, Jürgens G. A conserved domain of Arabidopsis ARF GEF mediates subunit interaction and cyclophilin 5 binding. Plant Cell. 2000;12:343–356. doi: 10.1105/tpc.12.3.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann T, Mayer U, Jürgens G. The auxin-insensitive bodenlos mutation affects primary root formation and apical–basal patterning in the Arabidopsis embryo. Development. 1999;126:1387–1395. doi: 10.1242/dev.126.7.1387. [DOI] [PubMed] [Google Scholar]
- Hartmann MA, Benveniste P. Plant membrane sterols: Isolation, identification and biosynthesis. Methods Enzymol. 1987;148:632–650. [Google Scholar]
- Holmer L, Pezhman A, Worman HJ. The human lamin B receptor/sterol reductase multigene family. Genomics. 1998;54:469–476. doi: 10.1006/geno.1998.5615. [DOI] [PubMed] [Google Scholar]
- Husselstein T, Schaller H, Gachotte D, Benveniste P. Delta7-sterol-C5-desaturase: Molecular characterization and functional expression of wild-type and mutant alleles. Plant Mol Biol. 1999;39:891–906. doi: 10.1023/a:1006113919172. [DOI] [PubMed] [Google Scholar]
- Jürgens G, Mayer U. Arabidopsis. In: Bard J, editor. Embryos: A colour atlas of development. London, UK: Wolfe Publications; 1994. pp. 7–21. [Google Scholar]
- Kauschmann A, Jessop A, Koncz C, Szekeres M, Willmitzer L, Altmann T. Genetic evidence for an essential role of brassinosteroids in plant development. Plant J. 1996;9:701–713. [Google Scholar]
- Klahre U, Noguchi T, Fujioka S, Takatsuto S, Yokota T, Nomura T, Yoshida S, Chua NH. The Arabidopsis DIMINUTO/DWARF1 gene encodes a protein involved in steroid synthesis. Plant Cell. 1998;10:1677–1690. doi: 10.1105/tpc.10.10.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
- Lai MH, Bard M, Pierson CA, Alexander JF, Goebl M, Carter GT, Kirsch D. The identification of a gene family in the Saccharomyces cerevisiae ergosterol biosynthesis pathway. Gene. 1994;140:41–49. doi: 10.1016/0378-1119(94)90728-5. [DOI] [PubMed] [Google Scholar]
- Laux T, Jürgens G. Embryogenesis: A new start in life. Plant Cell. 1997;9:989–1000. doi: 10.1105/tpc.9.7.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees ND, Bard M, Kirsch DR. Biochemistry and molecular biology of sterol synthesis in Saccharomyces cerevisiae. Crit Rev Biochem Mol Biol. 1999;34:33–47. [PubMed] [Google Scholar]
- Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- Liu YG, Whittier RF. Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics. 1995;25:674–681. doi: 10.1016/0888-7543(95)80010-j. [DOI] [PubMed] [Google Scholar]
- Long JA, Moan EI, Medford JI, Barton MK. A member of the KNOTTED class of homeodomain proteins encoded by the STM gene of Arabidopsis. Nature. 1996;379:66–69. doi: 10.1038/379066a0. [DOI] [PubMed] [Google Scholar]
- Lorenz TR, Parks LW. Cloning, sequencing, and disruption of the gene encoding sterol C-14 reductase in Saccharomyces cerevisiae. DNA Cell Biol. 1992;11:685–692. doi: 10.1089/dna.1992.11.685. [DOI] [PubMed] [Google Scholar]
- Marcireau C, Guyonnet D, Karst F. Construction and growth properties of a yeast strain defective in sterol 14-reductase. Curr Genet. 1992;22:267–272. doi: 10.1007/BF00317919. [DOI] [PubMed] [Google Scholar]
- Mayer KF, Schoof H, Haecker A, Lenhard M, Jürgens G, Laux T. Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell. 1998;95:805–815. doi: 10.1016/s0092-8674(00)81703-1. [DOI] [PubMed] [Google Scholar]
- Mayer U, Torres-Ruiz RA, Berleth T, Misera S, Jürgens G. Mutations affecting the body organization in the Arabidopsis embryo. Nature. 1991;353:402–407. [Google Scholar]
- Mayer U, Büttner G, Jürgens G. Apical–basal pattern formation in the Arabidopsis embryo: Studies on the role of the gnom gene. Development. 1993;117:149–162. [Google Scholar]
- Mayer U, Herzog U, Berger F, Inze D, Jürgens G. Mutations in the PILZ group genes disrupt the microtubule cytoskeleton and uncouple cell cycle progression from cell division in Arabidopsis embryo and endosperm. Eur J Cell Biol. 1999;78:100–108. doi: 10.1016/S0171-9335(99)80011-9. [DOI] [PubMed] [Google Scholar]
- Meyer KM, Leube P, Grill E. A protein phosphatase 2C involved in ABA signal transduction in Arabidopsis thaliana. Science. 1994;264:1452–1455. doi: 10.1126/science.8197457. [DOI] [PubMed] [Google Scholar]
- Moussian B, Schoof H, Haecker A, Jürgens G, Laux T. Role of the ZWILLE gene in the regulation of central shoot meristem cell fate during Arabidopsis embryogenesis. EMBO J. 1998;17:1799–1809. doi: 10.1093/emboj/17.6.1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozo T, Fischer S, Meier-Ewert S, Lehrach H, Altmann T. Use of the IGF BAC library for physical mapping of the Arabidopsis thaliana genome. Plant J. 1998;16:377–384. doi: 10.1046/j.1365-313x.1998.00299.x. [DOI] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- Olszewski NE, Martin FB, Ausubel FM. Specialized binary vector for plant transformation: Expression of the Arabidopsis thaliana AHAS gene in Nicotiana tabacum. Nucleic Acids Res. 1988;16:10766–10782. doi: 10.1093/nar/16.22.10765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ourisson G. Pecularities of sterol biosynthesis in plants. J Plant Physiol. 1994;143:434–439. [Google Scholar]
- Patterson GW, Hugly S, Harrison D. Sterols and phytyl esters of Arabidopsis thaliana under normal and chilling temperatures. Phytochemistry. 1993;33:1381–1383. [Google Scholar]
- Prakash A, Sengupta S, Aparna K, Kasbekar P. The erg-3 (sterol Δ14-15-reductase) gene of Neurospora crassa: Generation of null mutants by repeat-induced point mutation and complementation by proteins chimeric for human lamin B receptor sequences. Microbiology. 1999;145:1443–1451. doi: 10.1099/13500872-145-6-1443. [DOI] [PubMed] [Google Scholar]
- Sakurai A, Fujioka S. Studies on biosynthesis of brassinosteroids. Biosci Biotechnol Biochem. 1997;61:757–762. doi: 10.1271/bbb.61.757. [DOI] [PubMed] [Google Scholar]
- Schaller H, Gondet L, Maillot-Vernier P, Benveniste P. Sterol overproduction is the biochemical basis of resistance to a triazole in calli from a tobacco mutant. Planta. 1994;194:295–305. [Google Scholar]
- Schmitt P, Scheid F, Benveniste P. Accumulation of delta8,14 sterols in suspension cultures of bramble cells cultured with an azasterol antimycotic agent (A25822B) Phytochemistry. 1980;19:525–530. [Google Scholar]
- Schuler I, Milon A, Nakatani Y, Ourisson G, Albrecht AM, Benveniste P, Hartmann MA. Differential effects of plant sterols on water permeability and on acyl chain ordering of soybean phosphatidylcholine bilayers. Proc Natl Acad Sci. 1991;88:6926–6930. doi: 10.1073/pnas.88.16.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Silve S, Dupuy PH, Ferrara P, Loison G. Human lamin B receptor exhibits sterol C14-reductase activity in Saccharomyces cerevisiae. Biochim Biophys Acta. 1998;1392:233–244. doi: 10.1016/s0005-2760(98)00041-1. [DOI] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes & Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- Taton M, Benveniste P, Rahier A. Microsomal delta 8,14-sterol delta 14-reductase in higher plants. Characterization and inhibition by analogues of a presumptive carbocationic intermediate of the reduction reaction. Eur J Biochem. 1989;185:605–614. doi: 10.1111/j.1432-1033.1989.tb15156.x. [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-n. [DOI] [PubMed] [Google Scholar]