Abstract
Remarkable progress has been made in the field of G protein coupled receptor (GPCR) structural biology during the past four years. Several obstacles to generating diffraction quality crystals of GPCRs have been overcome by combining innovative methods ranging from protein engineering to lipid-based screens and microdiffraction technology. The initial GPCR structures represent energetically stable inactive-state conformations. However, GPCRs signal through different G protein isoforms or G protein-independent effectors upon ligand binding suggesting the existence of multiple ligand-specific active states. These active-state conformations are unstable in the absence of specific cytosolic signaling partners representing new challenges for structural biology. Camelid single chain antibody fragments (nanobodies) show promise for stabilizing active GPCR conformations and as chaperones for crystallogenesis.
Introduction (journal format)
G protein-coupled receptors –GPCRs– are the largest class of receptors in the human genome and are the most commonly targeted membrane protein class for medicinal therapeutics. Over the past three decades, great progress has been made in characterizing the pharmacology, cellular physiology and in vivo function of many members of this family. The paradigm of GPCR signaling involves activation of heterotrimeric G proteins (Gαβγ). The inactive Gαβγ heterotrimer is composed of two principal elements, Gα•GDP and the Gβγ heterodimer. Gβγ sequesters the switch II element on Gα such that it is unable to interact with other proteins in the second messenger systems. Activated GPCRs catalyze the release of GDP from Gα, allowing GTP to bind and liberate the activated Gα-GTP subunit. In this state, switch II forms a helix stabilized by the γ-phosphate of GTP allowing it to interact with effectors such as adenylyl cyclase. Although much progress has been made in understanding how G α subunits interact with and regulate the activity of their downstream targets, it is not clear how activated GPCRs initiate this process by catalyzing nucleotide exchange on Gαβγ.[1].
In the classical models, signaling by the activated GPCR is terminated by phosphorylation of the cytoplasmatic loops and/or tail of the receptor by GPCR kinases (GRKs). This results in the binding of arrestins that mediate receptor desensitization and internalization via clathrin-coated pits. This classical model is both oversimplified and incomplete. Over the past decade, we learned that arrestins not only act as regulators of GPCR desensitization but also as multifunctional adaptor proteins that have the ability to signal through multiple effectors such as MAPKs, SRC, NF-kB and PI3K [2]. In this revised model, β-arrestins are interacting with and recruiting intracellular signaling molecules, as well as mediating desensitization. It is still unclear whether the same receptor conformations that result in arrestin-mediated signal transduction also lead to receptor desensitization. For a number of different receptor systems, it has been found that the G protein dependent and the arrestin dependent signaling events are pharmacologically separable [3]. In other words, a class of ligands referred to as biased agonists selectively trigger signaling towards one pathway over the other; that is, they preferentially signal through either the G protein- or arrestin-mediated pathway [4]. It thus appears that GPCRs, despite their small size, are sophisticated allosteric machines with multiple signaling outputs. Characterizing these functionally distinct structures is challenging, but essential for understanding the mechanism of physiologic signaling and for developing more effective drugs.
Active-state GPCR structures
Polytopic membrane proteins such as GPCRs, transporters and channels are dynamic proteins that exist in an ensemble of functionally distinct conformational states [5]. Crystallogenesis typically traps the most stable low energy states, making it difficult to obtain high-resolution structures of other less stable but biologically relevant functional states. The first structures of rhodopsin covalently bound to 11-cis-retinal represent a completely inactive state with virtually no basal activity [6–7]. Similarly, the first crystal structures of GPCRs for hormones and neurotransmitters were bound to inverse agonists and represent inactive conformations. These include the human β2AR [8–10], the avian β1AR [11], the human D3 dopamine [12], the human CXCR4 [13] receptor, the human adenosine A2A receptor [14] and the human histamine H1 receptor [15].
As summarized above, there is a growing body of evidence that GPCRs are conformationally complex and can signal through different pathways in a ligand specific manner. The functional complexity suggests multiple active states. For the purpose of this review, we will focus on G protein activation and define an “active-state” structure is one that is competent to couple to and catalyze nucleotide exchange on a G protein. The first active-state GPCR structure was that of opsin, the retinal-free form of rhodopsin [16]. Upon light activation, retinal isomerizes and initiates a series of conformational changes leading to the formation of metarhodopsin II, the conformational state capable of activating the G protein tranducin [17]. Following the formation of metarhodopsin II, the Schiff base is hydrolyzed and retinal dissociates to generate opsin (the retinal-free form of rhodopsin). Under physiologic pH opsin is a very weak activator of transducin, but at reduced pH (5–6) it assumes a more active conformation that is nearly identical to metarhodopsin II as determined by FTIR spectroscopy [18]. This is in agreement with previous studies demonstrating a role of protonation in the process of rhodopsin activation [19]. In 2008, Hofmann, Ernst and colleagues reported the structure of opsin obtained from crystals grown at pH5 [16] as well as the structure of opsin bound to the C terminal peptide of transducin [20], the G protein activated by rhodopsin. Recently two new active-state structures of rhodopsin have been obtained: metarhodopsin II [21], and a constitutively active mutant of opsin bound to trans-retinal [22]. Both of these structures also include the C-terminnal peptide of transducin. All these active-state rhodopsin structures have in common that they were obtained from crystals grown at a pH between 4.5 and 6.0, and all show the same overall structural changes observed originally in the first opsin structure (lacking both trans-retinal and the transducin peptide), suggesting that the pH plays the most important role in stabilizing the active conformation of this protein.
Efforts to obtain active-state structures of other GPCRs has been more challenging. Recent crystal structures of the β2AR bound to a covalent agonist [23] and a thermostabilized avian β1AR bound to several agonists and partial agonists [24] are inactive conformations. This is consistent with previous studies suggesting that, under physiologic conditions (pH, ionic strength) agonist alone is not sufficient to stabilize a fully active conformation of the β2AR [25]. Like rhodopsin, the β2AR becomes more active at reduced pH; however, it also becomes less stable and denatures below pH 6.5 [26].
Nanobodies as G protein surrogates
For GPCRs that do not tolerated acidic conditions, stabilization of an active conformation can be achieved in different ways. The most physiologic approach is to use a native signaling partner such as a G protein or arrestin. Unfortunately, interactions of GPCRs with G proteins or arrestins are highly sensitive to pH, detergents and nucleotides used during the solubilization and purification of these proteins. It has therefore been difficult to form complexes of sufficient stability for crystallography. An alternative approach is to apply mutagenesis [27] to enhance the stability of the active conformation. Constitutively active mutants have been described for many GPCRs including the β2AR [28]. These mutations lead to a high level of basal, agonist independent signaling. However, for the β2AR, these mutations are also associated with reduced expression and structural instability [29].
An alterative to using a G protein or arrestin is to identify another binding protein that can stabilize the same conformational state stabilized by a native signaling partner. A characteristic feature of the active state of many GPCRs in a GPCR-G protein complex is an increase in agonist affinity relative to the GPCR alone [30]. For example, the β2AR couples preferentially to Gs, the stimulatory G protein for adenylyl cyclase. The affinity of the agonist isoproterenol for the β2AR-Gs complex is approximately 100 fold higher that its affinity for the β2AR alone [31]. The requirement for Gs to stabilize the β2AR in an active conformation is consistent with the cooperative nature of the agonist-β2AR-Gs complex. Arrestin has been shown to have a similar effect on β2AR affinity for agonists [32–33]. We therefore attempted to identify G protein surrogates that would exhibit similar properties.
Antibodies evolved to bind to a diverse array of protein structures with high affinity and specificity, and are therefore logical candidates for stabilizing specific GPCR conformations. Nanobodies are antibody-derived single domain proteins that contain the unique structural and functional properties of heavy chain only antibodies that naturally occur in Camelids [34]. Nanobodies are small (15kD) and stable single domain fragments harboring the full antigen-binding capacity of the original heavy chain only antibodies [35]. Nanobodies are encoded by single genes and are efficiently produced in prokaryotic and eukaryotic hosts including bacteria and yeast [36]. They exhibit a superior stability compared to conventional antibodies and derivatives thereof like FABs or scFvs [37]. Due to their unique 3-dimensional structure, nanobodies have access to cavities or clefts on the surface of proteins [38–39•]. These cryptic epitopes are largely inaccessible to conventional antibodies but can be readily recognized by a long and protruding CDR3 loop of the nanobody (Fig.1). The nanobody platform has the competitive advantage to other recombinant scaffold libraries in that large numbers (109) of fragments harboring the full antigen-binding capacity of genuine in vivo matured antibodies can be screened for high affinity binders in a couple of days, allowing one to fully exploit the humoral response of large mammals against native antigens. The applications of nanobodies in structural biology are numerous. Nanobodies can trap unstable structural intermediates along the fibrillation pathway of amyloidogenic proteins [40•]. A multidomain protein is more rigid in a complex with a nanobody than the multidomain protein by itself [41]. In complex with a nanobody, the total amount of structured polypeptide increases, thus providing a much better starting point for the crystallization of intrinsically unfolded proteins [42]. Nanobodies can also be used to stabilize the protomers of larger protein assemblies [43] in one-to-one heterodimers. With the exception of one case [44], all nanobodies that have been characterized in complex with an antigen recognize discontinuous amino acids that come together in native protein conformations (i.e. conformational epitopes), making them ideal tools to selectively stabilize specific conformational states of (membrane) proteins.
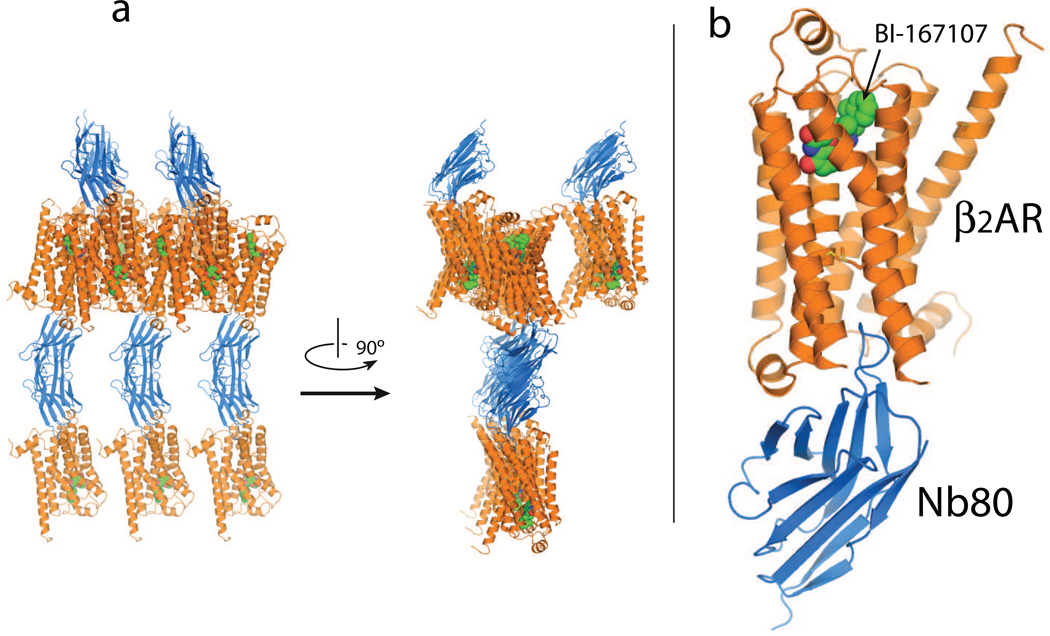
Figure 1.
Agonist-β2AR-T4L-Nb80 complex in crystals formed in lipidic cubic phase [45••]. (a) Two different views of the crystal packing: β2AR indicated in orange and Nb80 in blue. The β2AR-nanobody complexes are arranged with the lipid bilayers approximately parallel to the bc plane of the crystal. Twofold symmetry related nanobody molecules interact along the a axis to generate a tightly packed lattice in this direction. (b) Nb80 binds to the cytoplasmic end of the β2AR, with the third complementarity determining region loop (CDR3) projecting 14Å into the core of the receptor.
The greatest challenge to generating a nanobody that recognizes and stabilizes an active-state GPCR structure is preparing the active-state antigen. Most commercially available β2AR agonists are relatively low affinity and would dissociate rapidly after immunization. Therefore, as a first step we screened over 50 proprietary β2AR agonists provided by several pharmaceutical companies and identified a full agonist with an affinity of 84 pM and a dissociation half-life of approximately 30 hours (BI-167107, Boehringer Ingelheim) [45••]. β2AR was purified by antibody and ligand affinity chromatography to guarantee a high specific activity [45••]. Receptor was loaded with the high affinity agonist and reconstituted at high protein to lipid ratio. The high affinity and slow off-rate of the agonist increased the probability of maintaining the β2AR in an active conformation following immunization. Under the conditions of reconstitution, receptors were oriented randomly in the lipid bilayer of the vesicle.
One llama (Lama glama) received six weekly administrations of 100µg of the reconstituted agonist-bound receptor. Lymphocytes were isolated from the blood of the immunized llama and total RNA was prepared from these cells. The coding sequences of the nanobody repertoire were amplified by RT-PCR and cloned into a phage display vector [46]. β2AR specific phages were enriched in vitro by bio-panning on the immobilized receptor. Antigen bound phages were recovered from antigen-coated wells by the addition of freshly grown E. coli cells. After 2 rounds of panning, 96 individual colonies were randomly picked and the nanobodies produced as a soluble His-tagged protein in the periplasm of E. coli. The initial solid-phase ELISA screen identified 16 nanobodies that recognized native, but not heat denatured β2AR. Of these, 7 bound preferentially to agonist-bound β2AR as determined by size exclusion chromatography. One of these was selected based on its effect on β2AR agonist binding affinity. When bound to nanobody 80 (Nb80) the β2AR affinity for the catecholamine agonist isoproterenol increased by 100 fold, nearly identical to the effect observed when the β2AR is complexed with Gs [31].
Nanobody-assisted crystallography of GPCRs
The β2AR-T4L-Nb80 complex was crystallized in lipidic cubic phase [47]. Diffraction data were collected using minibeam technology [48] and the solution determined by molecular replacement [45••]. Fig. 1a shows the crystallographic packing of the β2AR-T4L-Nb80 complex. Crystallographic contacts are primarily mediated by Nb80. As shown in Fig. 1b, the long CDR3 loop of the nanobody projects into the transmembrane core occupying a position nearly identical to the transducin peptide in opsin [20].
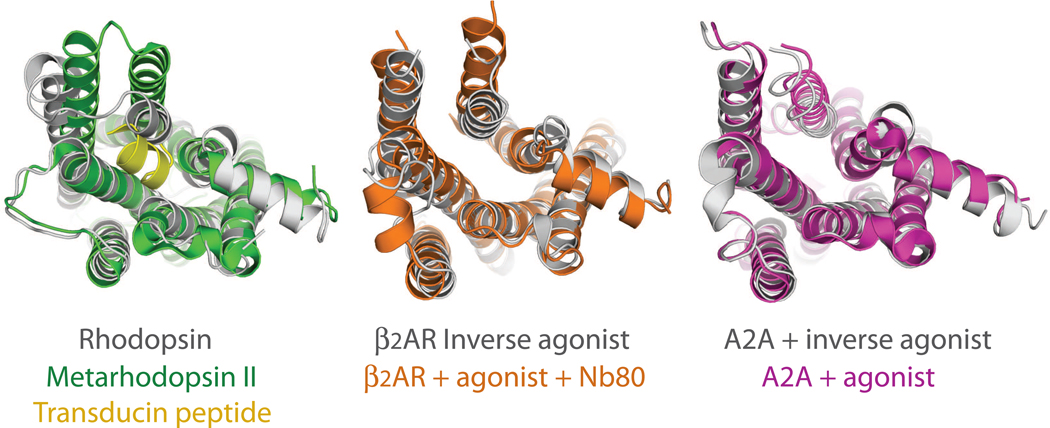
Fig. 2 compares the inactive and active state structures of metarhosopsin II [6,20] and the β2AR [9,45••], as well as the recent agonist and antagonist bound structures of the adenosine receptor [14,49]. In metarhodopsin II, the largest change is observed in the cytoplasmic end of TM6, where an approximately 6Å outward movement allows the docking of the carboxyl terminal peptide of transducin (Fig. 2a shown in yellow). The conformational changes in the nanobody-stabilized β2AR structure are similar to those observed in metarhodoopsin II, except for a larger (11Å) outward movement of the cytoplasmic end of TM6. The agonist-bound adenosine receptor structure (Fig. 2c) shows some of the conformational changes observed in opsin; however, the magnitude of the conformational change in TM6 would not accommodate the carboxyl terminal peptide of a G protein as observed in metarhodopsin II. Like opsin, crystals of the agonist-bound adenosine receptor were grown at pH 5.0 – 5.5 [49], suggesting that protonation may have played a role in stabilizing the partial activation state. The relatively large agonist used (UK-432097, 778 Daltons) may also have contributed to the observed differences with the antagonist structure. The T4 lysozyme fusion used to obtain the Adenosine receptor crystals may have restricted movements of TMs 5 and 6, preventing it from assuming a fully active conformation. The larger conformational change observed in the β2AR-Nb80 complex may raise concern that the nanobody has trapped the β2AR in a nonnative conformation. In such a case however, the nanobody would pay a substantial energetic penalty for distorting the β2AR structure into a conformation that does not appreciably exist in the absence of the bound nanobody. In the case of the β2AR, the Nb80 stabilized state has an increased agonist binding affinity that is identical to that observed for the β2AR-Gs protein complex [45••]. This would not be expected if Nb80 bound to and stabilized a non-physiologic receptor conformation.
Figure 2.
Cytoplasmic view of the active- and inactive-state rhodopsin and β2AR structures, compared to the antagonist and agonist bound structures of the adenosine receptor. (a) rhodopsin [6] compared to metarhodopsin II in complex with the transducing peptide [20] (b) β2AR-TL4 bound to the inverse agonist carazolol [9] overlayed on β2AR-TL4-Nb80 bound to the agonist BI-167107 [45••] (c) and superposition of the antagonist ZM241385 bound [14] on the agonist UK-432097 bound adenosine receptor [49].
In summary, nanobodies represent a new tool for membrane protein structural biology. They efficiently rigidify flexible regions and are able to stabilize specific conformations of polytopic membrane proteins. Nanobodies should facilitate obtaining structures of non-engineered hormone receptors in different functional states, providing new insights into the structural basis of ligand efficacy and biased signaling.
Highlights.
Last four years, several inactive-state GPCR structures have been solved
Active-state structures may be unstable without a native signaling partner
Nanobodies act as surrogates of GPCR signaling partners
Nanobody 80 has G protein-like properties and stabilizes an agonist activated state of the β2AR
Acknowledgements
The work in JS’s laboratory is supported by Vrije Universiteit Brussel (VUB, GOA65), Vlaams Instituut Biotechnologie (VIB), the Fund for Scientific Research of Flanders (FWOAL551), the Hercules Foundation (HERC2) and and the Institute for the encouragement of Scientific Research and Innovation of Brussels (ISRIB). BKK receives support from National Institutes of Health Grants NS028471 and GM083118 and the Mathers Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors declare that they have no conflict of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Tesmer JJG. The quest to understand heterotrimeric G protein signaling. Nat Struct ol Biol. 2010;17:650–652. doi: 10.1038/nsmb0610-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kovacs JJ, Hara MR, Davenport CL, Kim J, Lefkowitz RJ. Arrestin development: merging roles for beta-arrestins in developmental signaling pathways. Developmental cell. 2009;17:443–458. doi: 10.1016/j.devcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whalen EJ, Rajagopal S, Lefkowitz RJ. Therapeutic potential of β-arrestin- and G protein-biased agonists. Trends in Molecular medicine. 2011;17:126–139. doi: 10.1016/j.molmed.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajagopal S, Rajagopal K, Lefkowitz RJ. Teaching old receptors new tricks: biasing seven-transmembrane receptors. Nat Rev Drug discov. 2010;9:373–386. doi: 10.1038/nrd3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deupi X, Kobilka BK. Energy Landscapes as a Tool to Integrate GPCR Structure, Dynamics, and Function. Physiology. 2010;25:293–303. doi: 10.1152/physiol.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 7.Li J, Edwards P, Burghammer M, Villa C, Schertler GFX. Structure of bovine rhodopsin in a trigonal crystal form. J mol biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 8.Rasmussen SGF, Choi H-J, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VRP, Sanishvili R, Fischetti RF, et al. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 9.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Yao X-J, Weis WI, Stevens RC, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 10.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H-J, Kuhn P, Weis WI, Kobilka BK, et al. High-resolution crystal structure of an engineered human 2-adrenergic G protein coupled receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warne T, Serrano-Vega MJ, Baker JG, Moukhametzianov R, Edwards PC, Henderson R, Leslie AGW, Tate CG, Schertler GFX. Structure of a β1-adrenergic G-protein-coupled receptor. Nature. 2008;454:486–491. doi: 10.1038/nature07101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chien EYT, Liu W, Zhao Q, Katritch V, Won Han G, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V, et al. Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science. 2010;330:1091–1095. doi: 10.1126/science.1197410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010;1066 doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaakola V-P, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, IJzerman AP, Stevens RC. The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW, et al. Structure of the human histamine H1 receptor complex with doxepin. Nature. 2011 doi: 10.1038/nature10236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JH, Scheerer P, Hofmann KP, Choe H-W, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 17.Hofmann KP. Signalling states of photoactivated rhodopsin. Novartis Foundation symposium. 1999;224:158–175. doi: 10.1002/9780470515693.ch10. discussion 175–180. [DOI] [PubMed] [Google Scholar]
- 18.Vogel R, Siebert F. Conformations of the active and inactive states of opsin. The Journal biol chem. 200;276:38487–38493. doi: 10.1074/jbc.M105423200. [DOI] [PubMed] [Google Scholar]
- 19.Arnis S, Fahmy K, Hofmann KP, Sakmar TP. A conserved carboxylic acid group mediates light-dependent proton uptake and signaling by rhodopsin. J Biol Chem. 1994;269:23879–23881. [PubMed] [Google Scholar]
- 20.Scheerer P, Park JH, Hildebrand PW, Kim YJ, Krauss N, Choe H-W, Hofmann KP, Ernst OP. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 21.Choe H-W, Kim YJ, Park JH, Morizumi T, Pai EF, Krauβ N, Hofmann KP, Scheerer P, Ernst OP. Crystal structure of metarhodopsin II. Nature. 2011:1–6. doi: 10.1038/nature09789. [DOI] [PubMed] [Google Scholar]
- 22.Standfuss J, Edwards PC, D’Antona A, Fransen M, Xie G, Oprian DD, Schertler GFX. The structural basis of agonist-induced activation in constitutively active rhodopsin. Nature. 2010;45:656–660. doi: 10.1038/nature09795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rosenbaum DM, Zhang C, Lyons JA, Holl R, Aragao D, Arlow DH, Rasmussen SGF, Choi H-J, DeVree BT, Sunahara RK, et al. Structure and function of an irreversible agonist-β2 adrenoceptor complex. Nature. 2011;469:236–240. doi: 10.1038/nature09665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Warne T, Moukhametzianov R, Baker JG, Nehmé R, Edwards PC, Leslie AGW, Schertler GFX, Tate CG. The structural basis for agonist and partial agonist action on a β1-adrenergic receptor. Nature. 2011;469:241–244. doi: 10.1038/nature09746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whorton MR, Rasmussen SGF, Devree BT, Deupi X, Jie X, Ve G, Sunahara RK, Kobilka BK. The effect of ligand efficacy on the formation and stability of a GPCR-G protein complex. Proc Natl Acad Sci U S A. 2009;106:1–6. doi: 10.1073/pnas.0811437106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghanouni P, Schambye H, Seifert R. The Effect of pH on β2 Adrenoceptor Function. J Biol Chem. 2000;275:3121–3127. doi: 10.1074/jbc.275.5.3121. [DOI] [PubMed] [Google Scholar]
- 27.Tate CG, Schertler GFX. Engineering G protein-coupled receptors to facilitate their structure determination. Curr Opin Struct Biol. 2009;19:386–395. doi: 10.1016/j.sbi.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 28.Parnot C, Miserey-Lenkei S, Bardin S, Corvol P, Clauser E. Lessons from constitutively active mutants of G protein-coupled receptors. Trends in endocrinology and metabolism. 2002;13:336–343. doi: 10.1016/s1043-2760(02)00628-8. [DOI] [PubMed] [Google Scholar]
- 29.Gether U, Seifert J. Structural instability of a constitutively active G protein-coupled receptor. J Biol Chem. 1997;272:2587–2590. doi: 10.1074/jbc.272.5.2587. [DOI] [PubMed] [Google Scholar]
- 30.Delean A, Stadel JM, Lefkowitz RJ. A ternary complex model explains the egonist-specific binding properties of the adenylate cyclase-coupled β-adrenergic Receptor. J Biol Chem. 1980;255:7108–7117. [PubMed] [Google Scholar]
- 31.Whorton MR, Bokoch MP, Rasmussen SGF, Huang B, Zare RN, Kobilka BK, Sunahara RK. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gurevich V, Pals-Rylaarsdam R. Agonist-receptor-arrestin, an alternative ternary complex with high agonist affinity. J Biol Chem. 1997;272:28849–28852. doi: 10.1074/jbc.272.46.28849. [DOI] [PubMed] [Google Scholar]
- 33.Martini L, Hastrup H, Holst B, Fraile-Ramos A, Marsh M, Schwartz TW. NK1 receptor fused to beta-arrestin displays a single-component, high-affinity molecular phenotype. Molecular pharmacology. 2002;62:30–37. doi: 10.1124/mol.62.1.30. [DOI] [PubMed] [Google Scholar]
- 34.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 35.Muyldermans S, Cambillau C, Wyns L. Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends in biochemical sciences. 2001;26:230–235. doi: 10.1016/s0968-0004(01)01790-x. [DOI] [PubMed] [Google Scholar]
- 36.Frenken LGJ, van der Linden RHJ, Hermans PWJJ, Ruuls RC, De Geus B, Verrips CT. Isolation of antigen specific Llama VHH antibody fragments and their high level secretion by Saccharomyces cerevisiae. Journal of biotechnology. 2000;78:11–21. doi: 10.1016/s0168-1656(99)00228-x. [DOI] [PubMed] [Google Scholar]
- 37.Muyldermans S. Single domain camel antibodies: current status. Journal of biotechnology. 2001;74:277–302. doi: 10.1016/s1389-0352(01)00021-6. [DOI] [PubMed] [Google Scholar]
- 38.Lauwereys M, Arbabi GM, Desmyter A, Kinne J, Holzer W, De Genst E, Wyns L, Muyldermans S. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 1998;17:3512–3520. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Genst E, Silence K, Decanniere K, Conrath K, Loris R, Kinne J, Muyldermans S, Wyns L. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci U S A. 2006;103:4586–4591. doi: 10.1073/pnas.0505379103. This paper shows that Nanobodies have a prolate shape and present a large convex paratope that is predominantly formed by a long CDR3. Such single domain antibodies have a clear structural advantage over conventional dimeric formats for targeting clefts on antigenic surfaces.
- 40. Domanska K, Vanderhaegen S, Srinivasan V, Pardon E, Dupeux F, Marquez JA, Giorgetti S, Stoppini M, Wyns L, Bellotti V, et al. Atomic structure of a nanobody-trapped domain-swapped dimer of an amyloidogenic β2-microglobulin variant. Proc Natl Acad Sci U S A. 2011;108:1314–1319. doi: 10.1073/pnas.1008560108. This paper illustrates one of several uses of nanobodies in structural biology. Here, a nanobody was used to trap and characterize a dimeric intermediate of β2-microglobulin amyloidogenesis by X-ray crystallography.
- 41.Korotkov KV, Pardon E, Steyaert J, Hol WGJ. Crystal structure of the N-terminal domain of the secretin GspD from ETEC determined with the assistance of a nanobody. Structure. 2009;17:255–265. doi: 10.1016/j.str.2008.11.011. 10.1016/j.str.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loris R, Marianovsky I, Lah J, Laeremans T, Engelberg-Kulka H, Glaser G, Muyldermans S, Wyns L. Crystal structure of the intrinsically flexible addiction antidote MazE. J Biol Chem. 2003;278:28252–28257. doi: 10.1074/jbc.M302336200. [DOI] [PubMed] [Google Scholar]
- 43.Wu M, Park Y-J, Pardon E, Turley S, Hayhurst A, Deng J, Steyaert J, Hol WGJ. Structures of a key interaction protein from the Trypanosoma brucei editosome in complex with single domain antibodies. Journal of structural biology. 2011;174:124–136. doi: 10.1016/j.jsb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Genst EJ, Guilliams T, Wellens J, O’Day EM, Waudby CA, Meehan S, Dumoulin M, Hsu S-TD, Cremades N, Verschueren KHG, et al. Structure and Properties of a Complex of alpha-Synuclein and a Single-Domain Camelid Antibody. J Mol biol. 2010;402:326–343. doi: 10.1016/j.jmb.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 45. Rasmussen SGF, Choi HJ, Fung JJ, Pardon E, Casarosa P, Chae PS, DeVree BT, Rosenbaum DM, Thian FS, Kobilka TS, et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. 2011;469:175–180. doi: 10.1038/nature09648. This paper describes the first active-state structure of a GPCR activated by a diffusible ligand solved in complex with a nanobody that acts as a G protein surrogate.
- 46.Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Caffrey M. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Annual review of biophysics. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- 48.Sanishvili R, Nagarajan V, Yoder D, Becker M, Xu S, Corcoran S, Akey DL, Smith JL, Fischetti RF. A 7µm mini-beam improves diffraction data from small or imperfect crystals of macromolecules. Acta crystallographica. Section D, Biological crystallography. 2008;64:425–435. doi: 10.1107/S0907444908001741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu F, Wu H, Katritch V, Han GW, Jacobson KA, Gao Z-G, Cherezov V, Stevens RC. Structure of an Agonist-Bound Human A2A Adenosine Receptor. Science. 2011;322:322–327. doi: 10.1126/science.1202793. [DOI] [PMC free article] [PubMed] [Google Scholar]