Abstract
T cells engineered to express T cell receptors (TCRs) specific for tumor antigens can drive cancer regression. The first TCRs used in cancer gene therapy, DMF4 and DMF5, recognize two structurally distinct peptide epitopes of the melanoma-associated MART-1/Melan-A protein, both presented by the class I MHC protein HLA-A*0201. To help understand the mechanisms of TCR cross-reactivity and provide a foundation for the further development of immunotherapy, we determined the crystallographic structures of DMF4 and DMF5 in complex with both of the MART-1/Melan-A epitopes. The two TCRs use different mechanisms to accommodate the two ligands. Whereas DMF4 binds the two with a different orientation, altering its position over the peptide/MHC, DMF5 binds them both identically. The simpler mode of cross-reactivity by DMF5 is associated with higher affinity towards both ligands, consistent with the superior functional avidity of DMF5. More generally, the observation of two diverging mechanisms of cross-reactivity with the same antigens and the finding that TCR binding orientation can be determined by peptide alone extend our understanding of the mechanisms underlying TCR cross-reactivity.
Introduction
The identification of tumor associated antigens preferentially presented by human cancers has led to the development of immunotherapeutic strategies for cancer such as peptide vaccines and adoptive T cell transfer. In adoptive T cell transfer, tumor-antigen specific T cells are activated ex vivo and transplanted back into a lymphodepleted patient. Although clinical trials with adoptive transfer have been promising (reviewed in 1), a liability is that variation in T cell repertoires impacts the likelihood of any individual producing a highly avid T cell receptor (TCR) specific for a given tumor antigen. That most tumor antigens are nonmutated self-antigens against which T cells will likely be negatively selected compounds this liability. A recent development that can address these concerns is the transfer of T cells genetically engineered to express tumor-antigen specific TCRs with defined recognition properties.
The first two trials examining the use of genetically engineered T cells in humans were recently published (2, 3). Both trials targeted the MART-1 protein (also referred to as Melan-A), upregulated in the majority of melanomas. The two trials used different class I MHC-restricted TCRs, DMF4 and DMF5. The two receptors are unrelated, utilizing different Vα and Vβ segments and possessing different CDR3 loops (Table I). In functional assays DMF5 T cells are more avid than DMF4, and DMF5 T cells are more efficiently stained with MART-1/HLA-A2 tetramers (4). Although the clinical trials were small, use of DMF5 resulted in an improved rate of cancer regression (13% with DMF4 vs. 30% with DMF5). Use of DMF5 was also associated with incidences of eye, ear, and skin autoimmune toxicity not reported with DMF4. Expanded trials with DMF5-engineered T cells are underway, and DMF5 continues to be exploited as a model receptor for the development of T cell-based gene therapy of cancer (5–7).
Table 1.
Gene usage and CDR loop sequences of DMF4 and DMF5
| DMF4 | DMF5 | |
|---|---|---|
| Vα | 35 | 12-2 |
| CDR1 | SSSIFNTW | YSDRGSQSF |
| CDR2 | YKAGELT | YSNGDK |
| CDR3 | AGGTGNQFYF | AVNFGGGKLIF |
| HV4 | GITRKDS | NKASQYV |
| Vβ | 10-3 | 6-4 |
| CDR1 | QTENHRY | QDMRHNA |
| CDR2 | YSYGVKDTD | YSNTAGTT |
| CDR3 | AISEVGVGQPQHF | ASSLSFGTEAFF |
Despite this progress, antigen recognition by MART-1-specific TCRs in general is complex and poorly understood. Most MART-1 specific TCRs examined cross-react between the decameric epitope spanning residues 26–35 (EAAGIGILTV) as well as the nonameric epitope spanning residues 27–35 (AAGIGILTV), both presented by the class I MHC HLA-A*0201 (HLA-A2). Compared to the nonamer, the additional amino acid in the decamer forces the peptide to bulge and zig-zag in the HLA-A2 peptide binding groove, resulting in the presentation of different surfaces to the T cell repertoire (8). In addition to highlighting the capacity for TCRs to cross-react with structurally diverse ligands (9), nonamer/decamer cross-reactivity is also likely to be important in melanoma immunotherapy. The nonamer is believed to be the clinically relevant antigen in HLA-A2+ individuals (10–13). Yet due to poor binding of the nonamer to HLA-A2 and the inability to generate a superior “heteroclitic” nonamer that maintains the nonameric conformation in the HLA-A2 peptide binding groove (8, 14), the majority of efforts targeting MART-1 have made use of the stronger binding decamer or a decameric variant modified at position 2 (ELAGIGILTV) to select, assay, and activate MART-1 specific T cells. The decamer (or its anchor-modified variant) was among the first peptides to be used in clinical trials of peptide-based cancer vaccines and remains a component of many candidate cancer vaccine formulations (e.g., ref. 15).
Here we studied MART-1 nonamer/decamer recognition by the DMF4 and DMF5 TCRs, determining the structural basis for cross-reactivity between the nonamer and decamer peptide/HLA-A2 complexes. We found that the two receptors cross-react via fundamentally different mechanisms. DMF4 cross-reacts with a complex mechanism, altering its orientation over the peptide/MHC complex in order to accommodate the differences in the peptides. In contrast, DMF5 binds the two ligands identically, accommodating the differences through the use of a permissive architecture that is pre-formed in the free receptor. The simpler mode of cross-reactivity for DMF5 is associated with higher affinity towards both ligands, helping to explain DMF5’s stronger functional avidity. In addition to providing a foundation for further developments in cancer immunotherapy, the results contribute to our understanding of the mechanisms underlying TCR binding cross-reactivity, demonstrating that different TCRs can utilize different mechanisms to cross-react with the same two ligands and that TCR binding orientation can be determined by peptide alone.
Materials & Methods
Proteins and peptides
Recombinant soluble TCRs and peptide/HLA-A2 molecules were refolded from bacterially expressed inclusion bodies using established procedures (44). Peptides were purchased from Genscript or synthesized locally using an ABI 433A instrument and verified by LC-MS. All structure and binding experiments with the MART-1 decamer used the anchor-modified ELAGIGILTV variant. Recombinant DMF4 and DMF5 utilized an engineered disulfide bond in the constant domains to enhance stability (45).
X-ray crystallography
Crystals of the DMF4-peptide/HLA-A2 complexes were grown from 15% PEG4000, 0.2 M MgCl2 buffered with 0.1 M Tris at pH 8.5 at 25 °C. Crystals of the DMF5-peptide/HLA-A2 complexes were grown from 20% PEG4000 buffered with 0.1 M HEPES at pH 7.5 with the addition of 10% propanol at 25 °C. Crystals of free DMF5 were grown from 15% PEG 3350, 0.2 M MgCl2 buffered with 0.1 M Tris at pH 8.5 at 25 °C. Crystallization was performed using sitting drop/vapor diffusion. Streak seeding was used to obtain higher quality crystals. For cryo-protection, crystals were transferred into 20% glycerol/80% mother liquor for 30 seconds and immediately frozen in liquid nitrogen. Diffraction data were collected at the 19BM, 19ID, 21ID, and 31ID beamlines at the Advanced Photon Source, Argonne National Laboratories. Data reduction was performed with HKL2000 (46). The ternary complexes were solved by molecular replacement using MOLREP or Phaser using PDB entry 2GJ6 (47) as a search model with the coordinates of peptides, solvent, and CDR loops removed. The structure of free DMF5 was solved using the coordinates of the TCR from PDB entry 1AO7 (25) as a search model with solvent and CDR loops removed. Rigid body refinement followed by TLS refinement and multiple steps of restrained refinement were performed with Refmac5 (48). TLS groups were chosen as previously described (47). Once defined, TLS parameters were included in all subsequent steps of the refinement. Anisotropic and bulk solvent corrections were taken into account throughout refinement. After TLS refinement, it was possible to unambiguously trace the position of peptides and TCR CDR loops in all structures against σA weighted 2Fo-Fc maps. Waters were added using ARP/wARP (49). Evaluation of models and fitting to maps was performed using Coot (50) and XtalView (51). Procheck (52), the template structure check in WHATIF (53), and MolProbity (54) were used to evaluate the structures during and after refinement. Hydrogen bonds were determined with the PISA web server and validated with distance and geometry criteria (55). Intermolecular contacts were tabulated using a cutoff of 4 Å. Measurements of TCR docking angle followed recommended procedure (16). Surface complementarities are the Sc statistic of Lawrence and Colman (56). Note that the peptides in the decamer complexes are numbered from 1 to 10, in contrast with our previous structure of the decamer/HLA-A2 complex, in which the peptide was numbered from 0 to 9 (8). PDB entries for the structures are listed in Table II.
Table 2.
X-ray data collection and refinement statistics
| DMF4- nonamer/HLA-A2 |
DMF4- decamer/HLA-A2 |
DMF5- nonamer/HLA-A2 |
DMF5- decamer/HLA-A2 |
DMF5 | |
|---|---|---|---|---|---|
| Data collection | 21ID | 19BM | 19ID | 21ID | 31ID |
| Space group | P212121 | P212121 | C2 | C2 | C2 |
| Cell dimensions | |||||
| a, b, c (Å) | 59.7, 73.7, 225.3 | 56.0, 69.8, 227.1 | 227.8, 46.3, 85.9 | 228.4, 46.6, 86.0 | 184.2, 86.5, 66.5 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 106.6, 90.0 | 90.0, 106.7, 90.0 | 90.0, 104.0, 90.0 |
| Resolution (Å) | 20–2.60 (2.64-2.60) | 20–2.80 (2.85-2.80) | 20–2.30 (2.34-2.30) | 20–2.70 (2.75-2.70)* | 30-2.10 (2.14-2.10) |
| Rmerge | 0.08 (0.28) | 0.15 (0.86) | 0.07 (0.41) | 0.08 (0.27) | 0.05 (0.29) |
| I / σI | 25.8 (5.6) | 13.7 (1.9) | 19.1 (2.0) | 18.2 (3.4) | 20.7 (3.2) |
| Completeness (%) | 99.6 (98.8) | 92.9 (89.8) | 97.0 (81.5) | 90.3 (62.2) | 99.7 (99.7) |
| Redundancy | 6.6 (6.1) | 5.2 (4.3) | 3.6 (2.8) | 3.1 (2.7) | 3.7 (3.4) |
| Refinement | |||||
| Resolution (Å) | 20.00-2.60 | 20.00-2.80 | 20.00-2.30 | 20.00-2.70 | 29.79-2.09 |
| No. reflections | 31550 | 21058 | 37477 | 22059 | 59136 |
| Rwork / Rfree | 0.23/0.27 | 0.21/0.28 | 0.24/0.30 | 0.22/0.28 | 0.21/0.27 |
| No. atoms | |||||
| Protein | 6576 | 6602 | 6598 | 6610 | 6863 |
| Water | 103 | 33 | 48 | 27 | 498 |
| B-factors | |||||
| Protein | 22.4 | 23.5 | 47.4 | 45.6 | 42.1 |
| Water | 19.7 | 14.0 | 39.9 | 35.1 | 43.4 |
| RMS deviations | |||||
| Bond lengths (Å) | 0.013 | 0.01 | 0.012 | 0.009 | 0.014 |
| Bond angles (°) | 1.589 | 1.476 | 1.562 | 1.354 | 1.668 |
| PDB entry | 3QEQ | 3QDM | 3QDJ | 3QDG | 3QEU |
Data in parenthesis is for the highest resolution shell.
Surface plasmon resonance
Surface plasmon resonance experiments were performed using a Biacore 3000 instrument as previously described (44). The TCR was coupled to the sensor surface using amine coupling. Data were corrected for bulk solvent effects using a blank flow cell. For experiments with the nonamer, improved accuracy was obtained by fixing the activity of the surface at values pre-determined with the decamer (57). Flow rates were 5 µL/min. All injections were repeated twice and affinity measurements reflect simultaneous fits to both datasets. Solution conditions were 10 mM HEPES, 150 mM NaCl, 3 mM EDTA, 0.005% surfactant P-20, pH 7.4, 25 °C. Data were processed with Biaevaluation 4.1 (GE Healthcare) and fit with OriginPro 7.5 (OriginLabs).
Results
Structures of the DMF4 and DMF5 TCRs bound to the nonameric and decameric MART-1/HLA-A2 complexes
The structures of the DMF4 and DMF5 TCRs bound to the MART-1 27–35 nonamer (AAGIGILTV) and anchor-modified 26–35 decamer (ELAGIGILTV) were determined at resolutions between 2.3 and 2.8 Å (Table II). All four ternary complexes displayed the diagonal docking mode traditionally seen in TCR recognition of foreign antigens. This and other structural descriptors, such as buried surface area and shape complementarity, were within the range seen for other TCR-pMHC interactions (16) and are summarized in Table III. Electron density images for key regions of each structure are shown in Supplemental Fig. 1. The structures are described and compared in detail below, beginning with the more complex DMF4 structures.
Table 3.
Structural descriptors of the DMF4 and DMF5 ternary complexes
| DMF4- nonamer/HLA-A2 |
DMF4- decamer/HLA-A2 |
DMF5- nonamer/HLA-A2 |
DMF5- decamer/HLA-A2 |
|
|---|---|---|---|---|
| Docking angle | 44° | 29° | 31° | 31° |
| Surface complementarity | 0.72 | 0.64 | 0.64 | 0.65 |
| TCR-MHC hydrogen bonds / salt-bridges | 6 | 2 | 5 | 4 |
| TCR-peptide hydrogen bonds / salt-bridges | 2 | 5 | 6 | 8 |
| Buried surface area (Å2) | ||||
| Total | 1890 | 1712 | 2201 | 2137 |
| CDR1α/CDR2α/HV4α/CDR3α | 78/157/52/137 | 78/138/76/130 | 344/129/41/140 | 324/135/35/116 |
| CDR1βCDR2β/CDR3β | 62/144/313 | 68/68/327 | 50/171/232 | 49/178/238 |
| αhelix/α2 helix/peptide | 426/289/230 | 355/191/280 | 471/361/261 | 449/320/296 |
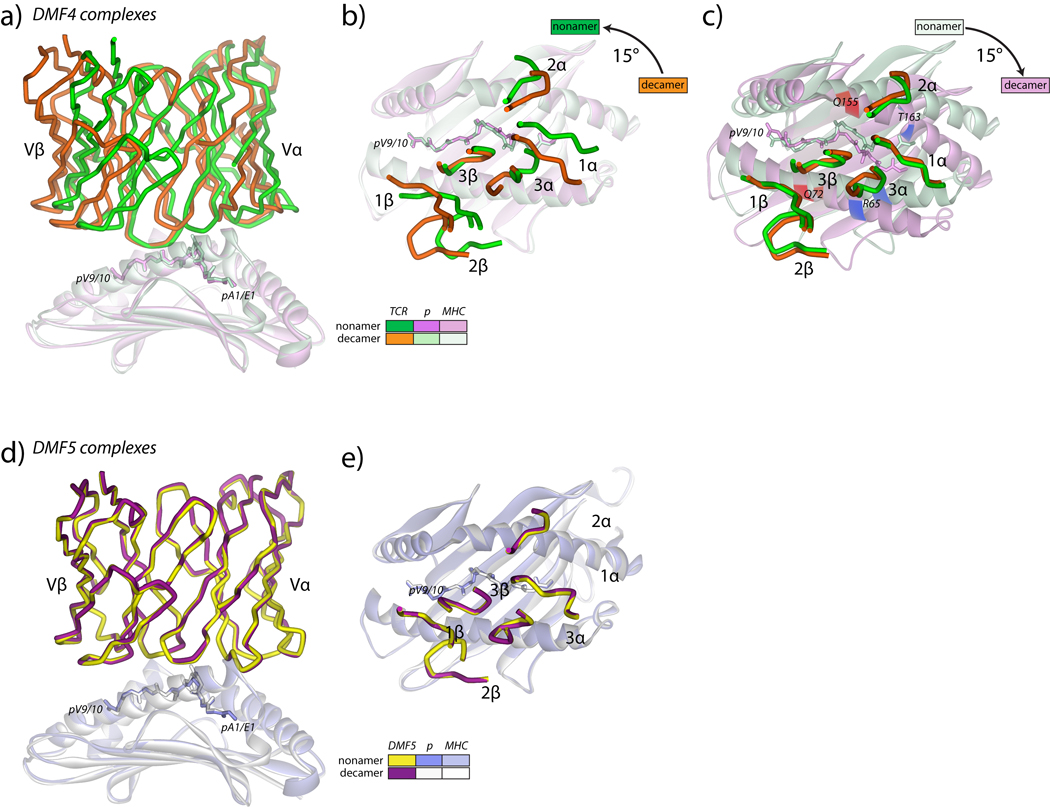
The DMF4 TCR is oriented differently over the MART-1 nonamer and decamer peptide/HLA-A2 complexes
The structures of the DMF4 TCR bound to the MART-1 nonamer and decamer peptide/HLA-A2 complexes showed that the TCR binds the two ligands differently, as shown in Fig. 1A. When the HLA-A2 peptide binding domains in the two structures are superimposed, the RMSD between the TCR variable domains is 5.1 Å. Viewed from the top through the TCR, each loop except CDR3β is arranged differently over the pMHCs (Fig. 1B). This is reflected in a 15° difference in docking angle, with the TCR positioned more diagonally over the nonamer (44°) than the decamer (29°). Other than CDR3α though, each CDR loop remains in the same conformation. DMF4 thus engages the nonameric and decameric MART-1/HLA-A2 complexes with geometries that differ predominantly by a rigid-body rotation over the pMHC, with the “pivot point” of rotation centered on CDR3β (Fig. 1C).
Figure 1.
Overview of the DMF4 and DMF5 MART-1 nonamer and decamer peptide/HLA-A2 ternary complexes. A) Side view of the two DMF4 complexes, showing the differences in the TCR variable domains when the HLA-A2 peptide binding domains are superimposed. The color scheme is indicated in the inset and maintained in panels B and C. B) Top view of the superimposition in panel A showing the positions of the DMF4 CDR loops over the peptide/HLA-A2 complexes. The differences in the TCR are attributable to a 15° rotation of the TCR over HLA-A2, with CDR3β as the pivot point. C) Same as panel B, but with the variable domains of the TCR used for superimposition. The positions of Arg65 and Thr163 are highlighted in blue. The positions of Gln72 and Gln155 are highlighted in red. D) Side view of the two DMF5 complexes, showing the identical binding mode of the TCR. The color scheme is indicated in the inset. E) Top view of the superimposition in panel D showing the positions of the DMF5 CDR loops over the peptide/HLA-A2 complexes.
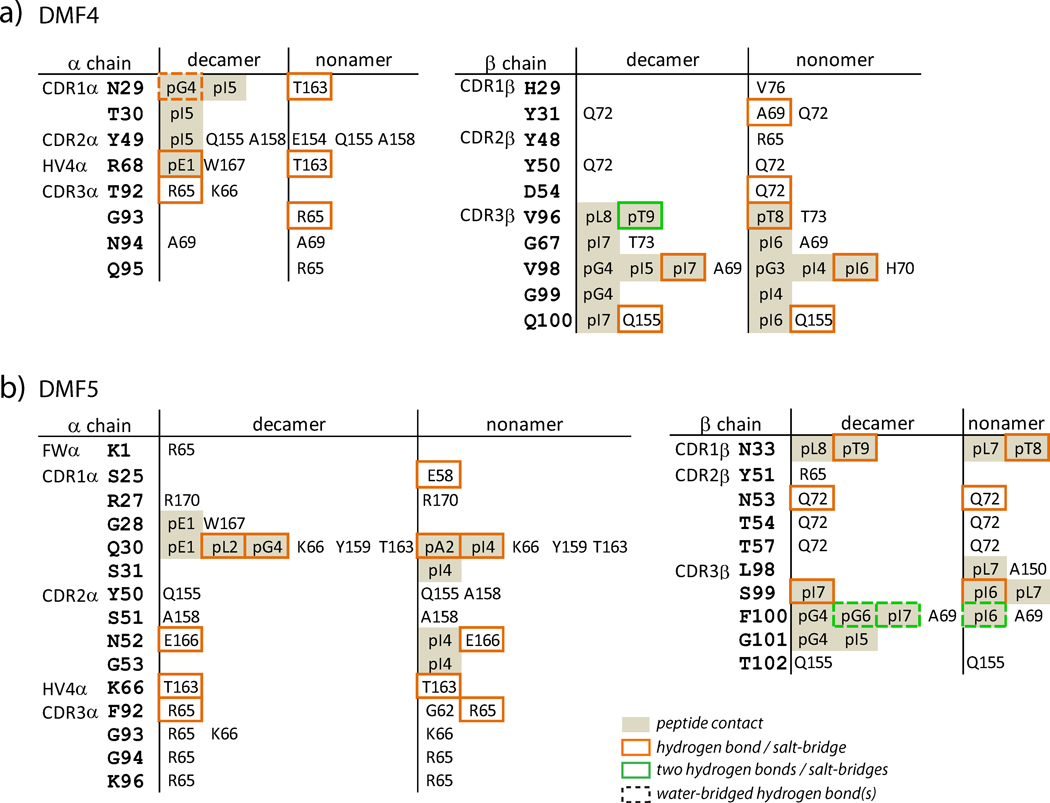
The different geometries by which DMF4 binds the nonamer and decamer complexes result in different contacts made by various CDR loop amino acids to positions on HLA-A2 (Fig. 2, a more detailed list is in Supplemental Fig. 2). One illustration of these differences is in the TCR-HLA-A2 hydrogen bonding patterns: only two hydrogen bonds are formed to HLA-A2 in the DMF4-decamer complex. In contrast, six TCR-HLA-A2 hydrogen bonds are formed in the nonamer complex.
Figure 2.
Amino acids on HLA-A2 involved in key intermolecular contacts in the DMF4 (A) and DMF5 (B) ternary complexes with the MART-1 nonamer and decamer. Key contacts are defined as those with interatomic distances less than or equal to 3.75 Å. More expanded lists of contacts are in Supplemental Fig. 2
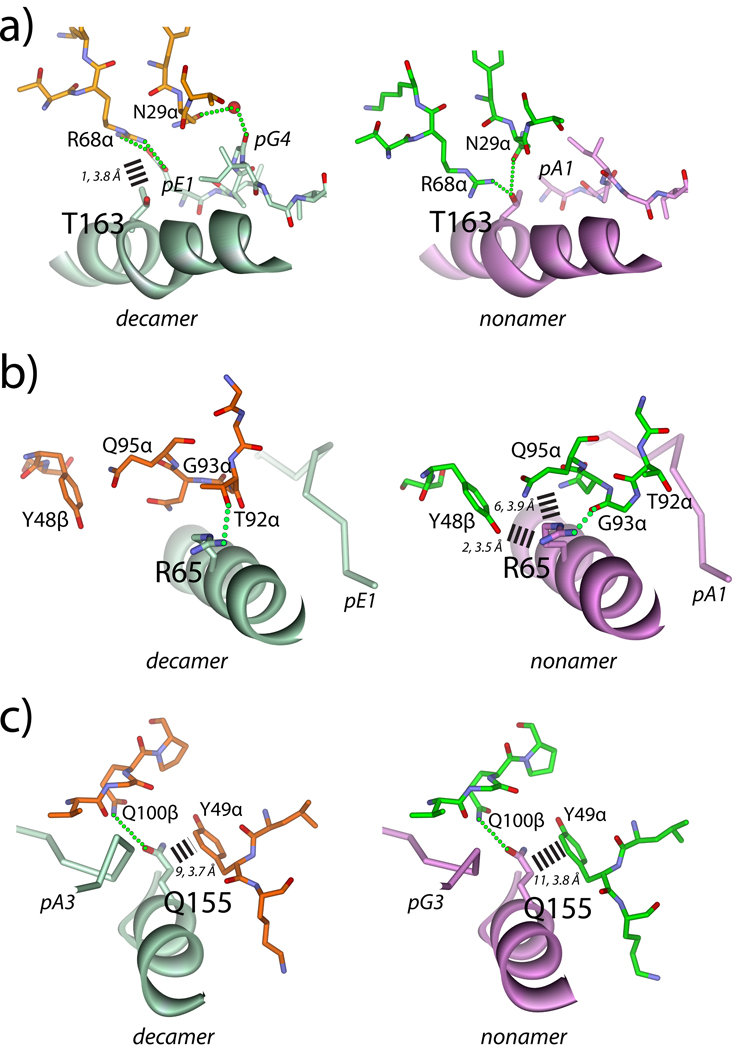
Examining the DMF4-HLA-A2 interfaces in more detail, the differences in environments due to the change in TCR orientation can be broken down into three general classes: 1) placement of TCR and HLA-A2 atoms into different environments with the formation of wholly new interatomic interactions; 2) a mimicking of the general chemical environment around HLA-A2 residues but using atoms from different TCR amino acids; and 3) retention of environment with only small changes in interatomic interactions. Instances of each class are shown in Fig. 3. The most dramatic change in environment occurs with Thr163 in the HLA-A2 α2 helix. In the nonamer structure, Thr163 hydrogen bonds with Asn29 of CDR1α and Arg68 of HV4α. In the decamer structure, however, Thr163 forms only a single long range van der Waals contact with Asn29α, with Asn29α and Arg68α instead interacting with the peptide (Fig. 3A).
Figure 3.
Molecular environments around HLA-A2 contact positions in the two DMF4 ternary complexes. For all panels, dotted green lines indicate hydrogen bonds. Dashed bars indicate interatomic van der Waals contacts, with the number and average length in Å indicated. A) Environment around Thr163, showing the switch in hydrogen bonding patterns between DMF4 recognition of decamer and nonamer. B) Environment around Arg65, showing the switch in van der Waals and hydrogen bonding patterns. Of note is the conformational change in CDR3α, which occurs in order for Arg65 to hydrogen bond with Thr92α in the decamer complex and Gly93sα in the nonamer complex. C) Environment around Gln155, showing the conserved van der Waals interactions with Tyr49 of CDR2α and the hydrogen bond to Gln100 of CDR3β
An example where the HLA-A2 chemical environment is mimicked using different TCR amino acids is seen with Arg65 in the HLA-A2 α1 helix, which hydrogen bonds with Thr92α of CDR3α in the decamer complex but with Gly93 of CDR3α in the nonamer complex (Fig. 3B). In addition to rotation of the DMF4 TCR, the change in environment around Arg65 is also driven by a shift in CDR3α conformation as shown in Fig. 1C. This conformational change appears to occur solely in order for the TCR to hydrogen bond with Arg65, as CDR3α forms no contacts to the peptide in either the nonamer or decamer structure and there are no steric clashes that would force a conformational change in the loop if DMF4 were to bind the nonamer with a decamer-like orientation. The importance of position 65 in TCR recognition of class I MHC, and HLA-A2 in particular, has been previously noted (17–19), likely reflected here in how the need to hydrogen bond with the TCR forces a conformational change CDR3α.
Thr163 and Arg65 of HLA-A2 lie towards the N-terminal end of the peptide in the HLA-A2 binding groove, where the differences in environment are magnified as they are most distant from the CDR3β pivot point (blue highlights in Fig. 1C). Positions on HLA-A2 closer to CDR3β thus retain more of their chemical environments in the two complexes. Indeed, the DMF4-HLA-A2 contacts near CDR3β are the only ones shared in the two DMF4 structures. Of particular interest are shared contacts between germline CDR loops and HLA-A2. Gln155 maintains the greatest number of these (Fig. 3C), forming eight with Tyr49 in CDR2α. Both Gln155 and tyrosines in CDR2α have been suggested to play a key role in TCR recognition of class I MHC (17, 20), and the close alignment of Gln155 with Tyr49α despite the different docking angle could indicate such a role here. Notably though, Gln155 also forms a hydrogen bond with Gln100 of CDR3α in both structures (Fig. 3C), complicating such an interpretation.
DMF4 cross-reactivity between the MART-1 nonamer and decamer is attributable to different binding orientations, nonamer conformational changes, and shared CDR3β-peptide interactions
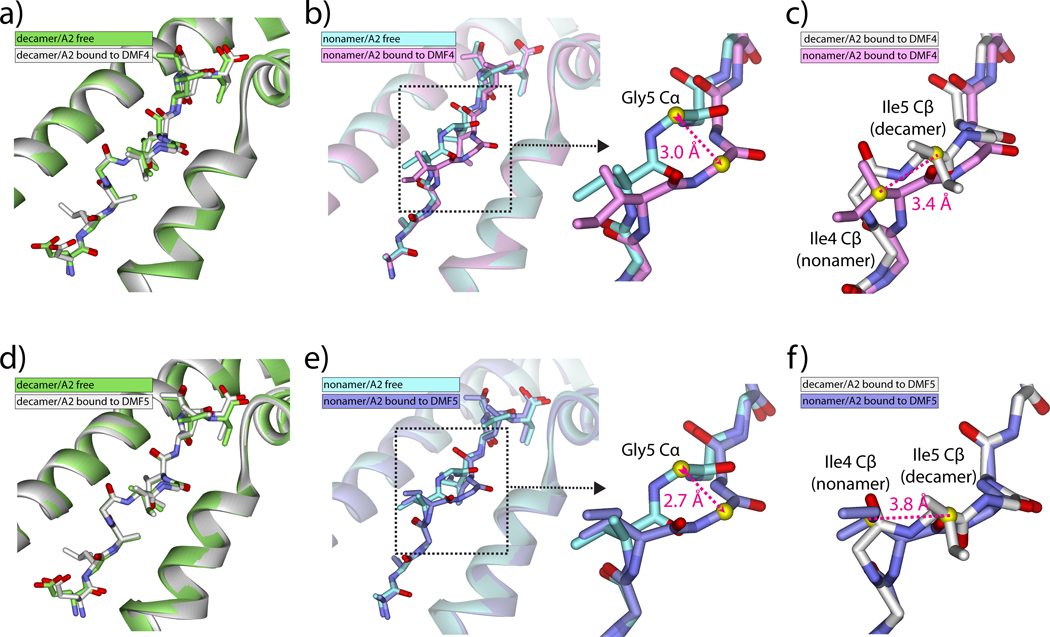
We next compared the structures of the DMF4-bound pMHC complexes with those of the previously solved free pMHCs (8, 21). No changes occur in either peptide or MHC upon TCR recognition of the decamer (Fig. 4A). Upon recognition of the nonamer though, a large shift occurs in the center of the peptide, bringing the conformation of the center closer to that of the decamer (Fig. 4B). The shift extends from the carbonyl oxygen of Ile4 to the amide nitrogen of Ile6, and is maximal at the amide nitrogen of Gly5, which moves 2.7 Å towards the HLA-A2 α2 helix. The shift in the nonamer is similar to a recent description of “induced molecular mimicry” upon TCR binding. (22). However, due to the presence of the additional amino acid in the decamer there are still conformational differences between the nonamer and decamer, with the peptides out of alignment and register at Ile4 (nonamer) and Ile5 (decamer) (Fig. 4C).
Figure 4.
DMF4 and DMF5 recognize the MART-1 decamer without changes in peptide conformation but force a shift in the center of the nonamer. A) The conformation of the decamer is unchanged upon DMF4 binding. RMSD for all atom peptide superimposition is 0.6 Å. B) The center of the nonamer undergoes a conformational change upon DMF4 binding, best summarized as a 3.0 Å shift at the αcarbon of Gly5. RMSD for all atom peptide superimposition is 1.3 Å. C) Although the shift in the nonamer brings the backbone conformation closer to that of the decamer, the nonamer and decamer are still out of register and alignment, with the β carbons (yellow spheres) of Ile4 (nonamer) and Ile5 (decamer) offset by 3.4 Å. D) The conformation of the decamer is unchanged upon DMF5 binding. RMSD for all atom superimposition is 0.4 Å. E) As with DMF4, the center of the nonamer undergoes a conformational change upon DMF5 binding, best summarized as a 2.7 Å shift at the α carbon of Gly5. RMSD for all atom superimposition is 1.0 Å. F) As with DMF4, although the backbone conformations are closer, the nonamer and decamer peptides are still out of register and alignment, with the β carbons (yellow spheres) of Ile4 (nonamer) and Ile5 (decamer) offset by 3.8 Å.
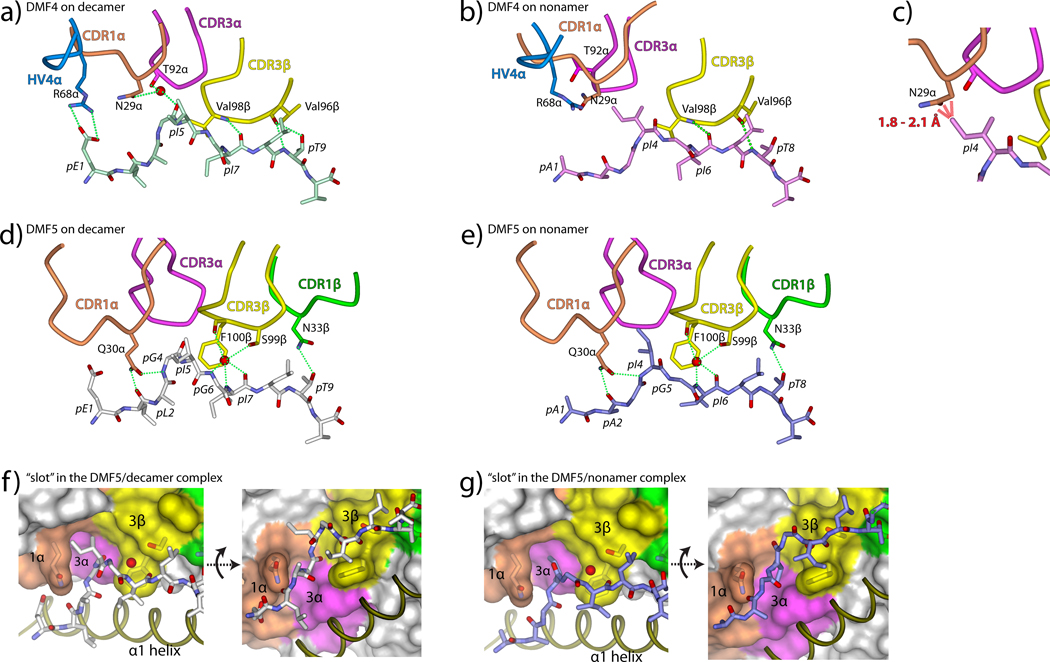
Closer examination of the DMF4-peptide/HLA-A2 interfaces shows how the repositioning of DMF4 over the two pMHC molecules allows the TCR to accommodate the remaining structural differences in the peptides. Beginning with the peptide N-terminus, multiple electrostatic interactions link DMF4 to the decamer (Fig. 5A): Arg68 of the HV4α loop forms a salt-bridge with the N-terminal glutamate and an interfacial water links Asn29 of CDR1α and Thr92 of CDR3α to the carbonyl oxygen of Gly4. None of these interactions is present in the structure with the nonamer (Fig. 5B): without a hydrogen bonding partner the water molecule is absent, and most importantly, the side chain of Ile4 in the nonamer complex occupies the position of the Gly4 backbone in the decamer complex, forcing a repositioning of the CDR1α loop. Without movement of CDR1α, steric clashes would occur between the side chains of Ile4 and Asn29α (Fig. 5C). These clashes are avoided by the more diagonal placement of DMF4 over the nonamer, which moves CDR1α out of the way of the Ile4 side chain. The clashes between Ile4 and Asn29α are the only clashes that occur when the pMHC from the nonameric complex is superimposed onto that of the decameric complex. As the conformation of CDR1α is unchanged despite the different position of the TCR, the surprising conclusion is that the energetic cost for the TCR to bind in a different orientation is less than that for moving CDR1α out of the way via a conformational change.
Figure 5.
Mechanisms of peptide engagement in the DMF4 and DMF5 ternary complexes. A) DMF4 engages the decamer through a salt-bridge to Glu1 from HV4α and water-bridged hydrogen bonds to Ile5 from CDR1α and CDR3α. CDR3β aligns alongside the C-terminal half of the peptide, hydrogen bonding to Ile7 and Thr9 and forming van der Waals contacts using Val96 and Val98. Dotted green lines represent hydrogen bonds or salt-bridges in this and all subsequent panels. B) The rotation of the DMF4 over HLA-A2 moves the HV4α, CDR1α, and CDR3α loops away from the N-terminal half of the nonamer. Peptide engagement is only through CDR3β, the pivot point of DMF4 rotation, which mimics its role in recognition of the decamer. C) Without rotation of the TCR, the side chain of Asn29 of CDR1α would clash sterically with the side chain of Ile4 of the nonamer (red dashed lines). D–E) DMF5 engages the decamer (D) and nonamer (E) via hydrogen bonds from Glu30 of CDR1α, water-bridged hydrogen bonds from CDR3β, and a hydrogen bond from CDR1β. E–F) DMF5 accommodates the structural differences in the nonamer and decamer through the use of a wide “slot,” with sides formed by the side chains of Gln30 (CDR1α) and Phe100 (CDR3β) and a roof formed by the backbone of CDR3α.
After Ile4/5, the nonamer and decamer peptides begin to move into alignment and are superimposable at Ile6/7. At this point, both peptides interact with CDR3β, which as the “pivot point” for the TCR maintains its position in the two structures. CDR3β is aligned parallel to the C-terminal halves of the peptides, forming a motif similar to that of an anti-parallel β-sheet (Fig. 5A,B). A hydrogen bond is formed between the amide nitrogen of Val98β and the carbonyl oxygen of Ile6/7 in both DMF4 complexes, and the Val98 side chain forms several van der Waals interactions with the peptides. The position of Val98β appears to drive the conformational change that occurs in the nonamer peptide, as steric clashes would occur between Val98β and the backbone of Gly5 of the nonamer if the peptide did not move. Two residues down the CDR3β loop Val96 hydrogen bonds with Thr8/9.
The DMF5 TCR engages the MART-1 nonamer and decamer pMHC complexes identically
Unlike DMF4, the DMF5 TCR binds the MART-1 nonamer and decamer peptide/HLA-A2 complexes identically (Fig. 1D). In the two DMF5 complexes, the backbones of the TCR Vα/Vβ domains, common residues of the peptides, and the HLA-A2 peptide binding domains superimpose with an RMSD of only 0.5 Å, and the conformations of the CDR loops are the same (Fig. 1E). The key inter-residue contacts within the DMF5-peptide/HLA-A2 interfaces are listed in Fig. 2; a more detailed list of contacts is given in Supplemental Fig. 2. As expected from the near-identical structures, the participation of HLA-A2 amino acids in the two DMF5 interfaces is essentially the same.
DMF5 uses an open architecture and interfacial water to accommodate the structural differences in the peptides
We next compared the structures of the DMF5-bound pMHC complexes with those of the free. As with DMF4, no changes occur in either peptide or MHC upon DMF5 recognition of the decamer (Fig. 4D). Upon recognition of the nonamer though, a shift occurs in the center of the peptide, bringing the conformation of the center closer to that of the decamer (Fig. 4E). The shift is nearly identical to that seen with DMF4: it extends from the carbonyl oxygen of Ile4 to the amide nitrogen of Ile6 and is maximal at the amide nitrogen of Gly5, which moves 2.7 Å towards the HLA-A2 α2 helix. Again, although the backbones are closer, the peptides remain out of alignment and register at Ile4/5 (Fig. 4F).
A close inspection of the two DMF5-peptide/HLA-A2 interfaces reveals how DMF5 is able to recognize the decamer and the shifted nonamer without requiring the changes in TCR binding orientation or CDR loop conformation required for DMF4. Beginning with the N-termini of the peptides, the side chain of Gln30 of CDR1α hydrogen bonds to the carbonyl oxygen of Leu2 in the decamer and Ala2 in the nonamer (Figs. 5D,E). Although the conformations of the peptides begin to diverge after this hydrogen bond, they are close enough to permit the side chain of Gln30α to form a second hydrogen bond to the amide nitrogen of Gly5 (decamer) and Ile4 (nonamer). The DMF5 TCR does not form a hydrogen bond or salt-bridge with the N-terminal glutamate in the decamer, making only long-range van der Waals contacts to the glutamate side chain.
The structural differences between the nonamer and decamer become more significant following the second hydrogen bond made by Gln30 of CDR1α. After this hydrogen bond, the backbone of the decamer bulges up towards the TCR. This bulge does not occur in the nonamer, but the βcarbon of Ile4 of the nonamer occupies the same position as the carbonyl carbon of Gly4 of the decamer. Both the bulge in the decamer and the side chain of Ile4 in the nonamer are accommodated by a wide “slot” in the TCR that is walled by the side chains of Gln30 of CDR1α and Phe100 of CDR3β and roofed by the triple glycine motif in the center of CDR3α(Figs. 5E,F). The slot is large enough that it can accommodate both peptides without any compensatory adjustments. Indeed, the CDR3α “roof” is high enough such that CDR3α forms no contacts to the decamer and only three, long-range van der Waals contacts to the nonamer (Supplemental Fig. 3). An accommodating architecture is thus one component of how DMF5 recognizes both the MART-1 nonamer and decamer.
After exiting the slot, the side chain of Ile5 in the decamer extends towards the HLA-A2 α2 helix, occupying space at the periphery of the interface that is empty in the nonamer structure. At this point, the peptide backbones are closer in alignment and are linked to the TCR via a water molecule that serves as the hub of a network of hydrogen bonds between CDR3β and the centers of the peptides. In both structures, the water links the backbone of Ile6 (nonamer) and Ile7 (decamer) with the backbone of Phe100β and the side chain of Ser99β (Figs. 5C,D). An additional hydrogen bond is made to Gly6 in the decamer, but not to Gly5 in the nonamer due to lingering structural differences in the peptides. This network of hydrogen bonds explains the need for the structural shift that occurs in the center of the nonamer upon binding: if the nonamer did not adopt a conformation closer to the decamer at this point, there would be no room for the bridging water molecule, preventing the formation of the hydrogen bonds between CDR3β and the peptide.
Following the water-bridged hydrogen bonds to the centers of the peptides, the conformations of the nonamer and decamer peptides are identical. In both structures, a final TCR-peptide hydrogen bond is made by the side chain of Asn33 of CDR1β to side chain of Thr8/9 (Figs. 5D,E).
Only minor conformational adaptations are needed for the DMF5 TCR to engage peptide
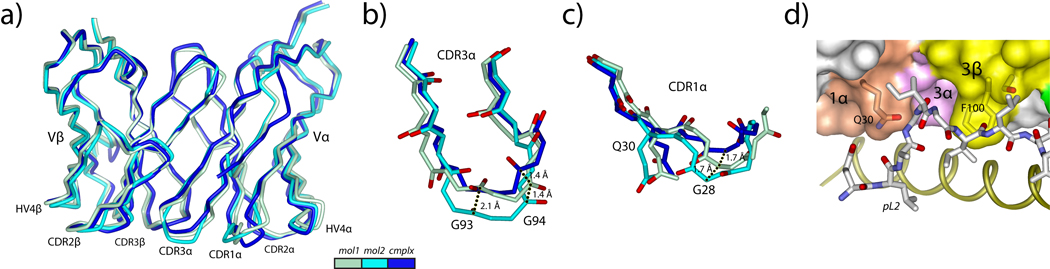
We next determined the structure of the free DMF5 TCR to 2.1 Å resolution in a crystal form with two molecules per asymmetric unit (Table II; electron density images are in Supplemental Fig. 1). The two copies of the molecule superimpose closely (RMSD for superimposition of the backbones of the variable domains is 0.8 Å). Each CDR loop adopts the same overall conformation in the two copies of the molecule (Fig. 6A). The positions at the tip of CDR3α however differ by 2.1 Å and 1.4 Å displacements at the backbone carbonyls of Gly93 and Gly94, respectively (Fig. 6B), indicating the triple-glycine motif of Gly93, Gly94, and Gly95 imparts a degree of flexibility to CDR3α. The conformation of CDR3α in the first molecule in the asymmetric unit is closest to the conformation seen in the bound state of the receptor.
Figure 6.
The structure of the free DMF5 TCR indicates that only minor conformational changes are needed to bind. A) Superimposition of the variable domains for the two molecules in the asymmetric unit of the free DMF5 structure onto the variable domain from the ternary complex with the decamer. The color scheme is given in the inset and maintained in panels B and C. B) Conformational diversity in CDR3α is centered on Gly93 and Gly94, with differences of 2.1 Å at the carbonyl carbon of Gly93 and 1.4 Å at the carbonyl carbon of Gly94. The conformation of the loop in the first molecule in the asymmetric unit most closely resembles that in the ternary complex. C) Conformational diversity for CDR1α is centered on Gly28, which is displaced by 1.7 Å in the two copies of the free TCR, and displaced a further 1.7 Å upon binding. D) Despite the conformational adjustments needed in CDR1α and CDR3α, the open architecture in bound DMF5 is largely present in free DMF5, evident when the structure of the free TCR is superimposed onto that in the complex with the decamer.
The position of CDR1α also differs slightly in the two molecules in the asymmetric unit of the free DMF5 structure, with a rotation around Gly28 that impacts the path and position of C-terminal end of the loop (Fig. 6C). The C-terminal end of CDR1α in the bound state of the receptor is displaced slightly away from the two conformations seen in the unbound state. The N-terminal half of CDR1α is largely the same in the bound and unbound conformations, although modest changes are needed in the Gln30 side chain torsion angles in order for it to engage peptide.
Although the structure indicates some flexibility for CDR1α and CDR3α, the conformational adaptations needed to bind ligand are small in context, less than the average seen for CDR1α and CDR3α in a recent comparison of bound and free TCRs (23). The major elements of DMF5 used to bind ligand thus appear largely pre-configured in the free receptor. This is illustrated in Fig. 6D, which shows how free DMF5 sits over the decamer peptide when superimposed onto the TCR in the DMF5-decamer/HLA-A2 complex, emphasizing the “slot” needed to accommodate the peptide.
DMF5 binds both the nonamer and decamer with higher affinity than DMF4
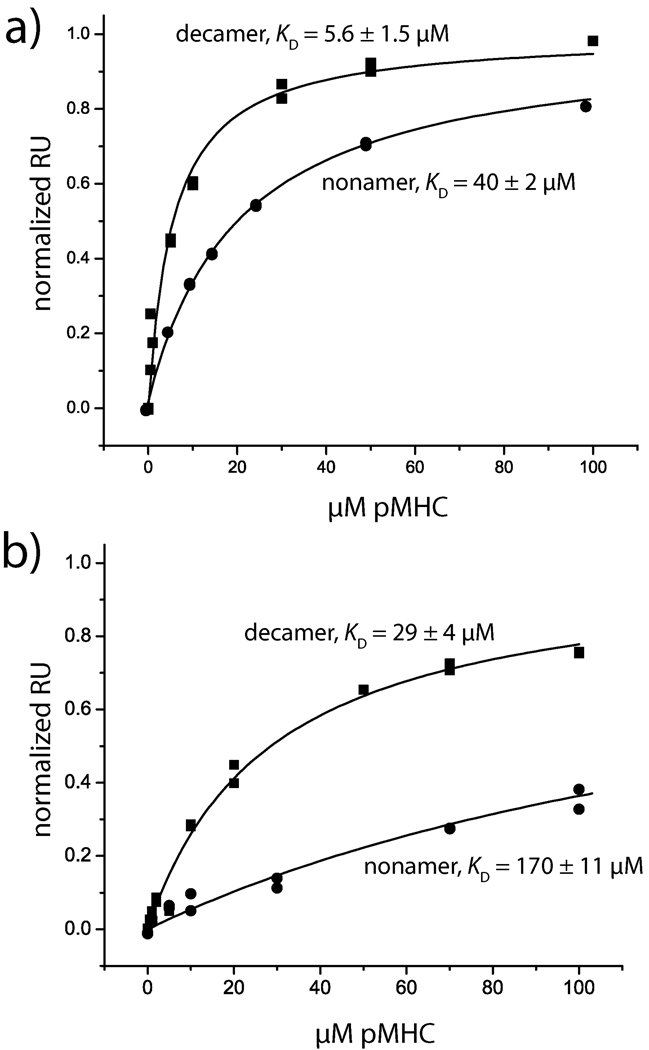
We next examined the interactions of the DMF4 and DMF5 TCRs with the two MART-1 ligands using surface plasmon resonance (Fig. 7A,B). The affinity of DMF5 towards the decamer and nonamer ligand was 6 µM and 40 µM, respectively. The affinity of DMF4 towards the decamer and nonamer was 29 µM and 170 µM, respectively. Both TCRs thus bind the decamer more strongly than the nonamer, with DMF5 possessing stronger affinity for both. The greater affinity towards decamer is consistent with the need to for the nonamer to undergo a structural shift upon binding of both TCRs.
Figure 7.
Surface plasmon resonance binding data define the hierarchy of DMF4/DMF5 nonamer/decamer recognition. A) Steady state equilibrium data for DMF5 recognition of the decamer and nonamer peptide/HLA-A2 complexes. Solid lines show fits to a single site binding model. Affinities are indicated. B) Steady state equilibrium data for DMF4 recognition of the decamer and nonamer peptide/HLA-A2 complexes. Affinities are indicated.
Interestingly, although DMF5 binds both the nonamer and decamer more tightly than DMF4, the difference in binding free energy between recognition of the nonamer and decamer is identical within error for the two TCRs (ΔΔG° = 1.2 ± 0.3 kcal/mol for decamer, 1.1 ± 0.1 kcal/mol for nonamer). Thus, from a free energy perspective, although DMF5 binds both nonamer and decamer with higher affinity, the DMF5’s mechanism of cross-reactivity is not superior to DMF4’s. Lastly, although we attempted kinetic measurements, dissociation rates for all cases were fast (> 0.5 s−1), precluding accurate measurements of association and dissociation rates.
Discussion
Recent clinical trials have demonstrated that the adoptive transfer of genetically re-directed T cells can lead to cancer regression in humans (2, 3). The first two T cell receptors used in this approach, DMF4 and DMF5, both recognize the overlapping but structurally diverging 26–35 (decamer) and 27–35 (nonamer) epitopes from the MART-1/Melan-A protein presented by HLA-A2 (4). Although the trials were small in size, clinical outcomes differed with the two receptors. Use of DMF4 led to a 13% rate of cancer regression, whereas use of DMF5 led to a 30% rate of regression and associated eye, ear, and skin toxicity. The DMF5 TCR is currently in use in larger clinical trials, and continues to be used as a model TCR for improvements in T cell-based gene therapy of cancer (5–7).
Early work assumed the MART-1 nonamer and decamer were structurally equivalent due to the high frequency of cross-reactive T cells in HLA-A2+ individuals (24). However, comparative structures of the two peptide/HLA-A2 complexes demonstrated that this is not the case, with the decamer adopting a bulged conformation due to the presence of the additional amino acid (8). Although the mechanisms underlying nonamer/decamer cross-reactivity are of interest given the fundamental role T cell cross-reactivity plays in cellular immunity (9), MART-1 nonamer/decamer cross-reactivity may also be important in immunotherapy: the nonamer is believed to be the physiologically relevant epitope in HLA-A2+ individuals (10–13), yet due to the poor binding of the nonamer to HLA-A2, the decamer or its anchor-modified variant is regularly used to identify and activate MART-1-specific T cells. The decamer or its variant also continues to be used as a chief component of many cancer vaccine formulations.
In cross-reacting between the MART-1 nonamer and decamer, both DMF4 and DMF5 require the nonamer to shift its backbone into a more decamer-like conformation, explaining the higher affinity towards decamer for both TCRs. Binding-induced conformational changes in peptide backbones have been observed previously in TCR recognition (e.g., 25–27), but it is interesting here that the changes are observed in the nonamer rather than the longer and more extensively bulged decamer. An earlier analysis of MART-1 bound to HLA-A2 suggested the nonamer possesses greater intrinsic flexibility than the decamer (8). Both DMF4 and DMF5 apparently utilize this flexibility in engaging nonamer, albeit for different reasons: DMF4 in order to avoid steric clashes, DMF5 in order to form hydrogen bonds.
After shifting the nonamer, the methods of DMF4 and DMF5 cross-reactivity diverge. The DMF4 TCR utilizes a complex mechanism, alternating between a binding orientation that, judging by hydrogen bonds, optimizes interactions with the peptide at the expense of the MHC (decamer), or optimizes interactions with the MHC at the expense of the peptide (nonamer). In contrast, DMF5 engages both ligands almost identically, utilizing an open architecture apparently pre-formed in the free TCR. Although the structures do not readily indicate why, DMF5’s simpler mode of cross-reactivity is associated with improved affinity towards both ligands.
The differences in affinity between the DMF4 and DMF5 TCRs are relatively modest, 5-fold for the decamer and 4-fold for the nonamer. These results are consistent with the differences in functional avidity with the original DMF4 and DMF5 T cell clones (4), and could help explain the reported differences in clinical outcomes with the two TCRs. Enhancing TCR affinity has been suggested as a means for improving immunotherapy (28), and TCRs with 1000-fold or more gains in affinity have been generated through molecular evolution techniques (e.g., 29 – 30). The differences in affinity between DMF4 and DMF5 raise the possibility that such large enhancements may not be needed to impact clinical results. This may be significant given that losses in peptide specificity have been observed with some very high affinity TCRs (31, 32), an outcome that would clearly be detrimental for antigen-specific immunotherapy (again though, we emphasize that the clinical trials with DMF4 and DMF5 were small, and other factors such as differences in α/β chain pairing in the transduced TCRs could have contributed to the outcomes).
Structure-guided computational design has also been used to enhance TCR affinity (34), in principle providing a degree of control absent in molecular evolution and reducing the risk of losses in antigen specificity. The structures with the DMF5 TCR provide a starting point for pursuing such an approach. The fact that DMF5 engages both the MART-1 nonamer and decamer identically using a pre-formed architecture raises the likelihood of successfully enhancing affinity towards both nonamer and decamer without losses in specificity, reducing concerns that a MART-1-specific TCR with enhanced affinity towards the nonamer would have reduced affinity towards the decamer or vice versa, either of which would be negatively impact immunotherapy utilizing decamer-based peptides to identify or activate nonamer-specific T cells. A natural concern is that higher affinity TCRs targeting MART-1 may induce even stronger autoimmune toxicity if used clinically, requiring more significant interventions than previously used (2).
The finding that the binding orientation of DMF4 differed with the nonamer and decamer was surprising. The switch in binding orientation appears attributable to two structural features: the need to avoid steric clashes between CDR1α and the center of the nonamer and the ability of HV4α to form a salt-bridge with the N-terminal glutamate of the decamer. Different orientations of a single TCR over two different ligands have been observed with the murine 2C TCR in complex with the dEV8 and the QL9 peptides (35). In that case though, the two peptides were presented by different MHC proteins (H-2Ld and H-2Kb, respectively). The results with DMF4 extend this finding by demonstrating that TCR binding orientations can be dictated by peptide alone. This peptide-determined binding mode necessitates a structurally and energetically permissive relationship between the germline elements of DMF4 and HLA-A2. Such permissiveness may be a fundamental mechanism of TCR cross-reactivity, and is likely further illustrated in the observation of unusual TCR binding modes seen with TCRs bound to self-antigens associated with autoimmunity (reviewed in 36).
Although the changes in binding orientation seen with DMF4 alters the interactions between the TCR germline elements and HLA-A2, some interactions are conserved, most notably those between Tyr49 of CDR2α and Gln155 of HLA-A2. Both tyrosines in TCR CDR2 loops and Gln155 in class I MHC molecules have been proposed to play key roles in TCR binding, with tyrosines in particular implicated in encoding a genetic bias of TCRs towards MHC proteins (17, 20, 37). The two DMF4 structures, with different binding orientations despite the same variable domains and MHC, provide a new opportunity to test this hypothesis with structure-guided mutations. It is notable that in addition to interacting with Tyr49 of CDR2α, Gln155 also hydrogen bonds with Gln100 of CDR3α, highlighting possible cooperativity in the interactions of the germline and non-germline elements with HLA-A2.
Recognition of MART-1 antigens in HLA-A2+ individuals is characterized by a strong bias towards TCRs utilizing the Vα 12-2 variable domain (38, 39). Based on the structure of the Vα 12-2 Mel5 TCR with the MART-1 nonamer presented by HLA-A2, Cole et al. proposed that this bias was attributable to interactions between the germline CDR1α loop and the peptide, describing this as “innate-like” recognition of antigen (40). The DMF5 TCR forms the same CDR1α -peptide interactions as does Mel5, utilizing Gln30 to form two hydrogen bonds to the peptide backbone (Supplemental Fig. 5A). Interestingly, CDR1α of the well-characterized TCR A6, which also utilizes Vα 12-2, forms similar interactions with the Tax, Tel1p, and HuD peptides (25, 41, 42). Although these peptides are unrelated to those of MART-1, their N-terminal conformations when bound to HLA-A2 are very similar. CDR1α of Vα 12-2 thus appears optimally positioned to interact with this peptide conformation. However, the extent to which CDR1α-peptide interactions underlies the Vα 12-2 bias in MART-1 specific TCRs remains uncertain, as there are conserved patterns of van der Waals interactions between the Vα 12-2 germline loops and HLA-A2 in the various structures (Supplemental Fig. 5B). Determining the energetic balance between these two sets of interactions will again require more probing investigations. Lastly, MART-1-specific Vα 12-2 TCRs also show a weak conservation in the length and sequence of CDR3α and CDR3β (43). The two CDR3 loops of Mel5 form a similar “slot” as in DMF5 in order to accommodate the bulge in the MART-1 decamer (Supplemental Fig. 6); the weak bias in CDR3α/CDR3β composition may reflect that only a subset of possible CDR3 loops are compatible with this architecture.
Supplementary Material
Acknowledgements
We thank Cynthia Piepenbrink for outstanding technical assistance.
Footnotes
Supported by grants GM067079 from NIGMS, NIH, RR025761 from NCRR, NIH, and RSG-05-202-01-GMC from the American Cancer Society. SMS was supported by a fellowship from the Walther Cancer Center. Results shown in this report are derived from work performed at the Structural Biology Center, LS-CAT, and LRL-CAT at the Advanced Photon Source, Argonne National Laboratory. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy under contract DE-AC02-06CH11357. Use of LS-CAT at APS Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). Use of the LRL-CAT at APS Sector 31 was provided by Eli Lilly & Company who operates the facility.
References
- 1.Rosenberg SA, Restifo NP, Yang JC, Morgan RA, Dudley ME. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat Rev Cancer. 2008;8:299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, Kammula US, Royal RE, Sherry RM, Wunderlich JR, Lee CC, Restifo NP, Schwarz SL, Cogdill AP, Bishop RJ, Kim H, Brewer CC, Rudy SF, VanWaes C, Davis JL, Mathur A, Ripley RT, Nathan DA, Laurencot CM, Rosenberg SA. Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood. 2009;114:535–546. doi: 10.1182/blood-2009-03-211714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, Zheng Z, Nahvi A, de Vries CR, Rogers-Freezer LJ, Mavroukakis SA, Rosenberg SA. Cancer Regression in Patients After Transfer of Genetically Engineered Lymphocytes. Science. 2006;314:126–129. doi: 10.1126/science.1129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson LA, Heemskerk B, Powell DJ, Jr, Cohen CJ, Morgan RA, Dudley ME, Robbins PF, Rosenberg SA. Gene Transfer of Tumor-Reactive TCR Confers Both High Avidity and Tumor Reactivity to Nonreactive Peripheral Blood Mononuclear Cells and Tumor-Infiltrating Lymphocytes. J Immunol. 2006;177:6548–6559. doi: 10.4049/jimmunol.177.9.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goff S, Johnson L, Black M, Xu H, Zheng Z, Cohen C, Morgan R, Rosenberg S, Feldman S. Enhanced receptor expression and in vitro effector function of a murine-human hybrid MART-1-reactive T cell receptor following a rapid expansion. Cancer Immunology, Immunotherapy. 59:1551–1560. doi: 10.1007/s00262-010-0882-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ray S, Chhabra A, Chakraborty NG, Hegde U, Dorsky DI, Chodon T, von Euw E, Comin-Anduix B, Koya RC, Ribas A, Economou JS, Rosenberg SA, Mukherji B. MHC-I-restricted melanoma antigen specific TCR-engineered human CD4+ T cells exhibit multifunctional effector and helper responses, in vitro. Clinical Immunology. 136:338–347. doi: 10.1016/j.clim.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jazirehi AR, Baritaki S, Koya RC, Bonavida B, Economou JS. Molecular Mechanism of MART-1+/A*0201+ Human Melanoma Resistance to Specific CTL-Killing Despite Functional Tumor-CTL Interaction. Cancer Research. doi: 10.1158/0008-5472.CAN-10-1296. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Borbulevych OY, Insaidoo FK, Baxter TK, Powell DJ, Jr, Johnson LA, Restifo NP, Baker BM. Structures of MART-1(26/27–35) Peptide/HLA-A2 Complexes Reveal a Remarkable Disconnect between Antigen Structural Homology and T Cell Recognition. J Mol Biol. 2007;372:1123–1136. doi: 10.1016/j.jmb.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mason D. A very high level of crossreactivity is an essential feature of the T-cell receptor. Immunology Today. 1998;19:395–404. doi: 10.1016/s0167-5699(98)01299-7. [DOI] [PubMed] [Google Scholar]
- 10.Skipper JCA, Gulden PH, Hendrickson RC, Harthun N, Caldwell JA, Shabanowitz J, Engelhard VH, Hunt DF, Craig J, Slingluff L. Mass-spectrometric evaluation of HLA-A*0201-associated peptides identifies dominant naturally processed forms of CTL epitopes from MART-1 and gp100. International Journal of Cancer. 1999;82:669–677. doi: 10.1002/(sici)1097-0215(19990827)82:5<669::aid-ijc9>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 11.Derre L, Ferber M, Touvrey C, Devevre E, Zoete V, Leimgruber A, Romero P, Michielin O, Levy F, Speiser DE. A novel population of human melanoma-specific CD8 T cells recognizes Melan-AMART-1 immunodominant nonapeptide but not the corresponding decapeptide. J Immunol. 2007;179:7635–7645. doi: 10.4049/jimmunol.179.11.7635. [DOI] [PubMed] [Google Scholar]
- 12.Held G, Wadle A, Dauth N, Stewart-Jones G, Sturm C, Thiel M, Zwick C, Dieckmann D, Schuler G, Hoogenboom HR, Levy F, Cerundolo V, Pfreundschuh M, Renner C. MHC-peptide-specific antibodies reveal inefficient presentation of an HLA-A*0201-restricted, Melan-A-derived peptide after active intracellular processing. European Journal of Immunology. 2007;37:2008–2017. doi: 10.1002/eji.200636545. [DOI] [PubMed] [Google Scholar]
- 13.Michaeli Y, Denkberg G, Sinik K, Lantzy L, Chih-Sheng C, Beauverd C, Ziv T, Romero P, Reiter Y. Expression hierarchy of T cell epitopes from melanoma differentiation antigens: unexpected high level presentation of tyrosinase-HLA-A2 Complexes revealed by peptide-specific, MHC-restricted, TCR-like antibodies. J Immunol. 2009;182:6328–6341. doi: 10.4049/jimmunol.0801898. [DOI] [PubMed] [Google Scholar]
- 14.Valmori D, Fonteneau J-F, Lizana CM, Gervois N, Lienard D, Rimoldi D, Jongeneel V, Jotereau F, Cerottini J-C, Romero P. Enhanced Generation of Specific Tumor-Reactive CTL In Vitro by Selected Melan-A/MART-1 Immunodominant Peptide Analogues. J Immunol. 1998;160:1750–1758. [PubMed] [Google Scholar]
- 15.Ribas A, Comin-Anduix Ba, Chmielowski B, Jalil J, de la Rocha P, McCannel TA, Ochoa MT, Seja E, Villanueva A, Oseguera DK, Straatsma BR, Cochran AJ, Glaspy JA, Hui L, Marincola FM, Wang E, Economou JS, Gomez-Navarro J. Dendritic Cell Vaccination Combined with CTLA4 Blockade in Patients with Metastatic Melanoma. Clinical Cancer Research. 2009;15:6267–6276. doi: 10.1158/1078-0432.CCR-09-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rudolph MG, Stanfield RL, Wilson IA. How TCRs Bind MHCs, Peptides, and Coreceptors. Annu Rev Immunol. 2006;24:419–466. doi: 10.1146/annurev.immunol.23.021704.115658. [DOI] [PubMed] [Google Scholar]
- 17.Clements CS, Dunstone MA, Macdonald WA, McCluskey J, Rossjohn J. Specificity on a knife-edge: the [alpha][beta] T cell receptor. Current Opinion in Structural Biology. 2006;16:787–795. doi: 10.1016/j.sbi.2006.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Baxter TK, Gagnon SJ, Davis-Harrison RL, Beck JC, Binz A-K, Turner RV, Biddison WE, Baker BM. Strategic mutations in the class I MHC HLA-A2 independently affect both peptide binding and T cell receptor recognition. J. Biol. Chem. 2004;279:29175–29184. doi: 10.1074/jbc.M403372200. [DOI] [PubMed] [Google Scholar]
- 19.Gagnon SJ, Borbulevych OY, Davis-Harrison RL, Baxter TK, Clemens JR, Armstrong KM, Turner RV, Damirjian M, Biddison WE, Baker BM. Unraveling a Hotspot for TCR Recognition on HLA-A2: Evidence Against the Existence of Peptide-independent TCR Binding Determinants. Journal of Molecular Biology. 2005;353:556. doi: 10.1016/j.jmb.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 20.Marrack P, Scott-Browne JP, Dai S, Gapin L, Kappler JW. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu Rev Immunol. 2008;26:171–203. doi: 10.1146/annurev.immunol.26.021607.090421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sliz P, Michielin O, Cerottini J-C, Luescher I, Romero P, Karplus M, Wiley DC. Crystal Structures of Two Closely Related but Antigenically Distinct HLA-A2/Melanocyte-Melanoma Tumor-Antigen Peptide Complexes. J Immunol. 2001;167:3276–3284. doi: 10.4049/jimmunol.167.6.3276. [DOI] [PubMed] [Google Scholar]
- 22.Macdonald WA, Chen Z, Gras S, Archbold JK, Tynan FE, Clements CS, Bharadwaj M, Kjer-Nielsen L, Saunders PM, Wilce MCJ, Crawford F, Stadinsky B, Jackson D, Brooks AG, Purcell AW, Kappler JW, Burrows SR, Rossjohn J, McCluskey J. T Cell Allorecognition via Molecular Mimicry. 2009;31:897–908. doi: 10.1016/j.immuni.2009.09.025. [DOI] [PubMed] [Google Scholar]
- 23.Armstrong KM, Piepenbrink KH, Baker BM. Conformational changes and flexibility in T-cell receptor recognition of peptide-MHC complexes. Biochem J. 2008;415:183–196. doi: 10.1042/BJ20080850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pittet MJ, Valmori D, Dunbar PR, Speiser DE, Lienard D, Lejeune F, Fleischhauer K, Cerundolo V, Cerottini J-C, Romero P. High Frequencies of Naive Melan-A/MART-1-specific CD8+ T Cells in a Large Proportion of Human Histocompatibility Leukocyte Antigen (HLA)-A2 Individuals. J. Exp. Med. 1999;190:705–716. doi: 10.1084/jem.190.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garboczi DN, Ghosh P, Utz U, Fan QR, Biddison WE, Wiley DC. Structure of the complex between human T-cell receptor, viral peptide and HLA-A2. Nature. 1996;384:134–141. doi: 10.1038/384134a0. [DOI] [PubMed] [Google Scholar]
- 26.Tynan FE, Reid HH, Kjer-Nielsen L, Miles JJ, Wilce MCJ, Kostenko L, Borg NA, Williamson NA, Beddoe T, Purcell AW, Burrows SR, McCluskey J, Rossjohn J. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat Immunol. 2007;8:268–276. doi: 10.1038/ni1432. [DOI] [PubMed] [Google Scholar]
- 27.Lee JK, Stewart-Jones G, Dong T, Harlos K, Di Gleria K, Dorrell L, Douek DC, van der Merwe PA, Jones EY, McMichael AJ. T Cell Cross-Reactivity and Conformational Changes during TCR Engagement. J Exp Med. 2004;200:1455–1466. doi: 10.1084/jem.20041251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenberg SA, Dudley ME. Adoptive cell therapy for the treatment of patients with metastatic melanoma. Current Opinion in Immunology. 2009;21:233–240. doi: 10.1016/j.coi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holler PD, Holman PO, Shusta EV, O'Herrin S, Wittrup KD, Kranz DM. In vitro evolution of a T cell receptor with high affinity for peptide/MHC. Proc Natl Acad Sci U S A. 2000;97:5387–5392. doi: 10.1073/pnas.080078297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Moysey R, Molloy PE, Vuidepot A-L, Mahon T, Baston E, Dunn S, Liddy N, Jacob J, Jakobsen BK, Boulter JM. Directed evolution of human T-cell receptors with picomolar affinities by phage display. Nat Biotech. 2005;23:349–354. doi: 10.1038/nbt1070. [DOI] [PubMed] [Google Scholar]
- 31.Holler PD, Chlewicki LK, Kranz DM. TCRs with high affinity for foreign pMHC show self-reactivity. Nat Immunol. 2003;4:55–62. doi: 10.1038/ni863. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Bennett AD, Zheng Z, Wang QJ, Robbins PF, Yu LYL, Li Y, Molloy PE, Dunn SM, Jakobsen BK, Rosenberg SA, Morgan RA. High-affinity TCRs generated by phage display provide CD4(+) T cells with the ability to recognize and kill tumor cell lines. Journal of Immunology. 2007;179:5845–5854. doi: 10.4049/jimmunol.179.9.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chervin AS, Stone JD, Holler PD, Bai A, Chen J, Eisen HN, Kranz DM. The Impact of TCR-Binding Properties and Antigen Presentation Format on T Cell Responsiveness. J Immunol. 2009;183:1166–1178. doi: 10.4049/jimmunol.0900054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haidar JN, Pierce B, Yu Y, Tong W, Li M, Weng Z. Structure-based design of a T-cell receptor leads to nearly 100-fold improvement in binding affinity for pepMHC. Proteins: Structure, Function, and Bioinformatics. 2009;74:948–960. doi: 10.1002/prot.22203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colf LA, Bankovich AJ, Hanick NA, Bowerman NA, Jones LL, Kranz DM, Garcia KC. How a Single T Cell Receptor Recognizes Both Self and Foreign MHC. Cell. 2007;129:135–146. doi: 10.1016/j.cell.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 36.Wucherpfennig KW, Call MJ, Deng L, Mariuzza R. Structural alterations in peptide-MHC recognition by self-reactive T cell receptors. Curr Opin Immunol. 2009;21:590–595. doi: 10.1016/j.coi.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction 'codon'. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 38.Trautmann L, Labarriere N, Jotereau F, Karanikas V, Gervois N, Connerotte T, Coulie P, Bonneville M. Dominant TCR V alpha usage by virus and tumor-reactive T cells with wide affinity ranges for their specific antigens. Eur J Immunol. 2002;32:3181–3190. doi: 10.1002/1521-4141(200211)32:11<3181::AID-IMMU3181>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Dietrich PY, Le Gal FA, Dutoit V, Pittet MJ, Trautman L, Zippelius A, Cognet I, Widmer V, Walker PR, Michielin O, Guillaume P, Connerotte T, Jotereau F, Coulie PG, Romero P, Cerottini JC, Bonneville M, Valmori D. Prevalent role of TCR alpha-chain in the selection of the preimmune repertoire specific for a human tumor-associated self-antigen. J Immunol. 2003;170:5103–5109. doi: 10.4049/jimmunol.170.10.5103. [DOI] [PubMed] [Google Scholar]
- 40.Cole DK, Yuan F, Rizkallah PJ, Miles JJ, Gostick E, Price DA, Gao GF, Jakobsen BK, Sewell AK. Germline-governed recognition of a cancer epitope by an immunodominant human T-cell receptor. Journal of Biological Chemistry. 2009;284:27281–27289. doi: 10.1074/jbc.M109.022509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Borbulevych OY, Piepenbrink KH, Baker BM. Conformational Melding Permits a Conserved Binding Geometry in TCR Recognition of Foreign and Self Molecular Mimics. J Immunol. 2011;186:2950–2958. doi: 10.4049/jimmunol.1003150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borbulevych OY, Piepenbrink KH, Gloor BE, Scott DR, Sommese RF, Cole DK, Sewell AK, Baker BM. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity. 2009;31:885–896. doi: 10.1016/j.immuni.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wieckowski S, Baumgaertner P, Corthesy P, Voelter V, Romero P, Speiser DE, Rufer N. Fine Structural Variations of {alpha}{beta}TCRs Selected by Vaccination with Natural versus Altered Self-Antigen in Melanoma Patients. J Immunol. 2009;183:5397–5406. doi: 10.4049/jimmunol.0901460. [DOI] [PubMed] [Google Scholar]
- 44.Davis-Harrison RL, Armstrong KM, Baker BM. Two Different T Cell Receptors use Different Thermodynamic Strategies to Recognize the Same Peptide/MHC Ligand. Journal of Molecular Biology. 2005;346:533–550. doi: 10.1016/j.jmb.2004.11.063. [DOI] [PubMed] [Google Scholar]
- 45.Laugel B, Boulter JM, Lissin N, Vuidepot A, Li Y, Gostick E, Crotty LE, Douek DC, Hemelaar J, Price DA, Jakobsen BK, Sewell AK. Design of Soluble Recombinant T Cell Receptors for Antigen Targeting and T Cell Inhibition. J. Biol. Chem. 2005;280:1882–1892. doi: 10.1074/jbc.M409427200. [DOI] [PubMed] [Google Scholar]
- 46.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 47.Gagnon SJ, Borbulevych OY, Davis-Harrison RL, Turner RV, Damirjian M, Wojnarowicz A, Biddison WE, Baker BM. T Cell Receptor Recognition via Cooperative Conformational Plasticity. Journal of Molecular Biology. 2006;363:228–243. doi: 10.1016/j.jmb.2006.08.045. [DOI] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Perrakis A, Sixma TK, Wilson KS, Lamzin VS. wARP: improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D Biol Crystallogr. 1997;53:448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.McRee DE. XtalView/Xfit--A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 52.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst. 1993;26:283–291. [Google Scholar]
- 53.Rodriguez R, Chinea G, Lopez N, Pons T, Vriend G. Homology modeling, model and software evaluation: three related resources. Bioinformatics. 1998;14:523–528. doi: 10.1093/bioinformatics/14.6.523. [DOI] [PubMed] [Google Scholar]
- 54.Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 56.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 57.Piepenbrink KH, Gloor BE, Armstrong KM, Baker BM. Methods for Quantifying T Cell Receptor Binding Affinities and Thermodynamics. Methods Enzymol. 2009;466:359–381. doi: 10.1016/S0076-6879(09)66015-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.