Abstract
The Drosophila sticks-and-stones (sns) locus was identified on the basis of its mutant phenotype, the complete absence of body wall muscles and corresponding presence of unfused myoblasts. The genetic location of the mutation responsible for this apparent defect in myoblast fusion was determined by recombination and deficiency mapping, and the corresponding wild-type gene was isolated in a molecular walk. Identification of the SNS coding sequence revealed a putative member of the immunoglobulin superfamily (IgSF) of cell adhesion molecules. As anticipated from this homology, SNS is enriched at the membrane and clusters at discrete sites, coincident with the occurrence of myoblast fusion. Both the sns transcript and the encoded protein are expressed in precursors of the somatic and visceral musculature of the embryo. Within the presumptive somatic musculature, SNS expression is restricted to the putative fusion-competent cells and is not detected in unfused founder cells. Thus, SNS represents the first known marker for this subgroup of myoblasts, and provides an opportunity to identify pathways specifying this cell type as well as transcriptional regulators of fusion-specific genes. To these ends, we demonstrate that the presence of SNS-expressing cells is absolutely dependent on Notch, and that expression of SNS does not require the myogenic regulatory protein MEF2.
Keywords: Myoblast-fusion, IgSF, sticks-and-stones, Notch, somatic musculature, myogenesis
Myogenesis is a highly regulated process through which cells acquire a variety of specialized characteristics. Among the morphological changes that accompany myogenesis are the formation of the contractile apparatus and the fusion of myoblasts into multinucleate myotubes. Studies in both vertebrate and invertebrate systems have shown that molecules associated with the contractile apparatus are conserved (for review, see Bernstein et al. 1993). Moreover, regulatory proteins such as MEF2, which is critical for the transcriptional initiation of such genes, are also conserved (Lilly et al. 1994; Nguyen et al. 1994). One additional feature common to myogenesis in most metazoan animals is the migration of developing myoblasts to specific regions of the organism, and fusion to form an ordered array of multi-nucleate muscle fibers. However, by contrast to the proteins associated with the contractile apparatus, much less is known about the components associated with fusion or their regulation during myogenesis.
On the basis of the studies using vertebrate tissue culture systems, the process of fusion has been divided into a series of steps that includes differentiation, the acquisition of fusion competence, recognition, adhesion, and membrane breakdown (for review, see Knudsen 1992). Transmission electron microscopic studies (Engel et al. 1985) have revealed the presence of electron-opaque material localized at discrete points of cell-cell contact in fusing rat myoblast cell lines, suggesting the existence of specialized sites for fusion on the myoblast membrane surface. In addition, examination of myoblast-myotube fusion in cultured quail cells (Lipton and Konigsberg 1972) revealed that a single point of cytoplasmic continuity is present between apposing myoblasts, suggesting that fusion is initiated at a single site in this system. Finally, electron dense plaques have been observed in vertebrates as well (Rash and Fambrough 1973), and are thought to be associated with the fusion process.
Identification of molecules that inhibit myoblast fusion in vertebrate tissue culture systems has provided some biochemical insight into the fusion process. Essential components identified through these studies include, for example, cell adhesion molecules, calcium and calmodulin, metalloproteases, phospholipases, lipids, and others (for review, see Wakelam 1985; Knudsen 1992; Yagami-Hiromasa et al. 1995). However, an important caveat to the interpretation of results derived from blocking agents is the potential for indirect effects. By contrast, efforts to address the specific role of proteins through a loss-of-function phenotype can be complicated by the presence of molecules with redundant functions in the intact organism or lethality prior to the stage of interest (see, for example, Mege et al. 1992; Charlton et al. 1997; Radice et al. 1997).
Like vertebrates, the larval body wall muscles of Drosophila include a unique array of multinucleate muscle fibers. In the embryo, two distinct populations of myoblasts appear to be involved in formation of these fibers. The first, termed muscle founder cells, appear in characteristic and reproducible positions and contain information that specifies muscle identity, size, position, and attachment. Morphologically, these cells first appear as individual progenitors that divide asymetrically and then fuse into bi- and tri-nucleate clusters termed muscle precursors (Bate 1990; Dohrmann et al. 1990; Rushton et al. 1995). Several proteins have been identified that mark subsets of founder cells, and function in their specification and differentiation (Dohrmann et al. 1990; Michelson et al. 1990; Paterson et al. 1991; Williams et al. 1991; Bourgouin et al. 1992; Keller et al. 1998; Knirr et al. 1999; for review, see Frasch 1999). The second and more populous group of cells has been termed fusion-competent myoblasts. As defined, these cells are committed to myogenesis, but have no inherent fiber specificity. Rather, these cells are thought to take on the identity of the muscle precursors with which they fuse (Bate 1990; Dohrmann et al. 1990; Rushton et al. 1995).
Ultrastructural studies of Drosophila embryos have revealed a series of events associated with the formation of multinucleate syncytia that are reminiscent of those described above in vertebrate systems (Doberstein et al. 1997). This pathway begins with cell–cell recognition and adherence. Cells then elongate, align with each other, and establish multiple small zones of cytoplasmic continuity between the apposed plasma membranes. During this time, electron dense vesicles are found near the cytoplasmic face of the plasma membrane, at the contact point between myoblasts. These vesicles align with similar vesicles located in the apposing myoblasts, and have been referred to as the prefusion complex. Electron dense plaques thought to be formed from these vesicles extend for ∼500 nm, and fusion then occurs as the intervening cell membrane vesiculates. Whereas the composition of these vesicles and their role in fusion remain unclear, they are reminiscent of the electron opaque material seen in fusing rat myoblasts (Engel et al. 1985).
Whereas homologs of vertebrate factors associated with myoblast fusion have not been examined in detail in Drosophila, loss-of-function mutations that exhibit dramatic defects in myoblast fusion have been identified. Essential loci include rolling stone (rost; Paululat et al. 1995), myoblast city (mbc; Rushton et al. 1995), and blown fuse (blow; Doberstein et al. 1997). MBC, the Drosophila homolog of human DOCK180 and Caenorhabditis elegans ced-5, appears to be ubiquitously expressed and functions in the rac1 pathway (Erickson et al. 1997; Nolan et al. 1998). BLOW and ROST are muscle specific, but their protein coding sequences have not yet provided insight into their function (Doberstein et al. 1997; Paululat et al. 1997).
The present report describes the characterization of the sticks and stones (sns) gene, which is essential for the formation of multinucleate muscle fibers in Drosophila. Within the somatic mesoderm, it is the first marker specific to the fusion-competent class of myoblasts and is not detected in founder cells. SNS expression is not detected in Notch mutant embryos, suggesting that Notch is required for specification of the SNS-expressing population of cells. Surprisingly, the transcriptional regulator MEF2 is not required for SNS expression. Finally, sns encodes a protein in the Ig superfamily of cell adhesion molecules. Consistent with this identification, SNS is detected at the membrane and becomes localized to discrete sites that may be associated with contact between fusing myoblasts.
Results
Identification and genetic mapping of the sns locus
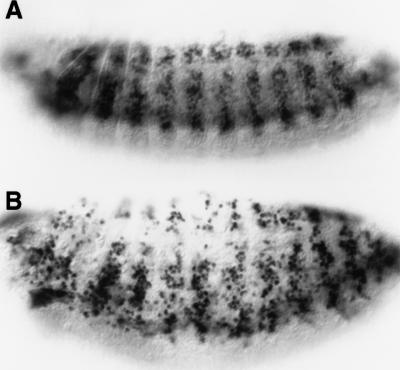
The sns locus, which is essential for myoblast fusion, was uncovered during an F2 lethal screen for EMS-induced point mutations in cytological region 95A on the third chromosome (Erickson et al. 1997; Keller et al. 1998). In this screen, the original mutagenized fly was later found to have contained two recessive lethal mutations, one in the region of interest on the third chromosome and one on the second chromosome. Genetic mapping revealed that the muscle defect segregated with the second chromosome, and the recovered mutant locus was named sticks and stones (sns). As shown in Figure 2B (see below), examination of the developing body wall muscles in snsA3.24 mutant embryos revealed an almost complete block in myoblast fusion.
Figure 2.
MHC, NAU, and MEF2 positive cells are present in sns mutant embryos but do not fuse to form muscle fibers. All embryos are oriented ventrolaterally with anterior to the left. Wild-type embryos are shown in A, C, and E. (B,D,F) Embryos that are genetically snsA3.24/Df(2R)BB1. Embryos homozygous for snsA3.24 exhibit the same mutant phenotype (data not shown) and Df(2R)BB1 completely removes the sns genomic region (Results, Materials and Methods). Therefore, by genetic criteria, snsA3.24 appears to represent the null phenotype for sns. (A,B) Stage 15 embryos immunostained with an MHC antibody to visualize the musculature. By comparison with wild-type in A, embryos mutant for sns exhibit unfused MHC expressing myoblasts in the place of mature muscle fibers. (C,D) Stage 13 embryos immunostained with antisera against NAU to visualize a subset of founder cells. In wild-type embryos (C), NAU-expressing cells are distributed in an array of muscle founders and precursors. (D) The pattern of NAU-expressing cells is not affected in sns mutant embryos, suggesting that founder cell specification does not require sns. (E,F) Stage 15 embryos immunostained with antisera against MEF2 to visualize the entire myoblast population. MEF2 expression can be seen in developing muscle fibers in wild-type embryos (E) and in the corresponding population of unfused myoblasts in sns mutant embryos (F).
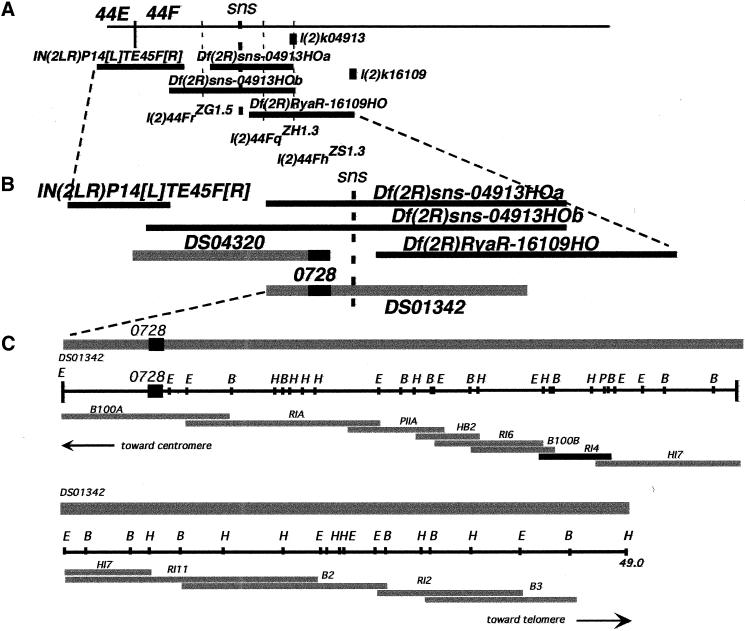
Genetic mapping (Materials and Methods) placed the sns locus between positions 58.2–61.5, corresponding roughly to cytological position 44–47. Deficiencies that deleted regions 43A–44DE, 44F2–45EF, and 45A–47 did not uncover sns. Deficiencies in region 44E–F were subsequently generated by gamma irradiation and imprecise excision of P-element insertions in the region (Materials and Methods). Select deficiencies, shown in Figure 1, uncovered sns and narrowed its location to cytological region 44F1–4, between the proximal breakpoints of deficiencies Df(2R)sns-04913HOa and Df(2R)RyaR-16109HO.
Figure 1.
Genetic and molecular map of the chromosomal region that includes sns. (A) Genetic map of cytological region 44F. (Horizontal bars) Deficiencies, (vertical dashed lines) lethal complementation groups, (gray boxes) P-element insertions. The genetic positions of lethal complementation groups in the region, including the sns locus, are indicated. As shown, sns maps between the proximal breakpoints of Df(2R)sns-04913HOa and Df(2R)RyaR-16109HO. (B) Enlargement of the genetic map shown in A onto which the positions of cloned sequences have been mapped. Black horizontal bars represent deficiencies as in A. The gray bars indicate the positions of two P1 clones (provided by the Berkeley Drosophila Genome Project) relative to the deficiencies. The STS 728 is indicated by the dark black box within each clone. As indicated, DS04320 begins at STS 728 and extends proximally. The proximal sequence of this clone is deleted in IN(2LR)P14[L]TE45F[R], whereas the distal end is deleted in both Df(2R)sns-04913HOa and Df(2R)sns-04913HOb. DS01342, which includes a significant portion of the sns genomic region, extends distally, and is deleted in Df(2R)RyaR-16109HO as shown. (C) Molecular walk within the P1 clone DS01342. The gray bar at the top represents the P1 clone used for a chromosomal walk to clone sns. The position of STS 728 is indicated. In the restriction map of the region, (E) EcoRI, (B) BamHI, (P) PstI, and (H) HindIII. Subclones are as indicated below the restriction map. A 2.9-kb EcoRI fragment (indicated RI4) is highlighted. This fragment includes sns coding sequence, and revealed the splicing defect in snsXB3 (refer to text).
The sns locus is essential for myoblast fusion
The muscle phenotype of embryos homozygous for the original snsA3.24 allele includes a large number of unfused myosin-expressing cells and a corresponding absence of differentiated muscle fibers. Embryos transheterozygous for this sns allele and Df(2R)BB1, which deletes the entire sns region (data not shown), exhibited the same mutant phenotype (Fig. 2B). Thus snsA3.24 behaves as a null allele by genetic criteria. The presence of founder cells was then assessed using an antibody directed against NAU (Keller et al. 1998). As shown in Figure 2, C and D, NAU-expressing cells are detected in their correct positions in sns mutant embryos, but do not appear to fuse. These cells are capable of producing myosin heavy chain (MHC) protein, and appear to be analogous to the muscle founders described in mbc mutant embryos (Rushton et al. 1995; data not shown). The entire myoblast population was also examined using a polyclonal antibody directed against MEF2, an early marker for most if not all cells of the somatic musculature (Lilly et al. 1994; Nguyen et al. 1994). MEF2 expression is detected in the myoblasts of sns mutant embryos, in numbers approximately equivalent to that observed in wild-type embryos. These results imply that the precursors of the somatic musculature begin their differentiation program in sns mutant embryos, but become blocked at the point of myoblast fusion.
In contrast to the severe defects in the somatic musculature, only subtle defects were observed in constrictions of the visceral musculature, and are under further investigation (data not shown). The monoclonal antibody 22C10 (Zipursky et al. 1984), which labels the nervous system, as well as antisera directed against Fasciclin II (FASII; Grenningloh et al. 1991), which labels the motorneurons, and Fasciclin III (FASIII; Patel et al. 1987), a glycoprotein on the surface of epidermal cells, revealed no obvious defects in these ectodermally-derived issues (data not shown).
Identification of the sns gene
The proximal breakpoints of deficiencies Df(2R)sns-04913HOa and Df(2R)RyaR-16109HO define the location of the sns gene (Fig. 1). Bacteriophage P1 clones DS04320 and DS01342, which had been localized to cytological region 44F by the Berkeley Drosophila Genome Project, were mapped molecularly to the region deleted by these deficiencies. Plasmid mini-libraries were generated from the P1 clones and STS0728, which is located at the end of DS04320, was used to initiate a chromosomal walk. Genomic fragments were evaluated for candidate transcripts using Northern blots and in situ hybridization. Fragment RI4 (Fig. 1) detected a muscle-specific transcript of roughly 8 kb (data not shown), and was used to isolate overlapping partial cDNA clones. The full-length cDNA sequence was then assembled from these clones.
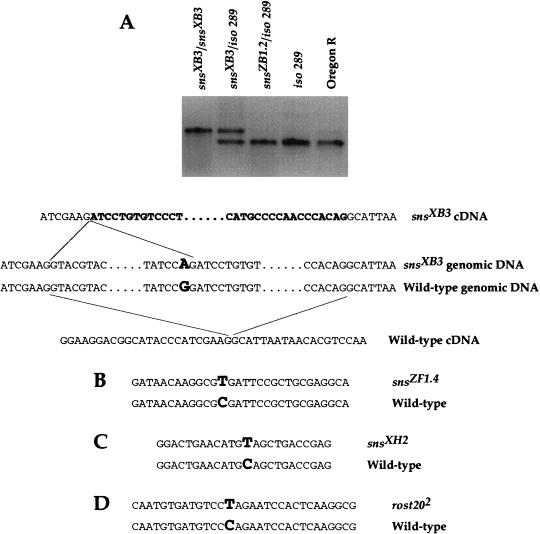
To confirm that this sequence corresponds to the sns gene, it was examined in mutant alleles of sns for aberrations. Small deletions and restriction site alterations were identified by Southern analysis. This approach uncovered a novel band in DNA from snsXB3, the result of a G to A transition that destroyed a BamHI site within an intron of the putative sns sequence (data not shown). This lesion created a novel 3′ splice acceptor site and leads to incorrect splicing of the transcript in snsXB3 mutant embryos, as revealed by RT–PCR (Fig. 3A). Incorrect splicing alters the reading frame of the transcript, such that a stop codon is introduced 67 base pairs from the splice junction.
Figure 3.
The putative sns sequence is altered in mutant alleles of sns. (A) A BamHI site located within an intron of the sns gene is altered in snsXB3 by a G to A transition in the first G of the BamHI recognition sequence. As confirmed by RT–PCR of mRNA from snsXB3 mutant embryos, this change introduces a splice acceptor site that results in a spliced product of 248 bp rather than the wild-type product of 195 bp. Direct sequencing of the 248 bp product confirmed the altered sequence indicated in A. (B–D) Non-isotopic RNase cleavage revealed mutations in the putative sns gene in other EMS induced alleles of sns (data not shown). Direct sequencing of these alleles revealed the indicated alterations. (B) snsZF1.4 contains a C to T transition that introduces a stop codon at amino acid position 356. (C) snsXH2 contains a C to T transition that introduces a stop codon at amino acid 465. (D) rost202 a putative mutant allele of the previously described rost gene (Paululat et al. 1995) contains a C to T transition that introduces a stop codon at amino acid position 367 of SNS.
Sequence aberrations were also identified using the Non-Isotopic RNase Cleavage Assay (NIRCA). This method utilizes the ability of RNase A to cleave an unpaired base in an RNA/RNA duplex. Coding regions of the putative sns gene were analyzed in various mutant alleles, and potential alterations were confirmed by direct DNA sequencing. As shown in Figure 3B and C, the presence of C to T transitions in snsZF1.4 and snsXH2 results in nonsense mutations at amino acid residues 356 and 465, respectively. Immunohistochemical staining of embryos homozygous for these alleles with antisera directed against the cytoplasmic domain of SNS revealed no SNS expression, consistent with the presence of stop codons in these positions.
Select mutations in rost are allelic to sns
The rost locus, located at cytological position 30, encodes another protein that was reported to be essential for myoblast fusion (Paululat et al. 1995, 1997). However, the original rostP20-containing chromosome was recently shown to contain two mutations that affect muscle development, a P-element insertion in the rost locus and a second mutation that maps to genetic position 43–49 (Paululat et al. 1999). Because the doubly-mutant chromosome was used in an EMS mutagenesis screen to isolate noncomplementing mutations (Paululat et al. 1995), the resulting alleles could be in either locus. Due to the localization of this second mutation to cytological region 43–49, it was of interest to determine whether any of the EMS alleles obtained in this screen were, in fact, allelic to sns. Indeed, all sns mutant alleles fail to complement the 43–49 mutation (data not shown). Recombinants of rost2023 that separated cytological regions 30 and 44 were also isolated and crossed to sns6.1. Recombinants that retained region 44 did not complement sns6.1, whereas recombinants that retained region 30 complemented sns6.1. These data strongly suggest that the rost2023 mutation is allelic to sns. In addition, the sns sequence was examined in several putative rost alleles by NIRCA (described above). Through this analysis, rost202 was found to contain a C to T transition that results in a nonsense mutation at amino acid residue 367 of the sns gene (Fig. 3D).
The structure of the SNS protein
The sns cDNA sequence, determined from overlapping clones, includes an apparent open reading frame (ORF) of 1483 amino acids (Fig. 4), and untranslated regions of 549 and 3480 nucleotides on the 5′ and 3′ ends, respectively. The predicted size of the SNS protein without modification is 162 kD. The amino acid sequence strongly suggests that sns encodes a cell-adhesion molecule in the immunoglobulin superfamily. SNS contains eight putative Ig domains in the amino-terminal region, a single fibronectin type III domain and a putative transmembrane domain. A potential signal anchor sequence characteristic of membrane bound proteins is present in the amino terminus. This sequence, centered around a glutamine-rich stretch, is composed of 13 contiguous uncharged polar residues flanked by basic residues. A similar sequence of unknown function is present in the COOH terminal portion of SNS. BLAST database searches (Altschul et al. 1990) for proteins with structural homology to SNS identified the human Nephrin protein (Kestila et al. 1998; Lenkkeri et al. 1999), which is expressed specifically in the kidney glomerulus and is associated with Congenital Nephrotic Syndrome. SNS has homology to Nephrin throughout the eight immunoglobulin domains and the fibronectin type III repeat. In SNS, this extracellular region includes 14 NXS and NXT consensus triplets for N-glycosylation. Like Nephrin, SNS also contains several SG doublets that are potential attachment sites for heparin sulfate. Homology to Nephrin continues through the putative transmembrane domain and into the cytosolic carboxy-terminal region. Although SNS has a longer cytoplasmic domain, the Nephrin-related region of SNS includes several tyrosine residues that may act as sites for phosphorylation. In addition to Nephrin, other genes with significant homology to SNS include the recently reported hibris gene of Drosophila (GenBank accession no. AF210316; Artero and Baylies 1999) and a single C. elegans ORF (GenBank accession no. CAB63432). These orthologs are included in the alignment shown in Figure 4.
Figure 4.
Amino acid sequence of the putative SNS protein and structurally-related sequences in multiple organisms. The putative SNS open reading frame is indicated in the top line (GenBank accession no. AF254867). Structural homologs include the hypothetical ORF of the Drosophila hibris gene (GenBank accession no. AF210316; Artero and Baylies 1999), human Nephrin (GenBank accession no. AF035385; Kestila et al. 1998; Lenkkeri et al. 1999), and a C. elegans ORF (GenBank accession no. CAB63432). Alignment was done using ClustalW and is presented using Boxshade. (Black boxes) Amino acid identity; (gray boxes) amino acid similarity; (horizontal black lines) immunoglobulin homology domains; (dashed horizontal line) the fibronectin type III domain; (thick black horizontal bar) the transmembrane domain. Asterisks highlight the residues altered in various mutant alleles of sns.
Of note, the available Drosophila genomic sequence has revealed the presence of at least 20 introns, and a transcription unit that covers a minimum of 50 kb. These preliminary findings suggest that the genomic region that corresponds to the sns locus is quite large, not unlike that of the human Nephrin gene, which includes 29 introns spanning a total of 26 kb.
Temporal and spatial patterns of sns expression
The embryonic expression pattern of the sns transcript was examined using a digoxigenin labeled cDNA fragment. The earliest expression of sns is seen during stage 11 in the visceral mesoderm, and in the somatic mesoderm prior to the onset of myoblast fusion (Fig. 5A, arrowhead and arrow, respectively). Although expression in the visceral mesoderm diminishes as germ band retraction proceeds, expression persists in the somatic mesoderm at high levels until stage 14, during which time myoblast fusion is taking place (Fig. 5B,C). No expression was evident in the visceral musculature or in the dorsal vessel at this time. By stage 15, transcript levels have also declined in the somatic musculature, and by stage 17 only faint expression could be detected (Fig. 5I). During stage 17, weak expression was also seen in the muscle attachment sites (Fig. 5I, arrows).
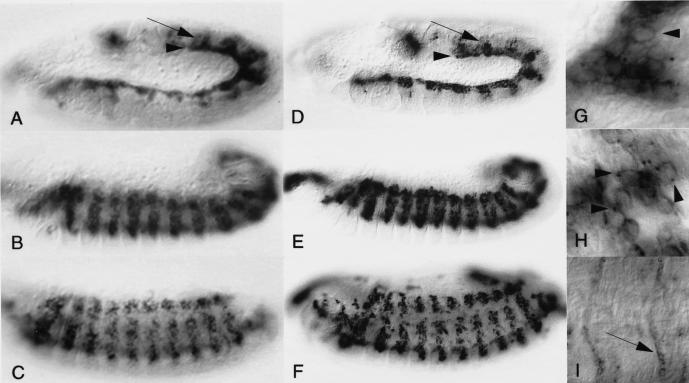
Figure 5.
Embryonic expression of the sns transcript and encoded protein are restricted to myogenic cells. (A–C,I) Localization of the sns transcript in wild-type embryos by in situ hybridization using a digoxigenin labeled probe. (D–H) Localization of the sns-encoded protein, detected by antisera generated against the cytoplasmic domain of SNS. All embryos are oriented laterally with anterior to the left. (A,D) Early Stage 12, (B,E) early Stage 13, (C,F) late Stage 14. Expression of SNS essentially mimics that of the transcript. (A,D) Expression at early stages is observed in both the developing somatic and visceral musculature (arrows and arrowheads, respectively). Expression persists in the somatic mesoderm throughout the time that myoblast fusion occurs. (G–I) High magnification of embryos at comparable stages to those shown in D–F. (G) SNS protein outlines the membrane of the entire cell initially (arrowhead). (H) As development proceeds, SNS becomes increasingly localized to distinct points in the membrane (arrowheads). (I) By Stage 17, expression of sns transcript has declined significantly in the muscles, but is observed at a low level in muscle attachment sites (arrow).
Antisera generated against the carboxy-terminal portion of the SNS protein confirmed that the pattern of protein expression is similar to that of the transcript (Fig. 5D–F). Confocal microscopy confirmed the co-expression of SNS and FASIII in the visceral musculature (data not shown). As anticipated for a putative cell adhesion molecule, both colorimetric and immunofluorescent confocal staining revealed the enrichment of the SNS protein at the cell membrane (note arrowheads in Fig. 5G,H; data not shown). In addition, SNS appears to become localized to discrete sites in the membrane as fusion progresses (note arrowheads in Fig. 5G,H). This membrane localization of SNS clusters has been confirmed by examination of serial confocal sections through the embryo. Like its transcript, SNS also appears to decline significantly as fusion progresses, such that little expression is observed in multinucleate syncitia.
SNS expression in the putative fusion competent cells of the somatic mesoderm
Three independent experimental approaches have been used to address whether SNS is expressed in both the founder cells and putative fusion competent cells of the somatic mesoderm. In the first approach, wild-type embryos were double-labeled with antisera against SNS and various founder cell markers, and examined by confocal microscopy. Markers included the enhancer trap rP298–lacZ (Nose et al. 1998; Fig. 6) and even-skipped (EVE; Frasch et al. 1987; Fig. 7). In brief, SNS expression was not detected in isolated cells that express rP298–lacZ (Fig. 6, arrowheads). These isolated cells were observed in many confocal sections, in embryos in several orientations (Fig. 6). Because most of these are not near the ventral nerve cord, we infer that these are unfused founder cells rather than rP298–lacZ expressing glial cells (Campos-Ortega and Hartenstein 1997; Nose et al. 1998). A similar analysis addressed co-expression of the EVE founder cell marker and SNS (Fig. 7). At the earliest appearance of the EVE-expressing founder cell, no SNS expressing cells are observed in its vicinity (Fig. 7A–C), consistent with the pattern of expression observed with rP298–lacZ. After a brief period of time, SNS-expressing cells are observed in close proximity (Fig. 7D–I), and begin to fuse almost immediately (Fig. 7J–L). These data suggest that SNS marks the fusion-competent cells shortly before fusion, and is not expressed in the founder cells.
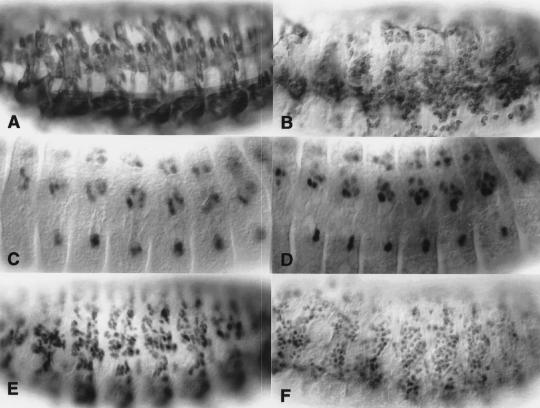
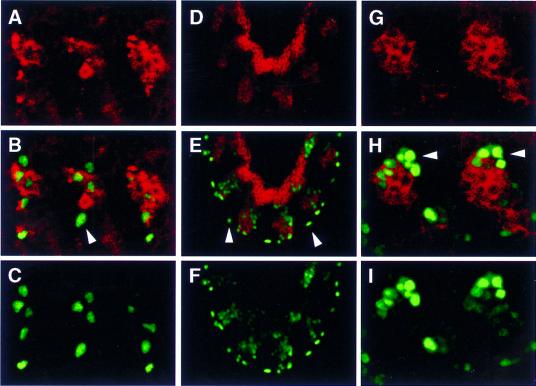
Figure 6.
SNS is not detected in unfused rP298–lacZ-expressing founder cells. Embryos were analyzed for coincident expression of SNS and the enhancer trap line rP298–lacZ by immunofluorescence and confocal microscopy. SNS expression is indicated in red (A,B,D,E,G,H), and rP298-driven expression of β-galactosidase is indicated in green (B,C,E,F,H,I). One micron confocal sections are shown. (A–C) A dorsolateral view of a Stage 13 embryo where anterior is to the left and dorsal to the top. (D–F) A lateral view of a Stage 12 embryo where anterior is to the top and dorsal to the right. (G–I) A dorsal view of a late Stage 12 embryo where anterior is to the left and the midline is to the top. Unfused β-galactosidase-expressing founder cells are indicated by arrowheads in B, E, and H. As evidenced by comparisons with the locations of the SNS-expressing cells, none of these founder cells co-express SNS.
Figure 7.
SNS is not detected in the unfused EVE-expressing founder cell. Embryos were analyzed for coincident expression of SNS and EVE by immunofluorescence and confocal microscopy. SNS expression is indicated in red (B,C,E,F,H,I,K,L), and EVE expression is indicated in green (A,B,D,E,G,H,J,K). (A–C) Stage 12, (D–I) Stage 13, and (J–L) Stage 14. A projection of 3 × 1 micron confocal sections are shown in all panels. All views are lateral, with anterior to the left and dorsal to the top. Unfused EVE-expressing founder cells are indicated by arrowheads in B and H.
Two additional analyses addressed whether SNS expression in the somatic musculature might be exclusive to the fusion competent cells. Both a non-null allele of sns and a null allele of mbc were used to determine whether SNS was expressed in morphologically distinct founder cells analogous to those described previously (Rushton et al. 1995). Elongated cells were observed in both snsXS5 and mbcD11.2 mutant embryos immunostained with MHC (note arrows; Fig. 8B,D) but not with SNS (Fig. 8A,C), consistent with the interpretation that SNS is not expressed in the founder cells. A second analysis relied on the hypothesis that Notch mediates a cell-fate decision between muscle progenitors (from which the founder cells arise) and the putative fusion competent cells (Corbin et al. 1991; Fuerstenberg and Giniger 1998; Rusconi and Corbin 1998). Notch NXK11 mutant embryos were double-labeled with vestigial (VG; Williams et al. 1991) another founder cell marker, and SNS. Wild-type embryos exhibit normal distribution patterns of both markers (Fig. 9A–C). By comparison, one observes a dramatic expansion in the number of VG-expressing founder cells in NXK11 mutant embryos (Fig. 9E,F), in agreement with studies using other founder cell markers (Bate 1993). Of most significance is a dramatic reduction in the number of SNS-expressing cells in these mutant embryos (Fig. 9D,E). This observation supports the hypothesis that Notch mediates a cell-fate decision between muscle progenitors and fusion-competent cells, that Notch is necessary for the selection of fusion-competent myoblasts, and that SNS specifically marks this population of myoblasts.
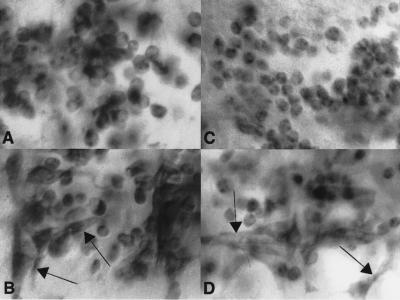
Figure 8.
SNS expression is not detected in morphologically distinct founder cells in embryos blocked for myoblast fusion. snsXS5 and mbcD11.2 mutant embryos were analyzed for the presence of morphologically distinct founder cells (Rushton et al. 1995) that express either MHC or SNS. (A,B) Lateral view of a Stage 16 embryo homozygous for the non-null sns allele snsXS5, immunostained for SNS (A) or MHC (B). (C,D) Lateral view of a Stage 16 embryo homozygous for mbcD11.2, immunostained for SNS (C) or MHC (D). Arrows in B and D indicate the positions of elongated founder cells that are visualized by their expression of MHC. By contrast, no such cells are revealed by expression of SNS.
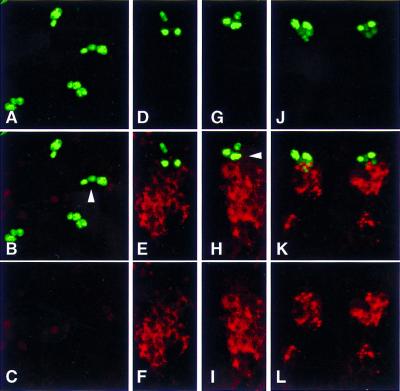
Figure 9.
SNS expression in the putative fusion competent cells is dependent on Notch. Wild-type and NXK11 mutant embryos were analyzed for expression of SNS and the founder cell marker VG (Williams et al. 1991). SNS expression is indicated in red (A,B,D,E), and VG expression is indicated in green (B,C,E,F). Both embryos are at Stage 13, and all views are ventral with anterior to the left. (A–C) Confocal projections of 22 × 1 micron sections of a wild-type embryo. As expected, both SNS- and VG-expressing cells are seen. (D–F) Confocal projections of 16 × 1 micron sections through an NXK11 mutant embryo at the same stage. As expected, the Notch mutant embryo exhibits a dramatically expanded population of VG-expressing founder cells compared to wild-type (compare B and C with E and F). By contrast, SNS expression is dramatically decreased (compare A and B with D and E), presumably due to the absence of fusion competent cells.
MEF2 independence of sns expression
SNS represents one of the first identified muscle-specific molecules that are absolutely essential for myoblast fusion. Consequently, it was of interest to examine whether genes associated with myoblast fusion are regulated by the same mechanisms as those associated with the contractile apparatus. Studies have established the role of Drosophila MEF2 in the regulation of muscle-specific genes that include myosin, tropomyosin I, and MLP60A (Bour et al. 1995; Lilly et al. 1995; Ranganayakulu et al. 1995; Lin et al. 1996, 1997; Stronach et al. 1999). As an entry point to identify potential regulators of sns, expression was examined in embryos mutant for the protein-null allele mef222-21. As shown in Figure 10, sns is detected at significant levels in these embryos. Thus, while one cannot conclude from these results that mef2 plays no role in sns regulation, it is clear that mef2 is not essential for its expression.
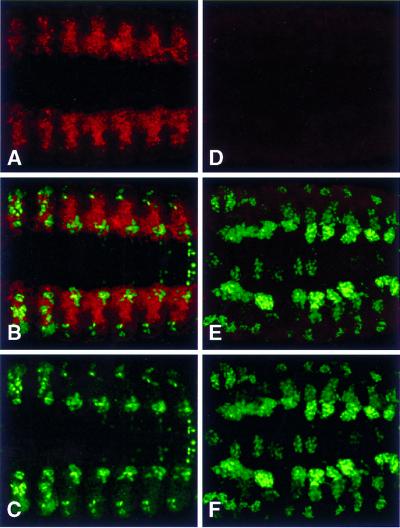
Figure 10.
SNS expression is detected in the absence of the myogenic transcriptional activator MEF2. Expression of the sns transcript was detected immunohistochemically, by in situ hybridization using a digoxigenin labeled probe. All views are lateral, with anterior to the left and dorsal to the top. (A) Wild-type Stage 14 embryo. (B) Stage 14 embryo homozygous for the mef2 mutation mef222–21 (Bour et al. 1995). As shown, levels of sns transcript appear to be relatively unaffected in mef2 mutant embryos, indicating that MEF2 is not essential for sns expression.
Discussion
The results reported herein describe the characterization of sns, a gene that plays an essential role in the fusion of myoblasts into multinucleate myotubes. In embryos mutant for sns, one observes a dramatic absence of multinucleate body wall muscles in the Drosophila embryo, and a correspondingly large number of unfused myosin-expressing myoblasts. Identification of the gene responsible for this defect has revealed that sns encodes a cell adhesion molecule in the immunoglobulin superfamily that, along with its closest human counterpart Nephrin (Kestila et al. 1998; Lenkkeri et al. 1999), defines a new organizational subdivision of this superfamily. As might be expected for a putative cell adhesion molecule, SNS is detected in the cell membrane, and becomes clustered at discrete sites as myoblast fusion progresses. Although it remains to be determined whether these sites are related to the paired vesicles or fusion plaques revealed in electron micrographs (Doberstein et al. 1997), SNS clearly serves as an entry point in identifying other molecules that are essential for myoblast fusion through the same pathway. Somewhat surprisingly, expression of sns is detected in progenitors of both the somatic and mononucleate visceral musculature. Of particular note, however, SNS expression within the somatic musculature is exclusive to the putative fusion competent cells and is not detected in founder cells. Because sns is the first gene to exhibit these characteristics, it also serves as an entry point for examining the specification of the fusion competent cells during subdivision of the mesoderm.
SNS is essential for myoblast fusion
The most apparent defect in embryos mutant for the sns gene is an inability of committed myosin-expressing myoblasts to fuse into muscle fibers. These myoblasts have clearly undergone significant progress in their myogenic differentiation, as evidenced by expression of proteins associated with the contractile apparatus. Consequently, sns appears to function at a later, or independent, step in the progression of cells from partially differentiated myoblasts to myotubes, most likely in the pathway through which these cells acquire features that mediate fusion. As already introduced, myoblast fusion can be divided into stages that include the acquisition of fusion competence, myoblast adhesion and plasma membrane union. This latter process is associated with changes in the composition of the plasma membrane, particularly in regard to lipid composition (for review, see Knudsen 1992). Although establishing the roles of particular molecules in vivo has been somewhat problematic, it seems prudent to assume that molecules identified in vertebrate tissue culture systems will be important for myoblast fusion in intact organisms. Nevertheless, as revealed by the isolation of sns, mbc, blow, and rost described in the introductory section, the use of genetic approaches in Drosophila has resulted in the identification of molecules that are absolutely essential.
The SNS protein establishes a new subgroup of the immunoglobulin superfamily
The sns gene encodes a protein with several features characteristic of the transmembrane cell adhesion molecules in the immunoglobulin superfamily (IgSF). Structurally, it contains several Ig domains, a fibronectin (Fn) III-like domain, transmembrane domain, and cytoplasmic tail. Other members of this broad family include N-CAM, L1-CAM, Neurofascin, and NrCam in vertebrates and FASII, Neuroglian and Roundabout in invertebrates (Brummendorf and Rathjen 1994; Kidd et al. 1998). As implied in this abbreviated list, many proteins in the IgSF have been identified on the basis of their role in neural development. One exception to this generalization is the recent identification of CDO, a roundabout-related molecule that can induce myogenic differentiation of mouse cells in culture (Kang et al. 1998).
The extracellular domain of several of the proteins described above includes a series of Ig domains followed by Fn III-like domains. SNS and its closest vertebrate counterpart Nephrin are distinct in having an 8 + 1 arrangement of these two domains in the extracellular region. Nephrin that is essential for filtration in the kidney glomerulus, was identified on the basis of its association with Congenital Nephrotic Syndrome (Kestila et al. 1998; Lenkkeri et al. 1999). Of note, while the homology between these two proteins extends beyond that found in the Ig and Fn III-like domains, it seems unlikely that Nephrin is a functional homolog of sns in vertebrates. The recent identification of hibris (Artero and Baylies 1999) indicates that this 8 + 1 subgroup of the IgSF includes multiple proteins in Drosophila. Therefore, one might also anticipate several structurally related mammalian proteins, one or more of which may have some functional relevance to myoblast fusion.
SNS is expressed in the fusion-competent, but not founder, cells of the somatic mesoderm
The body wall musculature of a Drosophila larva includes ∼30 distinct muscles per hemisegment that are generated by the fusion of founder cells and fusion-competent cells. The founder myoblasts appear to be specified with respect to their developmental identity whereas the fusion competent cells are thought to take on the characteristics of the founder cell with which they fuse (Bate 1990; Rushton et al. 1995; for review, see Frasch 1999). Although many genes have been identified that are expressed in, and required for, the specification or differentiation of founder cells (Dohrmann et al. 1990; Michelson et al. 1990; Paterson et al. 1991; Williams et al. 1991; Bourgouin et al. 1992; Keller et al. 1998; for review, see Frasch 1999; Knirr et al. 1999), a protein specific to the fusion competent class of myoblasts had remained elusive. The observation that SNS is expressed in a different subset of unfused myoblasts than rP298–lacZ and EVE, both markers for the founder cells, suggests that its expression is restricted in the somatic mesoderm to the putative fusion competent cells. The absence of SNS expressing cells in Notch mutant embryos (discussed below) further supports this conclusion. Therefore, not only does SNS serve as a marker to examine the transcriptional regulatory pathways associated with myoblast fusion, but also the pathways controlling specification of this class of cells.
The Notch pathway is essential for the presence of SNS-expressing cells in the somatic mesoderm
Numerous studies have established the involvement of the Notch pathway in the development of the somatic mesoderm, and support the hypothesis that Notch mediates a cell fate decision in this tissue (Corbin et al. 1991; Bate et al. 1993). Specifically, one observes a dramatic expansion in the number of founder cells in Notch mutant embryos, presumably at the expense of other mesodermal cells that might include the fusion competent cells. Studies have now identified domains of cells expressing lethal-of-scute (L'SC) which, through a process of lateral inhibition mediated by Notch, resolve into a single L'SC-expressing muscle progenitor (Carmena et al. 1995; for review, see Frasch 1999). It remains a formal possibility that the remaining cells, in which L'SC expression declines, then become fusion competent myoblasts. In the absence of Notch, lateral inhibition does not occur and all cells of the cluster appear to become muscle progenitors. If this expansion in muscle progenitors occurs at the expense of fusion competent cells, one might expect to observe a corresponding decrease in the number of fusion competent cells in a Notch mutant embryo. The finding that the population of SNS-expressing cells is severely reduced in Notch mutant embryos, in conjunction with the observation that SNS is not expressed in the founder cells, would seem to support this hypothesis. In addition, the absence of fusion competent myoblasts in Notch mutant embryos provides one explanation for the observation that these mutant embryos exhibit defects in myoblast fusion (Cross and Sang 1978a,b).
The transcription factor MEF2 is not essential for SNS expression
Drosophila mef2 encodes a highly conserved MADS-box domain-containing transcription factor that functions through specific sequences in the promoters of such muscle-specific genes as myosin, tropomyosin I, and the LIM-domain containing molecule MLP60A (Bour et al. 1995; Lilly et al. 1995; Ranganayakulu et al. 1995; Lin et al. 1996; Lin et al. 1997; Stronach et al. 1999). These genes are not expressed in embryos lacking MEF2, consistent with a direct role for MEF2 in their transcriptional initiation. By comparison, SNS is expressed at significant levels in mef2 mutant embryos, suggesting that MEF2 plays only a minor role, if any, in the transcriptional initiation of SNS. These data support a requirement for other transcriptional regulators in the transcriptional initiation of genes essential for myoblast fusion. It is worth noting that the myoblasts present in mef2 mutant embryos do not fuse, consistent with the possibility that MEF2 may be required for expression of some fusion-specific genes. Alternatively, MEF2 may play a role in establishing a state of differentiation that is permissive for, or a prerequisite to, fusion without having a direct role in the initiation of fusion-specific genes per se. This might be the case if, for example, cytoskeletal rearrangements that precede fusion are dependent on MEF2-regulated structural genes. Therefore, the definitive role of MEF2 in the transcriptional regulation of the fusion machinery awaits the identification of more genes specifically associated with fusion.
What is the function of the SNS protein in myoblast fusion?
In Drosophila, progress has been made in the last few years in the identification of molecules that are absolutely essential for myoblast fusion (Paululat et al. 1999). However, with few exceptions, these genes have encoded novel proteins of unknown function that have not yet yielded major insights. By virtue of its identification as a member of the Ig superfamily, one can more easily speculate that the function of SNS is to mediate cell–cell interactions or cell–cell communication essential for fusion.
It is enticing to consider that sns plays a role in the formation, recruitment, or processing of the paired-vesicles described by Doberstein et al. (Doberstein et al. 1997). The observation that SNS becomes clustered at specific sites within the membrane is consistent with this suggestion. However, one must also consider that SNS is expressed in the visceral mesoderm, which is not a syncitial tissue. Resolutions of this paradox include the possibilities that early expression of SNS in this tissue is of no consequence, that SNS serves a different purpose in the visceral musculature, or that SNS functions in cell–cell communication critical to myogenesis of both tissues. Although it is possible that SNS serves a purely adhesive function, the presence of an uncharacteristically long cytoplasmic region might be an indication that sns, like many of the neural cell adhesion molecules, plays a more complex role in signaling between cells. Resolution of these issues will require identification of the sequences essential to sns function, and molecules with which they interact.
Materials and methods
Drosophila stocks and P1 clones
All stocks were grown on standard cornmeal medium at 18°C or 25°C, as necessary. Stocks for recombinational mapping were obtained from the Bloomington Stock Center. The deficiency IN(2LR)P14[L]TE45F[R] was provided by Dr. J. Hooper (Hooper and Scott 1989). rost alleles were obtained from R. Renkawitz-Pohl (Paululat et al. 1995). The protein null alleles mef222–21 and mbcD11.2, and the Notch NXK11 allele, have been described (Bour et al. 1995; Erickson et al. 1997; Brennan et al. 1997). Deficiency Df(2R)BB1 was generated by gamma irradiation of flies bearing P-element insertion Pl(2){ry+cp70ZT}P44F,which was obtained from the Bloomington Stock Center. Df(2R)sns-04913HOa, Df(2R)sns-04913HOb, and Df(2R)sns-16109HO were generated by excision of P-element insertions Pl(2)k0491304913 or Pl(2)k1610916109, which have been described by Torok et al. (1993). For phenotypic studies, relevant stocks were balanced with CyO P[w+wgen11lacZ], which directs expression of β-galactosidase under the control of the wgen11 (Kassis et al. 1992).
Bacteriophage P1 clones DS04320 and DS01342, and primer sequences corresponding to sequence tagged sites (STSs), were obtained from the Berkeley Drosophila Genome Project (BDGP; Kimmerly et al. 1996).
Genetic mapping
The genetic location of sns was initially determined by recombinational mapping using dominant markers. Recombination occurred in females trans-heterozygous for snsA3.24 and a chromosome carrying Sp, Bl, L, and Pu. Progeny representing different recombination events were recovered over a CyO balancer chromosome and crossed to snsA3.24. One hundred sixty nine recombinants between the markers Bl and L were examined. Of these, 28 were Bl, sns, L+; 44 were Bl+, sns, L; 89 were Bl, sns+, L+; 8 were Bl+, sns+, L. The sns locus was also mapped relative to the recessive markers cn and either c or sca. Of the recombination events between cn and c, 355 were found to be cn sns c, whereas 110 were cn sns+ c. Of the recombination events between cn and sca, 163 were found to be cn sns sca, while 8 were cn sns+ sca.
The approximate genetic position of rost2023 was determined by isolation of recombinants between pr and cn, and subsequent complementation testing with sns6.1. Ten of ten rost2023 recombinants that contained the region distal to cn did not complement sns6.1, whereas 9 of 9 recombinants that contained the region proximal to pr complemented sns6.1.
Genetic screens
Df(2R)BB1 was isolated in a gamma irradiation screen using flies homozygous for P(2){ry+cp70ZT}44F. Males were irradiated with 4000 rads of X-rays, and mated to BcElp/Cyo; ry/ry virgin females. Approximately 20,000 progeny were screened for deletion of the P-element associated rosy+ eye color marker. Three deficiencies were recovered but only Df(2R)BB1 failed to complement snsA3.24. Smaller deletions in the region were generated by imprecise excision of either Pl(2)k0491304913 (an insert in the ryanodine receptor; data not shown) or Pl(2)k1610916109. Progeny containing one copy of the P element insertion and one copy of Sb delta 2-3 were mated to flies that were genetically yw and carried a CyO balancer chromosome. Excisions were revealed by loss of the white+ eye color marker, and potential deficiencies identified by noncomplementation of select lethal mutations in this region (shown in Fig. 1). These screens yielded Df(2R)sns-04913HOa, Df(2R)sns-04913HOb, and Df(2R)RyaR-16109HO.
EMS-induced mutations in the region were generated from an isogenic stock homozygous for b el pr cn or. Males were fed 25 mm EMS in 5% sucrose (Lewis and Bacher 1968), and the mutagenized chromosome was recovered over CyO. Mutant loci were identified by non-complementation of snsA3.24 (6200 mutagenized chromosomes) or Df(2R)BB1 (4300 mutagenized chromosomes). These genetic screens resulted in the isolation of 21 additional EMS induced alleles of sns, and 128 lethal mutations that fall in a minimum of 16 complementation groups uncovered by Df(2R)BB1 but not Df(2R)NP1 or IN(2LR)P14[L]TE45F[R].
Molecular analyses
Southern blots containing DNA enriched for the various deficiency chromosomes (Erickson et al. 1997) were probed with STS sequences to identify relevant P1 clones. DNA from P1 clones DS04320 and DS01342 was then used to create plasmid libraries by random subcloning of fragments into the pBSKII vector (Stratagene, La Jolla, CA). A chromosomal walk was initiated using STS 0728 as a molecular entry point. To isolate cDNA clones, a 9–12 hr Drosophila cDNA library (Zinn et al. 1988) was screened with genomic clone RI4 and portions of RI11 (Fig. 1). Three overlapping clones C1 (nucleotides 829–4890), K2 (nucleotides 1–2539), and O2 (nucleotides 4210–8035) were used to generate a full-length cDNA subclone. Both strands of the full-length cDNA were sequenced by the Penn State Nucleic Acid facility.
Immunohistochemical methods
The embryonic expression pattern of sns mRNA was determined using a digoxigenin-labeled cDNA fragment (nucleotides 1972–4383) (Tautz and Pfeifle 1989; Michelson et al. 1990). Colorimetric immunohistochemistry utilized a mouse monoclonal anti-MHC antibody (provided by D. Kiehart), a polyclonal rat antisera directed against NAU (Keller et al. 1998), and a polyclonal rabbit antisera directed against MEF2 (Bour et al. 1995). Primary antibodies were detected using the Vectastain ABC Elite kit (Vector Laboratories, Burlingame, CA). Balancer chromosomes were identified either by an enzymatic assay for β-galactosidase activity (Klambt et al. 1991) or by immunohistochemistry using a monoclonal anti-β-galactosidase antibody generated in mouse (Promega Corp., Madison, WI). Confocal studies utilized rabbit polyclonal antisera against VG (Williams et al. 1991) and EVE (Frasch et al. 1987). SNS-associated confocal microscopy utilized a polyclonal rat antiserum (Cocalico Biologicals) directed against a 258 amino acid fusion protein (amino acids 1225–1482) that was generated using the pET expression system (Novagen Inc., Madison, WI), and purified from inclusion bodies. The antisera was affinity-purified against the original antigen coupled to Affigel 15 (Bio-Rad), and used at a dilution of 1:5. It was detected using a Cy3 conjugated goat anti-rat antiserum (Vector Laboratories).
Molecular characterization of mutations
The snsXB3 mutation was originally detected in a Southern blot of 10 μg of DNA from heterozygous adults. DNA was prepared (Jowett 1986), digested with BamHI–EcoRI, and transferred to nylon membrane according to standard procedures. The blot was hybridized with a 2.9 kb genomic EcoRI fragment, RI4 (Fig. 1). Subsequent RT–PCR and sequence analysis of the snsXB3 transcript in this mutant allele utilized RNA from embryos enriched for the mutant chromosome by methods previously described (Erickson et al. 1997). Mutations in snsXS5, snsZF1.4, snsXH2, and rost202 were detected using the Non-Isotopic RNase Cleavage Assay (NIRCA) of the Mismatch Detect II kit (Ambion, Inc., Austin, TX) as described (Erickson et al. 1997). Appropriate regions were sequenced by the Penn State Nucleic Acid Facility.
Acknowledgments
We thank M. Kulp, M. Grill, D. Heyser, W. Williams, C. Keller, and M. Hazen for assistance with genetic screens, molecular characterization of deficiencies, and embryo immunohistochemistry. We are grateful to D. Kiehart, C. Goodman, H. Nguyen, M. Frasch, S. Carroll, and S. Benzer for gifts of antibodies, and E. Siegfried, R. Renkawitz-Pohl, J. Hooper, and H. Nguyen for fly strains. We thank C. Gay and J. Staines for assistance with confocal microscopy and B. Galletta for critical reading of the manuscript. This work has been supported by NIH grant RO1 AR44274 to S.M.A.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sma1@psu.edu; FAX (814) 863-7024.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Artero R, Baylies M. Characterization of genes involved in Drosophila myogenesis. 1999. GenBank accession no. AF210316. [Google Scholar]
- Bate M. The embryonic development of larval muscles in Drosophila. Development. 1990;110:791–804. doi: 10.1242/dev.110.3.791. [DOI] [PubMed] [Google Scholar]
- ————— . The mesoderm and its derivatives. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 1013–1090. [Google Scholar]
- Bate, M., E. Rushton, and M. Frasch. 1993. A dual requirement for neurogenic genes in Drosophila myogenesis. Development (Suppl.) 149–161. [PubMed]
- Bernstein SI, O'Donnell PT, Cripps RM. Molecular genetic analysis of muscle development, structure and function in Drosophila. Int Rev Cytol. 1993;143:63–152. doi: 10.1016/s0074-7696(08)61874-4. [DOI] [PubMed] [Google Scholar]
- Bour BA, O'Brien MA, Lockwood WL, Goldstein ES, Bodmer R, Taghert PH, Abmayr SM, Nguyen HT. Drosophila MEF2, a transcription factor that is essential for myogenesis. Genes & Dev. 1995;9:730–741. doi: 10.1101/gad.9.6.730. [DOI] [PubMed] [Google Scholar]
- Bourgouin C, Lundgren SE, Thomas JB. apterous is a Drosophila LIM domain gene required for the development of a subset of embryonic muscles. Neuron. 1992;9:549–561. doi: 10.1016/0896-6273(92)90192-g. [DOI] [PubMed] [Google Scholar]
- Brennan K, Tateson R, Lewis K, Martinez-Arias A. A functional analysis of Notch mutations in Drosophila. Genetics. 1997;147:177–188. doi: 10.1093/genetics/147.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brummendorf T, Rathjen FG. Cell Adhesion Molecules I: The Immunoglobulin Superfamily. Prot Profiles. 1994;1:951–1058. [PubMed] [Google Scholar]
- Campos-Ortega JA, Hartenstein V. The embryonic development of Drosophila melanogaster. Berlin, Germany: Springer-Verlag; 1997. [Google Scholar]
- Carmena A, Bate M, Jimenez F. lethal of scute, a proneural gene, participates in the specification of muscle progenitors during Drosophila embryogenesis. Genes & Dev. 1995;9:2373–2383. doi: 10.1101/gad.9.19.2373. [DOI] [PubMed] [Google Scholar]
- Charlton CA, Mohler WA, Radice GL, Hynes RO, Blau HM. Fusion competance of myoblasts rendered genetically null for N-cadherin in culture. J Cell Biol. 1997;138:331–336. doi: 10.1083/jcb.138.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin V, Michelson AM, Abmayr SM, Neel V, Alcamo E, Maniatis T, Young MW. A role for the Drosophila neurogenic genes in mesoderm differentiation. Cell. 1991;67:311–323. doi: 10.1016/0092-8674(91)90183-y. [DOI] [PubMed] [Google Scholar]
- Cross DP, Sang JH. Cell culture of individual Drosophila embyos I: Development of wild-type cultures. J Embryol Exp Morph. 1978a;45:161–172. [PubMed] [Google Scholar]
- ————— Cell culture of individual Drosophila embryos II: Culture of X-linked embryonic lethals. J Embryol Exp Morph. 1978b;45:173–187. [PubMed] [Google Scholar]
- Doberstein SK, Fetter RD, Mehta AY, Goodman CS. Genetic analysis of myoblast fusion: blown fuse is required for progression beyond the prefusion complex. J Cell Biol. 1997;136:1249–1261. doi: 10.1083/jcb.136.6.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohrmann C, Azpiazu N, Frasch M. A new Drosophila homeo box gene is expressed in mesodermal precursor cells of distinct muscles during embryogenesis. Genes & Dev. 1990;4:2098–2111. doi: 10.1101/gad.4.12a.2098. [DOI] [PubMed] [Google Scholar]
- Engel LC, Egar MW, Przybylski RJ. Morphological characterization of actively fusing L6 myoblasts. Eur J Cell Biol. 1985;39:360–365. [PubMed] [Google Scholar]
- Erickson MRS, Galletta BJ, Abmayr SM. Drosophila myoblast city encodes a conserved protein that is essential for myoblast fusion, dorsal closure and cytoskeletal organization. J Cell Biol. 1997;138:589–603. doi: 10.1083/jcb.138.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M. Controls in patterning and diversification of somatic muscles during Drosophila embryogenesis. Curr Opin Genet Dev. 1999;9:522–529. doi: 10.1016/s0959-437x(99)00014-3. [DOI] [PubMed] [Google Scholar]
- Frasch M, Hoey T, Rushlow C, Doyle H, Levine M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 1987;6:749–759. doi: 10.1002/j.1460-2075.1987.tb04817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerstenberg S, Giniger E. Multiple roles for Notch in Drosophila myogenesis. Dev Biol. 1998;201:66–77. doi: 10.1006/dbio.1998.8944. [DOI] [PubMed] [Google Scholar]
- Grenningloh G, Rehm EJ, Goodman CS. Genetic analysis of growth cone guidance in Drosophila: Fasciclin II functions as a neuronal recognition molecule. Cell. 1991;67:45–57. doi: 10.1016/0092-8674(91)90571-f. [DOI] [PubMed] [Google Scholar]
- Hooper JE, Scott MP. The Drosophila patched gene encodes a putative membrane protein required for segmental patterning. Cell. 1989;59:751–765. doi: 10.1016/0092-8674(89)90021-4. [DOI] [PubMed] [Google Scholar]
- Jowett T. Preparation of nucleic acids. In: Roberts DB, editor. Drosophila: A practical approach. Washington, D.C.: IRL press; 1986. pp. 275–286. [Google Scholar]
- Kang J-S, Mulieri P, Miller C, Sassoon DA, Krauss RS. CDO, A robo-related cell surface protein that mediates myogenic differentiation. J Cell Biol. 1998;143:403–413. doi: 10.1083/jcb.143.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassis JA, Noll E, VanSickle EP, Odenwald WF, Perrimon N. Altering the insertional specificity of a Drosophila transposable element. Proc Natl Acad Sci. 1992;89:1919–1923. doi: 10.1073/pnas.89.5.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller CA, Grill MA, Abmayr SM. A role for nautilus in the differentiation of muscle precursors. Dev Biol. 1998;202:157–171. doi: 10.1006/dbio.1998.9009. [DOI] [PubMed] [Google Scholar]
- Kestila M, Lenkkeri U, Mannikko M, Lamerdin J, McCready P, Putaala H, Ruotsalainen V, Morita T, Nissinen M, Peltonen L, et al. Positionally cloned gene for a novel glomerular protein-Nephrin-is mutated in congenital nephrotic syndrome. Mol Cell. 1998;1:572–582. doi: 10.1016/s1097-2765(00)80057-x. [DOI] [PubMed] [Google Scholar]
- Kidd T, Brose K, Mitchell KJ, Fetter RD, TessierLavigne M, Goodman CS, Tear G. Roundabout controls axon crossing of the CNS midline and defines a novel subfamily of evolutionarily conserved guidance receptors. Cell. 1998;92:205–215. doi: 10.1016/s0092-8674(00)80915-0. [DOI] [PubMed] [Google Scholar]
- Kimmerly W, Stultz K, Lewis S, Lewis K, Lustre V, Romero R, Benke J, Sun D, Shirley G, Martin C, Palazzolo M. A P1-based physical map of the Drosophila euchromatic genome. Genome Res. 1996;6:414–430. doi: 10.1101/gr.6.5.414. [DOI] [PubMed] [Google Scholar]
- Klambt C, Jacobs JR, Goodman C. The midline of the Drosophila central nervous system: A model for the genetic analysis of cell fate, cell migration, and growth cone guidance. Cell. 1991;64:801–815. doi: 10.1016/0092-8674(91)90509-w. [DOI] [PubMed] [Google Scholar]
- Knirr S, Azpiazu N, Frasch M. The role of the NK-homeobox gene slouch (S59) in somatic muscle patterning. Development. 1999;126:4525–4535. doi: 10.1242/dev.126.20.4525. [DOI] [PubMed] [Google Scholar]
- Knudsen KA. Fusion of myoblasts. In: Wilschut J, Hoekstra D, editors. Membrane fusion. New York, NY: Marcel Dekker, Inc.; 1992. pp. 601–626. [Google Scholar]
- Lenkkeri U, Mannikko M, McCready P, Lamerdin J, Gribouval O, Niaudet P, Antignac C, Kashtan CE, Holmberg C, Olsen A, et al. Structure of the gene for congenital nephrotic syndrome of the Finnish type (NPHS1) and characterization of mutations. Am J Hum Genet. 1999;64:51–61. doi: 10.1086/302182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis EB, Bacher F. Methods of feeding ethyl methane sulfonate (EMS) to Drosophila males. Dros Inf Ser. 1968;43:193. [Google Scholar]
- Lilly B, Galewsky S, Firulli AB, Schulz RA, Olson EN. D-MEF2: A MADS box transcription factor expressed in differentiating mesoderm and muscle cell lineages during Drosophila embryogenesis. Proc Natl Acad Sci. 1994;91:5662–5666. doi: 10.1073/pnas.91.12.5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly B, Zhao B, Ranganayakulu G, Paterson BM, Schulz RA, Olson EN. Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science. 1995;267:688–693. doi: 10.1126/science.7839146. [DOI] [PubMed] [Google Scholar]
- Lin M-H, Nguyen HT, Dybala C, Storti RV. Myocyte-specific enhancer factor 2 acts cooperatively with a muscle activator region to regulate Drosophila tropomyosin gene muscle expression. Proc Natl Acad Sci. 1996;93:4623–4628. doi: 10.1073/pnas.93.10.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MH, Bour BA, Abmayr SM, Storti RV. Ectopic expression of MEF2 in the epidermis induces epidermal expression of muscle genes and abnormal muscle development in Drosophila. Dev Biol. 1997;182:240–255. doi: 10.1006/dbio.1996.8484. [DOI] [PubMed] [Google Scholar]
- Lipton BH, Konigsberg IR. A fine-structural analysis of the fusion of myogenic cells. J Cell Biol. 1972;53:348–364. doi: 10.1083/jcb.53.2.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mege RM, Goudou D, Diaz C, Nicolet M, Garcia L, Geraud G, Rieger F. N-cadherin and N-CAM in myoblast fusion: Compared localisation and effect of blockade by peptides and antibodies. J Cell Sci. 1992;103:897–906. doi: 10.1242/jcs.103.4.897. [DOI] [PubMed] [Google Scholar]
- Michelson AM, Abmayr SM, Bate M, Martinez Arias A, Maniatis T. Expression of a MyoD family member prefigures muscle pattern in Drosophila embryos. Genes & Dev. 1990;4:2086–2097. doi: 10.1101/gad.4.12a.2086. [DOI] [PubMed] [Google Scholar]
- Nguyen HT, Bodmer R, Abmayr SM, McDermott JC, Spoerel NA, Nadal-Ginard B. D-mef2: A new Drosophila mesoderm-specific MADS box-containing gene with a bi-modal expression profile during embryogenesis. Proc Natl Acad Sci. 1994;91:7520–7524. doi: 10.1073/pnas.91.16.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan KM, Barrett K, Lu Y, Hu K-Q, Vincent S, Settleman J. Myoblast city, the Drosophila homologue of DOCK180/CED-5, is required in a Rac signaling pathway utilized for multiple developmental processes. Genes & Dev. 1998;12:3337–3342. doi: 10.1101/gad.12.21.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nose A, Isshiki T, Takeichi M. Regional specification of muscle progenitors in Drosophila: The role of the msh homeobox gene. Development. 1998;125:215–223. doi: 10.1242/dev.125.2.215. [DOI] [PubMed] [Google Scholar]
- Patel NH, Snow PM, Goodman CS. Characterization and cloning of Fasciclin III: A glycoprotein expressed on a subset of neurons and axon pathways in Drosophila. Cell. 1987;48:975–988. doi: 10.1016/0092-8674(87)90706-9. [DOI] [PubMed] [Google Scholar]
- Paterson BM, Walldorf U, Eldridge J, Dubendorfer A, Frasch M, Gehring WJ. The Drosophila homologue of vertebrate myogenic-determination genes encodes a transiently expressed nuclear protein marking primary myogenic cells. Proc Natl Acad Sci. 1991;88:3782–3786. doi: 10.1073/pnas.88.9.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paululat A, Burchard S, Renkawitz-Pohl R. Fusion from myoblasts to myotubes is dependent on the rolling stone gene (rost) of Drosophila. Development. 1995;121:2611–2620. doi: 10.1242/dev.121.8.2611. [DOI] [PubMed] [Google Scholar]
- Paululat A, Goubeaud A, Damm C, Knirr S, Burchard S, Renkawitz-Pohl R. The mesodermal expression of rolling stone (rost) is essential for myoblast fusion in Drosophila and encodes a potential transmembrane protein. J Cell Biol. 1997;138:337–348. doi: 10.1083/jcb.138.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paululat A, Holz A, Renkawitz-Pohl R. Essential genes for myoblast fusion in Drosophila embryogenesis. Mech Dev. 1999;83:17–26. doi: 10.1016/s0925-4773(99)00029-5. [DOI] [PubMed] [Google Scholar]
- Radice GL, Rayburn H, Matsunami H, Knudsen KA, Takeichi M, Hynes RO. Developmental defects in mouse embryos lacking N-cadherin. Dev Biol. 1997;181:64–78. doi: 10.1006/dbio.1996.8443. [DOI] [PubMed] [Google Scholar]
- Ranganayakulu G, Zhao B, Dokidis A, Molkentin JD, Olson EN, Schulz RA. A series of mutations in the D-MEF2 transcription factor reveal multiple functions in larval and adult myogenesis in Drosophila. Dev Biol. 1995;171:169–181. doi: 10.1006/dbio.1995.1269. [DOI] [PubMed] [Google Scholar]
- Rash JE, Fambrough D. Ultrastructural and electrophysiological correlates of cell coupling and cytoplasmic fusion during myogenesis in vitro. Dev Biol. 1973;30:166–186. doi: 10.1016/0012-1606(73)90055-9. [DOI] [PubMed] [Google Scholar]
- Rusconi J, Corbin V. Evidence for a novel Notch pathway required for muscle precursor selection in Drosophila. Mech Dev. 1998;79:39–50. doi: 10.1016/s0925-4773(98)00170-1. [DOI] [PubMed] [Google Scholar]
- Rushton E, Drysdale R, Abmayr SM, Michelson AM, Bate M. Mutations in a novel gene, myoblast city, provide evidence in support of the founder cell hypothesis for Drosophila muscle development. Development. 1995;121:1979–1988. doi: 10.1242/dev.121.7.1979. [DOI] [PubMed] [Google Scholar]
- Stronach BE, Renfranz PJ, Lilly B, Beckerle MC. Muscle LIM proteins are associated with muscle sarcomeres and require dMEF2 for their expression during Drosophila myogenesis. Mol Biol Cell. 1999;10:2329–2342. doi: 10.1091/mbc.10.7.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Torok T, Tick G, Alvarado M, Kiss I. P-lacW insertional mutagenesis on the second chromosome of Drosophila melanogaster: Isolation of lethals with different overgrowth phenotypes. Genetics. 1993;135:71–80. doi: 10.1093/genetics/135.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakelam MJO. The fusion of myoblasts. Biochem J. 1985;228:1–12. doi: 10.1042/bj2280001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JA, Bell JB, Carroll SB. Control of Drosophila wing and haltere development by the nuclear vestigial gene product. Genes & Dev. 1991;5:2481–2495. doi: 10.1101/gad.5.12b.2481. [DOI] [PubMed] [Google Scholar]
- Yagami-Hiromasa T, Sato T, Kurisaki T, Kamijo K, Nabeshima Y, Fujisawa-Sehara A. A metalloprotease-disintegrin participating in myoblast fusion. Nature. 1995;377:652–656. doi: 10.1038/377652a0. [DOI] [PubMed] [Google Scholar]
- Zinn K, McAllister L, Goodman CS. Sequence analysis and neuronal expression of Fasciclin I in Grasshopper and Drosophila. Cell. 1988;53:577–587. doi: 10.1016/0092-8674(88)90574-0. [DOI] [PubMed] [Google Scholar]
- Zipursky SL, Venkatesh TR, Teplow DB, Benzer S. Neuronal development in the Drosophila retina: Monoclonal antibodies as molecular probes. Cell. 1984;36:15–26. doi: 10.1016/0092-8674(84)90069-2. [DOI] [PubMed] [Google Scholar]