Abstract
Cdc42p, a Rho-related GTP-binding protein, regulates cytoskeletal polarization and rearrangements in eukaryotic cells, but the effectors mediating this control remain unknown. Through the use of the complete yeast genomic sequence, we have identified two novel Cdc42p targets, Gic1p and Gic2p, which contain consensus Cdc42/Rac interactive–binding (CRIB) domains and bind specifically to Cdc42p–GTP. Gic1p and Gic2p colocalize with Cdc42p as cell polarity is established during the cell cycle and during mating in response to pheromones. Cells deleted for both GIC genes exhibit defects in actin and microtubule polarization similar to those observed in cdc42 mutants. Finally, the interaction of the Gic proteins and Cdc42p is essential, as mutations in the CRIB domain of Gic2p that eliminate Cdc42p binding disrupt Gic2p localization and function. Thus, Gic1p and Gic2p define a novel class of Cdc42p targets that are specifically required for cytoskeletal polarization in vivo.
Keywords: Polarization, cytoskeleton, Rho GTPases, Saccharomyces cerevisiae
In polarized cells, the actin and microtubule cytoskeletons are highly asymmetric and serve to spatially regulate various cellular functions, including targeted secretion, signaling, and nuclear migration (Glotzer and Hyman 1995; Chant 1996; Drubin and Nelson 1996). The actin cytoskeleton maintains cell shape and plays a pivotal role in cell motility, cytokinesis, and phagocytosis. Reorganization of the actin cytoskeleton is regulated both through the cell cycle and in response to extracellular signals. Members of the Rho family of small GTPases have emerged as key regulators of actin filament dynamics and the assembly of focal adhesion contacts (Hall 1994; Ridley 1995). Rho GTPases are also important for maintaining cellular transformation (Symons 1995). GTPases act as molecular switches cycling between GTP- and GDP-bound conformations. When in the GTP-bound conformation, GTPases are thought to interact with target proteins that mediate their effects.
Cdc42p, a highly conserved Rho-type GTPase, has been shown to control polarized axis formation in both yeast and mammalian cells (Johnson and Pringle 1990; Hall 1994; Stowers et al. 1995). In fibroblasts, microinjection of activated Cdc42p triggers formation of filopodia or microspikes at the cell periphery (Kozma et al. 1995; Nobes and Hall 1995). In yeast, Cdc42p controls the polarization of actin and microtubules during both the vegetative cell cycle and mating. Polarity is initiated by choosing a site on the surface of the cell, and then growth is directed toward this site. During vegetative growth by budding, polarization is directed by a cell-type-specific program, which is controlled by a group of nonessential genes (BUD1-BUD9, AXL1, and BUD10/AXL2; Chant 1996). During mating, polarization is directed toward the mating partner by a mechanism involving the FAR1 gene (Dorer et al. 1995; Valtz et al. 1995). Remodeling of the actin cytoskeleton toward these sites during both budding and mating requires the products of multiple genes, including BEM1, CDC24, and CDC42. CDC24 encodes a GDP–GTP exchange factor for Cdc42p (Sloat and Pringle 1979; Zheng et al. 1994). Bem1p contains two SH3 domains and is thought to provide a scaffold by interacting directly with Cdc42p, Cdc24p, Ste20p, and Ste5p (Peterson et al. 1994; Leeuw et al. 1995; Lyons et al. 1996; Park et al. 1997). Cdc42p and Bem1p localize to the sites of polarized growth: the bud site during the cell cycle and the shmoo tip in cells exposed to pheromones (Ziman et al. 1993; Pringle et al. 1995). Cells lacking Cdc42p or Cdc24p function are unable to polarize their cytoskeleton and, as a consequence, arrest as large unbudded cells (Adams et al. 1990). In contrast, cells deleted for BEM1 are viable but exhibit morphological abnormalities (Chenevert et al. 1992).
In addition, Cdc42p has been shown to function upstream of mitogen-activated protein (MAP) kinase signal transduction pathways. In mammalian cells, Cdc42p triggers the Jun amino-terminal kinase/stress-activated protein kinase (JNK/SAPK) cascade (Bagrodia et al. 1995; Coso et al. 1995; Hill et al. 1995; Minden et al. 1995) and has also been implicated in the activation of the p70S6 kinase (Chou and Blenis 1996). In yeast, Cdc42p has been suggested to play a role in MAP kinase signaling pathways during mating (Simon et al. 1995; Zhao et al. 1995) and during pseudohyphal growth (Mösch et al. 1997). Members of the p21-activated kinase family (PAK) appear to be important effectors, mediating at least part of the signaling role of Cdc42p (Manser et al. 1994; Zhang et al. 1995; Brown et al. 1996; Peter et al. 1996; Leberer et al. 1997). Binding of Cdc42p to PAK-like kinases occurs through a short segment that is conserved among several Cdc42p targets and has been termed the Cdc42/Rac-interactive-binding (CRIB) domain (Burbelo et al. 1995). Activation of PAK kinases, however, is neither necessary nor sufficient for cytoskeletal polarization mediated by Cdc42p (Joneson et al. 1996; Lamarche et al. 1996).
The effectors of Cdc42p that control the cytoskeleton remain unknown. Although a number of candidate effectors have recently been described in yeast and mammals, none of these proteins can fully account for the effects of Cdc42p on cell polarization. The protein altered in patients suffering from Wiskott-Aldrich Syndrome (WASP) is important for some aspects of actin organization (Aspenstrom et al. 1996; Kolluri et al. 1996; Symons et al. 1996), but experiments in yeast have shown that the WASP-related molecule Las17/Bee1 is dispensible for cell polarization (Li 1997; D. Mitchell and G. Sprague, pers. comm). The formin-related proteins Bni1p and Bnr1p bind to several Rho-related GTPases, and thus may not act as specific Cdc42p targets (Kohno et al. 1996; Evangelista et al. 1997; Imamura et al. 1997). IQGAP family molecules, which are related by sequence to GTPase activating proteins (GAP), represent a possible class of Cdc42p effectors (Brill et al. 1996; Hart et al. 1996; Kuroda et al. 1996; McCallum et al. 1996), but our recent work suggests that yeast cells deleted for the gene, IQG1, encoding an IQGAP-related molecule are able to polarize (Epp and J. Chant, unpubl.). Finally, the ACK tyrosine kinase remains a largely unexplored mammalian target of Cdc42p (Manser et al 1994); however, ACK is not present in yeast and, therefore, seems an unlikely candidate for an ubiquitous Cdc42p effector involved in cytoskeletal polarization.
To identify Cdc42p effectors important for cytoskeletal polarization, we searched the complete Saccharomyces genomic sequence for additional proteins containing a CRIB domain. In this paper, we describe two novel Cdc42p effectors, Gic1p and Gic2p, which bind specifically to Cdc42p-GTP through their conserved CRIB domain. Importantly, Gic1p and Gic2p are required for cell polarization in vivo during the cell cycle and in response to extracellular signals, but they are dispensable for MAP kinase signal transduction. Our data suggest that Gic1p and Gic2p specifically link Cdc42p to dynamic rearrangements of the actin and microtubule cytoskeletons.
Results
Identification of GIC1 and GIC2
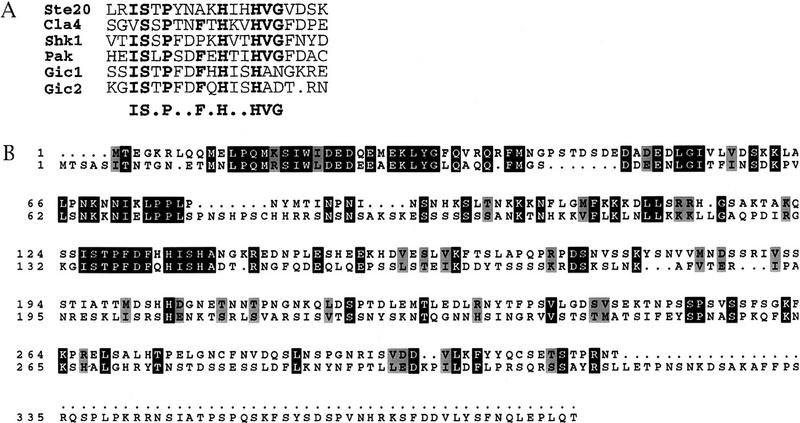
To identify potential Cdc42p targets, the complete genomic sequence of Saccharomyces cerevisiae was searched for gene products that contain the CRIB domain, which is shared by a number of known Cdc42p effectors (Manser et al. 1994; Burbelo et al. 1995) (Fig. 1A). Five proteins were identified: three PAK-related serine/threonine kinases (Ste20p, Cla4p, and Skm1p; Leberer et al. 1992; Ramer and Davis 1993; Cvrckova et al. 1995) and two uncharacterized open reading frames, which we denoted GIC1 and GIC2 (GTPase interactive components 1 and 2). Gic1p and Gic2p are related proteins of similar size with a highly conserved amino-terminal region followed by the CRIB domain and a less conserved carboxyl terminus (Fig. 1B). Further database searches indicated that Gic1p and Gic2p define a new class of CRIB domain proteins, and that there are only two Gic family members in yeast.
Figure 1.
Gic1p and Gic2p, two novel Cdc42p binding proteins. (A) Alignment of the CRIB domains of Gic1p and Gic2p to several Ste20/PAK-related kinases. Consensus amino acid residues are shown in bold (Burbelo et al. 1995). Sequences are from the following sources: Ste20p and Cla4p (Leberer et al. 1992; Ramer and Davis 1993; Cvrckova et al. 1995), Shk1p (Markus et al. 1995), and PAK1 (Manser et al. 1994). (B) Alignment of Gic1p (top) and Gic2p. Identical amino acid residues are indicated with black boxes. Similar amino acids are shaded.
Gic1p and Gic2p bind specifically to Cdc42p–GTP in vitro and in vivo
We tested whether the Gic proteins bind directly to Cdc42p by both biochemical and two-hybrid experiments. Columns containing immunoaffinity-purified Gic2 protein were assayed for their ability to retain recombinant Cdc42p. As illustrated in Figure 2A, Cdc42p preloaded with GTPγS readily bound to the Gic2p column, whereas Cdc42p–GDP exhibited no detectable affinity. Recombinant Cdc42p containing a T35A mutation in the GTPase effector domain, preloaded with GTPγS, failed to interact with Gic2p, suggesting that the interaction between Gic2p and Cdc42p occurs through the effector domain of Cdc42p.
Figure 2.
Gic1p and Gic2p bind Cdc42p-GTP. (A) Gic2p binds to Cdc42p-GTP. Sepharose beads containing purified yeast Gic2p were assayed for their ability to retain E. coli expressed Cdc42p preloaded with either GTPγS (lanes 1,3,4), or GDP (lane 2). (Lane 1) Control beads. (Lanes 2–4) Gic2-HAp beads. The following Cdc42 proteins were analyzed: (Lanes 1–3) wild-type Cdc42p. (Lane 4) Cdc42pT35A. Cdc42p was detected by immunoblotting. (B) Overproduction of an amino-terminal fragment of Gic2p inhibits cell growth (overexpressed from the GAL1 promoter). Cells accumulate with a large unbudded morphology. This growth defect could be suppressed by a high-copy-number plasmid carrying CDC42, but not by high-copy-number plasmids carrying no insert (VEC), CDC24, RHO1, CLA4, BEM1, or CDC42T35A.
The specificity of these interactions, particularly in relation to other Rho-type GTPases, was further examined by the two-hybrid system (Fields and Song 1989) (Table 1). Strong interactions were detected between the GTP-bound form of Cdc42p (G12V) and both Gic proteins, whereas no interaction was observed with the GDP-bound form of Cdc42p (D118A). Importantly, neither Bem1p nor any of the related Rho GTPases of yeast interacted with Gic2p, showing that Gic2p is a specific binding partner of Cdc42p. Mutating three conserved residues in the CRIB consensus sequence (Gic2pcrib−) to alanine residues abolished the interaction with Cdc42p, showing that the interaction between Gic2p and Cdc42p requires an intact CRIB domain.
Table 1.
Two-hybrid interactions between the Gic proteins, Bem1p, and Rho-family GTPases
| DNA-binding domain fusiona
|
Activation domain fusion b
|
lacZ expression (Miller units)c
|
|---|---|---|
| Cdc42p | vector | 49 |
| Cdc42p | Gic2p (full length) | 1503 |
| Cdc42p | Gic2p (1-208) | 1616 |
| Cdc42p | Gic1p (1-176) | 1530 |
| Cdc42p | Gic2p (Crib-) | 50 |
| Cdc42p (D118A) GDP bound | vector | 9 |
| Cdc42p (D118A) | Gic2p (full length) | 11 |
| Cdc42p (G12V) GTP bound | vector | 151 |
| Cdc42p (G12V) | Gic2p (full length) | 1372 |
| Rho1p | vector | 22 |
| Rho2p | vector | 24 |
| Rho3p | vector | 18 |
| Rho4p | vector | 27 |
| Rho1p | Gic2p (full length) | 17 |
| Rho2p | Gic2p (full length) | 14 |
| Rho3p | Gic2p (full length) | 15 |
| Rho4p | Gic2p (full length) | 7 |
| Bem1p | vector | 8 |
| Bem1p | Gic2p (full length) | 12 |
The LexA DNA binding domain fusions were carried on pEG202 and contain the carboxy-terminal Cys-Ser substitutions, which prevent prenylation (Stevenson et al. 1995).
The activation domain fusions were carried on pJG4-5, and pJG4-5 provided the vector control.
The average Miller units expressed of the lexA-lacZ reporter from three independent experiments are presented.
Further evidence showing an in vivo interaction between the Gic proteins and Cdc42p is illustrated in Figure 2B. Overproduction of a Gic2p fragment (Gic2p1–208), containing the CRIB domain, interferes in a dominant-negative fashion with cell growth. This growth defect was suppressed by overproduction of Cdc42p, but not by overproduction of Cdc42pT35A, a related GTPase (Rho1p), proteins involved in cell polarity (Cdc24p or Bem1p), or the established Cdc42p target, Cla4p. The simplest interpretation of this result is that overproduction of the Gic2p fragment competes for active Cdc42p within the cell, thereby preventing proper regulation of endogenous Cdc42p targets. Additional Cdc42p overcomes this deficiency. The restoration of growth by Cdc42p, but not by Cdc42pT35A, supports this view and suggests that Gic2p binds the Cdc42p effector domain. Taken together, the in vitro binding data, two-hybrid analysis, and genetic suppression experiments show that Gic1p and Gic2p interact specifically with GTP-bound Cdc42p in vivo.
Gic2p is produced in a cell cycle-dependent manner
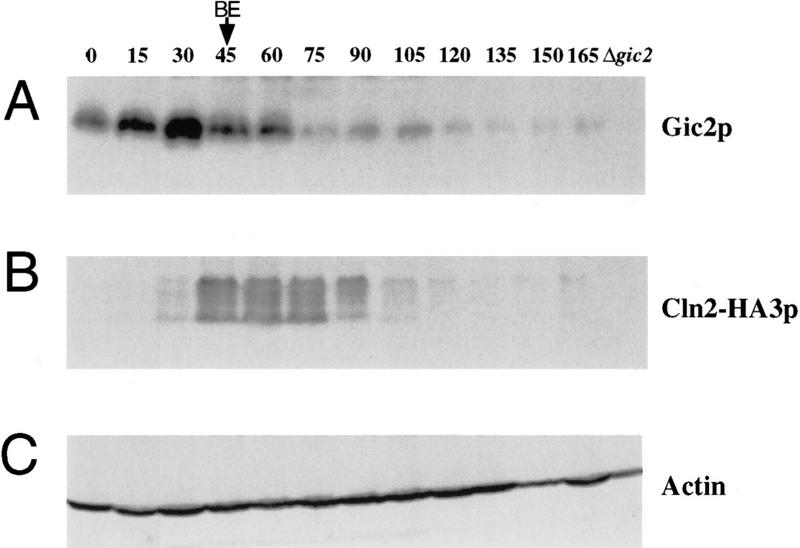
We examined whether the Gic proteins were present during the G1 phase of the cell cycle when Cdc42p directs cell polarity. Temperature-sensitive cdc15 cells were arrested in late mitosis and released from the cell cycle block by shifting the culture to the permissive temperature. As shown in Figure 3A, Gic2p accumulated throughout the G1-phase and peaked at the time of polarity establishment (15 min prior to bud emergence; BE in Fig. 3A). Gic2p rapidly disappeared at the time of bud emergence, concomitant with the appearance of Cln2p (Fig. 3B). No Gic2 protein was detected in G2 cells. In contrast, Cdc42p levels remained constant throughout the time course (data not shown; Ziman et al. 1993). We conclude that Gic2p is expressed in a cell cycle-dependent manner reaching maximal levels in late G1, consistent with a role in the establishment of cell polarity.
Figure 3.
Gic2p is expressed in a cell cycle-dependent manner. Cells were synchronized by releasing temperature-sensitive cdc15 cells from their block in late mitosis. Aliquots were collected every 15 min and analyzed by immunoblotting. The time of bud emergence is indicated as BE. (A) Gic2p; (B) Cln2p; (C) actin.
Gic1p and Gic2p colocalize with Cdc42p to regions of polarized cell growth
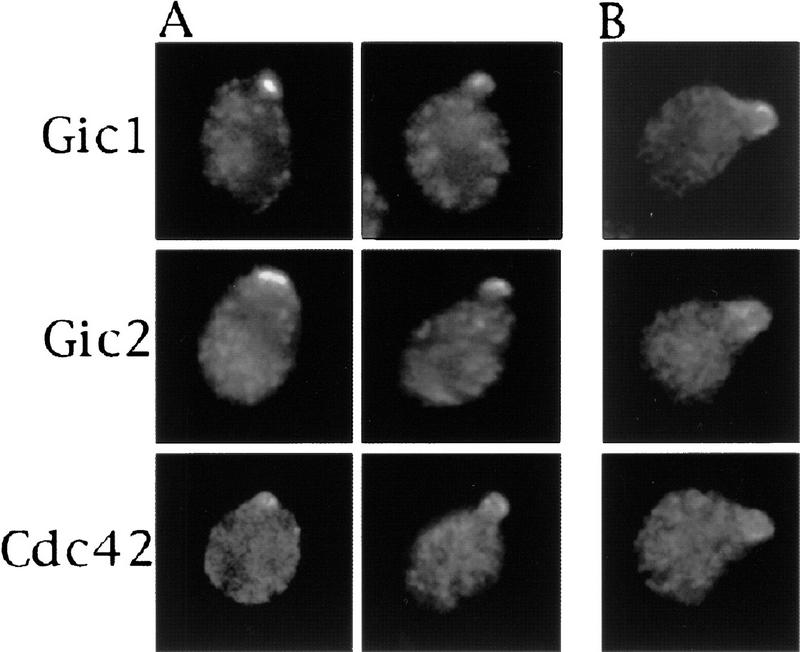
Next, we determined the subcellular localization of the Gic proteins (Fig. 4). As axes of cell polarity are being established, Gic1p and Gic2p were found to be asymmetrically distributed in patterns similar to that of Cdc42p. During cell division, Gic1p and Gic2p localized to the future bud site and later to the surfaces of small buds, as visualized by immunofluorescence microscopy (Fig. 4A). Cdc42p has been shown previously to localize to these same regions of polarized growth (Ziman et al. 1993). No localized Gic1p, Gic2p, or Cdc42p was observed in cells with medium to large buds; however, on occasion, some Cdc42p or Gic protein could be seen in the mother-bud neck very late in the cell cycle (data not shown). During mating, Gic1p, Gic2p, and Cdc42p colocalized to the tips of mating projections (shmoos) (Fig. 4B). Thus, Gic1p and Gic2p, together with Cdc42p, are located in regions that direct cytoskeletal polarization.
Figure 4.
Gic1p and Gic2p colocalize with Cdc42p to regions of polarized growth. Epitope-tagged Gic1p, Gic2p, and Cdc42p, expressed from their native promoters on centromeric plasmids, were detected by indirect immunofluorescence. Gic1p, Gic2p, and Cdc42p proteins localize to the incipient bud site and the surfaces of small buds (A), and to the tips of polarized mating projections (B).
Gic1p and Gic2p are functionally redundant proteins necessary for cell polarization in vivo
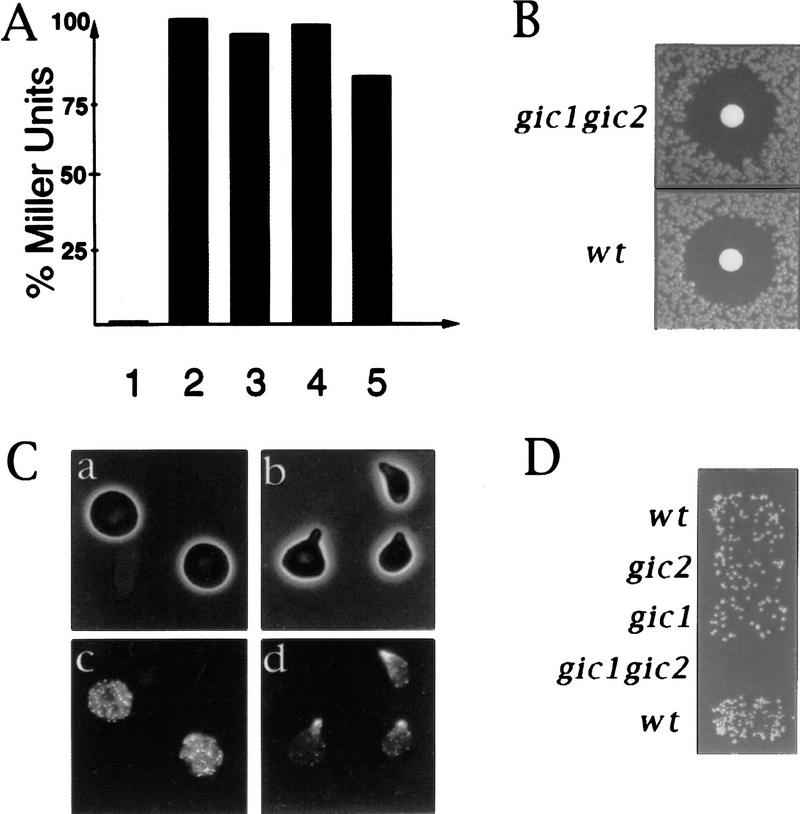
To determine whether the Gic proteins play a role in cell polarization, we studied the effects of GIC1 and GIC2 null mutations. As Gic1p and Gic2p are closely related by sequence, it seemed likely that the two genes would be functionally redundant. Consistent with this prediction, strains carrying either the GIC1 or GIC2 deletion grew normally, but strains deleted for both GIC1 and GIC2 were slow growing at 23°C and 30°C, and were dead at 37°C (Fig. 5A, data not shown). In a population of double mutants at 30°C, >80% of the cells accumulated as large, unbudded, multinucleate cells (Fig. 5D,I). Examination of both actin and microtubules in these mutants showed that the morphological defects reflected an underlying deficiency in cytoskeletal polarization. Whereas wild-type cells and the single gic mutant strains budded and polarized actin normally (Fig. 5B,C; data not shown), gic1Δ gic2Δ double mutants were deficient in actin polarization with actin patches distributed randomly at the cell cortex and actin cables either absent or misaligned (Fig. 5E). Spindle alignment in gic1Δ gic2Δ cells was also aberrant (Fig. 5H). Thus, cells lacking Gic proteins exhibit defects in cell polarization similar to those observed in cells lacking Cdc42p function (Adams et al. 1990).
Figure 5.
Gic1p and Gic2p encode redundant proteins required for efficient growth and cell polarization in vivo. (A) Strains harboring deletions in GIC1 or GIC2 grow at rates indistinguishable from the wild-type parental strain. Strains deleted for both GIC1 and GIC2 grow slowly at 30°C. (B–I) Wild-type (B,C,F,G) and gic1Δ gic2Δ cells (D,E,H,I) were observed by DIC (B,D) and epifluorescence microscopy (C,E,F–I). Actin (C,E), DNA (F,I), and microtubules (G,H) are shown.
Gic1p and Gic2p are required for polarized morphogenesis but not MAP kinase signal transduction during mating
Cdc42p has been implicated in both cell polarization and MAP kinase signal transduction during yeast mating (Zhao et al. 1995; Simon et al. 1995; Stevenson et al. 1995). To determine whether the Gic proteins play a role in signal transduction, we measured transcriptional induction and cell cycle arrest in response to mating pheromones, both readouts of MAP kinase signaling (Herskowitz 1995). When exposed to pheromone, the gic1Δ gic2Δ strains induced wild-type levels of the transcriptional reporter FUS1–LacZ (Trueheart et al. 1987) and were unaffected for cell cycle arrest (Fig. 6A,B). Thus, the Gic proteins are not required to activate the MAP kinase pathway during yeast mating. In contrast to wild-type cells, however, gic1Δ gic2Δ cells did not form polarized mating projections (shmoos) in response to mating pheromones (Fig. 6C) and, consequently, exhibited a 100-fold decrease in mating efficiency (Fig. 6D). Thus, Gic1p and Gic2p are unlikely to mediate any effect of Cdc42p on MAP kinase signal transduction, but they are important effectors of Cdc42p for the establishment of cell polarity during mating, as well as during vegetative growth.
Figure 6.
Gic1p and Gic2p are required for polarized morphogenesis but not for MAP kinase signaling in response to pheromones. (A) Activity of the MAP kinase signaling pathway triggered by pheromones was measured by induction of the FUS1–LacZ reporter, and plotted as percent Miller Units relative to wild-type controls. (Lane 1) No α-factor added. (Lanes 2–5) α-factor added for 1 hr. The following strains were analyzed: (1 and 2) wild-type; (3) gic1Δ; (4) gic2Δ; 5: gic1Δ gic2Δ. (B) gic1Δ gic2Δ (top panel) and wild-type cells (bottom panel) are able to arrest their cell cycle in response to α-factor as assessed by halo assay. (C) gic double mutants (a and c), but not cells lacking GIC2 (b and d), are defective for polarized morphogenesis in response to α-factor. (a and b) Phase contrast photographs; (c and d) actin visualized by rhodamine-phalloidin. (D) Mating efficiencies of wild-type and gic mutant strains to a far1-c tester (Valtz and Peter 1997).
Binding of Gic2p to Cdc42p is essential to Gic2p function in vivo
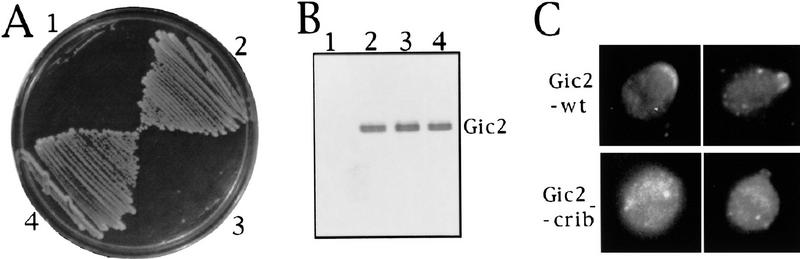
To assess the importance of Cdc42p binding for Gic2p function in vivo, we analyzed the function of a Gic2 protein containing CRIB domain mutations that abolish detectable Cdc42p interaction (Manser et al. 1994; Burbelo et al. 1995; Peter et al. 1996; Leberer et al. 1997) (Table 1). Although the mutant protein was produced at wild-type levels (Fig. 7B), the gic2crib− allele was unable to complement the growth defect of the gic1Δ gic2Δ double mutant (Fig. 7A), showing that binding of Gic2p to Cdc42p is essential for Gic2p function in vivo. Additionally, Gic2crib− protein failed to concentrate efficiently at sites of polarized growth and, instead, was distributed throughout the cytoplasm (Fig. 7C). Thus, the interaction between Cdc42p and Gic2p is required for proper Gic2p function and localization. Recently, it has been shown that Cdc42p also directs the localization of the protein kinase Ste20p (Peter et al. 1996; Leberer et al. 1997), suggesting that Cdc42p might generally target effectors to sites of polarized growth.
Figure 7.
Interaction between Gic2p and Cdc42p is essential for Gic2p function and localization. (A) The growth defect of gic1Δ gic2Δ cells could be complemented by plasmids expressing wild-type GIC2 but not gic2crib−. (1) gic1Δ gic2Δ strain carrying vector. (2) gic1Δ gic2Δ carrying vector with a GIC2 insert. (3) gic1Δ gic2Δ carrying vector with a gic2crib− insert. (4) A gic1 strain carrying no plasmid for comparison to sector 2. (B) Wild-type and Gic2pcrib− proteins are present at equal levels. Lane numbers correspond to those of panel A. Gic2 protein was detected by immunoblotting with specific Gic2p-antibodies. (C) Asymmetric localization of Gic2p is dependent on a functional CRIB domain. Localization of epitope-tagged wild-type Gic2p (top panels) is contrasted with the localization of epitope-tagged Gic2pcrib− (bottom panels), both expressed from a high-copy-number plasmid.
Genetic interactions between the GIC genes and other factors involved in cell polarization
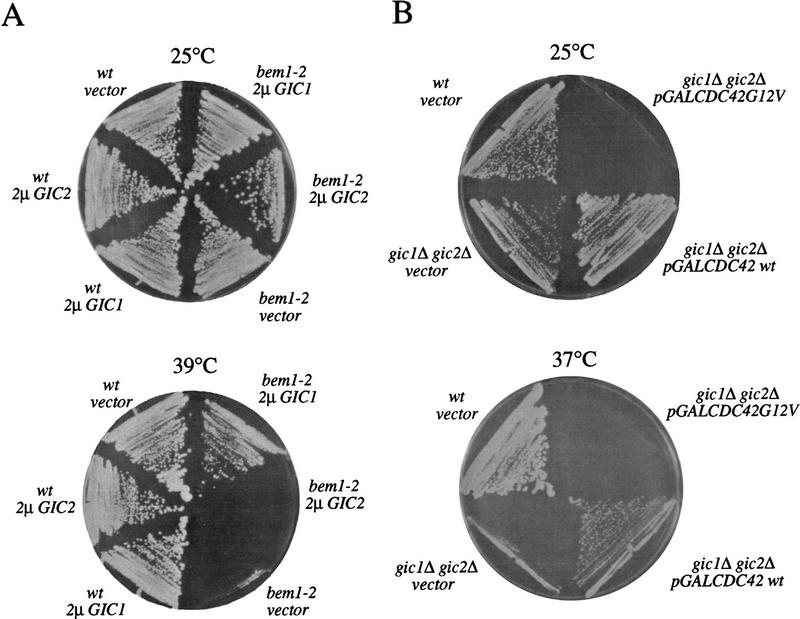
In addition to Cdc42p, Bem1p and Bem2p are also involved in the establishment of cell polarity. Bem2p functions as a GAP for Rho-type GTPases (Zheng et al. 1993; Peterson et al. 1994), whereas Bem1p may provide a scaffold for several polarity establishment proteins. Because the phenotype of gic1Δ gic2Δ cells resembles that of cells lacking Bem1p and Bem2p, we tested whether overproduction of Gic1p and Gic2p could rescue the growth defect of these cells. We found that high-copy-number vectors carrying GIC1, but not GIC2, partially restored growth at 37°C to bem1Δ or bem2Δ cells (Fig. 8A, and data not shown). Conversely, a multicopy plasmid carrying BEM1 was able to partially rescue the growth defect of gic1Δ gic2Δ cells (data not shown). These results further support the view that Gic1p and Gic2p are involved in polarity establishment.
Figure 8.
Genetic interaction between the Gic proteins, Bem1p, and Cdc42p. (A) Overexpression of Gic1p but not Gic2p is able to partially restore growth of cells lacking Bem1p function at 39°C. (B) Overexpression of wild-type Cdc42p but not Cdc42p in its active GTP-bound form (G12V) is able to suppress the growth defect of gic1Δgic2 mutants at 37°C.
Interestingly, the growth defect of gic1Δ gic2Δ cells could also be suppressed by overproduction of wild-type Cdc42p (Fig. 8B). Overproduction of Cdc42p in its GTP-bound form interfered with cell proliferation of gic1Δ gic2Δ cells, indicating that at least some of the lethal effects of Cdc42p-GTP are not dependent on the presence of the Gic proteins (Fig. 8B). Finally, multicopy plasmids carrying either GIC1 or GIC2 were not able to restore growth of a temperature-sensitive cdc42 mutant (data not shown). The simplest interpretation of these genetic suppression results is that Cdc42p controls multiple targets that coordinate cytoskeletal polarization, and that overproduction of Cdc42p can compensate, at least partially, for the lack of some effectors.
Discussion
Gic1p and Gic2p are Cdc42p-specific effectors required for establishment of cell polarity
Our results identify two novel effectors of Cdc42p that are required for cellular polarization during polarized cell division and mating chemotropism. A number of other Cdc42p-interacting proteins have been identified previously, including PAK kinases, WASP-related factors, and formins (Symons 1996; Frazier and Field 1997). Although these molecules are suggested or shown to be Cdc42p targets, they do not appear to account for the effect of Cdc42p on cell polarization. Available evidence suggests that members of the PAK kinase family are involved in the activation of MAP kinase signaling cascades rather than affecting cytoskeleton polarization (Cvrckova et al. 1995; Joneson et al. 1996; Lamarche et al. 1996). WASP-related molecules are not necessary for actin polarization in yeast, although they do affect some aspects of cortical actin morphology (Li 1997; D. Mitchell and G.F. Sprague, pers. comm.). Finally, recent work has implicated Bni1p, a formin-related molecule, as a Cdc42p effector that contributes to actin polarization during mating (Evangelista et al. 1997). BNI1, however, is dispensable for polarization of actin during budding (Jansen et al. 1997), and furthermore, Bni1p does not bind specifically to Cdc42p but also interacts with the related GTPases Rho1p, Rho3p, and Rho4p (Evangelista et al. 1997; Imamura et al. 1997; Kohno et al. 1997).
In contrast, we have presented four lines of evidence that argue strongly that Gic1p and Gic2p are Cdc42p effectors critical for controlling cell polarity. First, Gic1p and Gic2p bind specifically to Cdc42p-GTP with no detectable affinity for Cdc42p–GDP or related Rho proteins. Second, Gic1p and Gic2p colocalize with Cdc42p to the future bud site or mating projection at times when cytoskeletal polarization is being established. Third, the cell polarization defects exhibited by gic1Δ gic2Δ cells are similar to those of cdc42 mutants. Finally, and importantly, Gic2p function correlates with Cdc42p binding: Disruption of the Cdc42p–Gic2p interaction eliminates Gic protein function in vivo.
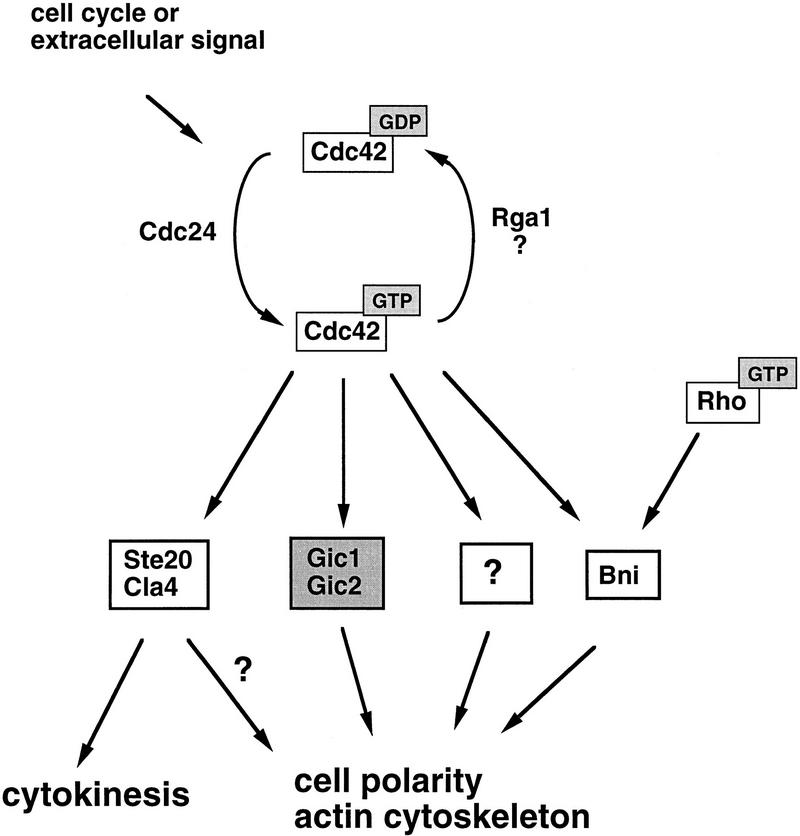
On the basis of these observations, we propose that during the cell cycle or in response to extracellular signals, Cdc42p is converted to the GTP-bound form in a spatially restricted manner. In turn, Cdc42p–GTP binds to Gic1p and Gic2p, which contribute toward cytoskeletal polarization (Fig. 9). It is unclear, at present, how Gic1p and Gic2p exert these effects, and the GIC sequences provide no obvious clues. Gic proteins may bind cytoskeletal elements directly, or they may link Cdc42p to key actin or microtubule-binding factors. Although no Gic homologs are yet known from other organisms, we consider it likely that Gic-like proteins link Cdc42p to cell polarity and cytoskeletal organization in higher eukaryotes, as, to date, all known Cdc42-binding proteins are conserved broadly.
Figure 9.
A pathway for cytoskeletal polarization mediated by Cdc42p in yeast. Cell cycle progression or extracellular signals lead to the conversion of GDP to GTP on Cdc42p, possibly by activation of the exchange factor Cdc24p. GTP–Cdc42p then binds and thereby localizes the effectors Gic1p and Gic2p to sites of polarized growth, which in turn contribute to polarization of the actin cytoskeleton. Cdc42p might exert some of its effects on the cytoskeleton not only through Gic1p and Gic2p but also through other targets including Bni1p, Ste20p, and Cla4p. Bni1p is known to interact also with other members of the Rho-GTPase family.
The effects of Cdc42p on the cytoskeletal organization and MAP kinase signaling are mediated through distinct effectors
Evidence presented here and elsewhere confirms the view that Cdc42p mediates its effects on the cytoskeleton and signal transduction pathways through distinct effectors. PAK-related kinases function upstream of MAP kinase signaling cascades (Leberer et al. 1992), and Cdc42p has been shown to bind and thereby activate these kinases in vitro (Manser et al. 1994; Martin et al. 1995; Simon et al. 1995). Available evidence, however, suggests that PAK-like kinases are not sufficient to account for cytoskeletal polarization mediated by Cdc42p, as a mutant form of Cdc42p that is unable to bind PAK kinases and fails to activate the JNK/SAPK kinase pathway fully promotes actin rearrangements when injected into fibroblasts (Joneson et al. 1996; Lamarche et al. 1996). Consistent with these observations, yeast cells lacking the two PAK-related kinases Ste20p and Cla4p are able to polarize their actin cytoskeleton (Cvrckova et al. 1995). In contrast, we show here that Gic1p and Gic2p are essential for cell polarization in yeast, but that Gic1p and Gic2p are dispensible for pheromone-induced signal transduction, indicating that these Cdc42p-targets are specific for mediating cytoskeletal rearrangements.
Cdc42p may regulate the cytoskeleton through multiple effectors
The evidence presented here firmly establishes that the Gic proteins are Cdc42p targets controlling cytoskeletal polarization. The Gic proteins, however, are not likely to be the sole cytoskeletal effectors of Cdc42p. First, cdc42 mutant cells are nonviable, whereas cells lacking both Gic1p and Gic2p are slow growing. Second, overexpression of either Cdc42p or Bem1p was able to at least partially restore the growth defect of gic1Δ gic2Δ cells, showing that increased levels of Cdc42p and Bem1p can reduce the need for the Gic proteins. In contrast, overexpression of either Gic protein failed to restore growth of cells lacking functional Cdc42p. Taken together, these results raise the possibility that Cdc42p and related GTPases direct cytoskeletal polarization, not through a single effector, but by signaling through a combination of effectors, each of which controls a parameter of actin or microtubule dynamics (Fig. 9). Together, Gic proteins and other factors would account for the essential role of Cdc42p in establishing axes of cell polarization.
Materials and methods
Genetic, recombinant, and database search methods
Standard yeast growth conditions and genetic manipulations were used (Rose and Fink 1990). Yeast transformations were performed by the lithium acetate method (Ito et al. 1983). Standard procedures were used for recombinant DNA manipulations (Sambrook et al. 1989; Ausubel et al. 1991). PCR reactions were performed by use of the Expand polymerase kit as recommended by the manufacturer (Boehringer Mannheim). Products were purified with the Wizard PCR purification kit according to the instructions of the manufacturer (Promega). Database searches were performed by use of the SGD (Stanford University) and the NCBI BLAST programs (National Institutes of Health).
GIC cloning and deletions
The GIC1 and GIC2 genes were cloned by PCR of genomic DNA from a haploid S288C-derived yeast strain (MATa ade2-101, ura3-52, trp1-Δ1, his3Δ200, leu2Δ, lys2-801) and the following oligonucleotides:
GIC1: oTP358 (5′-ATTTGCGGCCGCCGTCATCAGGAGTTCGAGGTCCAGAGGCATTGTTATCGG-3′) and oTP359 (5′-ATAAAGCTTCATTACGAGGAAACTATGGTGAAGATTACTGGG-3′); GIC2: oTP356 (5′-GCATTGCGGCCGCTTATAATTTTGGCGTTCAGCAAGCGCGCGG-3′) and oTP357 (5′-GTTAATTCGAAGATATAATAAACGAATGTATGGGATAACGCC-3′). An internal BamH1–SphI fragment of the GIC1 gene was replaced with the URA3 gene to generate the plasmid pMJ33. The GIC2 deletion construct was produced by removing an internal SpeI–BamHI fragment of the GIC2 gene, and replacing it with the full length LEU2 gene to generate the pBSK+ based plasmid, pMJ43. The resulting disruption constructs were excised from pMJ43 and pMJ33 and used to generate heterozygous diploid gic2Δ::LEU2 and gic1Δ::URA3 strains in the W303 background (K700: MAT a/α ade2-1, trp1-1, leu2-3, 112, his3-11, 15, ura3, GAL, psi+) by single step gene replacement (Rothstein 1991). Haploid gic1::URA3 and gic2::LEU2 segregants of opposite mating type were obtained by sporulation and used to generate a diploid strain (YMP1053), which was then sporulated to isolate gic1 gic2 double deletion mutants. Tetrads were dissected on YPD medium with 1 m sorbitol and grown at 25°C.
Construction of the crib− and truncation mutations of GIC2
The gic2crib− allele was generated by mutating three consensus CRIB amino acids (I134, S135, and P137) to alanine residues by two-step PCR with the following oligonucleotide combinations: oTP356 and oTP434 (5′-GTGAAATATGTTGAAAATCAAATGCTGTGGCG-3′); oTP429 (5′-GGTGCCGCCACAGCATTTGATTTTCAACATATTTCAC-3′) and oTP357. The mutated Crib− GIC2 gene was sequenced, and subcloned into pJG4-5 for two hybrid experiments (generating pMJ177), as well as several other yeast vectors (pRS314, pRS424, YEp352; Sikorski and Hieter 1989).
The amino-terminal fragment of GIC2 (amino acid residues 1–208) was amplified by PCR with oligonucleotides oTP409: (5′-ACTGGAATTCAATATGACTAGTGCAAGTATTACC-3′) and oTP423: (5′-ACGCTCGAGTCATAGTCTTGATGTCTTATTTTCGTGCG-3′), and subcloned into the two hybrid vector pJG4-5 (generating pMJ64) and a GAL1 expression vector to generate pMJ127.
Construction of epitope-tagged Gic1p, Gic2p, and Cdc42p
Epitope-tagged versions of Gic1p and Gic2p containing the influenza hemagglutinin (HA) epitope (YPYDVPDYA) fused to the carboxyl terminus of each protein were constructed by use of PCR and the following oligonucleotide pairs: GIC1: (5′-CGCGAATTCGCGAAAAGACAACAAC-3′) and (5′-GCTCTAGATTAAGCGTAGTCTGGGACGTCGTATGGGTAGGTATTTCGAGGAGTACTAGTTTC-3′); GIC2: (5′-CGAGATCTAGATGTTGCCTATTTCTCG-3′), and (5′-GCTCTAGATTAAGCGTAGTCTGGGACGTCGTATGGGTAAGTTTGCAGGGGCTCGAGCTGG-3′). Isolated PCR products containing 300 bp of the endogenous GIC1 and GIC2 promoters were derived directly from SEY6210 genomic DNA and cloned into pRS313, pRS314, YEp351, and Yep352-based plasmids as EcoRI–XbaI or XbaI fragments. The integrity of the constructs was confirmed by sequencing and Western blotting. Plasmids expressing either HA–Gic1p or HA–Gic2p were found to fully complement both the growth and morphology defects associated with gic1Δ gic2Δ mutants. Construction of an amino-terminal epitope-tagged Cdc42p in a pRS315-based plasmid was generated by use of PCR and oligonucleotides (5′-CGGGATCCTATTAGCTCTTCCACAAAATGTACCCATACGACGTCCCAGACTACGCTCAAACGCTAAAGTGTGTTGTTGTCGG-3′) and (5′-GCTCTAGACGGGCATATACTAATATGACTACA-3′). PCR products were cloned into the pRS315 vector containing 500 bp of the endogenous CDC42 promoter as BamHI/XbaI fragments, and analyzed by Western blotting. Centromeric plasmids expressing HA–Cdc42p fully complemented a cdc42::TRP1 disrupted strain in plasmid shuffle experiments.
Production of polyclonal Anti-Gic2p antisera
Polyclonal anti-Gic2p antibodies were generated against the full length Gic2p coding sequence cloned into pGEX-4T (Pharmacia) and expressed as a GST-fusion protein. Soluble GST–Gic2p, purified with glutathione Sepharose (Pharmacia) was used to immunize rabbits (Elevage Scientifique des Dombes, France). Antibodies were affinity-purified against GST–Gic2p as described (Harlow and Lane 1988). Standard procedures were used for yeast cell extract preparation and immunoblotting (Peter et al. 1993). Antibodies are specific to Gic2p as no signal is detected in extracts from gic2Δ cells (data not shown).
Cell synchronization
To release cells from a cdc15-2 block, cells were grown to exponential phase in YPD medium and then shifted to 37°C for 2 hr. Cells were released by shifting the culture to 25°C, and aliquots were taken at 15-min intervals. Protein extracts were prepared as described above. Cell cycle synchrony was monitored by fluorescence-activated cell sorting analysis and microscopic determination of the budding index.
Two-hybrid assays
Two-hybrid assays were performed in yeast strain EGY48 transformed with pEG202-based plasmids expressing LexA DNA-binding domain fusions, and pJG4-5-based plasmids containing transcriptional activation domain fusions (Gyuris et al. 1993). LexA–GTPase fusions all contain carboxy-terminal Cys to Ser substitutions that prevent prenylation. LacZ reporter activity was measured as described previously (Stern et al. 1984).
Cdc42p-binding assays
Gic2p affinity matrices were prepared as follows: Extracts of yeast expressing Gic2-HAp from the GAL1 promoter were prepared in TNE450 buffer (450 mm NaCl, 10 mm EDTA, 50 mm Tris-HCl at pH 7.5, 0.1% NP-40) and depleted of Gic2-HAp by use of 11HA monoclonal antibodies (Babco, Berkeley, CA) covalently coupled to protein G-Sepharose (Peter et al. 1996) (Pharmacia). Gic2-HAp columns were washed three times with TMT (10 mm Tris at pH 7.5, 10 mm MgCl2, 1 mm DTT, 0.1 % Triton X-100) prior to Cdc42p binding. Cdc42p and Cdc42pT35A proteins were produced in Escherichia coli strain NB42 as 6His fusion proteins by use of the pTrcHis vector (Invitrogen) and purified on Co2+ Sepharose-6B columns coupled with iminodiacetic acid (Sigma). Purified Cdc42p was preloaded with GTP-γS or GDP, as described (Park et al. 1993), and incubated for 1 hr at 4°C with affinity matrix in 200 μl of buffer B (10 mm Tris at pH 7.5, 85 mm NaCl, 6 mm MgCl2, 10% glycerol) containing 0.6 mm GTP-γS, or 0.6 mm GDP. After four washes with TMT buffer, Cdc42p was eluted with 100 μl of gel sample buffer and detected by immunoblotting with Cdc42p-specific antisera (Peter et al. 1996).
Immunofluorescence
Actin and microtubule staining was performed on cells fixed and treated by standard methods (Pringle et al. 1991). Actin was visualized by use of rhodamine-conjugated phalloidin (Molecular Probes) at a concentration of 1 mg/ml, and microtubules were detected with the YOL1/34 monoclonal antibody (Accurate) and FITC-conjugated goat anti-rat secondary antibodies (Jackson Labs) at 1/1000 dilution. The DNA stain Hoechst (Sigma) was used at a final concentration of 0.01 mg/ml.
Immunofluorescence microscopy of the Gic proteins was performed in W303 cells on fully complementing carboxy-terminally HA epitope-tagged versions expressed under their own promoters in gic1Δ gic2Δ cells from the pRS313 or pRS314-based centromeric plasmids, respectively. Amino-terminally-tagged Cdc42p was expressed from its own promoter in the pRS315 plasmid in wild-type haploid strain 1241-2D (Chant and Hersowitz 1991). HA-tagged proteins were visualized (Pringle et al. 1991; Brown et al. 1994) with the 11 HA monoclonal (Babco) and CY3-conjugated goat anti-mouse secondary antibodies (Jackson Labs) at 1/1000 dilutions. Pheromone treated cells were exposed to 4 mg/ml of synthetic α-factor (Sigma) for 1.5 hr. For Figure 7C, HA-tagged Gic2p and Gic2pcrib− were produced from multicopy YEp351-based plasmids in a wild-type strain.
Mating assays
Mating assays were performed with the tester strains IH1793 (MATα, lys1) and IH2625 (MATα, lys1, far1-c) as described (Valtz and Peter 1997). Shmoo morphology was examined after addition of 10−6 m α-factor to 3 ml of log phase cultures for 3 hr at 25°C. For cell cycle arrest (halo) assays, 103–104 cells were plated on YPD–sorbitol plates (1m sorbitol). Ten micrograms of α-factor in 20 μl of 0.01 m HCl was spotted on a sterile filter disc (Schleicher and Schuell) and placed on plates, which were then incubated for 3 days at 25°C.
Acknowledgments
The authors would like to thank the members of each lab for helpful discussions, M. van Lohuizen and C. Boone for strains and plasmids, K. Hofmann for help with computer analysis, and A. Rushforth for help with early aspects of this work. We acknowledge D. Mitchell and G. Sprague for sharing unpublished results, and J. Philips, B. Amati, and V. Simanis for critical reading of the manuscript. J.B. received support from a National Institutes of Health (NIH) Postdoctoral Fellowship, and J.C. has operating grants from the NIH and the Searle Family/Chicago Community Trust. M.P. is supported by the Swiss National Science Foundation, the Swiss Cancer League, and a Helmut Horten Incentive Award.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL matthias.peter@isrec.unil.ch; FAX (41) 21 652-6933.
References
- Adams AEM, Johnson DI, Longnecker RM, Sloat BF, Pringle JR. CDC42 and CDC43, two additional genes involved in budding and the establishment of cell polarity in the yeast Saccharomyces cerevisiae. J Cell Biol. 1990;111:131–142. doi: 10.1083/jcb.111.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspenström P, Lindberg U, Hall A. Two GTPases, Cdc42 and Rac, bind directly to a protein implicated in the immunodeficiency disorder Wiskott-Aldrich syndrome. Curr Biol. 1996;6:70–75. doi: 10.1016/s0960-9822(02)00423-2. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: Greene Publishing Associates/Wiley-Interscience; 1991. [Google Scholar]
- Bagrodia S, Derijard B, Davis RJ, Cerione RA. Cdc42 and PAK-mediated signalling leads to Jun kinase and p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:27995–27998. doi: 10.1074/jbc.270.47.27995. [DOI] [PubMed] [Google Scholar]
- Brill S, Li S, Lyman CW, Church DM, Wasmuth JJ, Weissbach L, Bernards A, Snijders AJ. The Ras GTPase-activating-protein-related human protein IQGAP2 harbors a potential actin binding domain and interacts with calmodulin and Rho family GTPases. Mol Cell Biol. 1996;16:4869–4878. doi: 10.1128/mcb.16.9.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Bussey H, Stewart RC. Yeast Skn7p functions in a eukaryotic two-component regulatory pathway. EMBO J. 1994;13:5186–5194. doi: 10.1002/j.1460-2075.1994.tb06849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Stowers L, Baer M, Trejo JA, Coughlin S, Chant J. Human Ste20 homologue hPAK1 links GTPases to the JNK MAP kinase pathway. Curr Biol. 1996;6:598–605. doi: 10.1016/s0960-9822(02)00546-8. [DOI] [PubMed] [Google Scholar]
- Burbelo PD, Drechsel D, Hall A. A conserved binding motif defines numerous candidate target proteins for both Cdc42 and Rac GTPases. J Biol Chem. 1995;270:29071–29074. doi: 10.1074/jbc.270.49.29071. [DOI] [PubMed] [Google Scholar]
- Chant J. Generation of cell polarity in yeast. Curr Opin Cell Biol. 1996;8:557–565. doi: 10.1016/s0955-0674(96)80035-4. [DOI] [PubMed] [Google Scholar]
- Chant J, Herskowitz I. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 1991;65:1203–1212. doi: 10.1016/0092-8674(91)90015-q. [DOI] [PubMed] [Google Scholar]
- Chenevert J, Corrado K, Bender A, Pringle J, Herskowitz I. A yeast gene (BEM1) necessary for cell polarization whose product contains two SH3 domains. Nature. 1992;356:77–79. doi: 10.1038/356077a0. [DOI] [PubMed] [Google Scholar]
- Chou MM, Blenis J. The 70KDa S6 kinase complexes with and is activated by the Rho family G proteins Cdc42 and Rac1. Cell. 1996;85:573–583. doi: 10.1016/s0092-8674(00)81257-x. [DOI] [PubMed] [Google Scholar]
- Coso OA, Chiariello M, Yu JC, Teramoto H, Crespo P, Xu N, Miki T, Gutkind JS. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signalling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- Cvrckova F, De Virgilio C, Manser E, Pringle JR, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes & Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- Dorer R, Pryciak PM, Hartwell LH. Saccharomyces cerevisiae cells execute a default pathway to select a mate in the absence of pheromone gradients. J Cell Biol. 1995;131:845–861. doi: 10.1083/jcb.131.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin DG, Nelson WJ. Origins of cell polarity. Cell. 1996;84:335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Evangelista M, Blundell K, Chow CJ, Adames N, Pringle JR, Peter M, Boone C. Bni1p, a yeast formin linking Cdc42p and the actin cytoskeleton during polarized morphogenesis. Science. 1997;276:118–122. doi: 10.1126/science.276.5309.118. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Frazier JA, Field CM. Are FH proteins local organizers? Curr Biol. 1997;7:414–417. doi: 10.1016/s0960-9822(06)00205-3. [DOI] [PubMed] [Google Scholar]
- Glotzer M, Hyman AA. The importance of being polar. Curr Biol. 1995;5:1102–1105. doi: 10.1016/s0960-9822(95)00221-1. [DOI] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Hall A. Small GTP-binding proteins and the regulation of the actin cytoskeleton. Annu Rev Cell Biol. 1994;10:31–54. doi: 10.1146/annurev.cb.10.110194.000335. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hart MJ, Callow MG, Souza B, Polakis P. IQGAP1, a calmodulin-binding protein with a RasGAP-related domain, is a potential effector for Cdc42Hs. EMBO J. 1996;15:2997–3005. [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. MAP kinase pathways in yeast: For mating and more. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]
- Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- Imamura H, Tanaka K, Hihara T, Umikawa M, Kamei T, Takahashi K, Sasaki T, Takai Y. Bni1p and Bnr1p: Downstream targets of the Rho family small G-proteins which interact with profilin and regulate actin cytoskeleton in Saccharomyces cerevisiae. EMBO J. 1997;16:2745–2755. doi: 10.1093/emboj/16.10.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RP, Dowzer C, Michaelis C, Galova M, Nasmyth K. Mother cell-specific HO expression in budding yeast depends on the unconventional myosin myo4p and other cytoplasmic proteins. Cell. 1996;84:687–697. doi: 10.1016/s0092-8674(00)81047-8. [DOI] [PubMed] [Google Scholar]
- Johnson DI, Pringle JR. Molecular characterization of CDC42, a Saccharomyces cerevisiae gene involved in the development of cell polarity. J Cell Biol. 1990;111:143–152. doi: 10.1083/jcb.111.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joneson T, McDonough M, Bar-Sagi D, Van Aelst L. Rac regulation of actin polymerization and proliferation by a pathway distinct from Jun kinase. Science. 1996;274:1374–1376. doi: 10.1126/science.274.5291.1374. [DOI] [PubMed] [Google Scholar]
- Kolluri R, Tolias KF, Carpenter CL, Rosen FS, Kirchhausen T. Direct interaction of the Wiskott-Aldrich syndrome protein with the GTPase Cdc42. Proc Natl Acad Sci. 1996;93:5615–5618. doi: 10.1073/pnas.93.11.5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Fujita Y, Hotta K, Qatoda H, Watanabe T, Ohya Y, Takai Y. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- Kozma R, Ahmed S, Best A, Lim L. The Ras-related protein Cdc42Hs and bradykinin promote formation of peripheral actin mikrospikes and filopodia in Swiss 3T3 fibroblasts. Mol Cell Biol. 1995;15:1942–1952. doi: 10.1128/mcb.15.4.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda S, Fukata M, Kobayashi K, Nakafuku M, Nomura N, Iwamatsu A, Kaibuchi K. Identification of IQGAP as a putative target for the small GTPases, Cdc42 and Rac1. J Biol Chem. 1996;271:23363–23367. doi: 10.1074/jbc.271.38.23363. [DOI] [PubMed] [Google Scholar]
- Lamarche N, Tapon N, Stowers L, Burbelo PD, Aspenstrom P, Bridges T, Chant J, Hall A. Rac and Cdc42 induce actin polymerization and G1 cell cycle progression independently of p65PAK and the JNK/SAPK MAP kinase cascade. Cell. 1996;87:519–529. doi: 10.1016/s0092-8674(00)81371-9. [DOI] [PubMed] [Google Scholar]
- Leberer E, Dignard D, Harcus D, Thomas DY, Whiteway M. The protein kinase homologue Ste20p is required to link the yeast pheromone response G-protein beta gamma subunits to downstream signalling components. EMBO J. 1992;11:4815–4824. doi: 10.1002/j.1460-2075.1992.tb05587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Wu C, Leeuw T, Fourest-Lieuvin A, Segall JE, Thomas DY. Functional characterization of the Cdc42 binding domain of yeast Ste20p protein kinase. EMBO J. 1997;16:83–97. doi: 10.1093/emboj/16.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeuw T, Fourest-Lieuvin A, Wu C, Chenevert J, Clark K, Whiteway M, Thomas DY, Leberer E. Pheromone response in yeast: Association of Bem1p with protein of the MAP kinase cascade and actin. Science. 1995;270:1210–1213. doi: 10.1126/science.270.5239.1210. [DOI] [PubMed] [Google Scholar]
- Li R. Bee 1, a yeast protein with homology to Wiscott-Aldrich syndrome protein, is critical for the assembly of cortical actin cytoskeleton. J Cell Biol. 1997;136:649–658. doi: 10.1083/jcb.136.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DM, Mahanty MC, Choi K-Y, Manandhar M, Elion EA. The SH3-domain protein Bem1p coordinates mitogen-activated protein kinase cascade activation with cell cycle control in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4095–4106. doi: 10.1128/mcb.16.8.4095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L. A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature. 1994;367:40–46. doi: 10.1038/367040a0. [DOI] [PubMed] [Google Scholar]
- Markus S, Polverino A, Chang E, Robbins D, Cobb MH, Wigler MH. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signalling module in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci. 1995;92:6180–6184. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin GA, Bollag G, McCormick F, Abo A. A novel serine kinase activated by Rac1/CDC42Hs-dependent autophosphorylation is related to PAK65 and STE20. EMBO J. 1995;14:1970–1978. doi: 10.1002/j.1460-2075.1995.tb07189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum SJ, Wu WJ, Cerione RA. Identification of a putative effector for Cdc42Hs with high sequence similarity to the RasGAP-related protein IQGAP1 and a Cdc42Hs binding partner with similarity to IQGAP2. J Biol Chem. 1996;271:21732–21737. doi: 10.1074/jbc.271.36.21732. [DOI] [PubMed] [Google Scholar]
- Minden A, Lin A, Claret FX, Abo A, Karin M. Selective activation of the JNK signalling cascade and c-Jun transcriptional activity by the small GTPases Rac and Cdc42Hs. Cell. 1995;81:1147–1157. doi: 10.1016/s0092-8674(05)80019-4. [DOI] [PubMed] [Google Scholar]
- Mösch H-U, Roberts RL, Fink GR. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1996;93:5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobes CD, Hall A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 1995;81:53–62. doi: 10.1016/0092-8674(95)90370-4. [DOI] [PubMed] [Google Scholar]
- Park HO, Chant J, Herskowitz I. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 1993;365:269–274. doi: 10.1038/365269a0. [DOI] [PubMed] [Google Scholar]
- Park HO, Bi E, Pringle JR, Herskowitz I. Two active states of the Ras-related Bud1/Rsr1 protein bind to different effectors to determine yeast cell polarity. Proc Natl Acad Sci. 1997;94:4463–4468. doi: 10.1073/pnas.94.9.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter M, Gartner A, Horecka J, Ammerer G, Herskowitz I. FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell. 1993;73:747–760. doi: 10.1016/0092-8674(93)90254-n. [DOI] [PubMed] [Google Scholar]
- Peter M, Neiman AM, Park H-O, van Lohuizen M, Herskowitz I. Functional analysis of the interaction between the small GTP binding protein Cdc42 and the Ste20 protein kinase in yeast. EMBO J. 1996;15:7046–7059. [PMC free article] [PubMed] [Google Scholar]
- Peterson J, Zheng Y, Bender L, Meyers R, Cerione R, Bender A. Interactions between the bud emergence proteins Bem1p and Bem2p and Rho-type GTPases in yeast. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle JR, Adams AEM, Drubin DG, Haarer BK. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Pringle JR, Bi E, Harkins HA, Zahner JE, De Virgilio C, Chant J, Corrado K, Fares H. Establishment of cell polarity in yeast. Cold Spring Harbor Symp Quant Biol. 1995;60:729–744. doi: 10.1101/sqb.1995.060.01.079. [DOI] [PubMed] [Google Scholar]
- Ramer SW, Davis RW. A dominant truncation allele identifies a gene, STE20, that encodes a putative protein kinase necessary for mating in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1993;89:452–456. doi: 10.1073/pnas.90.2.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. Rho-related proteins: Actin cytoskeleton and cell cycle.Curr. Opin Genet Dev. 1995;5:24–30. doi: 10.1016/s0959-437x(95)90049-7. [DOI] [PubMed] [Google Scholar]
- Rose MD, Fink GR. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. pp. 119–187. [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: Integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MN, De Virgilio C, Souza B, Pringle JR, Abo A, Reed SI. Role for the Rho-family GTPase Cdc42 in yeast mating-pheromone signal pathway. Nature. 1995;376:702–705. doi: 10.1038/376702a0. [DOI] [PubMed] [Google Scholar]
- Sloat BF, Pringle JR. A mutant of yeast defective in cellular morphogenesis. Science. 1978;200:1171–1173. doi: 10.1126/science.349694. [DOI] [PubMed] [Google Scholar]
- Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- Stevenson BG, Ferguson B, De Virgilio C, Bi E, Pringle JR, Ammerer G, Sprague GF., Jr Mutation of RGA1, which encodes a putative GTPase-activating protein for polarity-establishment protein Cdc42, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes & Dev. 1995;9:2949–2963. doi: 10.1101/gad.9.23.2949. [DOI] [PubMed] [Google Scholar]
- Stowers L, Yelon D, Berg LJ, Chant J. Regulation of the polarization of T cells towards antigen-presenting cells by Ras-related GTPase CDC42. Proc Natl Acad Sci. 1995;92:5027–5031. doi: 10.1073/pnas.92.11.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symons M. The Rac and Rho pathways as a source of drug targets for Ras-mediated malignancies. Curr Opin Biotech. 1995;6:668–674. doi: 10.1016/0958-1669(95)80110-3. [DOI] [PubMed] [Google Scholar]
- ————— Rho family GTPases: The cytoskeleton and beyond. Trends Biochem Sci. 1996;21:178–181. [PubMed] [Google Scholar]
- Symons M, Derry JMJ, Karlak B, Jiang S, Lemahieu V, McCormick F, Francke U, Abo A. Wiskott-Aldrich syndrome protein, a novel effector for the GTPase CDC42Hs, is implicated in actin polymerization. Cell. 1996;84:723–734. doi: 10.1016/s0092-8674(00)81050-8. [DOI] [PubMed] [Google Scholar]
- Trueheart J, Boeke JD, Fink GR. Two genes required for cell fusion during yeast conjugation: Evidence for a pheromone-induced surface protein. Mol Cell Biol. 1987;7:2316–2328. doi: 10.1128/mcb.7.7.2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtz N, Peter M. Functional analysis of FAR1 in yeast. In Cell cycle control. Methods Enzymol. 1997;283:350–365. doi: 10.1016/s0076-6879(97)83029-7. [DOI] [PubMed] [Google Scholar]
- Valtz N, Peter M, Herskowitz I. FAR1 is required for oriented polarization of yeast cells in response to mating pheromones. J Cell Biol. 1995;131:863–873. doi: 10.1083/jcb.131.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Han J, Sells MA, Chernoff J, Knaus UG, Ulevitch RJ, Bokoch GM. Rho family GTPases regulate p38 mitogen-activated protein kinase activation. J Biol Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- Zhao Z S, Leung T, Manser E, Lim L. Pheromone signalling in Saccharomyces cerevisiae requires the small GTP-binding protein Cdc42 and its activator CDC24. Mol Cell Biol. 1995;15:5246–5257. doi: 10.1128/mcb.15.10.5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Hart MJ, Shinjo K, Evans T, Bender A, Cerione RA. Biochemical comparison of the Saccharomyces cerevisiae Bem2 and Bem3 proteins. J Biol Chem. 1993;268:24629–24634. [PubMed] [Google Scholar]
- Ziman M, Preuss D, Mulholland J, O’Brien JM, Botstein D, Johnson DI. Subcellular localization of Cdc42, a Saccharomyces cerevisiae GTP-binding protein involved in the control of cell polarity. Mol Cell Biol. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]