Abstract
Relatively little is known about the molecular mechanisms involved in transcriptional repression, despite its importance in development and differentiation. Recent evidence suggests that some transcriptional repressors act by way of adaptor molecules known as corepressors. Here, we use in vivo functional assays to test whether different repressor activities are mediated by the Groucho (Gro) corepressor in the Drosophila embryo. Previously, Gro was proposed to mediate repression by the Hairy-related family of basic helix–loop–helix proteins. Our results indicate not only that repression by Hairy requires Gro, but that a repressor domain from the Engrailed (En) homeodomain protein is also Gro dependent. The latter result correlates with an ability of this En domain to bind to Gro in vitro. In contrast, repressor regions from the Even-skipped, Snail, Krüppel, and Knirps transcription factors are effective in the absence of Gro. These results show that Gro is not generally required for repression, but acts as a specific corepressor for a fraction of negative regulators, including Hairy and En.
Keywords: Groucho, corepressor, repression, Hairy, Engrailed, Drosophila
Transcriptional repression plays a key role in establishing precise patterns of gene expression during development (Gray and Levine 1996b; Ip and Hemavathy 1997). For example, most transcriptional regulators that control pattern formation in the early Drosophila embryo behave as repressors. Although mechanisms of repression are less well understood than those leading to gene activation, two major modes of repression have been identified (Cowell 1994; Johnson 1995; Hanna-Rose and Hansen 1996). In one case, repressors function by interfering passively with binding of activators to target promoters. This can involve the formation of repressor/activator complexes that are unable to bind DNA, or the occlusion of activator DNA-binding sites by binding of the repressor to overlapping target sequences. A second class of repressor binds to specific DNA sites, and blocks transcription actively through protein–protein interactions with other promoter factors.
Like transcriptional activators, “active” repressors of the second class appear to have a modular nature. In addition to a DNA-binding domain, they include discrete protein domains that can impose repressor activity on heterologous DNA-binding regions. Repressor modules can share certain structural similarities, including a preponderance of alanine residues [e.g., those from the Drosophila Engrailed (En), Even-skipped (Eve), and Krüppel (Kr) proteins; Han and Manley 1993a,b; Licht et al. 1994; for review, see Hanna-Rose and Hansen 1996], but the role of such structural features is not understood. Some repressor domains are likely to establish direct interactions with components of the transcriptional machinery (e.g., those from Eve and Kr; Sauer et al. 1995; Um et al. 1995) or with promoter-bound activator factors. However, others probably function by recruiting accessory proteins (“corepressors”) that themselves effect repression.
The best-characterized example of corepression involves a complex formed by the yeast proteins TUP1 and SSN6, which are required for repression by the α2 homeodomain protein (Keleher et al. 1992). These two proteins do not bind inherently to DNA, but appear to be recruited to promoters through interactions with α2. Once at the promoter, the TUP1/SSN6 complex mediates repression, possibly by binding downstream proteins such as basal transcription factors or histones (Edmondson et al. 1996). Other repressor–corepressor models include Mad-mSin3 and nuclear hormone receptor–N-CoR/mSin3 associations in mammalian cells (Horlein et al. 1995; Kurokawa et al. 1995; Alland et al. 1997; Heinzel et al. 1997).
The Drosophila Groucho (Gro) protein, a maternally contributed nuclear factor (Hartley et al. 1988; Delidakis et al. 1991), is a putative corepressor (Paroush et al. 1994; Fisher et al. 1996). Gro lacks a recognizable DNA-binding domain but, like TUP1, contains repeated WD motifs that are believed to mediate protein–protein recognition (for review, see Neer et al. 1994). Previously, we have proposed that Gro functions as a corepressor for the Hairy family of basic helix–loop–helix (bHLH) transcription factors (Paroush et al. 1994), which includes the Hairy, Deadpan (Dpn), and Enhancer of split [E(spl)] proteins that act in segmentation, sex determination, and neurogenesis, respectively (Klämbt et al. 1989; Rushlow et al. 1989; Bier et al. 1992; Delidakis and Artavanis-Tsakonas 1992; Knust et al. 1992; Schrons et al. 1992; Younger-Shepherd et al. 1992). The Hairy/Gro corepression model is supported by the finding that Gro binds in yeast and in vitro to a conserved carboxy-terminal tetrapeptide (WRPW) present in the above bHLH proteins, which functions as an active repressor domain in cultured cells (Paroush et al. 1994; Fisher et al. 1996). In addition, Gro activity is required maternally for the processes regulated by the Hairy-related proteins. In embryos lacking maternal gro (hereafter referred to as gro mutant embryos), hairy (h) expression overlaps that of one of its targets of repression, fushi tarazu (ftz) (Carroll and Scott 1986; Howard and Ingham 1986; Ish-Horowicz and Pinchin 1987; Paroush et al. 1994). Also, the female-specific gene Sex-lethal (Sxl), which is normally repressed by Dpn, is expressed ectopically in male embryos (Younger-Shepherd et al. 1992; Paroush et al. 1994). Finally, the mutant embryos show neural hyperplasia attributable to excessive selection of neural precursors, a process that is normally inhibited by E(spl) (Delidakis et al. 1991; Schrons et al. 1992; Paroush et al. 1994).
Despite the emergence of an increasing number of corepression models, limited information is still available on the specificity of corepressors, that is, whether they mediate the activity of unique repressors/repressor families, or whether they serve as common effectors for different classes of repressors (or even all or most repressors). In the case of TUP1/SSN6, a number of studies suggest that this complex mediates repression by several classes of repressors (Keleher et al. 1992; Treitel and Carlson 1995). In this paper, we analyze the specificity of corepression in a higher eukaryote by examining whether different repressor activities require Gro for their function in Drosophila embryos. Our results indicate that the carboxy-terminal region of Hairy and a repressor domain from the En homeodomain protein both need Gro to function in vivo. In contrast, repressor domains from the Snail (Sna), Eve, Kr, and Knirps (Kni) proteins can act independently of Gro. These results suggest that Gro mediates repression by a subset of negative regulators in Drosophila.
Results
Gro is required for Hairy repression of Sxl transcription
Gro appears to play several roles during early development and it can be difficult to distinguish between its direct and indirect effects in vivo. For example, segmentation in gro mutant embryos is altered dramatically before the action of Hairy (Paroush et al. 1994, 1997), therefore Hairy’s requirement for Gro could be indirect. To partially circumvent these problems we have explored the role of Gro in repression using an assay in which ectopic Hairy interferes with expression of the Sxl sex determination gene (Parkhurst et al. 1990). Sxl transcription is initiated selectively in early female embryos by a set of activator proteins encoded on the X chromosome (Cline 1980, 1988; Bell et al. 1988; Keyes et al. 1992; for review, see Parkhurst and Meneely 1994). In male embryos, Sxl remains off because these activators are outcompeted successfully by autosomal repressors, which include the Hairy-related factor Dpn (Younger-Shepherd et al. 1992; Barbash and Cline 1995). Although Hairy is not normally involved in sex determination, its misexpression at early blastoderm stages mimics Dpn’s activity and inhibits Sxl transcription (Parkhurst et al. 1990). Thus, ectopic Hairy expression driven by the hunchback (hb) gap gene promoter, which is active in the anterior half of the blastoderm embryo at the time of Sxl initiation (nuclear cycles 11–13), leads to repression of Sxl in the anterior of female embryos. This can be monitored by a monoclonal antibody specific for active, full-length Sxl protein, which normally stains only female embryos (Fig. 1A). The hb–h assay offers inherent advantages for exploring the role of Gro in repression by Hairy. First, regulation of Sxl is among the first transcriptional switches in Drosophila, and therefore, is probably independent of other Gro roles in embryonic development. Also, zygotic activity of the hb promoter is largely normal in gro mutant embryos (Fig. 1C), therefore it drives equivalent transgene expression in gro and wild-type embryos.
Figure 1.
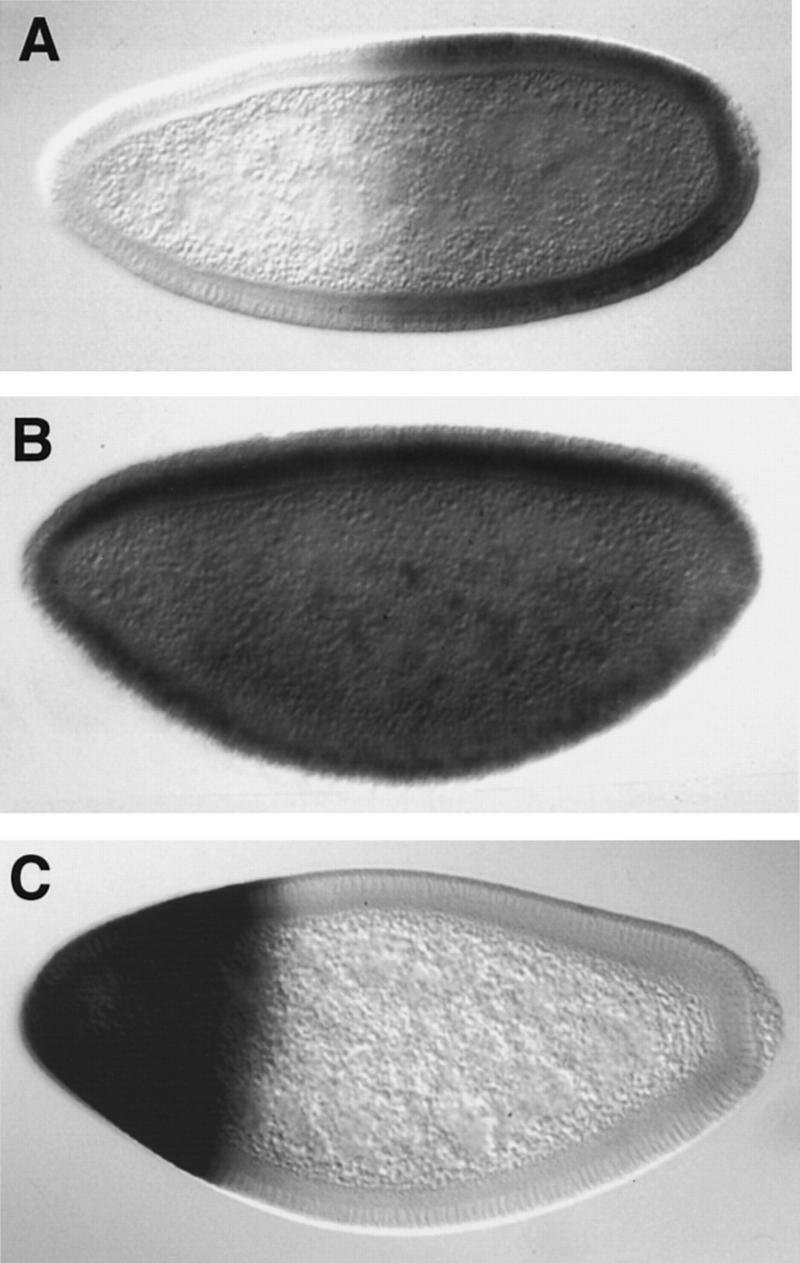
Hairy requires Gro for repression of Sxl. Effect of the hb–h construct in otherwise wild-type (A) or groE48 (B) female blastoderm embryos. Efficient repression at the anterior is observed in the presence but not in the absence of Gro. Embryos were stained with an antibody against active Sxl protein. (C) Pattern of lacZ mRNA expression directed by the hb promoter in gro embryos; efficient activity of the promoter is observed in the anterior region of the embryo. In this and subsequent figures anterior is to the left and dorsal is up.
To examine whether Hairy requires Gro to repress Sxl, we analyzed the effects of ectopic h transcription in embryos deprived of maternal Gro activity. As gro homozygous females are lethal, such embryos were obtained from mosaic females in which all embryos laid derive from gro− germ cells (see Materials and Methods). These experiments involved the groE48 and groE75 alleles, which behave as nulls in genetic tests (Preiss et al. 1988; Delidakis et al. 1991) and Df(3R)BX22, a deficiency that removes gro-coding sequences and adjacent E(spl) genes that are not expressed maternally, and whose loss should not affect early transcriptional regulation (Preiss et al. 1988; Shepard et al. 1989). In general, all three gro alleles yielded equivalent results. If repression of Sxl by Hairy depends on Gro, anterior Sxl expression should be restored in hb–h embryos from gro mutant mothers (hb–h, gro embryos). Indeed, all such embryos express Sxl both anteriorly and posteriorly (Fig. 1B). Thus, Hairy is unable to repress Sxl transcription in gro embryos.
Replacement of the carboxyl terminus of Hairy by the Eve repressor domain generates a Gro-independent repressor
The above experiment argues that Gro is necessary for Hairy to repress Sxl, and is consistent with previous evidence that such repression depends on the WRPW Gro-binding domain (Dawson et al. 1995). This result also offers an assay to test the requirement of Gro for the activity of other repressor domains. We reasoned that replacement of the Hairy carboxy-terminal domain by heterologous repressor domains would generate Hairy repressor derivatives whose repression of Sxl might be either dependent or independent of Gro. To test this idea, we made a HairyEve fusion derivative by replacing the carboxy-terminal 69 amino acids of Hairy by a repression domain from the Eve segmentation protein (EveR; amino acids 140–247; Fig. 2A) that mediates repression in cultured cells when fused to a heterologous DNA-binding domain (Han and Manley 1993a). EveR is believed to act by direct interactions with the basal transcription complex (Austin and Biggin 1995; Um et al. 1995) and therefore is a good candidate for acting independently of Gro.
Figure 2.
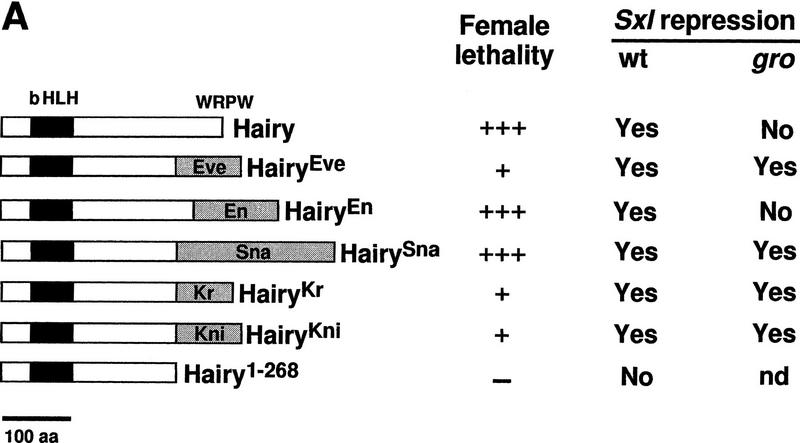
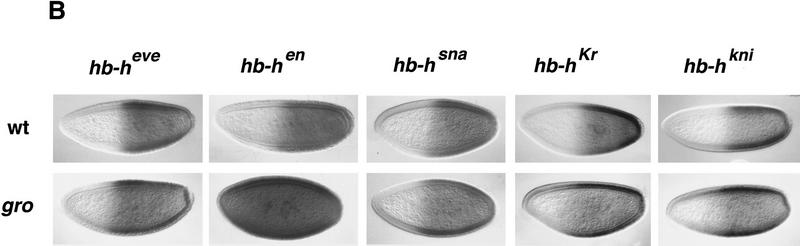
Effects of Hairy repressor chimeras in otherwise wild-type or gro embryos. (A) Diagram of Hairy derivatives expressed under the control of the hb promoter and their effects on female viability and Sxl expression. The degree of female lethality is represented by crosses: (+++) most lines show >80% lethality; (+) most lines show <30% lethality; (−) no lethality detected. (B) Effects on Sxl expression of hb–heve, hb–hen, hb–hsna, hb–hKr, and hb–hkni in otherwise wild-type or groE48 embryos. Repression of Sxl is observed in all cases, except for hb–hen in gro embryos.
We first examined the effects of hb–heve in otherwise wild-type embryos. hb–heve does not cause the high levels of female lethality normally associated with hb–h. Viability of transformant females in several independent lines is typically >80% (Fig. 2A; data not shown). However, three different hb–heve lines examined show effective repression of anterior Sxl (Fig. 2B), similar to that caused by hb–h. In these lines, Sxl expression recovers partially as development proceeds (mainly after stage 9; data not shown), suggesting that residual, undetectable Sxl transcription in early hb–heve embryos is sufficient to trigger Sxl maintenance and rescue most females to adulthood. Repression by HairyEve is dependent on EveR because the equivalent truncated Hairy protein lacking the carboxy-terminal 69 amino acids (Hairy1–268) is inactive in the hb assay (Fig. 2A; data not shown).
To test whether HairyEve is dependent on Gro activity, we monitored its effects on Sxl expression in gro mutant embryos. Anterior Sxl expression is repressed efficiently in hb–heve, gro embryos (Fig. 2B), showing that EveR functions independently of Gro. This result also provides direct evidence that the failure of Hairy to repress Sxl in gro embryos results from a loss of activity of the carboxy-terminal WRPW domain, strongly suggesting that the latter acts through Gro in vivo.
A repression domain from En is Gro dependent
We then tested HairyEn (Fig. 2A), a chimeric protein including Hairy1–286 fused to a repressor domain from the En segmentation protein (amino acids 168–298). This domain (EnR) has been shown previously to mediate potent repression in Drosophila cells and in other cell and embryonic systems (Jaynes and O’Farrell 1991; Han and Manley 1993b; Badiani et al. 1994; John et al. 1995; Conlon et al. 1996; Smith and Jaynes 1996). Indeed, hb–hen behaves as a strong repressor of Sxl; in most hb–hen lines, viability of females drops to <10% of that of males (Fig. 2A; data not shown), and efficient repression is observed in the anterior half of all transformant embryos (Fig. 2B). Strikingly, hb–hen is unable to repress Sxl in groE48 (Fig. 2B), groE75, or BX22 embryos (data not shown). These results indicate that EnR is a second Gro-dependent domain.
We also examined three further repressor domains, from the Sna (amino acids 1–244), Kr (amino acids 26–114), and Kni (amino acids 159–257) proteins that function as negative regulators of transcription in blastoderm embryos (Gerwin et al. 1994; Licht et al. 1994; Arnosti et al. 1996; Gray and Levine 1996a; Fig. 2A). All three fusions have detrimental effects on female viability when expressed from the hb promoter in a wild-type background: HairySna leads to high levels of female lethality (>80%); HairyKr and HairyKni have weaker effects, similar to that of HairyEve (Fig. 2A; data not shown). Nevertheless, all three constructs repress Sxl efficiently in blastoderm embryos (Fig. 2B). All three fusions also repress Sxl in gro mutant embryos (Fig. 2B), indicating that, at least in our assay, none of these repressor domains requires Gro.
Repression of ftz by HairyEn is also Gro dependent
The above experiments indicate that EnR mediates repression of Sxl in a Gro-dependent manner. To test whether such a requirement is specific to Sxl regulation or shared by other promoters we examined the ability of HairyEn to repress ftz. Previous results suggest that ftz is a direct target of repression by Hairy/Gro (Carroll and Scott 1986; Howard and Ingham 1986; Ish-Horowicz and Pinchin 1987; Paroush et al. 1994; Jiménez et al. 1996). We find that ectopic Hairy expression under the control of a heat-shock promoter (hs–h) leads to rapid and efficient repression of ftz transcription in wild-type, but not gro embryos (Fig. 3; Materials and Methods). In contrast, Hairy derivatives lacking the carboxy-terminal WRPW motif are unable to repress ftz in this assay (Jiménez et al. 1996).
Figure 3.
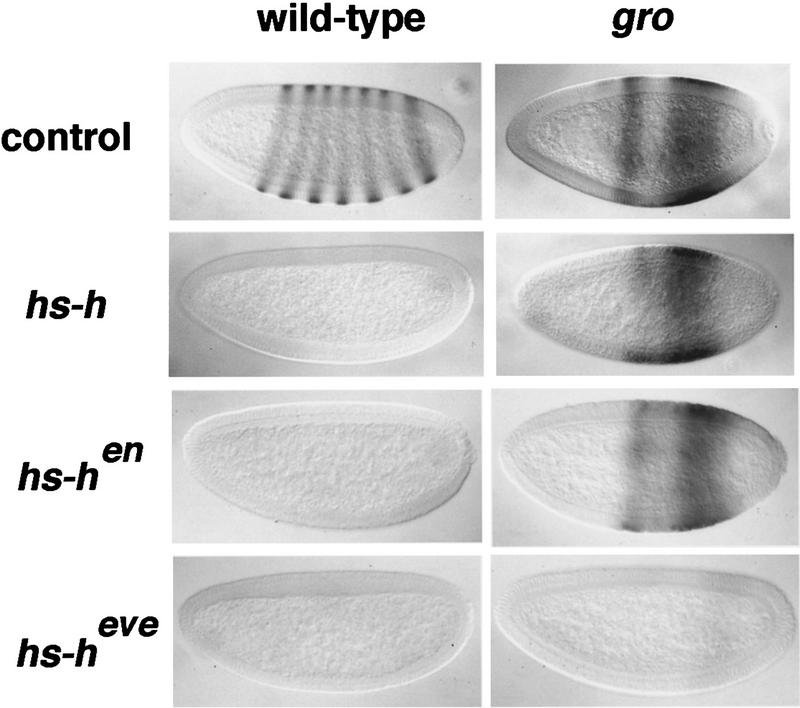
Effects of Hairy, HairyEn, and HairyEve on ftz transcription in the presence and absence of Gro. The Hairy derivatives were expressed in blastoderm embryos under the control of a heat-shock promoter (see Materials and Methods). The normal patterns of ftz mRNA in control wild-type and gro embryos are also shown. Because of the effects of gro on gap gene expression, ftz is not expressed in stripes in gro embryos, but occupies a broad domain in the trunk region. All three heat-shock constructs repress ftz in wild-type embryos, but only hs–heve leads to significant repression in gro embryos. The gro embryos were derived from groBX22 (control and hs–h) or groE48 (hs–hen and hs–heve) mosaic females.
We generated transformant hs–hen lines and monitored ftz expression after heat-shock induction (Materials and Methods). In embryos carrying the hs–hen transgene, ftz expression is completely abolished within 30 min of the start of a 10-min heat shock (Fig. 3), as occurs when wild-type Hairy is expressed ectopically (Ish-Horowicz and Pinchin 1987). However, ftz transcription is unaffected by hs–hen in gro mutant embryos (Fig. 3), indicating that EnR requires Gro to repress both ftz and Sxl. Failure to repress ftz is not attributable to inefficient induction of HairyEn in gro embryos, because the chimeric protein can be detected readily using an antibody against Hairy (data not shown).
In contrast, expression of HairyEve under heat-shock control results in efficient repression of ftz both in wild-type and gro embryos (Fig. 3). These results are consistent with those obtained in the hb assay, and show that the Eve domain can repress different promoters independently of Gro (see Discussion).
Repression of eve by En protein requires Gro
The above results indicate that Gro mediates the activity of EnR. Therefore, we asked whether Gro is required for repression by native En. The En protein functions as a regulator of anteroposterior pattern formation both in the embryo and the adult (for review, see Cohen 1993; Martinez Arias 1993). Although the activity of En as a repressor has been mainly characterized in cultured cells, there is also evidence that it inhibits transcription of various target genes in vivo (Sanicola et al. 1995; Schwartz et al. 1995). eve is one such target because ectopic en expression in blastoderm embryos causes rapid repression of eve (John et al. 1995). Also, after the blastoderm stage eve expression persists for longer than normal in en mutant embryos (Harding et al. 1986). Thus, En is likely to act as a negative regulator of late eve transcription.
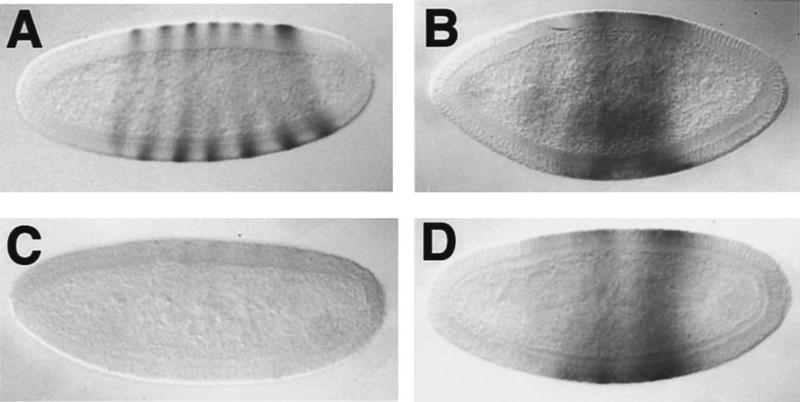
As shown in Figure 4C, ectopic en expression under the control of a heat-shock promoter clearly represses eve in embryos derived from wild-type females (see Materials and Methods). Repression is particularly efficient for eve stripes 2, 5, and 7 and even the more resistant stripe 4 is fully repressed in ∼60% of embryos (101 of 166). In gro embryos, eve transcripts accumulate in one or two broad central domains, not in stripes (Fig. 4B). This expression is not repressed after hs-en induction (Fig. 4D), although efficient accumulation of ectopic En protein is readily detected in those embryos (data not shown). Therefore, Gro is required for En to repress eve transcription.
Figure 4.
Ectopic En represses eve expression in wild-type but not in gro embryos. Wild-type pattern of eve mRNA (A) and its repression in a heat-shocked hs–en embryo (C). Pattern of eve in a control groE48 mutant embryo (B) and in a heat-shocked groE48 embryo carrying the hs–en construct (D); no significant repression is observed. As in the case of ftz, eve transcripts are not expressed in stripes in gro embryos, but accumulate in one or two broad central domains.
HairyEn binds to Gro in vitro
One mechanism whereby En might direct Gro-dependent repression is if the EnR repression domain binds Gro directly and recruits it to target promoters. Therefore, we compared the ability of full-length Hairy and HairyEn to bind to Gro in vitro. The Hairy derivatives were expressed in bacteria as glutathione S-transferase (GST) fusions, immobilized on glutathione–Sepharose beads, and tested for their ability to retain radiolabeled Gro protein. Both GST–Hairy and GST–HairyEn bind Gro, whereas control GST fusions (two different Hairy carboxy-terminal truncations lacking the WRPW tetrapeptide and the HairyEve and HairySna chimeras), show little or no binding (Fig. 5A). These results suggest that the EnR domain inhibits transcription by direct interactions with Gro.
Figure 5.
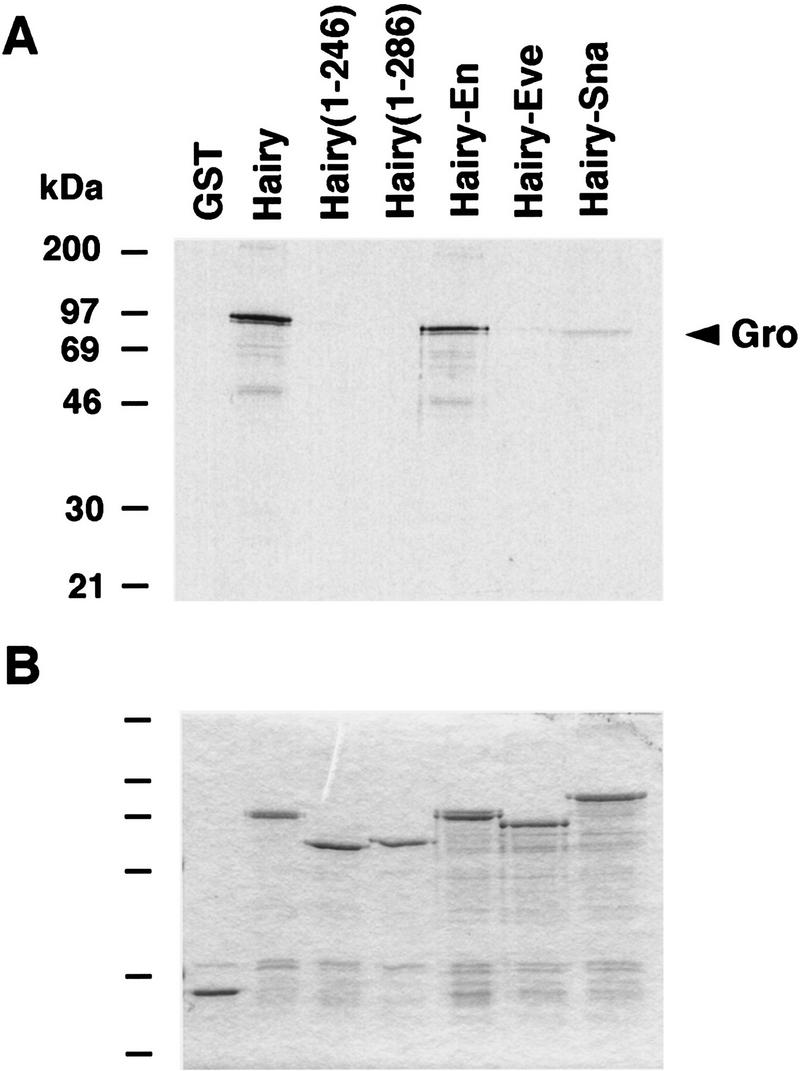
Gro binds in vitro to HairyEn. (A) Radiolabeled Gro protein was incubated with various GST–Hairy derivatives bound to glutathione–Sepharose beads. After washing the beads, the retained Gro protein was visualized by SDS-PAGE and autoradiography. (B) Coomassie staining showing the integrity of the different GST fusions after incubation with Gro.
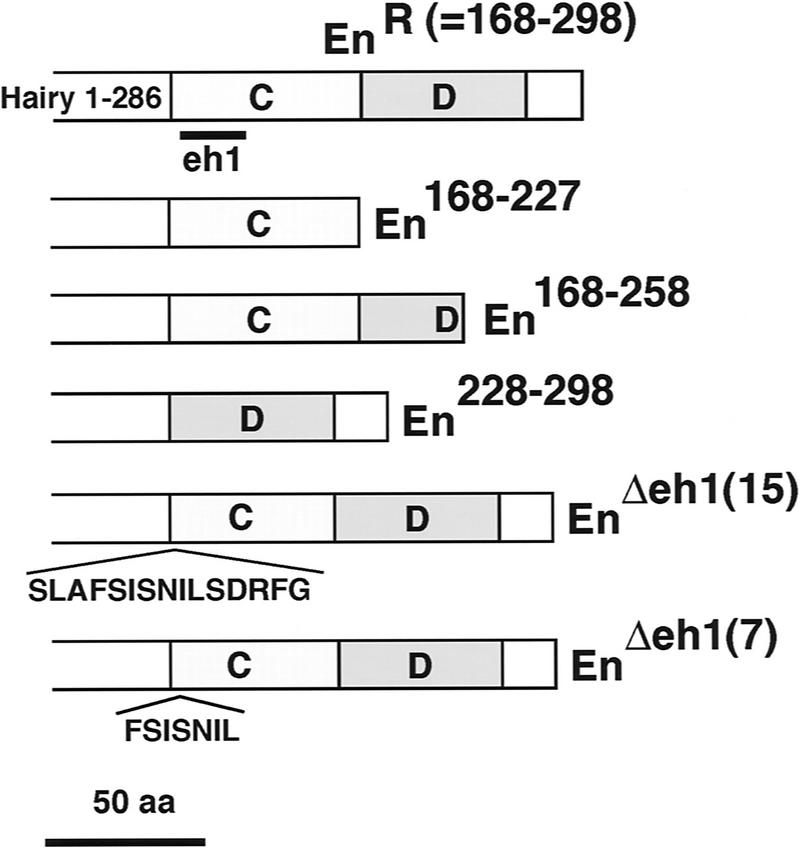
EnR does not contain a recognizable WRPW-like sequence, suggesting that its binding to Gro is mediated by other protein motifs. To begin the identification of such motifs, we tested several HairyEn derivatives with deletions in the EnR domain for their ability to bind to Gro in vitro. The original EnR domain used in our experiments contains two subdomains (regions C and D, see Fig. 6) that were initially characterized by Han and Manley (1993b) in cultured cells. These researchers found a stronger repressor activity for region D than for region C. However, recent experiments using an assay in blastoderm embryos indicate that region C is more active than region D in vivo (Smith and Jaynes 1996). We find that HairyEn proteins lacking region D are able to bind to Gro with the same affinity as the intact HairyEn chimera (Fig. 6). In contrast, deletion of region C results in a fourfold decrease in binding activity. These results indicate that although region D can contribute to the interaction, most of the binding activity resides within region C. Because the study of Smith and Jaynes (1996) suggested that most of the function of the C domain resides in a conserved element (called eh1), present in other homeodomain proteins such as Goosecoid, we examined the effects of removing this element in our binding assay. As shown in Figure 6, two different deletions of the eh1 motif result in a decrease in Gro binding comparable to that caused by removal of the whole C region. These results point to a important role of the eh1 region in the ability of EnR to bind to Gro. Moreover, the degree of Gro binding for the different constructs correlates well with their in vivo repressor activity as determined by Smith and Jaynes (1996), suggesting that the eh1/Gro interaction is indeed functionally significant.
Figure 6.
Mapping of sequences within EnR responsible for binding to Gro. A diagram of different EnR deletion mutants is shown. These mutations were introduced in the original GST–HairyEn construct and examined for their ability to bind full-length Gro. Deletions of region D (constructs En168–227 and En168–258) do not affect the interaction with Gro. In contrast, deletion of region C (construct En228–298) causes a fourfold decrease in the binding. A similar result is obtained after eliminating 15 or 7 amino acids [EnΔeh1(15) or EnΔeh1(7), respectively] that constitute the conserved eh1 motif (Smith and Jaynes 1996).
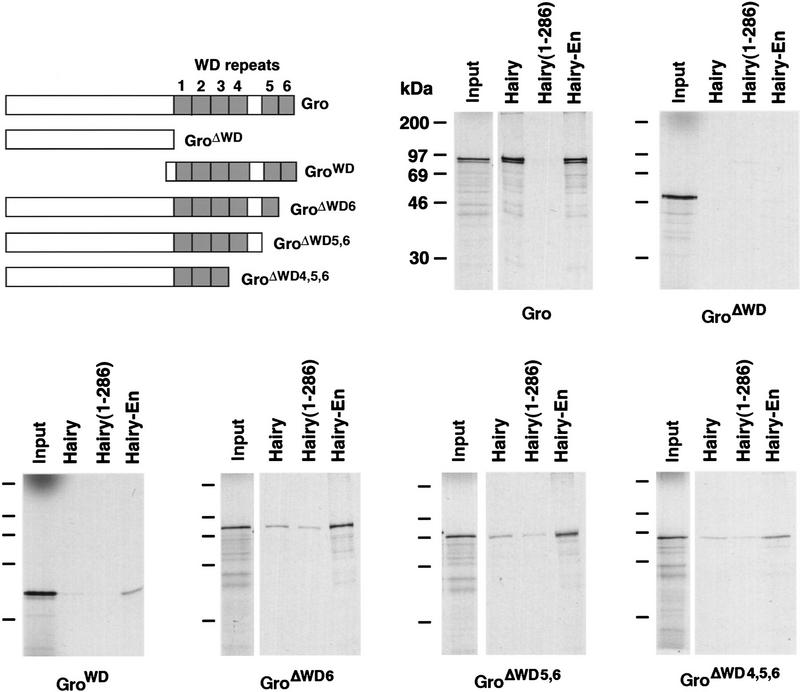
The ability of Gro to bind to the Hairy and En repressor domains raises the question of whether these associations are mediated by a single or different regions of Gro. Therefore, we compared the binding in vitro of various Gro deletion derivatives to Hairy and HairyEn. We first examined two Gro mutants lacking either the carboxy-terminal half of the protein (including the WD repeats) or the complementary amino-terminal domain (GroΔWD and GroWD, respectively; see Fig. 7). Both mutants show a dramatic loss of binding activity to Hairy and HairyEn, although we still detect weak binding of GroWD to HairyEn (Fig. 7). These results suggest that both halves of Gro are required for full interaction with both repressor domains. These findings differ from our previous result that a central domain of Gro, lacking all WD sequences, is sufficient for the interaction with Hairy in yeast (Paroush et al. 1994). Although we cannot explain this difference, the stronger and more specific binding of Gro to Hairy in vitro than in yeast (Paroush et al. 1994) favors a role for the WD repeats in this interaction (see below).
Figure 7.
Binding of Hairy and HairyEn to Gro deletion mutants. A diagram of the different Gro derivatives analyzed is shown. These derivatives were assayed for binding to Hairy, Hairy1–286 (a truncation similar to HairyEn but lacking EnR), and HairyEn. Equivalent aliquots of labeled Gro derivatives are shown as a control for the strength of the interactions (Input lanes). GroΔWD and GroWD show weak or no binding to either Hairy or HairyEn. Gro derivatives with progressive WD repeat deletions also show very weak binding to Hairy, but GroΔ6 and GroΔ5,6 interact significantly with HairyEn.
To test further the requirement of the WD region for binding to Hairy and HairyEn, we generated three additional Gro derivatives with progressive deletions of carboxy-terminal WD repeats (Fig. 7). All three derivatives show very weak interactions with full-length Hairy, similar to that of the Hairy1–286 negative control, again suggesting that the integrity of the WD region is essential for binding to Hairy. In contrast, GroΔWD6 and GroΔWD5,6 (lacking the sixth, and the fifth and sixth WD repeats, respectively) retain significant binding to HairyEn. These results suggest that these two repeats contribute toward, but are not essential for binding to EnR, and are consistent with the existence of different structural requirements for binding of Gro to Hairy and En repressor domains.
Discussion
A few studies suggest that active repression often involves corepressors. These proteins are believed to act as molecular bridges between repressor domains and target proteins such as components of the basal transcription complex. A prevailing question is whether corepressors act in concert with many or only a few repressors. In this paper, we provide the first systematic attempt to assess the specificity of a corepressor in higher eukaryotes. We find that Gro is not only required for repression by Hairy, but that it also mediates specifically the activity of a domain from En. In contrast, repressor domains from the Sna, Eve, Kr, and Kni proteins are active in the absence of Gro. These results argue that Gro is required for the activity of a subset of repressors in Drosophila.
Our results show that ectopic Hairy fails to repress either Sxl or ftz transcription in gro mutant embryos. In contrast, Hairy chimeric proteins in which the Gro-binding WRPW motif is substituted by alternative repressor domains can act independently of Gro. Thus, Hairy is able to direct repression through the Eve, Sna, Kr, or Kni repressor domains in gro mutant embryos, but not through its own WRPW motif. These results render unlikely the explanation that Hairy requires Gro directly or indirectly to bind to target promoters (see also Jiménez et al. 1996). Together with the in vitro results showing direct interactions between the WRPW motif and Gro, our results indicate that Gro functions as an authentic corepressor for Hairy (Paroush et al. 1994).
Recently, Aronson et al. (1997) have provided evidence that Runt, a transcriptional regulator unrelated to Hairy but that contains at its carboxyl terminus a WRPY sequence reminiscent of the WRPW motif, also interacts with Gro through this sequence. Thus, Gro also mediates some of Runt’s repressor activities during segmentation.
The Eve, Sna, Kr, and Kni domains must have a mechanism of action that is independent of Gro. In contrast, EnR appears functionally linked to Gro, as HairyEn represses transcription efficiently in wild-type but not gro mutant embryos. Indeed, native En requires Gro to repress eve in vivo and we have shown that EnR is able to bind Gro in vitro. These results suggest that, like the WRPW motif, EnR functions by recruiting Gro to target promoters. This association depends on the eh1 motif, which is conserved within En homologous genes, essential for repression in vivo, and also present in other homeodomain proteins from various organisms (see Smith and Jaynes 1996). Thus, these proteins may be transcriptional repressors that interact with Gro or its homologs in other animals.
Our results indicate that Gro acts as a multifunctional protein that integrates signals from different transcriptional repressors. These repressors can be unrelated structurally, as there are no significant similarities between Hairy and En, either within or outside their repressor domains. This illustrates that repressor domains classified apart according to their structure may have a common mechanism of action. Similarly, TUP1 appears to mediate repression by different classes of repressors such as α2 and MIG1 (Keleher et al. 1992; Treitel and Carlson 1995). Conversely, repressor domains with shared features may have different targets; En and Eve are both homeodomain proteins with alanine-rich repressor domains, yet only the former acts through Gro.
The eh1 and WRPW motifs appear unrelated, suggesting that the associations of these repressor domains with Gro involve different molecular interfaces. We addressed this issue by examining which regions in Gro mediate its binding to Hairy and HairyEn. Our results show that sequences both within and outside the WD repeats are important for each of these interactions. However, Hairy and EnR appear to show different requirements for the WD repeats. The most carboxy-terminal fifth and sixth WD repeats are necessary for binding to Hairy, but are partially dispensable for the interaction with EnR. Thus, Hairy must either bind the sixth repeat or require the integrity of the whole WD region. Crystal structure analyses of other WD proteins have shown that this region adopts a complex conformation called a β-propeller (Sondek et al. 1996; Wall et al. 1996), and perhaps Hairy recognizes features characteristic of such a structure. In contrast, EnR may target Gro motifs that do not necessarily need to be assembled into a full β-propeller.
Gro is probably only essential for some En functions. Thus, reduced or absent gro activity in the posterior compartment of imaginal discs does not seem to recapitulate the phenotype of en loss-of-function mutations (de Celis and Ruiz-Gómez 1995; Heitzler et al. 1996), suggesting that Gro may be dispensable for repression by En in some adult tissues. Similarly, Runt appears to repress at least one target gene independently of Gro (Aronson et al. 1997).
How might Gro mediate repression? We favor a model in which Gro is recruited to target promoters by specific DNA-binding proteins and then represses transcription (Paroush et al. 1994). This view is supported by recent experiments showing that Gro can mediate significant levels of repression in cultured cells when targeted to a promoter through a heterologous DNA-binding domain (Fisher et al. 1996). An alternative model is that Gro is a component of the basal transcriptional machinery and functions as a target for repressors such as Hairy or En. This model appears plausible in light of the finding that dTAF80, a Drosophila WD repeat protein that may be required for repression, is a component of the general TFIID complex (Dynlacht et al. 1993; Kokubo et al. 1993). However, studies on polytene chromosomes show that Gro localizes to a subset of chromosome bands, suggesting that it is not present at all promoters (Bettler et al. 1996). Although these results do not exclude the possibility that Gro is a component of the transcription complex assembled only in a subset of promoters, they favor the recruitment model of Gro action.
Once recruited to target promoters, Gro could interfere with transcription in several ways. The simplest possibility is that Gro interacts directly with components of the basal transcriptional machinery to disrupt their assembly, structure, or accessibility. A particular aspect of this model is that Gro might interfere with components of the transcriptional machinery specifically required for transcriptional activation [e.g., the TATA-binding protein (TBP)-associated factors (TAFs); for review, see Verrijzer and Tjian 1996]. It has been shown that in cultured cells EnR appears to mediate strong repression of promoters stimulated by activator proteins, while being unable to inhibit transcription of basal promoters (Han and Manley 1993b). Thus, these effects may reflect the mechanism of Gro action, and may also be a feature of repression by Hairy. Alternatively, Gro could inhibit transcription by “quenching” (i.e., by interacting with a promoter-bound activator and interfering with its ability to contact the basal machinery). Finally, it is possible that Gro acts by influencing chromatin organization directly. Another WD-containing protein, Extra-sex-combs, is thought to act as a repressor by altering chromatin structure (Gutjahr et al. 1995; Sathe and Harte 1995; Simon et al. 1995), and repression by TUP1 appears to rely on interactions with histones H3 and H4 (Edmondson et al. 1996). Moreover, a series of recent experiments suggest that the corepressor complex mRPD/mSin3/N-CoR modulates chromatin conformation through histone deacetylation (for review, see Pazin and Kadonaga 1997; Wolffe 1997). Future biochemical approaches will address whether Gro uses these or other activities to modify chromatin behavior.
Previous work has led to a distinction between repressors that need to lie close (<150 bp) to activators or the basal complex to function (short-range repressors) and those that can act over longer distances (long-range repressors; Gray and Levine 1996b). It has been suggested that this difference may result in distinct modes of regulation of a complex promoter. Short-range repressors could act locally to inhibit a particular enhancer, while leaving other enhancers free to activate expression; in contrast, long-range repressors would act dominantly to ensure that the promoter is repressed completely (Gray et al. 1994; Arnosti et al. 1996; Gray and Levine 1996a; Barolo and Levine 1997). We find that three repressor domains that come from short-range repressors (Sna, Kr, and Kni) all act independently of Gro. In contrast, recent experiments show that Hairy behaves as a long-range repressor on synthetic promoters (Barolo and Levine 1997), an effect that should depend on Gro. These results suggest that Gro may be a long-range, dominant corepressor. However, the HairySna, HairyKr, and HairyKni chimeras (in which Hairy has presumably been converted from a long- to short-range repressor) are still able to repress the endogenous Sxl promoter. These results indicate that short- and long-range repressors are to some extent interchangeable and do not necessarily exert different modes of promoter regulation.
In conclusion, our results indicate that Gro is not generally required for repression, but acts as a dedicated corepressor for a subset of negative regulators, including Hairy and En. The evolutionary conservation of Gro, Hairy, and En suggests that similar repressor/corepressor associations will also operate in vertebrates. Indeed, EnR is much used as a portable repressor domain in a variety of heterologous vertebrate systems (for example, see Badiani et al. 1994; Conlon et al. 1996), and it will be interesting to determine whether its activity in vertebrates is also Gro dependent. In yeast, the TUP1 protein is necessary for repression of a large number of genes, suggesting that it also acts in concert with different classes of negative regulators. These observations raise the possibility that at least a fraction of the repressors present in eukaryotes act through a relatively small number of corepressors. The characterization of repressor/corepressor interactions should be particularly fruitful in the search for common themes in transcriptional repression.
Materials and methods
Plasmids
Plasmid manipulations were performed according to standard protocols (Ausubel et al. 1987–97; Sambrook et al. 1989). The sequences encoding the different Hairy derivatives (except HairyEn) were first prepared in pBluescript (Stratagene) by inserting PCR fragments encoding the repressor domains as BamHI–XbaI fragments downstream of the unique BamHI site in the h cDNA. The hen fusion was made by first cloning a HindII–BamHI fragment from the en cDNA upstream of a synthetic BamHI–XbaI linker containing an in-frame stop codon. This was followed by the insertion of a HindIII–PvuII h cDNA fragment upstream of the en HindII site. All chimeric sequences were recovered as BstEII–XbaI fragments for cloning into pCaSpeR4. The hb–h plasmid is described in Parkhurst et al. (1990); all other hb–h derivatives were derived from it but constructed in the pCaSpeR4 “mini”-white vector (Pirrotta 1988). These plasmids include a 750-bp NheI–XbaI fragment from the hb gene (which includes the proximal promoter and all but 10 bp from the 5′-untranslated region), a 230-bp XbaI–BstEII fragment from the h 5′ leader, Hairy fusion sequences (as BstEII–XbaI fragments), and finally a 1.6-kb XbaI–EcoRV fragment from the h 3′-untranslated sequence. The XbaI sites in the hb and h 5′ leaders were lost during cloning (see Parkhurst et al. 1990). The unique XbaI site between the h coding and the 3′-end sequences was created during the cloning procedure. Final constructs were generated by assembling hb–h sequences between the NheI and HpaI sites of pCaSpeR4.
The hs–heve construct was made by cloning the heve sequences as a HindII–XbaI fragment in pCaSpeR-hs (Pirrotta 1988) digested with HpaI and XbaI. hs–hen was constructed by replacing a NotI–XbaI fragment in hs–h1–268-VP16 (containing Hairy amino acids 246–268 and the VP16 activation domain; Jiménez et al. 1996) by the equivalent NotI–XbaI hen fragment.
Plasmids encoding GST–Hairy and GST–Hairy1–246 (=HairyΔNotI) were described in Paroush et al. (1994). Plasmids for all other GST chimeras were made by cloning in-frame the relevant Hairy-derived sequences into pZEX, a modification of pGEX-2T (Smith and Johnston 1988) containing the following polylinker cloning sites: EcoRI, SmaI, BamHI, and XhoI.
pET–Gro plasmids were made by cloning full-length or partial gro sequences into pET-17b and pET-3a.
Additional details on the construction of the plasmids are available on request.
Germ-line transformation
P-element-mediated transformation was performed as described previously (Steller and Pirrotta 1985; Spradling 1986), selecting for G418-resistance (hb–h) or rescue of w eyes (all other constructs). In general, two or more independent lines were analyzed for each construct.
In the case of hb constructs, insertions on the X chromosome were maintained in males using an attached X chromosome [C(1)M3]; insertions on the autosomes were kept as unbalanced stocks selecting each generation for transformant males and nontransformant females. To analyze the effects of hb–h and hb–hen on gro embryos, we crossed mosaic gro females to males carrying the hb construct on the X chromosome, therefore all gro female embryos inherit the transgene.
For the experiments with hs–hen and hs–heve, we recombined two independent insertions of each construct into the same chromosome. Males carrying these recombinant chromosomes were then crossed to wild-type or mosaic gro females. To test the effects of hs–h, we used males carrying an insertion on the X chromosome, HSH33, and homozygous for another insertion on the second, HSH21 (Ish-Horowicz and Pinchin 1987).
The hs-en flies were a kind gift of A. Jacinto and P. Ingham (University of Sheffield, UK).
Germ-line clones and embryo analysis
Embryos deprived of maternal gro function were obtained using the ovoD-FLP-FRT system (Chou et al. 1993). Briefly, males of the genotype hs-FLP1/Y; FRT[82B] ovoD1/Sb were mated to FRT[82B] gro/TM3 females, and 2- to 4-day-old progeny heat-shocked daily for 4 hr at 37°C for the following 3–4 days. Eclosed TM3+ Sb+ virgin females were crossed to males carrying the hb and hs constructs. The progeny of these crosses was examined to confirm the expected lethality of gro mutant embryos.
In the experiments examining repression of Sxl, embryos were dechorionated 130–190 min after egg laying (AEL), and fixed for 12–15 min in heptane/4% formaldehyde/PBS. The embryos were stained with a monoclonal antibody specific for the active form of Sxl (Bopp et al. 1991).
For the heat-shock experiments, embryos between 140 and 170 min AEL were heat shocked for 10–12 min at 36.5°C on wet tissue inside a prewarmed container in a water bath, a protocol that does not cause significant effects on wild-type embryos (Ish-Horowicz and Pinchin 1987). After heat shock, the embryos were allowed to develop for 20–25 min, dechorionated, and fixed as above for 15–20 min. In the case of stainings with antibodies against Hairy (gift of S.M. Pinchin, this laboratory) and En (Patel et al. 1989), embryos were fixed for 12–15 min, 15 min after the heat shock. Whole-mount in situ hybridizations were performed using digoxygenin- and fluorescein-labeled RNA probes (Boehringer) as described (Tautz and Pfeifle 1989; Klingler and Gergen 1993). In all cases, signals were detected using secondary antibodies coupled to alkaline phosphatase (Jackson Immunoresearch Laboratories, Boehringer), and embryos were mounted in methacrylate (JB-4, Polyscience) and examined under Nomarski optics.
In vitro protein–protein interactions
GST fusions were expressed and purified essentially as described previously (Paroush et al. 1994) in the protease-deficient Escherichia coli strain SRP84 (gift of C. Higgins, ICRF). Binding assays were performed with equal amounts of fusion proteins (30 μl of beads supplemented when necessary with beads from a blank bacterial extract) and 20–30 μl of 35S-labeled Gro protein synthesized using the TNT-coupled rabbit reticulocyte lysate system (Promega), in 180 μl of binding buffer [20 mm HEPES-KOH (pH 7.9), 50 mm KCl, 2.5 mm MgCl2, 10% glycerol, 1 mm DTT, 0.2% NP-40] supplemented with 3 μl of rabbit serum and 3 μl of a 100 mm PMSF stock. Binding reactions were rolled overnight at 4°C, and washed four times with 1 ml of RIPA buffer [10 mm Tris-HCl (pH7.5), 150 mm NaCl, 1 mm EDTA, and 0.2% NP-40]. The beads were boiled for 1 min in sample buffer and aliquots examined by electrophoresis, followed by Coomassie staining to confirm the integrity of the GST fusions, and autoradiography to detect bound Gro protein. The extent of Gro binding to mutant EnR domains was quantified by a PhosphorImager (Molecular Dynamics).
Acknowledgments
We thank members of the Developmental Genetics Laboratory (especially M. Wainwright and S. Pinchin) for their help and encouragement. We are also grateful to P. Verrijzer for helpful comments on the manuscript, and to A. Jacinto, P. Ingham, D. Hartley, and M. Levine for fly stocks and plasmids. G.J. was supported by the European Community Human Capital and Mobility Program, the Imperial Cancer Research Fund, and the European Molecular Biology Organization. Z.P. was supported by the European Science Foundation, a Research Career Development Award of the Israel Cancer Research Fund, and by a research grant from the Israel Academy of Sciences and Humanities. This work was supported by the Imperial Cancer Research Fund. D.I.-H. is a Howard Hughes International Research Scholar.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL d.horowicz@icrf.icnet.uk; FAX +44-171-269-3417.
References
- Alland L, Muhle R, Hou H, Potes J, Chin L, Schreiberagus N, Depinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Arnosti DN, Gray S, Barolo S, Zhou J, Levine M. The gap protein Knirps mediates both quenching and direct repression in the Drosophila embryo. EMBO J. 1996;15:3659–3666. [PMC free article] [PubMed] [Google Scholar]
- Aronson BD, Fisher AL, Blechman K, Caudy M, Gergen J P. Groucho-dependent and -independent repression activities of Runt domain proteins. Mol Cell Biol. 1997;17:5581–5587. doi: 10.1128/mcb.17.9.5581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin RJ, Biggin MD. A domain of the Even-skipped protein represses transcription by preventing TFIID binding to a promoter: Repression by cooperative blocking. Mol Cell Biol. 1995;15:4683–4693. doi: 10.1128/mcb.15.9.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel, F.J., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl, eds. 1987–97. In Current protocols in molecular biology. Greene Publishing Associates and Wiley-Interscience, New York, NY.
- Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes & Dev. 1994;8:770–782. doi: 10.1101/gad.8.7.770. [DOI] [PubMed] [Google Scholar]
- Barbash DA, Cline TW. Genetic and molecular analysis of the autosomal component of the primary sex determination signal of Drosophila melanogaster. Genetics. 1995;141:1451–1471. doi: 10.1093/genetics/141.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barolo S, Levine M. Hairy mediates dominant repression in the Drosophila embryo. EMBO J. 1997;16:2883–2891. doi: 10.1093/emboj/16.10.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell LR, Maine EM, Schedl P, Cline TW. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific splicing and sequence similarity to RNA binding proteins. Cell. 1988;55:1037–1046. doi: 10.1016/0092-8674(88)90248-6. [DOI] [PubMed] [Google Scholar]
- Bettler D, Pearson S, Yedvobnick B. The nuclear protein encoded by the Drosophila neurogenic gene mastermind is widely expressed and associates with specific chromosomal regions. Genetics. 1996;143:859–875. doi: 10.1093/genetics/143.2.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E, Vässin H, Younger-Shepherd S, Jan LY, Jan YN. deadpan, an essential pan-neural gene in Drosophila, encodes a helix–loop–helix protein similar to the hairy gene product. Genes & Dev. 1992;6:2137–2151. doi: 10.1101/gad.6.11.2137. [DOI] [PubMed] [Google Scholar]
- Bopp D, Bell LR, Cline TW, Schedl P. Developmental distribution of female-specific Sex-lethal proteins in Drosophila melanogaster. Genes & Dev. 1991;5:403–415. doi: 10.1101/gad.5.3.403. [DOI] [PubMed] [Google Scholar]
- Carroll SB, Scott MP. Zygotically active genes that affect the spatial expression of the fushi tarazu segmentation gene during early Drosophila embryogenesis. Cell. 1986;45:113–126. doi: 10.1016/0092-8674(86)90543-x. [DOI] [PubMed] [Google Scholar]
- Chou TB, Noll E, Perrimon N. Autosomal P[ovoD1] dominant female-sterile insertions in Drosophila and their use in generating germ-line chimeras. Development. 1993;119:1359–1369. doi: 10.1242/dev.119.4.1359. [DOI] [PubMed] [Google Scholar]
- Cline TW. Maternal and zygotic sex-specific gene interactions in Drosophila melanogaster. Genetics. 1980;96:903–926. doi: 10.1093/genetics/96.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Evidence that sisterless-a and sisterless-b are two of several discrete “numerator elements” of the X/A sex determination signal in Drosophila that switch Sxl between two alternative stable expression states. Genetics. 1988;119:829–862. doi: 10.1093/genetics/119.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen SM. Imaginal disc development. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 747–842. [Google Scholar]
- Conlon FL, Sedgwick SG, Weston KM, Smith JC. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development. 1996;122:2427–2435. doi: 10.1242/dev.122.8.2427. [DOI] [PubMed] [Google Scholar]
- Cowell IG. Repression versus activation in the control of gene-transcription. Trends Biochem Sci. 1994;19:38–42. doi: 10.1016/0968-0004(94)90172-4. [DOI] [PubMed] [Google Scholar]
- Dawson SR, Turner DL, Weintraub H, Parkhurst SM. Specificity for the Hairy/Enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol Cell Biol. 1995;15:6923–6931. doi: 10.1128/mcb.15.12.6923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Celis JF, Ruiz-Gómez M. groucho and hedgehog regulate engrailed expression in the anterior compartment of the Drosophila wing. Development. 1995;121:3467–3476. doi: 10.1242/dev.121.10.3467. [DOI] [PubMed] [Google Scholar]
- Delidakis C, Artavanis-Tsakonas S. The Enhancer-of-split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc Natl Acad Sci. 1992;89:8731–8735. doi: 10.1073/pnas.89.18.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delidakis C, Preiss A, Hartley DA, Artavanis-Tsakonas S. Two genetically and molecularly distinct functions involved in early neurogenesis reside within the Enhancer-of-split locus of Drosophila melanogaster. Genetics. 1991;129:803–823. doi: 10.1093/genetics/129.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynlacht BD, Weinzierl RO, Admon A, Tjian R. The dTAFII80 subunit of Drosophila TFIID contains β-transducin repeats. Nature. 1993;363:176–179. doi: 10.1038/363176a0. [DOI] [PubMed] [Google Scholar]
- Edmondson DG, Smith M, Roth SY. Repression domain of the yeast global repressor TUP1 interacts directly with histones H3 and H4. Genes & Dev. 1996;10:1247–1259. doi: 10.1101/gad.10.10.1247. [DOI] [PubMed] [Google Scholar]
- Fisher AL, Ohsako S, Caudy M. The WRPW motif of the Hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol Cell Biol. 1996;16:2670–2677. doi: 10.1128/mcb.16.6.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwin N, La Rosee A, Sauer F, Halbritter HP, Neumann M, Jackle H, Nauber U. Functional and conserved domains of the Drosophila transcription factor encoded by the segmentation gene knirps. Mol Cell Biol. 1994;14:7899–7908. doi: 10.1128/mcb.14.12.7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray S, Levine M. Short-range transcriptional repressors mediate both quenching and direct repression within complex loci in Drosophila. Genes & Dev. 1996a;10:700–710. doi: 10.1101/gad.10.6.700. [DOI] [PubMed] [Google Scholar]
- ————— Transcriptional repression in development. Curr Opin Cell Biol. 1996b;8:358–364. doi: 10.1016/s0955-0674(96)80010-x. [DOI] [PubMed] [Google Scholar]
- Gray S, Szymanski P, Levine M. Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes & Dev. 1994;8:1829–1838. doi: 10.1101/gad.8.15.1829. [DOI] [PubMed] [Google Scholar]
- Gutjahr T, Frei E, Spicer C, Baumgartner S, White RA, Noll M. The Polycomb-group gene, extra sex combs, encodes a nuclear member of the WD-40 repeat family. EMBO J. 1995;14:4296–4306. doi: 10.1002/j.1460-2075.1995.tb00104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Manley JL. Transcriptional repression by the Drosophila even-skipped protein: Definition of a minimal repression domain. Genes & Dev. 1993a;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- ————— Functional domains of the Drosophila Engrailed protein. EMBO J. 1993b;12:2723–2733. doi: 10.1002/j.1460-2075.1993.tb05934.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna-Rose W, Hansen U. Active repression mechanisms of eukaryotic transcription repressors. Trends Genet. 1996;12:229–234. doi: 10.1016/0168-9525(96)10022-6. [DOI] [PubMed] [Google Scholar]
- Harding K, Rushlow C, Doyle HJ, Hoey T, Levine M. Cross regulatory interactions among pair-rule genes in Drosophila. Science. 1986;233:953–959. doi: 10.1126/science.3755551. [DOI] [PubMed] [Google Scholar]
- Hartley DA, Preiss A, Artavanis-Tsakonas S. A deduced gene product from the Drosophila neurogenic locus, Enhancer-of-split, shows homology to mammalian G-protein beta subunit. Cell. 1988;55:785–795. doi: 10.1016/0092-8674(88)90134-1. [DOI] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG. A complex containing N-CoR, Msin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and Achaete-Scute complexes are required for a regulatory loop between Notch and Delta during lateral signaling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- Horlein AJ, Naar AM, Heinzel T, Torchia J, Gloss B, Kurokawa R, Ryan A, Kamei Y, Soderstrom M, Glass CK, Rosenfeld MG. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature. 1995;377:397–404. doi: 10.1038/377397a0. [DOI] [PubMed] [Google Scholar]
- Howard K, Ingham P. Regulatory interactions between the segmentation genes fushi tarazu, hairy and engrailed in the Drosophila blastoderm. Cell. 1986;44:949–957. doi: 10.1016/0092-8674(86)90018-8. [DOI] [PubMed] [Google Scholar]
- Ip YT, Hemavathy K. Drosophila development—delimiting patterns by repression. Curr Biol. 1997;7:R216–R218. doi: 10.1016/s0960-9822(06)00104-7. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D, Pinchin SM. Pattern abnormalities induced by ectopic expression of the Drosophila gene hairy are associated with repression of fushi tarazu transcription. Cell. 1987;51:405–415. doi: 10.1016/0092-8674(87)90636-2. [DOI] [PubMed] [Google Scholar]
- Jaynes JB, O’Farrell PH. Active repression of transcription by the engrailed homeodomain protein. EMBO J. 1991;10:1427–1433. doi: 10.1002/j.1460-2075.1991.tb07663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez G, Pinchin SM, Ish-Horowicz D. In vivo interactions of the Drosophila Hairy and Runt transcriptional repressors with target promoters. EMBO J. 1996;15:7088–7098. [PMC free article] [PubMed] [Google Scholar]
- John A, Smith ST, Jaynes JB. Inserting the ftz homeodomain into engrailed creates a dominant transcriptional repressor that specifically turns off ftz target genes in vivo. Development. 1995;121:1801–1813. doi: 10.1242/dev.121.6.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AD. The price of repression. Cell. 1995;81:655–658. doi: 10.1016/0092-8674(95)90524-3. [DOI] [PubMed] [Google Scholar]
- Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- Keyes LN, Cline TW, Schedl P. The primary sex determination signal of Drosophila acts at the level of transcription. Cell. 1992;68:933–943. doi: 10.1016/0092-8674(92)90036-c. [DOI] [PubMed] [Google Scholar]
- Klämbt C, Knust E, Tietze K, Campos-Ortega JA. Closely related transcripts encoded by the neurogenic gene complex Enhancer of split of Drosophila melanogaster. EMBO J. 1989;8:203–210. doi: 10.1002/j.1460-2075.1989.tb03365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingler M, Gergen JP. Regulation of runt transcription by Drosophila segmentation genes. Mech Dev. 1993;43:3–19. doi: 10.1016/0925-4773(93)90019-t. [DOI] [PubMed] [Google Scholar]
- Knust E, Schrons H, Grawe F, Campos-Ortega JA. Seven genes of the Enhancer of split complex of Drosophila melanogaster encode helix-loop-helix proteins. Genetics. 1992;132:505–518. doi: 10.1093/genetics/132.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo T, Gong DW, Yamashita S, Takada R, Roeder RG, Horikoshi M, Nakatani Y. Molecular cloning, expression, and characterization of the Drosophila 85-kilodalton TFIID subunit. Mol Cell Biol. 1993;13:7859–7863. doi: 10.1128/mcb.13.12.7859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld MG, Glass CK. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- Licht JD, Hanna-Rose W, Reddy JC, English MA, Ro M, Grossel M, Shaknovich R, Hansen U. Mapping and mutagenesis of the amino-terminal transcriptional repression domain of the Drosophila Krüppel protein. Mol Cell Biol. 1994;14:4057–4066. doi: 10.1128/mcb.14.6.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Arias A. Development and patterning of the larval epidermis of Drosophila. In: Bate M, Martinez Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 517–608. [Google Scholar]
- Neer EJ, Schmidt CJ, Nambudripad R, Smith TF. The ancient regulatory-protein family of WD-repeat proteins. Nature. 1994;371:297–300. doi: 10.1038/371297a0. [DOI] [PubMed] [Google Scholar]
- Parkhurst SM, Meneely PM. Sex determination and dosage compensation: Lessons from flies and worms. Science. 1994;264:924–932. doi: 10.1126/science.8178152. [DOI] [PubMed] [Google Scholar]
- Parkhurst SM, Bopp D, Ish-Horowicz D. X:A ratio, the primary sex determining signal in Drosophila, is transduced by helix-loop-helix proteins. Cell. 1990;63:1179–1191. doi: 10.1016/0092-8674(90)90414-a. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Finley RLJ, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation and sex-determination, and interacts directly with Hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Paroush Z, Wainwright SM, Ish-Horowicz D. Torso signalling regulates terminal patterning in Drosophila by antagonising Groucho-mediated repression. Development. 1997;124:3827–3834. doi: 10.1242/dev.124.19.3827. [DOI] [PubMed] [Google Scholar]
- Patel NH, Martin-Bianco E, Coleman KG, Poole SJ, Ellis MC, Kornberg TB, Goodman CS. Expression of engrailed protein in arthropods, annelids and chordates. Cell. 1989;58:955–968. doi: 10.1016/0092-8674(89)90947-1. [DOI] [PubMed] [Google Scholar]
- Pazin MJ, Kadonaga JT. SWI2/SNF2 and related proteins—ATP-driven motors that disrupt protein-DNA interactions. Cell. 1997;88:737–740. doi: 10.1016/s0092-8674(00)81918-2. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. Vectors for P-mediated transformation in Drosophila. In: Rodriquez RL, Denhardt DT, editors. Vectors, a survey of molecular cloning vectors and their uses. Boston, MA: Butterworth; 1988. pp. 437–456. [Google Scholar]
- Preiss A, Hartley DA, Artavanis-Tsakonas S. The molecular genetics of Enhancer of split, a gene required for embryonic neural development in Drosophila. EMBO J. 1988;7:3917–3927. doi: 10.1002/j.1460-2075.1988.tb03278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushlow CA, Hogan A, Pinchin SM, Howe KR, Lardelli MT, Ish-Horowicz D. The Drosophila hairy protein acts in both segmentation and bristle patterning and shows homology to N-myc. EMBO J. 1989;8:3095–3103. doi: 10.1002/j.1460-2075.1989.tb08461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sanicola M, Sekelsky J, Elson S, Gelbart WM. Drawing a stripe in Drosophila imaginal disks: Negative regulation of decapentaplegic and patched expression by engrailed. Genetics. 1995;139:745–756. doi: 10.1093/genetics/139.2.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe SS, Harte PJ. The Drosophila Extra sex combs protein contains WD motifs essential for its function as a repressor of homeotic genes. Mech Dev. 1995;52:77–87. doi: 10.1016/0925-4773(95)00392-e. [DOI] [PubMed] [Google Scholar]
- Sauer F, Fondell JD, Ohkuma Y, Roeder RG, Jäckle H. Control of transcription by Kruppel through interactions with TFIIB and TFIIE beta. Nature. 1995;375:162–164. doi: 10.1038/375162a0. [DOI] [PubMed] [Google Scholar]
- Schrons H, Knust E, Campos-Ortega JA. The Enhancer of split complex and adjacent genes in the 96F region of Drosophila melanogaster are required for segregation of neural and epidermal progenitor cells. Genetics. 1992;132:481–503. doi: 10.1093/genetics/132.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C, Locke J, Nishida C, Kornberg TB. Analysis of cubitus interruptus regulation in Drosophila embryos and imaginal disks. Development. 1995;121:1625–1635. doi: 10.1242/dev.121.6.1625. [DOI] [PubMed] [Google Scholar]
- Shepard SB, Broverman SA, Muskavitch M. A tripartite interaction among alleles of Notch, Delta, and Enhancer-of-split during imaginal development of Drosophila melanogaster. Genetics. 1989;122:429–438. doi: 10.1093/genetics/122.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Bornemann D, Lunde K, Schwartz C. The extra sex combs product contains WD40 repeats and its time of action implies a role distinct from other Polycomb group products. Mech Dev. 1995;53:197–208. doi: 10.1016/0925-4773(95)00434-3. [DOI] [PubMed] [Google Scholar]
- Smith D, Johnston K. Single-step purification of polypeptides expressed in E. coli as fusions with glutathione S-transferase. Gene. 1988;76:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Smith ST, Jaynes JB. A conserved region of Engrailed, shared among all En-, Gsc-, Nk1-, Nk2- and Msh-class homeoproteins, mediates active transcriptional repression in vivo. Development. 1996;122:3141–3150. doi: 10.1242/dev.122.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a G-protein βγ dimer at 2.1Å resolution. Nature. 1996;379:369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- Spradling AC. P element-mediated transformation. In: Roberts DB, editor. Drosophila: A practical approach. Oxford, UK: IRL Press; 1986. pp. 175–197. [Google Scholar]
- Steller H, Pirrotta V. A transposable P vector that confers G418 resistance to Drosophila larvae. EMBO J. 1985;4:167–171. doi: 10.1002/j.1460-2075.1985.tb02332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Treitel MA, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Um M, Li C, Manley JL. The transcriptional repressor Even-skipped interacts directly with TATA-binding protein. Mol Cell Biol. 1995;15:5007–5016. doi: 10.1128/mcb.15.9.5007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrijzer CP, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- Wall MA, Coleman DE, Lee E, Iñiguez-Lluhi JA, Posner BA, Gilman AG, Sprang SR. The structure of the G protein heterotrimer: Giα1β1γ2. Cell. 1996;83:1047–1058. doi: 10.1016/0092-8674(95)90220-1. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Sinful repression. Nature. 1997;387:16–17. doi: 10.1038/387016a0. [DOI] [PubMed] [Google Scholar]
- Younger-Shepherd S, Vässin H, Bier E, Jan LY, Jan YN. deadpan, an essential pan-neural gene encoding an HLH protein, acts as a denominator in Drosophila sex determination. Cell. 1992;70:911–922. doi: 10.1016/0092-8674(92)90242-5. [DOI] [PubMed] [Google Scholar]