Abstract
The human U2B” protein is one of the unique proteins that comprise the U2 snRNP, but it is also a representative of the U1A/U2B” protein family. In the U2 snRNP, it is bound to Stem-Loop IV (SLIV) of the U2 snRNA. We find that in vitro it binds not only to human SLIV, but also to Stem-Loop II (SLII) from human U1 snRNA and to Drosophila U2 snRNA SLIV. The thermodynamics of these binding interactions show a striking similarity, leading to the conclusion that U2B” has a relaxed specificity for its RNA targets. The binding properties of U2B” are distinct from those of human U1A and of Drosophila SNF, despite its high homology to those proteins, and so provide important new information on how this protein family has modulated its target preferences.
Keywords: RRM, RNA binding protein, U2B”, protein:RNA thermodynamics
Introduction
The spliceosomal proteins U1A and U2B” are highly conserved in eukaryotes, where they are components of the U1 and U2 snRNP, respectively. In the snRNPs, U1A protein is thought to bind exclusively to stem-loop II (SLII) of the U1 snRNA and U2B” to stem-loop IV (SLIV) of the U2 snRNA. These RNA hairpins are highly conserved, and their loop sequences are very similar to each other. Although they are components of snRNPs, the roles of U1A and U2B” in splicing remain unclear, and in fact, in vitro snRNP reconstitution in the absence of U1A has no effect on splicing [1]. Other experiments using mutations of the fly homologue, SNF, that exclude the protein from either the U1 or U2 snRNP resulted in relatively mild phenotypic consequences [2][3]. In contrast, knocking out both U1A and U2B” in C. elegans is embryoniclethal, as is the SNF knockout in Drosophila [4][5]. There is certainly a possibility that these proteins have alternative functions outside of the snRNPs.
U1A, U2B”, and SNF consist of two RNA recognition motifs (RRMs) connected by a variable, flexible linker. The RRM is the most commonly used RNA binding domain in eukaryotes {Maris et al., 2005, FEBS J, 272, 2118-31}, and can be identified by two amino acid sequences (RNP1 and RNP2) that are located in two of the four β strands on its β–sheet {Ghetti et al., 1989, FEBS Lett, 257, 373-6}. The canonical view of RNA-RRM interactions is that single-stranded RNA binds to the β-sheet surface. Favorable electrostatic interactions, hydrogen bonding, and stacking between RNA bases and aromatic residues located in the RNP motifs are regarded as the predominant determinants of RNA binding {Clery et al., 2008, Curr Opin Struct Biol, 18, 290-8}. U1A uses its N-terminal RRM to bind its in vivo U1 snRNA target, Stem-Loop II (SLII), with very high affinity and specificity [6][7]. There are crystal and solution structures of U1A RRM1 [8][9], and a co-crystal structure of U1A RRM1 bound to SLII [10] shows how the loop of the RNA hairpin is positioned on the surface of the β-sheet. A co-crystal of U2B” RRM1 bound to both SLIV and U2A’ (an auxiliary protein) positions the RRM in the middle of the complex, with the RNA bound on the β-sheet surface and U2A’ wrapping around the opposite face of the RRM, predominantly making contacts with RRM α1[11]. There is a solution structure of SNF RRM1 [12]. Not surprisingly, the structures of these three RRMs are similar to each other and for U1A, the structures of the free and bound proteins are also similar {Nagai et al., 1990, Nature, 348, 515-20}.
The U1A/U2B” family of proteins provides a valuable opportunity to understand determinants of RNA:protein affinity and specificity. RRM1 of U1A is ~75% identical to the N-terminal RRMs of U2B” and the Drosophila homologue, SNF. While SNF binds to both U1 snRNA SLII and U2 snRNA SLIV [13], there has been significant debate regarding how (and how well) U2B” binds to its target, U2 snRNA SLIV. Some data report that U2B” binds both SLII and SLIV [14] , and there are conflicting studies on whether the U2A’ protein, which is present in the U2 snRNP and which binds U2B”, is required for U2B” to bind SLIV [15]. Regardless, the studies do suggest that RNA binding by U2B” is much weaker than binding of SLII by U1A.
The co-crystal structure of U2B”:SLIV showed that many of the interactions between U2B” and SLIV were also present in the earlier U1A:SLII cocrystal structure [11]. Since both the RNA loops contain identical 5′ sequences (human SLII: A1UUGCA6CUCC and human SLIV A1UUGCA6GUA9CC) and the RNP sequences of the proteins are identical, preservation of these contacts is not unexpected. Stacking of the nucleobases with Tyr10 in RNP2 (β1) and Phe53 in RNP1 (β3) (U2B” numbering) occurs in both complexes. Despite the phylogenetic conservation of A9 on the 3′ side of the SLIV RNA loop (and its absence from stem-loop II sequences), the U2B” co-crystal showed no interactions between the protein and A9. Within the 3′ UA9CC sequence, U8 and C11 do pack against the VALKT amino acids of U2B” β2 and Loop 3; those amino acids are frequently conserved in U2B” proteins but are distinct from the corresponding residues of U1A. The authors noted that the loop amino acids R52 and T48 appeared to interact with the phosphate backbone of the RNA near its loop/stem junction [11]. It is worth noting that the RNA stem in the U2B” co-crystal is distorted from normal A-form, and those 3′ ACC loop nucleotides stack on each other, perhaps due to crystal packing. Considering the similar patterns of interactions between U1A and U2B”, we might anticipate that the proteins have comparable RNA binding affinities and specificities. However, we find that U1A and U2B” have different RNA binding preferences in vitro which could not be anticipated by the apparent similarities in their respective cocrystal structures.
The U1A:RNA interaction is characterized by complicated thermodynamics. More specifically, the U1A:SLII interaction has a large apparent heat capacity (ΔCP,obs) of −3 kcal/mol, and its enthalpy and entropy are both temperature-dependent [16][7]. Interpretation of ΔCP,obs is made more difficult by the conformational transitions of both RNA and protein upon complex formation. It is reasonable to anticipate that the U2B”:RNA interaction will also involve conformational changes of RNA and protein.
Measurements of thermodynamic pairwise coupling in the U1A protein and in the complex helped to identify a network of amino acid sidechains that span the RNA binding surface, including the conserved Tyr13, Phe56, and Gln54 (numbered as per U1A) on the surface of the β-sheet, residues in Loop 3 and in the C-terminal tail of RRM1 {Kranz and Hall, 1998, J Mol Biol, 275, 465-81}[17]. While the Loop 3 sequences in the two proteins are quite different, the other amino acids that have been implicated in this network are conserved between U1A and U2B”, leading to our expectation that the RNA binding surface of U2B” will also span the entire face of RRM1. A comprehensive thermodynamic study of U2B”:RNA interactions is essential to describe this protein’s binding mechanism and elucidate how and why it differs from the other members of this protein family. Most significantly, this analysis will help us understand how RRMs with such similar sequences and structures have such different RNA binding properties.
In this study, we use full-length wild-type human U2B” to assess the binding affinities of this protein for several RNA stemloops (hairpins). We also assess the salt and temperature dependence of these interactions. An important result that has implications for the protein’s biological functions is that human U2B” binds U1 snRNA SLII and U2 snRNA SLIV with almost equal affinity. In contrast, human U1A protein effectively binds only SLII. Within this protein family, binding affinities and specificities for RNA sequences have been exquisitely modulated, and biothermodynamics is the only way to compare them.
Results
The U2B” protein has two RNA recognition motifs, separated by a 40 amino acid linker. This linker is much shorter than the corresponding linker of U1A. Sequences of the N-terminal RRMs of human U1A, U2B”, and Drosophila SNF are compared in Figure 1, including some of the linker sequences. A structural depiction is shown that highlights the residues on the RNA binding surface that differ between U1A and U2B”. Differences in β2 are extensive, but the most significant difference with respect to RNA binding is in Loop 3, which contacts the RNA..
Figure 1.
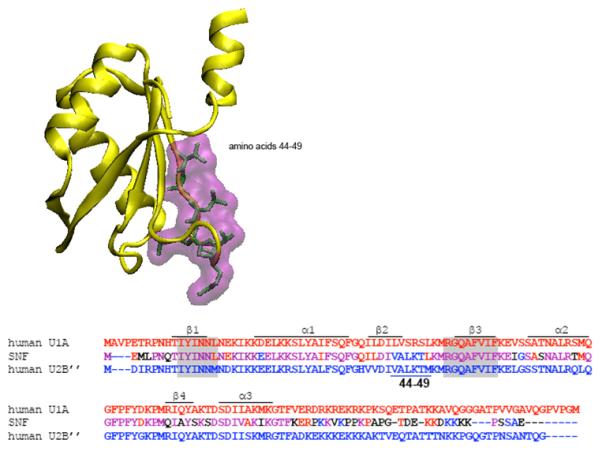
Structure of the U2B” protein taken from the co-crystal 1AVN. The β-sheet of the RRM faces out, and the residues on that RNA binding surface (44-49) are unique to U2B”. The sequences of U1A, SNF, and U2B” RRM1 are compared in the alignment, with the RNP sequences shaded. Amino acids 44-49 are noted below the U2B” sequence. Also included is the N-terminal part of the linker, which in U2B” contains many positively charged residues that could interact with the RNA stem.
Loop 3 of U1A, (SRSLKMRG) is a site that contributes to RNA:protein specificity; in both U1A:SLII and U2B”:SLIV co-crystals, the protein Loop 3 protrudes into the RNA loop where it is juxtaposed with the nucleotides on the bottom of the RNA loop, and so splays the RNA open. The sequence of this important protein loop is different in U2B” (LKTMKMRG) and Drosophila SNF (LKTLKMRG). For U1A, Loop 3 makes contacts with the RNA, including a hydrogen bond between the R47 amide and the backbone at the RNA loop-closing G. In the U2B”:SLIV co-crystal, Loop 3 contacts with the U-U appear to be minimal.
The RNA also needs to be considered in a discussion of U1A/U2B” binding. In the cocrystal of U2B” RRM1, Stemloop IV of U2 snRNA, and the U2A’ auxiliary protein, the RRM contacts the 5′ AUUGCAG sequence of the RNA loop. These interactions appear similar to those formed in the U1A:SLII complex, as expected based on the RNA sequence (AUUGCAC in SLII). One significant difference between SLII and SLIV is the loop-closing base pair, which in SLII is a C:G but in SLIV is a noncanonical U:U pair. In Drosophila, the U-U sequence has become U-G, and this difference appears to be important for recognition of the RNA by SNF {Williams and Hall, 2010, Biochemistry, 49, 4571-82}. The adjacent base pair in the stem is a G:C in human and fly SLIV (Figure 2), and it may be that this is the effective loop-closing base pair. Here, we refer to the U-U as either the loop-closing base pair or as inserted nucleotides at the bottom of the RNA loop. The difference in the loop-closing basepair may well be important for discrimination between the RNAs by the proteins.
Figure 2.
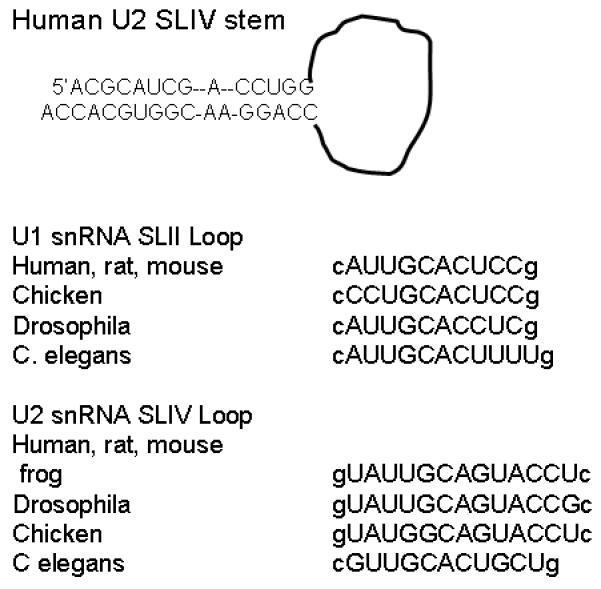
Sequences of the human U2 snRNA SLIV stem and SLIV and U1 snRNA SLII loops from several organisms. The loop sequences are capitalized; loop-closing base pair is lower case.
RNA Binding to U2B”
Stemloop IV, the binding site for U2B”, is located at the 3′ end of U2 snRNA. In vertebrates, the stem of this hairpin has eleven base pairs and an asymmetric internal A bulge in the middle of the stem (Figure 2). In most vertebrates, the loop sequence is conserved: 5′gUA1UUGC5AGUAC10CUc, where the loop-closing base pair is indicated in lower case. Much of this sequence is shared with U1 snRNA SLII: cA1UUGC5ACUCC10g. Notable differences include the ‘inserted’ U/U pair at the bottom of the SLIV loop, the C7-to-G7 substitution, and the longer SLIV loop. These phylogenetic differences could identify unique contacts with the corresponding protein, or indicate sites that are insensitive to mutation.
Binding of U2B” to several RNA sequences suggests some features recognized by the protein. As summarized in Table 1, the human SLII and SLIV RNAs are bound with equal affinity by the protein at room temperature in 100 mM KCl, 1 mM MgCl2, 10 mM sodium cacodylate pH 7.4. This rather surprising result indicates that the presence of the inserted U-U nucleotides does not limit protein binding. Indeed, the protein binds with equal affinity to Drosophila SLIV RNA with its U-G pair. In contrast, the human U1A protein does not detectably bind to the human SLIV RNA in nitrocellulose filter binding assays, but it is able to weakly bind to Drosophila SLIV (5 ±3 ×10−7 M in 100 mM KCl, 12 mM MgCl2, 10 mM sodium cacodylate pH 7, 22° C). The U-U pair in SLIV appears to function as a discriminator for U1A, but not for U2B”, pointing to a fundamental difference in their mechanisms of RNA recognition. Additionally, a C7-to-G7 substitution in SLII leads to a 10-fold weaker affinity of the U1A:RNA interaction (ΔΔG°=1.4 kcal/mol) [6]. In contrast, while the binding affinity of U2B” for SLII is significantly lower than the affinity of U1A for SLII, the protein tolerates the G7 in SLIV without a further loss of affinity (ΔΔG°=0), indicating different binding mechanisms.
Table I.
U2B” binding affinities to RNA hairpins.
| RNA | [KCl] mM | KD (M) | ΔG° (kcal/mol) |
|---|---|---|---|
| hSLIV 9 base pair stem | 100 | 1 (±2) ×10−9 | −12.2 (±0.3) |
|
| |||
| 6 base pair stem | 100 | 2 (±2) ×10−9 | −12 (±0.2) |
|
| |||
| 2AP9 | 100 | 2 (±2) ×10−9 | −12 (±0.2) |
|
| |||
| 250 | 2.6 (±0.6) ×10−8 | −10.3 (±0.1) | |
|
| |||
| hSLII | 100 | 2.1 (± 2) ×10−9 | −11.7 (±0.2) |
| 100/2* | 1.9 (±0.7) ×10−8 | −10.5 (±0.2) | |
|
| |||
| 250 | 1 (±2) ×10−7 | −10.1 (±0.2) | |
|
| |||
| dSLIV | 100 | 2 (±2) ×10−9 | −12 (±0.2) |
|
| |||
| 250 | 3.8 (±1.5) ×10−8 | −10.0 (±0.2) | |
Binding measured by nitrocellulose filter binding/fluorescence intensity at 22 °C in 100 or 250 mM KCl, 1 mM MgCl2 (*except for hSLII in 100/2 indicating 2 mM MgCl2), 10 mM sodium cacodylate pH 7.4 or 10 mM potassium phosphate pH 8. Free energies calculated from the KD.
Although the stem of SLIV in U2 snRNA contains an internal asymmetric bulge, here we use perfect duplexes for the hairpins. We find that stems of length 6 and 9 base pairs are bound with equal affinity by the protein. In the co-crystal, the RNA stem was a perfect 6 base pair duplex that appeared to make contacts with α1 of U2B” [11]. We posit that this contact arose through crystal packing that led to a buckling of the base pairs that unwound the stem, together with a twist of the RNA loop.
A significant difference between SLII and SLIV is the insertion of A9 on the 3′ side of the loop. While interactions with the protein were not observed in the cocrystal structure, this A9 could be used as a point of specific contact for U2B” recognition. To observe the structural consequence of U2B” binding to SLIV, A9 was replaced with 2-aminopurine (2AP), and its fluorescence was monitored with and without bound protein. The introduction of 2AP could potentially disrupt an RNA:protein contact, so U2B” affinity for 5′-32P-2AP-SLIV RNA was measured using nitrocellulose filter binding. The binding affinities for human and Drosophila SLIV RNAs were identical to affinities for the wild-type RNAs, giving us confidence that the fluorescence data report on normal binding events.
The steady state 2AP fluorescence intensity of the human SLIV RNA free and bound to U2B” is shown in Figure 3a. There is a 50% increase in the 2AP fluorescence intensity upon binding, indicating that stacking of the 2AP with neighboring bases (which significantly quenches 2AP fluorescence) has been disrupted. U2B” binding to Drosophila 2AP-SLIV results in a very similar increase in the fluorescence intensity, shown in Figure 3b. A feasible interpretation of these results, consistent with the binding data and with the U2B”:hSLIV crystal structure, is that the protein does not interact with the 2AP (or A9) base in the RNA but needs to open the loop to make specific contacts with the AUUGCAG sequence. Certainly this result indicates that binding of U2B” results in a large change in environment at the 2AP, and by extension, a large conformational change of the RNA.
Figure 3.
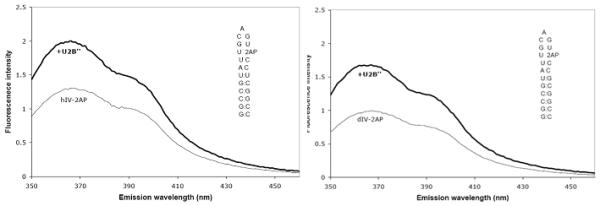
Fluorescence intensities of SLIV RNAs with 2-aminopurine at position A9 with and without U2B” protein. Fluorescence enhancement is 50 ±10% for hSLIV + U2B”, and 61 ±3% for dSLIV + U2B”. Experiments were performed at 23°C in 20 mM KCl, 10mM sodium cacodylate, pH 7. The concentration of RNA was 2μM. Protein was subsequently added to 2.5 μM. The RNA sequences are shown as insets within the spectra.
Salt dependence of binding
The salt concentration can affect the binding affinity of the protein for the RNA, and indeed, given the highly negative charge of RNA, one expects a large salt dependence for the interaction. On the basis of our experiments to date, we suspect electrostatic interactions do occur between the protein and the phosphates of the stem. U2B” binding to a dSLIV construct with a fluorescent probe at its 5′ end results in a significant enhancement of fluorescence intensity, as well as an increase in the anisotropy of the fluorophore (Figure 4). Protein binding results in increases to both the steady-state anisotropy and fluorescence intensity. When both anisotropy and fluorescence intensity were measured, fits yielded equivalent values for Kobs. However, data were less noisy for fluorescence enhancement. Similar fluorescence enhancements upon U2B” binding are seen in SLII and hSLIV constructs with 5′ fluorescein labels (data not shown).
Figure 4.
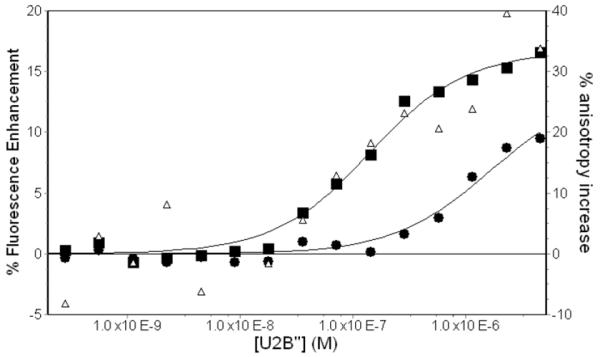
Representative titrations of U2B” into FAMN-dSLIV. Titrations were done at 23°C in 10 mM potassium phosphate, 2 mM MgCl2, pH 8. Fluorescence enhancement of FAMN are shown in 300 mM KCl (∎) and 500 mM KCl (● ). Increase in fluorescence anisotropy in 300 mM KCl is also shown (▵).
As with U2B”, SNF binding to the same RNAs results in a large enhancement of fluorescence intensity. With SNF, this enhancement is highly salt-dependent and is greatly decreased when RRM1 alone is used in titrations [13]. Such changes to the fluorescence intensity are hard to explain absent interactions between the protein and the RNA stem. A recent crystal structure of the U1 snRNP suggests that interactions between U1A and SLII occur well beyond the putative end of the RRM domain [18]. It therefore seems likely that the charged linker between the two RRMs interacts with the backbone of the RNA stem to increase the affinity of the interaction. An examination of the sequence of the U2B” linker shows that there are many lysines that follow α3 and might contribute to electrostatic association with the RNA. If the RRM1 tail is flexible and disordered, those lysines could reach to the stem of the RNA hairpin and interact with the negatively charged backbone. This is probably an important contributor to the salt dependence observed in the interactions.
As a first measure of the salt dependence, the KCl concentration was varied from 150 to 400 mM. The salt dependence of U2B” binding to hSLII, hSLIV, and dSLIV was determined by titrating 5′-FAM-RNAs (fluorescently labeled RNAs) with U2B” under various salt concentrations. To adequately probe the cation/anion response of the protein to RNA binding, these experiments should be repeated with a larger anion to remove the bias from the RNA; assessing anion effects to the protein:RNA interaction would also allow us to discriminate between the relative contributions of anions and cations to the salt dependence of these interactions [19]. As shown in Figure 5 and Table 2, the slope of the plot of ln(Kobs) vs ln[KCl] for U2B” binding to human SLII and SLIV indicates a net release of ions for all interactions studied. U2B” binding to SLII, hIV, and dIV occurs with a net release of 5.2 (±0.3), 4.6 (±0.3), and 4.8 (±0.2) ions, respectively.
Figure 5.
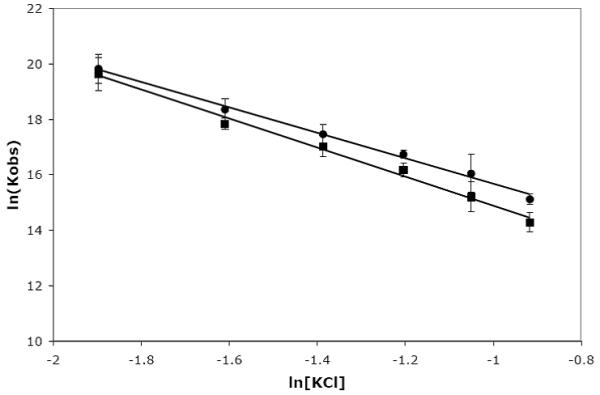
Salt dependence of U2B” binding to human SLII and SLIV. Binding was monitored by RNA fluorescence, and the salt concentration was varied between 150 to 400 mM KCl. The buffer also contained 10 mM potassium phosphate, pH 8, 2 mM MgCl2 and 20 μg/mL of BSA. All experiments were performed at 23°C. Data for hSLII are shown as (∎) and data for hSLIV are shown as (엯).
Table 2.
Salt dependence of RNA-Protein Interactions
| Protein:RNA | Slope |
|---|---|
| U1A:SLII | −6.7 (±1.1) |
| SNF:SLII | −5.7 (±0.2) |
| SNF:dSLIV | −4.0 (±0.2) |
| U2B”:SLII | −5.2 (±0.3) |
| U2B”:hSLIV | −4.6 (±0.3) |
| U2B”:dSLIV | −4.8 (±0.2) |
This analysis of the salt dependence and the interpretation of the data follows from studies undertaken by the Lohman lab. In a series of experiments to measure the interactions between oligopeptides and DNA and RNA strands, they defined a simple relationship between the change in the observed equilibrium constant with added monovalent salts: ∂logKobs/∂log[MX] = −(Δc + Δa) + 2[MX]·Δw/[H2O], where MX is the salt (KCl for our experiments), Δc is the change in cations bound, Δa is the change in anions bound, and Δw is the change in bound water. Preferential hydration effects can be ignored unless the salt concentration is very high (>0.5M), further simplifying the analysis [19]. A positive slope of the plot of log(Kobs) vs log[MX] indicates net uptake of anions and cations upon binding, whereas a negative slope indicates ion release. For the U2B” interaction with its RNAs, we find a net release of approximately 5 ions. By comparison, the value for SNF:dSLIV is −4, for SNF:SLII is −6, and U1A:SLII is −7 [13][6]. This indicates that the electrostatic contributions to binding are distinct in the various RNA:protein interactions and implies that electrostatics play an important role in distinguishing the different RNA binding mechanisms within this protein family. We found that SNF discriminates between the RNAs, and that electrostatics contribute to this discrimination. However, we find that U2B” binds with similar affinities to the three RNAs, and the overall electrostatic contributions to binding these RNAs appear to be similar.
Temperature dependence of binding
Although we anticipate from our previous work with U1A and SNF that only U2B” RRM1 is responsible for specific RNA recognition, the full length U2B” protein was used in these experiments. The protein is folded and stable to 35° C as determined by 15N/1H NMR experiments using 300 μM protein (data not shown). Binding to the human SLIV RNA was measured by nitrocellulose filter binding in 100 mM KCl, 1 mM MgCl2, 10 mM sodium cacodylate, pH 7.4, or by monitoring binding-induced changes in the fluorescence intensity of 5′-fluorescein-labeled RNA in 250 mM KCl. Where the measurements could be made using both nitrocellulose filter binding and protein titrations into fluorescein-labeled RNA, agreement was excellent, indicating that introduction of the fluorescein did not perturb the binding.
As anticipated, U2B” binding to the RNAs is both temperature- and salt-dependent. The van’t Hoff plot for binding to hSLIV in 250 mM KCl is linear and best fit by the van’t Hoff equation [ΔH°/T*R − ΔS°/R = ln(Kobs)] to give ΔH° = −16.4 (±1.0) kcal/mol and ΔS° = −21.2 (±3.5) cal/mol-K (Figure 6). In this salt, binding of hSLIV by U2B” is enthalpy-driven, possibly due to favorable stable stacking of the nucleobases with the aromatic amino acids on the β-sheet surface. In Figure 6, these data were also fit using the formalism from Ha et al., (1989), to give a small heat capacity of −414 (± 190) cal/mol, but we consider that the temperature-independent values of enthalpy and entropy give an adequate description of this interaction. Given the narrow temperature range for which we can perform these experiments, it is likely that at this salt concentration, we are sampling temperatures above the critical point, where a non-linear van’t Hoff relationship will nevertheless appear linear.
Figure 6.
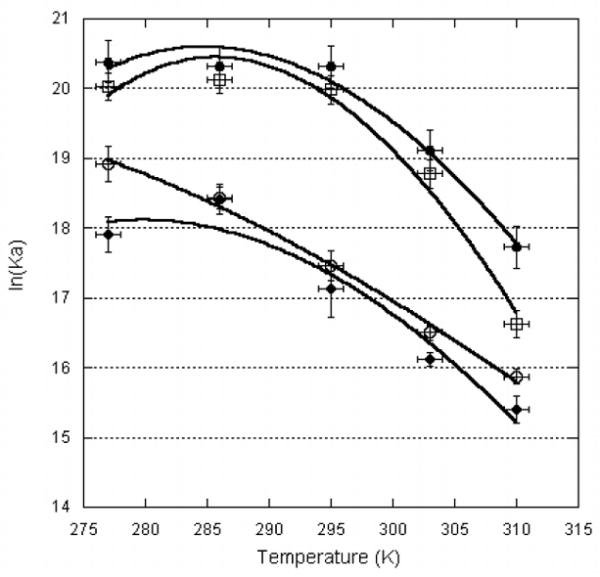
Temperature dependence of U2B” binding to RNA. (●) hSLIV in 250 mM KCl; (∎) hSLII in 100 mM KCl; (엯) hSLIV in 250 mM KCl; (▴) dSLIV in 250 mM KCl. Data were fit to ln(KA,obs) = ((ΔCP,obs)/R)(TH/T − ln(TS/T) −1), where R is the gas constant 1.98 cal-K-mol, T temperature in K, ΔCP,obs the heat capacity of binding, TH the temperature at which the enthalpy is zero, and TS the temperature at which the entropy is zero. These data were measured using nitrocellulose filter binding and/or fluorescence intensity of 5′-FAM-RNA.
In contrast, at a lower salt concentration (100 mM KCl), binding of U2B” to hSLIV is better fit by an expression that includes an apparent heat capacity, [ln(KA,obs) = (ΔCP,obs/R)[TH/T − ln(TS/T) −1] [20], where ΔCP, obs is the observed heat capacity change, R is the gas constant, T is the temperature in Kelvin, and TH and TS are values where the enthalpy and entropy of complex formation are zero. The result is that the enthalpy and entropy are temperature-dependent. The data do not fit perfectly, but we estimate ΔCP,obs for complex formation is −1.6 (±0.4) kcal/mol.
U2B” binding to Drosophila SLIV was measured by 5′-FAM-RNA fluorescence in 250 mM KCl. The binding data could be fit to the van’t Hoff relation to give ΔH° = −15.3 (±2.5) kcal/mol and ΔS° = −18.3 (±8.4) cal/mol-K. However, these data can also be fit by the expression ln(KA,obs) = (ΔCP,obs/R)[TH/T − ln(TS/T) −1] to give the apparent heat capacity of the association: here, ΔCP,obs = −1.1 (±0.4) kcal/mol. The only difference in the hSLIV and dSLIV loops is a U/U or U/G pair, respectively, at the junction with the stem. Since this is precisely where the protein Loop 3 is located in the complex, this apparently small change could have large consequences either through modulation of the RNA structure or RNA:protein contacts. However, binding affinity of U2B” for fly and human SLIV is nearly identical, suggesting that the protein does not make significant contacts with the base of the RNA loop.
U2B” also binds to human SLII. The data cannot be fit by the van’t Hoff equation, and instead leads to a ΔCP,obs = −2.2 (±0.4) kcal/mol, in 100 mM KCl, 1 mM MgCl2. As Figure 6 shows, the temperature dependence of U2B” binding to hSLIV and hSLII is nearly identical, suggesting that either the binding mechanism is the same, or that there are compensating interactions that result in similar thermodynamic signatures.
True heat capacity changes upon ligand binding are in general attributed to hydrophobic surface burial. We have observed these large apparent heat capacity changes in U1A, SNF, and now U2B”. However, one difficulty with interpretation of the heat capacity term for these RNA:protein complexes lies in the possibility that there are conformational changes of the components coupled to binding. It has been shown that apparent heat capacity changes can occur in the absence of hydrophobic surface burial, as a result of temperature-dependent conformational changes that are coupled to binding but that occur with a ΔCP=0 [21]. The 2AP fluorescence data show clearly that there is a large conformational change in the RNA upon binding, and it is likely that conformational changes also occur in the protein (particularly in Loop 3). Measurement of the temperature dependence of the calorimetric enthalpy of binding using isothermal titration calorimetry (ITC) would make interpretation clearer, as temperature dependence of this measurement would indicate contributions from a true ΔCP. These experiments are planned for U2B”.
Conclusions
These data show conclusively 1) that the human U2B” protein does not require an auxiliary protein for binding to RNA; 2) that U2B” binds to both U1 snRNA stemloop II as well as to U2 snRNA stemloop IV; and 3) U2B” shows no preference for SLIV or SLII on the basis of binding affinity. Temperature and salt dependence of these interactions are similar, which is distinct from recognition of the same RNAs by Drosophila SNF, suggesting that U2B” recognizes the different RNAs through a common mechanism. These results have important implications for the biology of snRNP assembly and composition, but also for development of a general model for RNA recognition by the U1A/U2B” family of RRM proteins.
snRNP composition
The U1 and U2 snRNPs mostly assemble in the cytoplasm, adding the Sm proteins to the snRNA after its transport out of the nucleus. Other protein components are added subsequently. The U1 snRNP is relatively simple, with only three U1-specific proteins in addition to the Sm cluster: U1A binds to SLII, the U1 70K protein binds to SLI, and the U1C protein binds to the tail of U1 70K [22][18]. The U2 snRNP is the most complex snRNP, and its composition is functionally regulated. Recent studies suggest that the U2B” protein is present in the particle throughout its life, even though it is located at the extreme 3′ end of the U2 snRNA which, in contrast to much of the rest of the snRNA, does not rearrange during splicing [23].
Both U1A and U2B” are found in the cytoplasm where they can bind to their snRNA targets, which for U2B” includes both SLII and SLIV. U1A would win the binding contest for its SLII target on the basis of its higher affinity. Comparing U1A and U2B” binding to SLII in vitro shows that U1A has (approximately) a 100-fold greater binding affinity. If cytoplasmic concentrations of each protein are equal, then U2B” would be effectively excluded from the U1 snRNP. Under conditions where the U1A concentration is limiting, U2B” could certainly bind to SLII, as indeed occurs in C. elegans when the U1A protein is knocked out [4]. However, in vertebrates, U1A could not replace U2B” since its affinity for the human SLIV is too weak. In this respect, the two vertebrate proteins are not interchangeable, and it seems that U2 SLIV may have evolved to exclude U1A.
RNA Recognition
Based on our in vitro results, it appears that the U2B” protein is quite tolerant of substitutions in the RNA loop, at least on the 3′ side and at the loop-closing base pair. One model is that only the loop AUUGCAG nucleotides constitute the recognition site for the protein, and all other interactions are nonspecific electrostatic contacts. Certainly the very similar binding affinity, temperature dependence, and salt dependence for hSLIV, hSLII, and dSLIV RNAs are consistent with this scheme. The protein T89D90S91 amino acids (numbered according to U1A) that recognize the G4CA6(G7/C7) sequences of SLIV and SLII are conserved in both U1A and U2B”. The T89D90S91 residues are located after the hinge that links β4 with the third α-helix, but it is their peptide backbone amide and carbonyl oxygen atoms that hydrogen bond with the nucleobases. This interaction appears to be modulated by the geometry of the protein that presents the TDS backbone for contacts, all of which are conserved in the two proteins.
The more interesting interactions are those that the thermodynamics show are not critical for RNA binding, but that involve Loop 3 of the protein, one of the phylogenetically conserved sequences in each U1A and U2B” sub-family. The construction of thermodynamic cubes that included RNA binding together with pairwise coupled sites on the protein [17] was inspired by the work of DiCera, which was presented at Gibbs Biothermodynamics meetings. Kranz’s novel application to a bimolecular system provided the first evidence of a network of interactions that together formed the RNA binding site of the protein. Thermodynamic pairwise coupling applied to U1A and the U1A:SLII complex helped to define a network of interactions that linked Loop 3 with the aromatic residues on the surface of the β-sheet.
One intriguing possibility is that in the U2B” protein, the pairwise coupling patterns are altered with the result that Loop 3 is no longer linked to the β sheet surface. The U1A protein is extremely sensitive to the nucleobases at the bottom of SLII through interactions with the C:G loop-closing base pair mediated by Loop 3. Insertion of a single U between that C:G base pair and the first A of SLII reduces the binding affinity of U1A by several orders of magnitude [7]. We proposed that the insertion causes a shift in the frame of RNA binding on the protein β-sheet through disruption of Loop 3/RNA interactions. Here, however, we observe that the U2B” protein seems insensitive to the nucleotides at the bottom of the loop, which implies a very different role for its Loop 3.
Although it is not apparent from the structures of the proteins alone or in their respective co-crystals, the thermodynamic analysis of U2B” binding to three RNA hairpins shows that its RNA binding surface is quite different from that of U1A. Its specificity is relaxed, and its mechanism of RNA recognition appears to rely on only a subset of the RNA loop nucleotides. Appreciation of the thermodynamic properties of this complex will direct further experiments that probe the atomic details of the interactions.
Materials and Methods
U2B” cloning and purification
The human U2B” clone was purchased from the American Type Culture Collection (ATCC). The full length cDNA was amplified using the following primers: Forward primer: GGTGGTCCATGGATATCAGACC; Reverse Primer: GGTGGTAAGCTTTTATTTCTTGGCATAGG and subcloned into the IPTG-inducible pTAC vector using the NcoI and HindIII sites. The plasmid was transformed into BL-21(DE3) cells for protein expression. Cells were grown in LB medium at 37°C to an optical density of 0.6-0.8 and were induced with 1mM IPTG for three hours. After addition of IPTG, the temperature was dropped to 28°C. Cells were harvested and stored at −70°C until lysis. Cells were resuspended in 30mM sodium acetate (pH 5.3), 200mM NaCl, 2mM EDTA, and 8.5% sucrose. PMSF, DNase II, and a protease inhibitor cocktail (Sigma) were added prior to French pressing the cells. The lysate was collected and spun down in an ultracentrifuge at 4°C, 45000g. The supernatant was filtered through a 0.22μm cellulose acetate membrane and loaded onto an SP Sepharose column pre-equilibrated in 50mM Tris (pH 7.5). U2B” was eluted over 170 minutes, using a 190 to 350mM NaCl gradient. EDTA and PMSF were added to the eluted fractions to a final concentration of 5mM and 20μg/mL, respectively. All column buffers were sterile-filtered through 0.45μm cellulose nitrate filters (Nalgene), and containers used in the purification were acid washed to remove RNases. Fractions containing U2B” were concentrated using a Vivaspin concentrator with a molecular weight cutoff of 10kDa and buffer-exchanged into 100mM KCl, 10mM cacodylate pH 7, 5mM EDTA. Concentration was determined spectrophotometrically using ε280=8960M−1cm−1.
RNA synthesis
Nitrocellulose filter binding experiments were performed with RNA transcripts that were enzymatically synthesized with T7 RNA polymerase, as described [24] [25]. The DNA oligonucleotides were obtained from IDT (Integrated DNA Technologies). The transcripts were internally labeled with [α-32P]UTP and [α-32P]CTP. Labeled RNAs were gel-purified before use in binding assays. The RNA product for d/hU1 SLII was: 5′GGGCUGCCAUUGCACCUCGGCGGUCC. The RNA product for dU2 SLIV was: 5′GGGCCCGGUAUUGCAGUACCGCCGGGUCC. Two hU2 SLIV constructs were: 5′GGACCUAUUGCAGUACCUGGUCC and 5′GGGCCCGGUAUUGCAGUACCUCCGGGUCC. The sequences corresponding to the loops are underlined.
2-aminopurine fluorescence experiments
RNA products internally labeled with 2AP were obtained from Dharmacon and IBA GmbH. The RNAs were: 5′GGCCGUAUUGCAGU2APCCUCGGCC (hSLIV) and 5′GGCCGUAUUGCAGU2APCCGCGGCC (dSLIV). An SLM 8000 instrument was used to perform fluorescence experiments. The temperature was set to 23°C and controlled by a circulating water bath. Cuvettes were blocked for at least one hour with 20 μg/mL of BSA. The cuvettes were subsequently rinsed with 1x buffer (20mM KCl, 10mM sodium cacodylate, pH 7). The RNA was folded as follows. The RNA was diluted to 2.2 μM in water, heated at 95°C for 5 minutes, and snap-cooled on ice for 2 minutes. 1/10 volume of 200 mM KCl, 10 mM sodium cacodylate pH 7 was then added. The final RNA concentration was 2 μM. At these salt and RNA concentrations, RNA hairpins were determined to be monomers. Emission scans were recorded for the buffer, RNA only, and RNA + 2.5μM U2B” from 350 to 460 nm. The excitation wavelength was set to 300 nm, and both excitation and emission bandwidths were 8 nm. The fluorescence enhancement upon RNA binding is given as the average of two values obtained from measurements on two separate samples of the fluorescence enhancement at 367nm, the emission maximum. The uncertainty is given as the standard deviation of those measurements.
For binding experiments, the synthesized RNA was 5′ end labeled with T4 polynucleotide kinase (NEB) and γ 32P ATP. Products were gel-purified to remove nucleases and degraded RNA.
Protein titrations with fluorescent RNA
The salt dependence of U2B” binding to dSLIV and temperature dependences of U2B” binding to dSLIV and hSLIV at 250mM KCl, 10mM potassium phosphate, 1mM MgCl2 pH 8 were performed using fluorescence-based assays. 5′-Fluorescein-labelled SLIV RNAs were obtained from IDT. The RNA sequences were: 5′-6-FAMN-GGGCCCGGUAUUGCAGUACCGCCGGGUCC (dSLIV) and 5′-6-FAM-GGGCCCGGUAUUGCAGUACCUCCGGGUCC (hSLIV). RNA samples were folded as described for the 2AP-labelled RNAs to a concentration of 2μM. Titrations were performed with a final RNA concentration between 0.1 and 10 nM. Cuvettes and stir bars were acid-washed to remove RNases. Cuvettes were subsequently blocked for one hour with buffer containing appropriate concentrations of KCl and MgCl2, 20 μg/mL of BSA and 10 mM potassium phosphate pH 8 to prevent sticking of U2B” to the cuvette walls. For initial experiments, steady-state anisotropy and fluorescence intensity were both measured throughout the protein titrations. While fits of the data yielded comparable Kds for both methods, fluorescence intensity proved to be more sensitive. Subsequent experiments were performed by measuring fluorescence intensity alone. Cuvette temperature was controlled using a circulating water bath. The excitation and emission wavelengths were set to 490 and 520nm, respectively. Temperature dependence experiments were performed using 250 mM KCl, 10 mM potassium phosphate, 1 mM MgCl2 pH 8. Salt dependence experiments were performed in variable amounts of KCl, 10 mM potassium phosphate, 2 mM MgCl2 pH 8. Binding curves were fit to a standard single-site binding model using Scientist (Micromath). Experiments were performed at least twice, and the reported values reflect the average of these experiments. Uncertainties are given as the larger value of either the standard deviation of these values or the propagated uncertainty.
RNA binding assays
Nitrocellulose filter binding assays were used to determine standard binding free energies for binding of RNA to different SNF protein constructs, as described [16]. A constant, picomolar concentration of RNA and variable protein concentrations were used. BSA (Roche) was added to a final concentration of 40 μg/mL. All experiments described in the text were done at pH 7.4. Solution conditions were otherwise variable and are indicated in the text and figures. All experiments were performed in duplicate, and binding curves were fit to a standard Langmuir isotherm using Scientist (Micromath). Kaleidegraph was used to fit van’t Hoff data.
Acknowledgments
This work is supported in part by NIH (SGW and KBH). Thanks to Dr Kim Delaney for cloning human U2B”. Thanks to many members of the Gibbs community who patiently schooled KBH in thermodynamics and appreciated its application to RNA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Will CL, Rumpler S, Gunnewiek J. Klein, van Venrooij WJ, Luhrmann R. In vitro reconstitution of mammalian U1 snRNPs active in splicing: the U1-C protein enhances the formation of early (E) spliceosomal complexes. Nucleic Acids Res. 1996;24:4614–4623. doi: 10.1093/nar/24.23.4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Salz HK, Flickinger TW. Both loss-of-function and gain-of-function mutations in snf define a role for snRNP proteins in regulating Sex-lethal pre-mRNA splicing in Drosophila development. Genetics. 1996;144:95–108. doi: 10.1093/genetics/144.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Stitzinger SM, Conrad TR, Zachlin AM, Salz HK. Functional analysis of SNF, the Drosophila U1A/U2B” homolog: identification of dispensable and indispensable motifs for both snRNP assembly and function in vivo. RNA. 1999;5:1440–1450. doi: 10.1017/s1355838299991306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Saldi T, Wilusz C, MacMorris M, Blumenthal T. Functional redundancy of worm spliceosomal proteins U1A and U2B”. Proc Natl Acad Sci U S A. 2007;104:9753–9757. doi: 10.1073/pnas.0701720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Flickinger TW, Salz HK. The Drosophila sex determination gene snf encodes a nuclear protein with sequence and functional similarity to the mammalian U1A snRNP protein. Genes Dev. 1994;8:914–925. doi: 10.1101/gad.8.8.914. [DOI] [PubMed] [Google Scholar]
- [6].Hall KB, Stump WT. Interaction of N-terminal domain of U1A protein with an RNA stem/loop. Nucleic Acids Res. 1992;20:4283–4290. doi: 10.1093/nar/20.16.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Williams DJ, Hall KB. RNA hairpins with non-nucleotide spacers bind efficiently to the human U1A protein. J Mol Biol. 1996;257:265–275. doi: 10.1006/jmbi.1996.0161. [DOI] [PubMed] [Google Scholar]
- [8].Nagai K, Oubridge C, Jessen TH, Li J, Evans PR. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- [9].Avis JM, Allain FH, Howe PW, Varani G, Nagai K, Neuhaus D. Solution structure of the N-terminal RNP domain of U1A protein: the role of C-terminal residues in structure stability and RNA binding. J Mol Biol. 1996;257:398–411. doi: 10.1006/jmbi.1996.0171. [DOI] [PubMed] [Google Scholar]
- [10].Oubridge C, Ito N, Evans PR, Teo CH, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- [11].Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B”-U2A’ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- [12].Hu J, Cui G, Li C, Liu C, Shang E, Lai L, Jin C, Wang J, Xia B. Structure and novel functional mechanism of Drosophila SNF in sex-lethal splicing. PLoS One. 2009;4:e6890. doi: 10.1371/journal.pone.0006890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Williams SG, Hall KB. Coevolution of Drosophila snf protein and its snRNA targets. Biochemistry. 2010;49:4571–4582. doi: 10.1021/bi100374d. [DOI] [PubMed] [Google Scholar]
- [14].Rimmele ME, Belasco JG. Target discrimination by RNA-binding proteins: role of the ancillary protein U2A’ and a critical leucine residue in differentiating the RNA-binding specificity of spliceosomal proteins U1A and U2B”. RNA. 1998;4:1386–1396. doi: 10.1017/s1355838298981171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Scherly D, Boelens W, Dathan NA, van Venrooij WJ, Mattaj IW. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B” and their cognate RNAs. Nature. 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- [16].Hall KB. Interaction of RNA hairpins with the human U1A N-terminal RNA binding domain. Biochemistry. 1994;33:10076–10088. doi: 10.1021/bi00199a035. [DOI] [PubMed] [Google Scholar]
- [17].Kranz JK, Hall KB. RNA binding mediates the local cooperativity between the beta-sheet and the C-terminal tail of the human U1A RBD1 protein. J Mol Biol. 1998;275:465–481. doi: 10.1006/jmbi.1997.1441. [DOI] [PubMed] [Google Scholar]
- [18].Weber G, Trowitzsch S, Kastner B, Luhrmann R, Wahl MC. Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010;29:4172–4184. doi: 10.1038/emboj.2010.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lohman TM, Mascotti DP. Thermodynamics of ligand-nucleic acid interactions. Methods Enzymol. 1992;212:400–424. doi: 10.1016/0076-6879(92)12026-m. [DOI] [PubMed] [Google Scholar]
- [20].Ha JH, Spolar RS, Record MTJ. Role of the hydrophobic effect in stability of site-specific protein-DNA complexes. J Mol Biol. 1989;209:801–816. doi: 10.1016/0022-2836(89)90608-6. [DOI] [PubMed] [Google Scholar]
- [21].Ferrari ME, Lohman TM. Apparent heat capacity change accompanying a nonspecific protein-DNA interaction. Escherichia coli SSB tetramer binding to oligodeoxyadenylates. Biochemistry. 1994;33:12896–12910. doi: 10.1021/bi00209a022. [DOI] [PubMed] [Google Scholar]
- [22].Krummel D.A. Pomeranz, Oubridge C, Leung AK, Li J, Nagai K. Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution. Nature. 2009;458:475–480. doi: 10.1038/nature07851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- [24].Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stump WT, Hall KB. SP6 RNA polymerase efficiently synthesizes RNA from short double-stranded DNA templates. Nucleic Acids Res. 1993;21:5480–5484. doi: 10.1093/nar/21.23.5480. [DOI] [PMC free article] [PubMed] [Google Scholar]


