Abstract
The TFIID complex interacts with at least three types of core promoter elements within protein-coding genes, including TATA, initiator (Inr), and downstream promoter elements. We have begun to explore the mechanism by which the TFIID–Inr interaction leads to functional synergy between TATA and Inr elements during both basal and activated transcription. In DNase I footprinting assays, GAL4–VP16 recruited TFIID–TFIIA to core promoters containing either a TATA box, an Inr, or both TATA and Inr elements, with synergistic interactions apparent on the TATA–Inr promoter. Appropriate spacing between the two elements was essential for the synergistic binding. Despite the sequence-specific TFIID–Inr interactions, gel shift experiments revealed that TFIID alone possesses similar affinities for the TATA–Inr and TATA promoters. Interestingly, however, recombinant TFIIA strongly and selectively enhanced TFIID binding to the TATA–Inr promoter, with little effect on binding to the TATA promoter. Studies of the natural adenovirus major late promoter confirmed these findings, despite the existence of specific but nonfunctional TFIID interactions downstream of the Inr in that promoter. These results suggest that a TFIIA-induced conformational change is essential for the sequence-specific TFIID–Inr interaction to occur with sufficient affinity to support the functional synergism between TATA and Inr elements.
Keywords: TFIID, TFIIA, TATA box, Inr, transcription, RNA polymerase II
The frequency of transcription initiation from a protein-coding gene is determined by an array of DNA sequence elements and by complex physical and functional interactions between factors that bind those elements. Nucleosomes and other chromatin-associated proteins contribute additional layers to the transcriptional regulation process. Because of this complexity, an understanding of gene regulation programs must begin with a dissection of the basic mechanisms of transcription initiation by RNA polymerase II and of the fundamental mechanisms of activation.
Fractionation of nuclear extracts that support accurate transcription initiation by RNA polymerase II led to the discovery of a set of essential general factors (Orphanides et al. 1996; Roeder 1996). In reactions reconstituted with these factors, initiation is preceded by the formation of a stable preinitiation complex nucleated by an interaction between the TATA box-binding protein (TBP) and a control element found in the promoters for most protein-coding genes, the TATA box. The TBP–TATA interaction is stabilized by TFIIA, with the final preinitiation complex including TFIIB, TFIIF, TFIIE, TFIIH, and RNA polymerase II.
The above factors constitute the minimal requirements for accurate transcription initiation, but they provide an oversimplification of this basic process as it occurs in vivo. First, within a cell, many of the general factors appear to be preassociated with the core RNA polymerase II in an RNA polymerase II holoenzyme, which contains several additional proteins of unknown function (Koleske and Young 1995). Second, TBP is not present as an isolated protein, but is instead a component of a large multiprotein complex known as TFIID, which contains other subunits referred to as TAFs (TBP-associated factors) (Burley and Roeder 1996; Verrijzer and Tjian 1996).
An additional complication relating to the mechanism of basal transcription is that many protein-coding genes contain important core promoter elements in addition to, or instead of, a TATA box (Smale 1997). Some TATA-containing promoters contain initiator (Inr) elements overlapping the transcription start site, with the two core elements strongly synergizing with each other. Many other promoters lack TATA boxes altogether; some of these TATA-less promoters contain Inr elements as functional analogs of TATA boxes (Smale 1997) or Inr elements combined with downstream promoter elements (DPEs; Burke and Kadonaga 1996).
Studies of Inr elements by several laboratories have provided evidence that their function depends on direct binding by a component of the TFIID complex. Specific binding of purified TFIID to consensus Inr elements has been observed in DNase I footprinting, electrophoretic mobility shift assay (EMSA), and binding site selection experiments (Zhou et al. 1992; Wang and Van Dyke 1993; Kaufmann and Smale 1994; Purnell et al. 1994; Bellorini et al. 1996). More recently, Burke and Kadonaga (1996) showed that Inr elements act in concert with a newly defined DPE to direct stable binding by purified Drosophila TFIID; stable binding required both elements and was not observed with either element alone. The TAF that recognizes the Inr has not been defined, but TAF250 is a leading candidate based on cross-linking studies and reconstitution experiments with partial TBP–TAF complexes (Verrijzer et al. 1995; Oelgeschäger et al. 1996).
TFIID-dependent Inr activity has not been reconstituted with a complete set of purified factors, but in addition to the factors required for TATA activity, a protein called TAF150 in Drosophila or CIF150 in humans is essential for an Inr to function when located downstream of a TATA box (Verrijzer et al. 1994, 1995; Kaufmann et al. 1996; Kaufmann et al. 1998). This protein, which is tightly associated with TFIID in Drosophila cells but not in human cells, binds DNA by itself and stabilizes TFIID binding to DNA (Verrijzer et al. 1994, 1995; Kaufmann et al. 1998). TAF150/CIF150 does not appear to be responsible for specific recognition of Inr elements, however, as mutations that disrupt Inr function do not disrupt its ability to bind DNA or to stabilize TFIID binding. Furthermore, human TFIID complexes lacking CIF150 retain their capacity to bind Inr elements in a sequence-specific manner (Kaufmann and Smale 1994, see below). Other proteins, including TFII-I, YY1, and RNA polymerase II, may contribute to the function of Inr elements in some promoters (Roy et al. 1991; Usheva and Shenk 1994; Weis and Reinberg 1997).
The knowledge of the basic initiation reaction has provided a framework for analyzing mechanisms of transcriptional activation. The precise activation mechanisms have not been established, but appear to include direct interactions between activator proteins and components of the general transcription machinery or adaptor proteins that interact with the general factors (Hori and Carey 1994; Roberts et al. 1995; Zawel and Reinberg 1995; Ge et al. 1996; Goodrich et al. 1996; Kaiser and Meisterernst 1996; Stargell and Struhl 1996; Struhl 1996; Tantin et al. 1996; Verrijzer and Tjian 1996; Ptashne and Gann 1997). One mechanism that appears to contribute to activation is the direct recruitment of the TFIID complex to the core promoter by activator proteins in a TAF- and TFIIA-dependent manner. Several in vitro experiments support this model, including DNase I footprinting and EMSAs, in which promoter-bound activators promoted the binding of TFIID and TFIIA to core promoters (Wang et al. 1992; White et al. 1992; Kaufmann and Smale 1994; Lieberman and Berk 1994; Chi et al. 1995; Kobayashi et al. 1995; Sauer et al. 1995; Shykind et al. 1995; Chi and Carey 1996; Struhl 1996; Shykind et al. 1997). The activator–TFIID–TFIIA interaction appears to be accompanied by a conformational change in TFIID that results in enhanced interactions downstream of the TATA box (Horikoshi et al. 1988a,b; Chi and Carey 1996). This conformational change may be necessary for activation, perhaps by facilitating the entry of TFIIB into the preinitiation complex (Horikoshi et al. 1988b; Chi and Carey 1996). In vivo studies in Saccharomyces cerevisiae have provided strong support for this recruitment model (Stargell and Struhl 1996; Struhl 1996).
Despite the evidence that both TATA and Inr elements are recognized by TFIID and that TFIID recruitment is an important event during transcriptional activation, little is known about the mechanisms of functional synergy between TATA and Inr elements and about the influence of core promoter architecture on TFIID recruitment. To provide insight into these issues, we compared the functional activities of synthetic and natural core promoters with their TFIID–TFIIA-binding and recruitment properties. The results provide a mechanism for the functional synergy between TATA and Inr elements during basal and activated transcription, and reveal a new function for TFIIA.
Results
GAL4–VP16 recruits TFIID–TFIIA to interact with heterogeneous core promoters
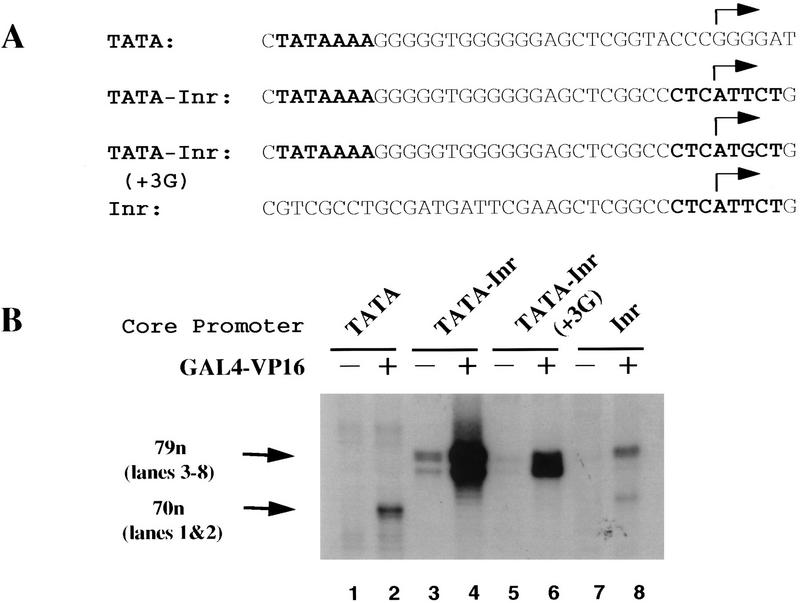
To investigate the influence of core promoter architecture on TFIID binding and recruitment, we performed DNase I footprinting experiments with three synthetic promoters containing either a consensus TATA box from the adenovirus major late (AdML) promoter, a consensus Inr from the terminal transferase (TdT) promoter, or both TATA and Inr elements separated by 25 bp (Fig. 1A; see also Emami et al. 1995). A fourth promoter was identical to TATA–Inr, but contained a single base pair substitution at nucleotide +3 (+3G) that reduces Inr activity more severely than any other single base pair change (Javahery et al. 1994; Lo and Smale 1996). These elements were located downstream of five binding sites for the yeast activator, GAL4 (see Materials and Methods). A recombinant fusion protein containing the GAL4 DNA-binding domain (amino acids 1–147) fused to the full-length transcriptional activation domain of the herpes simplex virus VP16 protein (amino acids 411 to 490; Carey et al. 1990) activates transcription from all four core promoters when added to HeLa nuclear extracts (Fig. 1B). By primer extension analysis of the RNA products, the TATA promoter yields cDNA products with an expected size of 70 nucleotides (lanes 1,2), whereas the TATA–Inr, TATA–Inr(+3G), and Inr promoters yield cDNA products with an expected size of 79 nucleotides (lanes 3–8). With both basal and activated transcription, the TATA and Inr elements act in a synergistic manner (Fig. 1B), with the activator stimulating transcription to a similar extent with each promoter. These results are qualitatively and quantitatively very similar to those obtained previously, both in vitro and in vivo, with the GAL4 DNA-binding domain fused to the amino-terminal half of the VP16 activation domain (Emami et al. 1995).
Figure 1.
Recruitment of TFIID by GAL4–VP16 to core promoters with distinct architectures. (A) Sequences of the TATA, TATA–Inr, TATA–Inr(+3G), and Inr core promoters. The location of the transcription start site is indicated (arrow). Boldface type denotes the AdML TATA and TdT Inr elements. (B) In vitro transcription reactions were performed in HeLa nuclear extracts with promoters containing multiple GAL4-binding sites upstream of the TATA (lanes 1,2), TATA–Inr (lanes 3,4), TATA–Inr(+3G) (lanes 5,6), and Inr (lanes 7,8) core promoters. Reactions were performed in the absence (lanes 1,3,5,7) or presence (lanes 2,4,6,8) of 600 ng GAL4–VP16. Arrows indicate the locations of the expected 70 (lanes 1,2) and 79 (lanes 3–8) nucleotide cDNA products following primer extension analysis. The TATA product is 9 nucleotides shorter than the other products because the TATA plasmid contains a 9-bp deletion of the Inr element, which reduces the distance from the transcription start site to the primer binding site. (C) DNase I footprinting experiments were performed with probes containing 5 GAL4-binding sites upstream of the TATA–Inr (lanes 1–7), Inr (lanes 8–14), and TATA (lanes 15–21) core promoters. The three probes were prepared simultaneously with the same radiolabeled primer and, therefore, are of the same specific activity. Binding reactions contained no protein or various combinations of purified TFIID (1 μl), purified TFIIA (0.1 μl), and GAL4–VP16 (600 ng), as indicated at the top. The locations of the GAL4-binding sites, TATA, and Inr elements are indicated to the right of each panel. Also indicated is a strong hypersenstive site at nucleotide +5 (arrow) and the protected region observed downstream of the Inr (DS).
DNase I footprinting assays were performed with radiolabeled probes spanning the TATA–Inr, Inr, and TATA promoters, by use of purified, recombinant GAL4–VP16, recombinant TFIIA, and epitope-tagged, immunoaffinity-purified TFIID (Zhou et al. 1992; Kaufmann and Smale 1994). It is noteworthy that the TFIID used for this study lacks a detectable homolog of the Drosophila TAF150 protein (Verrijzer et al. 1994, 1995). Although we cannot exclude the possibility that a TAF150 homolog is present at sub-stoichiometric amounts, subunits of TFIID other than TAF150 are likely to be responsible for the protein–DNA interactions described below.
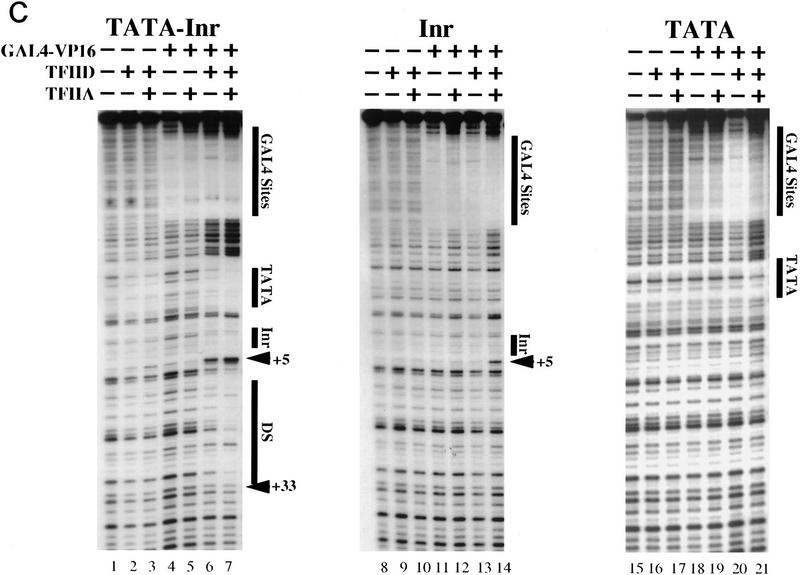
The footprinting results shown in the left panel of Figure 1C show that GAL4–VP16 is capable of recruiting TFIID to bind efficiently to the core promoter containing both TATA and Inr elements, with binding enhanced by TFIIA. Lanes 2 and 3 show that limiting amounts of TFIID in the absence of GAL4–VP16 and in the absence or presence of TFIIA result in weak protection of the TATA box, and very weak protection of the regions surrounding and downstream of the transcription start site. The degree to which TFIIA influenced TFIID binding in the DNase I footprinting assay varied, with the primary effects being slightly enhanced protection of the TATA box and the appearance of a hypersensitive site at nucleotide +5 (see also Fig. 2, lanes 2,3). Inclusion of GAL4–VP16 in the binding reactions, in the absence of TFIIA, also resulted in enhancement of the hypersensitive site at +5 and, in some experiments, enhanced the protection of the TATA box and downstream region (Fig. 1C, lane 6; see also Fig. 2, lane 6). In the presence of TFIIA, TFIID, and GAL4–VP16, strong protection of the TATA box and of the region downstream of the start site were observed, along with further enhancement of the hypersensitive site at +5 (Fig. 1C, lane 7; see also Fig. 2, lane 7). These results are largely consistent with those obtained with the Epstein–Barr virus Zebra (Zta) activator bound upstream of the adenovirus E4 (AdE4) core promoter (Lieberman and Berk 1994; Chi et al. 1995; Chi and Carey 1996), which appears to contain both TATA and Inr elements.
Figure 2.
A single base-pair substitution at nucleotide +3 disrupts TFIID interactions with the Inr and downstream region. DNase I footprinting experiments were performed as described in the legend to Fig. 1 and in Materials and Methods with probes derived from the TATA–Inr (lanes 1–7) and TATA–Inr(+3G) (lanes 8–14) promoters. The protein amounts used in the reactions are the same as indicated in the legend to Fig. 1, and the proteins added are indicated above each lane. The GAL4 sites, the TATA and Inr elements, the hypersensitive site at +5, and the region of downstream protection (DS) are indicated to the right.
The interactions detected with the Inr promoter and TATA promoter (Fig. 1C, lanes 8–21) were much less extensive than with the TATA–Inr promoter. With the Inr promoter (Fig. 1C, lanes 8–14), no protection of the core promoter was observed, even in the presence of GAL4–VP16, TFIID, and TFIIA (Fig. 1C, lane 14). However, the hypersensitive site at +5 provided evidence of a weak interaction at the Inr. With the TATA promoter (Fig. 1C, lanes 15–21), the core promoter interactions also appeared weak, with moderate protection of the TATA box observed only in the presence of GAL4–VP16, TFIID, and TFIIA (Fig. 1C, lane 21); no interactions were observed downstream of the TATA box. The synergistic interactions between TFIID–TFIIA and the TATA–Inr promoter correlate with the functional synergy between TATA and Inr, suggesting that these elements may influence promoter strength simply by modulating the affinity of TFIID–TFIIA for the promoter.
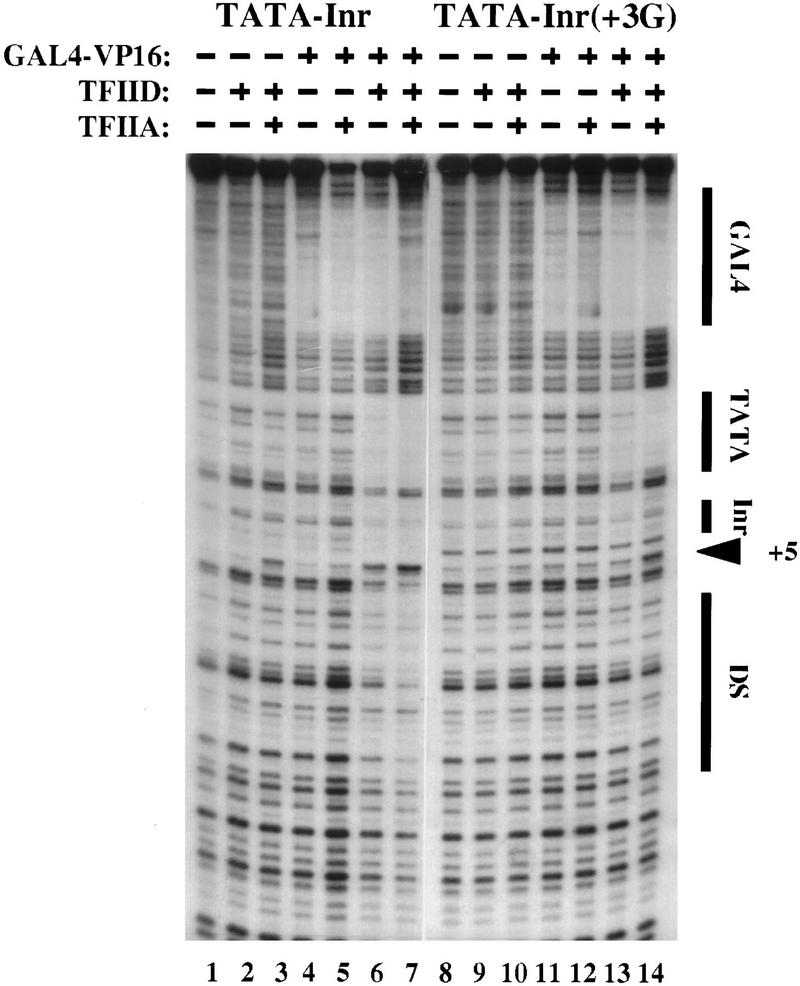
To provide a more rigorous test of the Inr-dependence of TFIID binding, the TATA–Inr(+3G) promoter was tested in the DNase I footprinting assay. The +3G mutation greatly weakened the TFIID interactions downstream of the TATA box (Fig. 2, cf. lanes 1–7 with lanes 8–14). This difference is most apparent when comparing the reactions performed with GAL4–VP16, TFIID, and TFIIA (Fig. 2, lanes 7,14). With the mutant promoter (Fig. 2, lane 14), no protection was observed at the Inr or downstream of the Inr, although a weak hypersensitive site remained. This hypersensitive site may be caused by the weak Inr activity that is retained with the mutant element. Alternatively, in the presence of a TATA box, the protein–DNA contact that yields the hypersensitive site may be Inr-independent, but enhanced by the presence of a consensus Inr. Regardless of the explanation, the data in Figure 2 show that TFIID–TFIIA can be recruited by GAL4–VP16 to interact with the TATA box within either the TATA–Inr or TATA–Inr(+3G) promoters. With the TATA–Inr(+3G) promoter, the interactions are largely restricted to the TATA box, whereas with the TATA–Inr promoter, the interactions are much more extensive.
As an important control for the above experiments, DNase I footprinting reactions were performed with the TATA–Inr promoter and purified, recombinant TBP in place of the purified TFIID complex. Previous studies had shown that the Inr does not function in transcription experiments performed with recombinant TBP (Kaufmann and Smale 1994; Kaufmann et al. 1996). Consistent with those data, concentrations of TBP and TFIIA that saturate the TATA box revealed no interactions at the Inr or downstream region (Fig. 3, lanes 1–6). Furthermore, GAL4–VP16 had no effect on the TBP–TFIIA interactions (data not shown). Thus, the interactions observed at the Inr and downstream region in Figures 1 and 2 are dependent on the TAFs within the TFIID complex.
Figure 3.
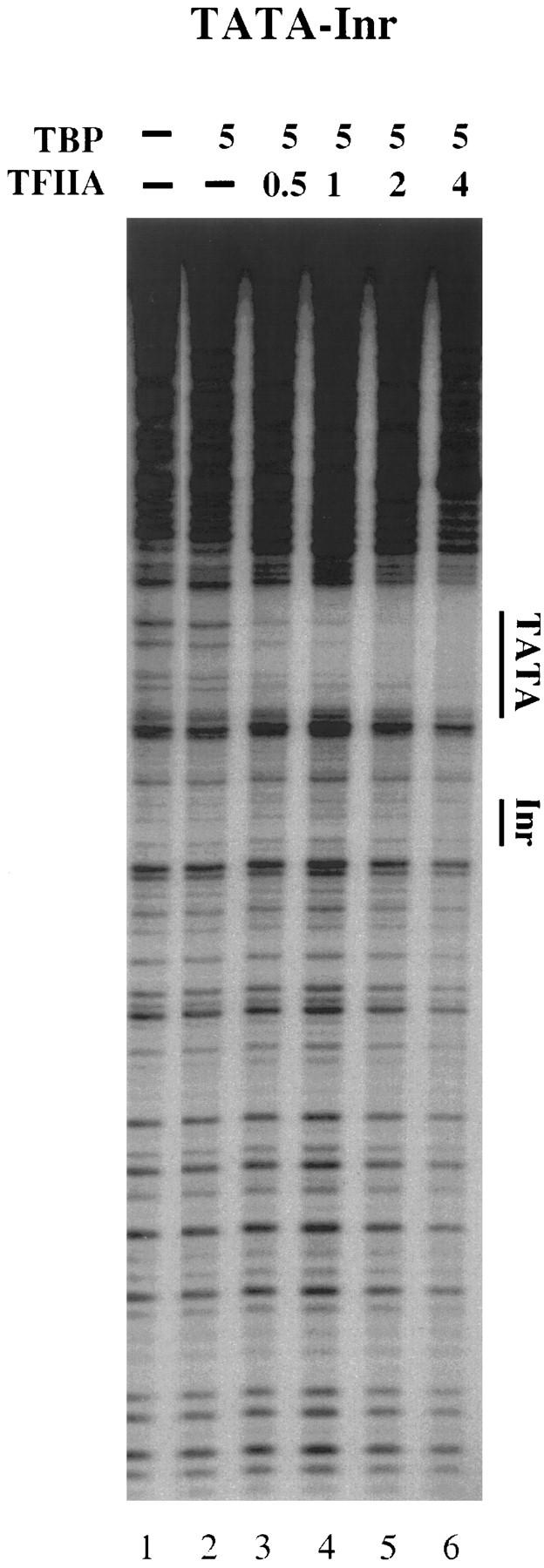
Binding of TBP to the TATA–Inr core promoter.DNase I footprinting reactions were performed with a probe containing the TATA–Inr core promoter. Reactions containing purified human TBP (5 μl of a 2 ng/μl solution; lanes 2–6) and 0.5, 1, 2, or 4 μl of a 10 ng/μl solution of TFIIA (lanes 3–6). The locations of the TATA box and Inr element are indicated to the right.
Spacing constraints of TFIID–TFIIA interactions with TATA–Inr core promoters
In the synthetic TATA–Inr promoter employed above, the TATA and Inr elements are separated by 25 bp (the distance from the final adenosine of the TATA sequence, TATAAAA, to the adenosine within the Inr). Previously, we reported that this spacing is critical for synergy between TATA and Inr elements during basal and Sp1-activated transcription (O’Shea-Greenfield and Smale 1992). When the distance between the TATA and Inr elements was increased to 30, 35, or 40 bp, promoter strength was greatly reduced. Interestingly, when the spacing was reduced to 20 or 15 bp, the TATA–Inr synergy was retained; promoter strength was comparable to that observed with the 25-bp spacing, but all of the transcripts initiated ∼25 bp downstream of the TATA box (O’Shea-Greenfield and Smale 1992). This result suggests that the Inr contributes to promoter strength when separated from the TATA box by 20 or 15 bp, but the TATA box is dominant in determining the transcription start site. Similar results were obtained when the core promoters with variable spacing were tested in the absence of activator (O’Shea-Greenfield and Smale 1992) or when activated by Sp1 (O’Shea-Greenfield and Smale 1992) or GAL4–VP16 (data not shown).
DNase I footprinting experiments were performed with probes containing these promoters to determine whether the TFIID-binding characteristics can explain the functional spacing constraints (Fig. 4). All binding reactions contained GAL4–VP16, TFIIA, and a more concentrated preparation of TFIID than that employed for Figures 1 and 2. Furthermore, all of the probes were prepared by PCR with the same radiolabeled primer (to insure comparable specific activities).
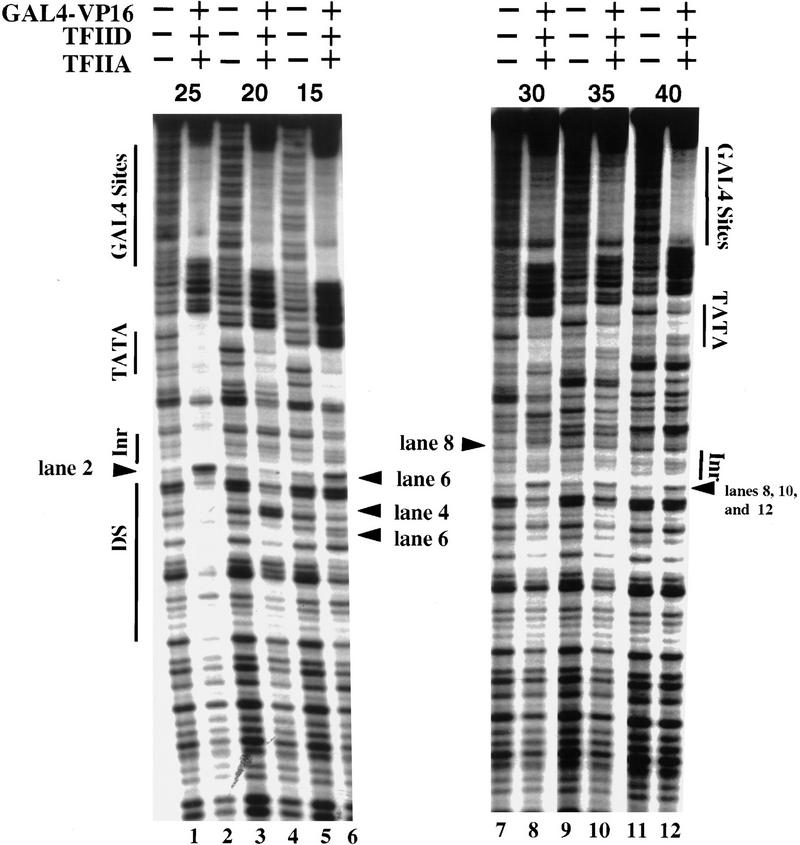
Figure 4.
Efficient protection of the Inr and downstream region within the TATA–Inr core promoter depend on a precise spacing between the TATA and Inr elements. DNase I footprinting reactions were performed with probes derived from the promoters containing 5 GAL4-binding sites upstream of TATA and Inr elements separated by variable spacing (indicated above each pair of lanes). The reactions contained either no protein (odd numbered lanes) or GAL4–VP16, TFIID (the TFIID preparation used for this experiment was ∼5 times more concentrated than the preparation used for Figs. 1 and 2), and TFIIA (even numbered lanes), as described in the legend to Fig. 1 and in Materials and Methods. All six probes were prepared by PCR with the same radiolabeled PCR primer to insure that the specific activities of the probes were similar. The left and right panels were from the same experiment and the same polyacrylamide gel. The locations of the GAL4-binding sites, the TATA and Inr elements, the hypersensitive site at +5 and the downstream protection observed in lane 2 (DS) are indicated to the left and right. Also indicated by arrows are the locations of hypersensitive sites observed in lanes 4, 6, and 8.
As expected, the concentrated TFIID bound very efficiently to the promoter in which the TATA and Inr elements were separated by 25 bp (Fig. 4, lane 2), with strong protection of the TATA box, the Inr, and the downstream region, and with a strong hypersensitive site at +5. In contrast, with all of the probes containing altered spacing, the interactions were reduced considerably. With each probe, TFIID binding to the TATA box was strong, but reduced relative to the wild-type TATA–Inr promoter. Furthermore, the contacts observed at the Inr and downstream region were greatly diminished. Interestingly, with TATA–Inr spacings of 15, 20, and 30 bp, new hypersensitive sites appeared ∼30 bp downstream of the TATA box (Fig. 4, lanes 4,6,8).
The TATA–Inr spacing constraints and the locations of the hypersensitive sites suggest that TFIID binds to the core promoters in a stereospecific manner and interacts, or induces a structural change in the DNA, ∼30 bp downstream of the TATA box. Presumably, if an Inr is located at an appropriate location relative to the TATA box (i.e., 25 bp), it enhances this interaction or structural change and also leads to more extensive protection of the start site and downstream regions. The stereospecific binding characteristics provide an attractive explanation for the greatly reduced functional activities of the promoters containing TATA–Inr spacings of 30, 35, and 40 bp. Given the binding data, however, the maintenance of functional synergy with TATA–Inr spacings of 20 and 15 bp is surprising. This apparent inconsistency suggests that the mechanistic basis of the functional synergy between TATA and Inr elements may actually be independent of the synergistic TFIID–TFIIA binding that is observed. A more likely explanation is that other components of the in vitro transcription reactions (e.g., topoisomerase I) may modify the DNA-binding properties of TFIID and TFIIA, leading to enhanced interactions despite the reduced TATA–Inr spacing.
The Inr influences the affinity of TFIID–TFIIA for a promoter, but not the affinity of TFIID alone
The reduced TFIID binding observed when the Inr is severely altered (Fig. 1C) or moved to a different location relative to the TATA box (Fig. 4) suggests that the primary function of the Inr in a TATA-containing promoter may be to increase the affinity of TFIID for the promoter, resulting in enhanced promoter strength. Relative TFIID affinities were difficult to determine with the DNase I footprinting assay, however, because of the large number of TFIID contacts and the lack of linearity when the concentration of TFIID in the binding reaction was titrated (data not shown).
As an alternative method for comparing the relative affinities of TFIID for the different core promoters, a Mg2+ agarose EMSA was employed (Zhou et al. 1992; Lieberman and Berk 1994). Radiolabeled probes were prepared by PCR with the same primers and templates used for the DNase I footprinting analysis. For each set of reactions shown, all probes were prepared with the same radiolabeled primers to insure similar specific activities. Activator was not used for these experiments because the mobilities of complexes containing and lacking activator are difficult to distinguish (Zhou et al. 1992; Lieberman and Berk 1994).
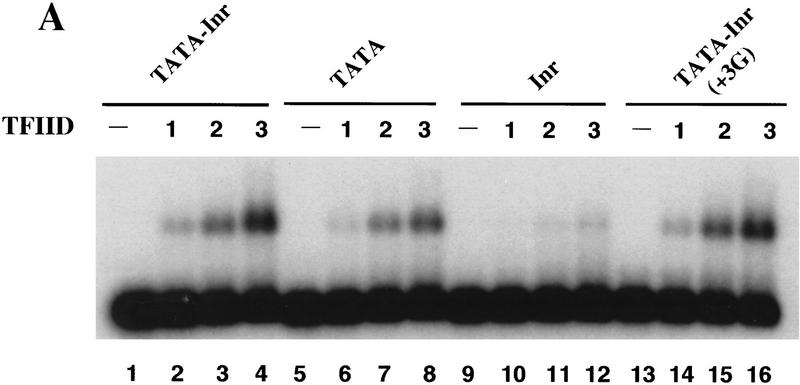
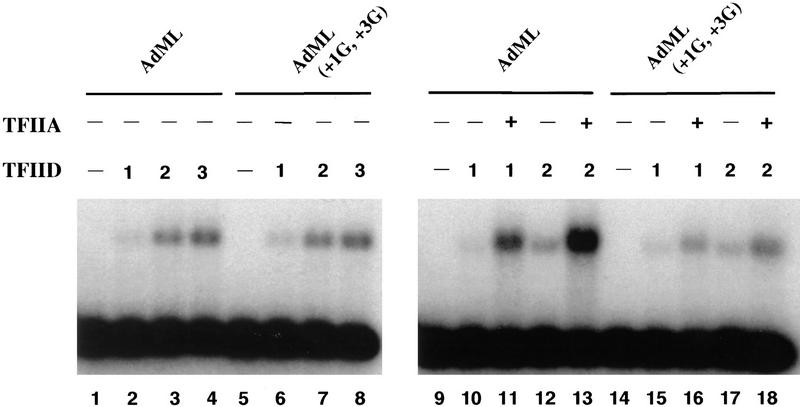
The results in Figure 5A compare the binding of TFIID alone with the TATA–Inr, TATA, Inr, and TATA–Inr(+3G) promoters. Three different concentrations of purified TFIID were tested with each probe. With the TATA–Inr promoter, a single protein–DNA complex was observed with a migration comparable to that observed in previous studies (Zhou et al. 1992; Lieberman and Berk 1994). With the TATA promoter, the abundance of the complex was only slightly reduced (by an average of 25%–40%) at each concentration of TFIID (Fig. 5A, cf. lanes 1–4 with lanes 5–8). Although this assay does not provide a precise measure of affinities, this result suggests that the affinity of TFIID for the TATA–Inr promoter is only slightly greater than its affinity for the TATA promoter. The affinity of TFIID for the Inr promoter appears to be reduced to a much greater extent, because the protein–DNA complex was barely detectable (Fig. 5, lanes 9–12). Finally, with the TATA–Inr(+3G) promoter, the amount of complex observed with each concentration of TFIID was routinely indistinguishable from that observed with the TATA–Inr promoter (Fig. 5, cf. lanes 1–4 with lanes 13–16). Thus, TFIID appears to bind with a similar affinity to the three TATA-containing promoters, TATA–Inr, TATA, and TATA–Inr(+3G), despite the striking differences in their functional activities.
Figure 5.
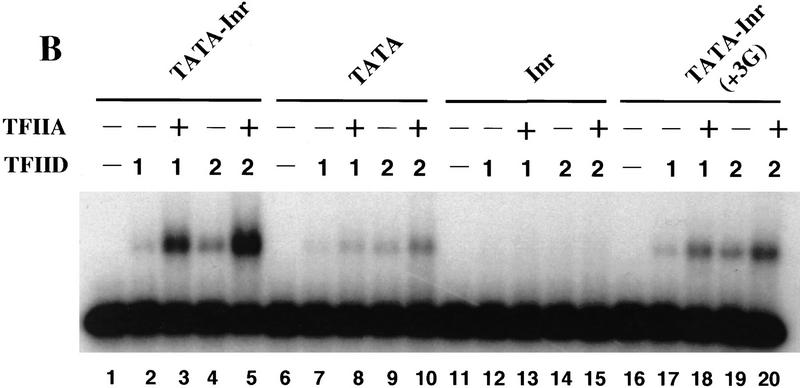
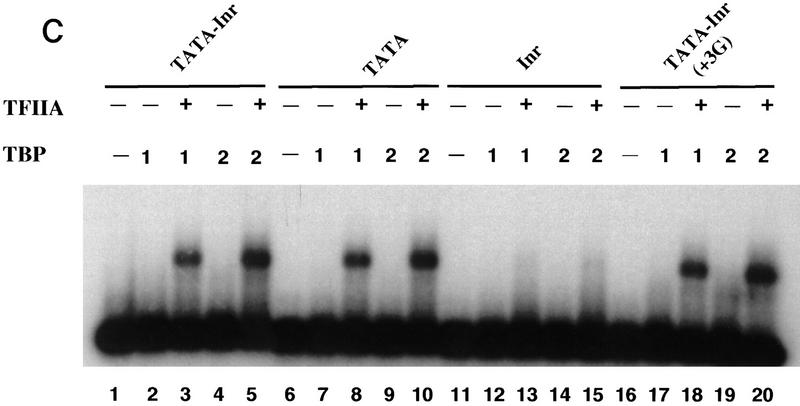
TFIIA selectively enhances TFIID binding to core promoters containing a functional Inr element as well as a TATA box. (A) Mg2+ agarose EMSAs (Zhou et al. 1992; Lieberman and Berk 1994) were performed with 0 (lanes 1,5,9,13), 1 (lanes 2,6,10,14), 2 (lanes 3,7,11,15), and 3 (lanes 4,8,12,16) μl of purified TFIID. Radiolabeled probes were prepared by PCR from plasmids containing the TATA–Inr (lanes 1–4), TATA (lanes 5–8), Inr (lanes 9–12), and TATA–Inr(+3G) (lanes 13–16) core promoters. All probes were prepared with the same radiolabeled primer, insuring that the probes were of similar specific activity. Complex abundances for lanes 1–16 relative to that in lane 2 (1.0) are 0, 1.0, 3.2, 4.6, 0, 0.8, 3.2, 4.5, 0, 0, 0.2, 0.3, 0, 1.0, 2.7, and 4.5, respectively. (B) Mg2+ agarose EMSAs were performed with the TATA–Inr (lanes 1–5), TATA (lanes 6–10), Inr (lanes 11–15), and TATA–Inr(+3G) (lanes 16–20) probes. Reactions contained 0 (lanes 1,6,11,16), 1 (lanes 2,3,7,8,12,13,17,18), or 2 (lanes 4,5,9,10,14,15,19,20) μl of purified TFIID and either 0 (lanes 1,2,4,6,7,9,11,12,14,16,17,19) or 200 (lanes 3,5,8,10,13,15,18,20) ng TFIIA. Complex abundances for lanes 1–20 relative to that in lane 2 (1.0) are 0, 1, 5.9, 2.7, 12.9, 0, 0.4, 1.3, 1.5, 2.5, 0, 0, 0, 0, 0, 0, 1.0, 3.6, 2.6, and 5.9, respectively. (C) Mg2+ agarose EMSAs were performed as in part B, but with 1 (lanes 2,3,7,8,12,13,17,18) or 2 (lanes 4,5,9,10,14,15,19,20) μl of a 2 ng/μl solution of TBP, and with 10 ng of TFIIA (lanes 3,5,8,10,13,15,18,20). The concentration of TFIIA that was optimal for TBP stabilization (10 ng) was greatly suboptimal for TFIID stabilization (200 ng optimum).
In contrast to the results described above, a very different picture emerged when TFIIA was included in the binding reactions (Fig. 5B). The results obtained with the TATA–Inr and TATA–Inr(+3G) promoters provide the most relevant comparison. As above, the protein–DNA complexes observed with TFIID alone are indistinguishable (Fig. 5B, cf. lanes 2–17 and lanes 4–19). However, TFIIA selectively enhanced binding to the TATA–Inr promoter, resulting in a significant difference in complex abundance between the TATA–Inr and TATA–Inr(+3G) promoters (Fig. 5B, cf. lanes 3–18 and lanes 5–20). Quantitation by PhosphorImager analysis revealed that the complexes obtained with TFIID–TFIIA on the TATA–Inr promoter are approximately twofold more abundant than the complexes obtained on the TATA–Inr(+3G) promoter (see figure legend). A more dramatic effect of TFIIA was observed when the TATA–Inr and TATA promoters were compared. With TFIID alone, the complexes observed with the TATA–Inr promoter were only 25%–40% more abundant than the complexes observed with the TATA promoter (Fig. 5B, cf. lanes 2 and 4 with lanes 7 and 9). With TFIID–TFIIA, however, the complexes observed with the TATA–Inr promoter were fivefold more abundant than observed with the TATA promoter (Fig. 5B, cf. lanes 3 and 5 with lanes 8 and 10).
These results do not provide a precise measure of the affinities or relative affinities of the protein–DNA interactions. However, TFIIA clearly is essential for a detectable affinity difference between the TATA–Inr and TATA–Inr(+3G) promoters. The twofold difference in complex abundance in the presence of TFIID and TFIIA can be compared with the 7-fold and 3.5-fold differences in the strengths of these promoters in vitro and in vivo, respectively (Fig. 1B; Lo and Smale 1996). For the TATA–Inr and TATA promoters, the 5-fold difference in complex abundance observed with TFIID and TFIIA can be compared with an ∼12-fold difference in promoter strength (Fig. 1B; Emami et al. 1995). These results suggest that the enhanced strength of a TATA-containing promoter by an Inr is manifested, at least in part, by the Inr’s influence on the affinity of the TFIID–TFIIA complex for the core promoter. TFIIA may be required in addition to TFIID because of its proposed ability to induce a conformational change in TFIID (Oelgeschläger et al. 1996; see Discussion).
Interestingly, TFIIA had little influence on the ability of TFIID to bind to the promoter containing a consensus Inr element, but lacking a TATA box (Fig. 5B, lanes 11–15). The weak complex that is often observed is not significantly above background. Therefore, we have been unable to determine if this complex involves specific interactions with the Inr or interactions with AT-rich sequences within the probe (see Discussion).
It is worth noting that the ability of TFIIA to stabilize TFIID binding to a TATA-containing promoter is small, unless the promoter contains an Inr element. The relative inability of TFIIA to stabilize TFIID binding to the TATA promoter is not caused by general inactivity of the recombinant, human TFIIA preparation used, as this same preparation dramatically stabilized binding of TBP to a TATA oligonucleotide (Fig. 5C). As expected, the Inr had no influence on the TBP-binding activity (Fig. 5C, cf. lanes 1–5 with lanes 6–10). Thus, the DNA-binding stabilization activity of TFIIA appears to be fundamentally distinct with TBP and TFIID. With TBP, TFIIA directly stabilizes the interaction between TBP and the TATA box. With TFIID, we hypothesize that the primary influence of TFIIA on binding affinity results from its ability to induce a conformational change in TFIID that allows it to contact the Inr more tightly, allowing the TFIID–Inr interaction to contribute to the overall affinity of TFIID for the core promoter.
Analysis of the natural AdML core promoter
The data in Figures 1C and 2 suggest that, with the synthetic promoters, the TATA and Inr elements are sufficient for TFIID protection of the TATA box, the transcription start site, and the region downstream of the start site. The downstream contacts do not appear to require elements other than the TATA and Inr. Nevertheless, studies of natural TATA-containing core promoters, including the AdML and Drosophila hsp70 promoters, have suggested that TFIID makes sequence-specific contacts downstream of the Inr (Nakajima et al. 1988; Chiang et al. 1993; Purnell and Gilmour 1993; Verrijzer et al. 1995). To study the significance of TFIID interactions with the Inr and downstream regions in greater detail, we performed DNase I footprinting, EMSA, and in vitro transcription experiments with the natural AdML promoter. The AdML promoter contains the same TATA box employed in the synthetic promoters described above and a strong Inr element that is almost identical to the TdT Inr (CCTCACTCT vs. CCTCATTCT). To simplify these experiments, they were performed in the absence of activator.
DNase I footprinting experiments with the AdML core promoter revealed the expected protection pattern (Nakajima et al. 1988), with strong protection of the TATA box, start-site region, and downstream region, and with multiple hypersensitive bands (Fig. 6A, lanes 7–9). These interactions appear to be stronger than those observed with the synthetic TATA–Inr promoter, as a comparable amount of TFIID yielded complete protection of sequences within the AdML downstream region, but only partial protection of sequences within the TATA–Inr downstream region (data not shown). Despite this apparent difference, a titration of TFIID with the two promoters, in the presence of a constant amount of TFIIA, revealed that a similar TFIID concentration was required for protection of both promoters (data not shown). This result suggests that TFIID–TFIIA binds with a comparable affinity to the two promoters (see below). The small contribution of the AdML downstream sequences to TFIID-binding affinity is consistent with previous DNase I footprinting studies, which revealed that the downstream interactions are extremely sensitive to competition by poly (dG.dC) (Oelgeschläger et al. 1996).
Figure 6.
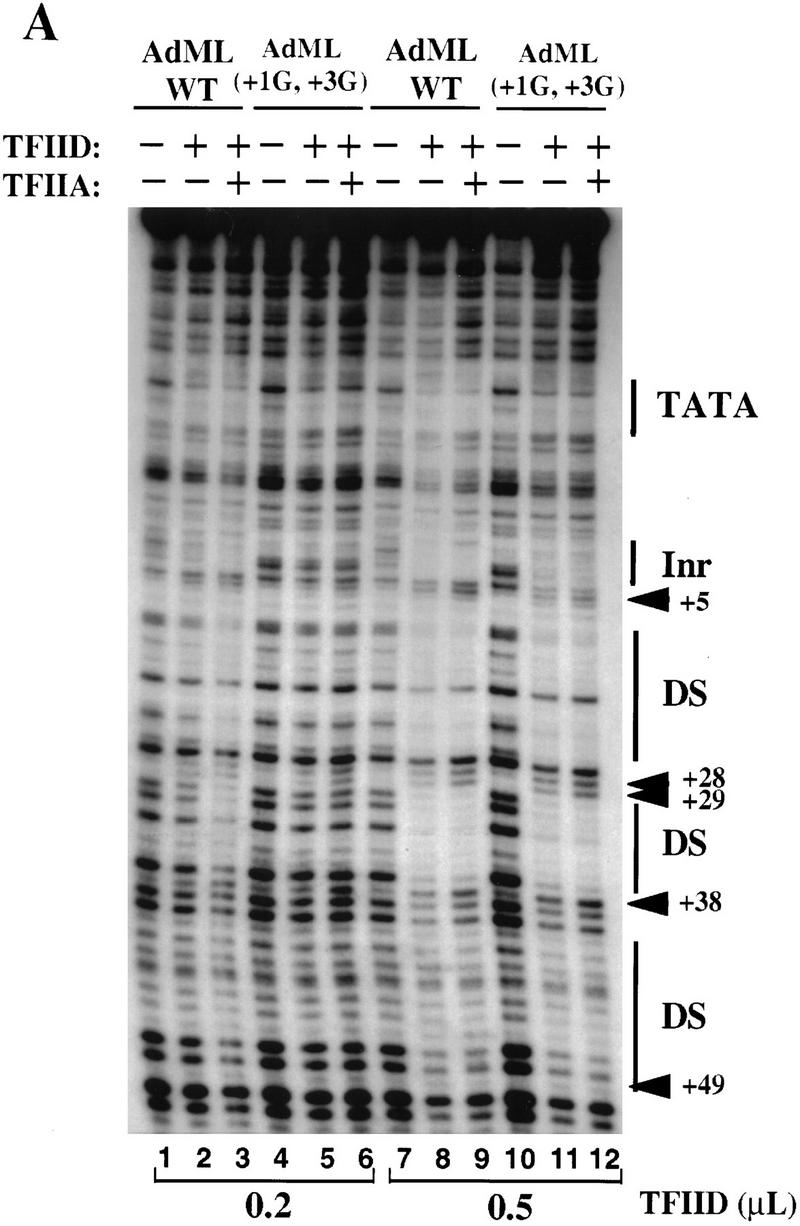
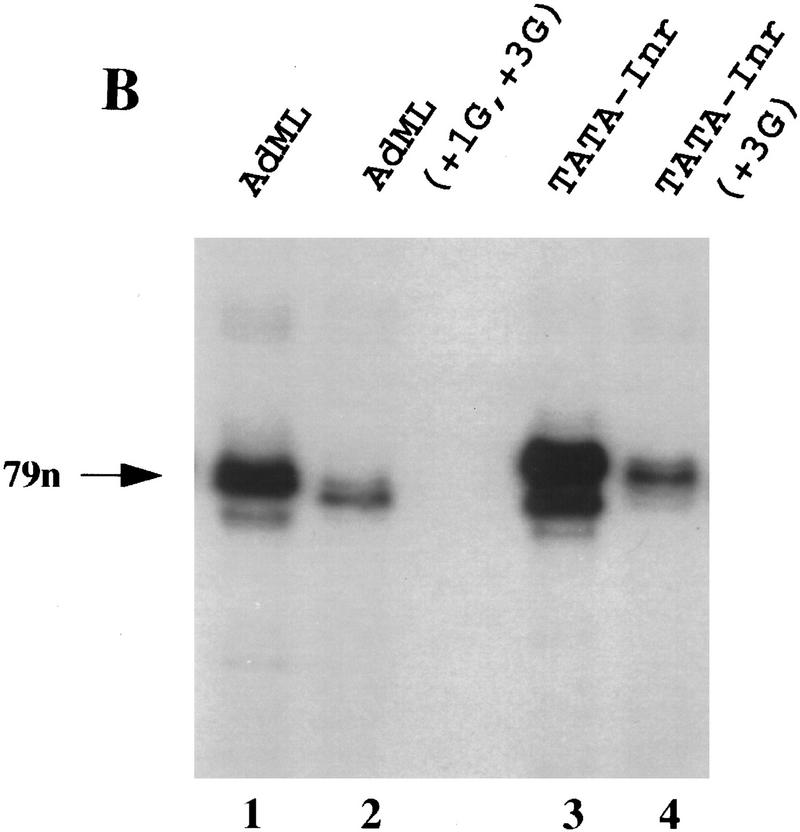
Analysis of TFIID–TFIIA binding to the natural AdML promoter. (A) DNase I footprinting reactions were performed with probes containing the AdML promoter (lanes 1–3,7–9) and the AdML(+1G,+3G) mutant promoter (lanes 4–6,10–12). Reactions contained no protein (lanes 1,4,7,10), 0.2 (lanes 2,3,5,6) or 0.5 (lanes 9,10,11,12) μl concentrated TFIID, and recombinant TFIIA (0.1 μl, lanes 3,6,9,12). The locations of the TATA and Inr elements, as well as regions of downstream protection (DS) and 4 hypersensitive sites (arrows) are depicted to the right. (B) In vitro transcription reactions were performed in HeLa nuclear extracts with supercoiled plasmids containing the AdML core promoter from −35 to +40 (lane 1; see Materials and Methods), the AdML core promoter containing two substitution mutations (+1G and +3G) within the Inr element (lane 2), the synthetic TATA–Inr core promoter (lane 3) and the synthetic TATA–Inr(+3G) core promoter (lane 4). Plasmid DNA (300 ng) and 100 μg HeLa nuclear extract were used for each reaction. RNA products were analyzed by primer extension, by use of the same radiolabeled SP6 primer to insure that the cDNA products possessed similar specific activities.
Interestingly, a mutation in the AdML Inr revealed that the downstream contacts observed with this promoter are largely Inr independent (Fig. 6A). With a high concentration of TFIID, no difference in binding to the wild-type and mutant promoters was observed (Fig. 6A, cf. lanes 7–9 and lanes 10–12). This result is similar to that reported previously with wild-type and mutant AdML promoters (Chiang et al. 1993). With a low concentration of TFIID, the wild-type promoter was protected to a slightly greater extent than the mutant promoter, but only when TFIIA was included in the binding reaction (Fig. 6A, cf. lanes 1–3 and lanes 4–6). These Inr-independent downstream contacts are in contrast to the Inr-dependent downstream contacts observed with the synthetic promoters (see Fig. 2). This difference is particularly striking, given that the mutant tested in the AdML promoter disrupts the Inr more severely than the mutant tested in the synthetic TATA–Inr promoter (+1G, +3G vs. +3G). These results suggest that sequence-specific contacts downstream of the AdML Inr compensate for (or mask) the loss of the TFIID–Inr interaction.
Despite the apparent ability of AdML downstream sequences to compensate for the loss of the Inr in the DNase I footprinting assay, the Inr remains critical for AdML promoter strength (Fig. 6B, lanes 1,2). In contrast, the region downstream of the AdML Inr, which apparently is responsible for retaining TFIID interactions on the Inr mutant promoter, does not contribute to promoter strength, as the AdML core promoter and the synthetic TATA–Inr promoter exhibit similar activities (Fig. 6B, lanes 1 and 3). Previous in vitro transcription experiments with AdML promoter deletion mutants also failed to show a functional role for the downstream sequences contacted by TFIID (Hu and Manley 1981). Furthermore, previous studies have shown that the TFIID complex is essential for the Inr activity observed with both the TATA–Inr and AdML promoters, with the TBP subunit insufficient (Kaufmann and Smale 1994; Martinez et al. 1994). Taken together, the results in Figure 6 suggest that both the AdML Inr and the AdML downstream region can promote TFIID binding to sequences downstream of the TATA box, but only the Inr enhances promoter strength.
To analyze the relative affinities of TFIID and TFIID–TFIIA for these promoters in greater detail, the Mg2+ agarose EMSA was employed. With TFIID alone, the amount of complex obtained with the wild-type AdML promoter was very similar to that obtained with the AdML(+1G, +3G) mutant (Fig. 7, cf. lanes 1–4 and lanes 5–8). This result is, therefore, similar to that observed with the synthetic promoters, where the Inr had no effect on TFIID affinity. In contrast, when TFIIA was included in the EMSA-binding reactions, the amount of complex observed with the wild-type AdML promoter was twofold greater than that observed with the AdML(+1G, +3G) promoter (Fig. 7, cf. lanes 9–13 and 14–18). Once again, this result is similar to that observed with the synthetic promoters. These results suggest that the putative TFIIA-induced conformational change in TFIID selectively enhances TFIID binding to the Inr. In the absence of the Inr, the downstream AdML sequences do not appear to support a similar TFIIA-dependent TFIID interaction.
Figure 7.
TFIIA selectively enhances TFIID binding to the AdML core promoter containing a functional Inr element. Mg2+ agarose EMSAs were performed with 0 (lanes 1,5,9,14), 1 (lanes 2,6,10,11,15,16), 2 (lanes 3,7,12,13,17, 18), or 3 (lanes 4,8) μl of purified TFIID, and 0 (lanes 1–10,12, 14,15,17) or 200 (lanes 11,13,16,18) ng TFIIA. Radiolabeled probes were prepared by PCR from plasmids containing the AdML (lanes 1–4,9–13) and AdML(+1G, +3G) (lanes 5–8,14–18) core promoters. All probes were prepared with the same radiolabeled primer, insuring that the probes are of similar specific activity. As determined by PhosphorImager analysis, complex abundances in lanes 1–18 relative to the complex abundance in lane 10 (1.0) are 0, 3.2, 10.9, 16.6, 0, 3.1, 6.8, 12.6, 0, 1.0, 8.2, 3.7, 17.2, 0, 1.1, 4.4, 2.6, and 9.4, respectively.
Discussion
The results presented in this manuscript show that GAL4–VP16 is capable of recruiting TFIID–TFIIA to core promoters containing either a TATA box, an Inr element, or both TATA and Inr elements. The TATA and Inr elements synergize with each other during activated transcription and TFIID–TFIIA recruitment, suggesting that the synergistic recruitment is responsible for the synergistic function. The synergistic binding of TFIID–TFIIA to the TATA–Inr core promoter is dependent on the precise spacing between the two elements, suggesting that TFIID binds in a stereospecific manner. Interestingly, EMSA experiments show that the affinity of TFIID for these core promoters is dictated primarily by the TATA box, with little contribution from the Inr element. TFIIA, however, enhances TFIID binding to the TATA–Inr promoter more strongly than to the TATA promoter, providing a correlation between binding affinity and promoter strength. Finally, experiments with the natural AdML promoter suggest that TFIID makes sequence-specific contacts downstream of the start site that are not observed with the synthetic promoters. These downstream contacts mask the effect of an Inr mutation in the DNase I footprinting assay, but they have little influence on promoter activity and they do not contribute significantly to TFIID–TFIIA affinity.
The TFIID–TFIIA recruitment experiments provide evidence of a weak interaction on the promoter containing only an Inr element. This interaction appears to be relatively unstable, as a significant protein–DNA complex was not observed by EMSA. In vitro transcription experiments have revealed that this promoter, which contains a GC-rich sequence between −25 and −30, does not direct detectable levels of transcription in the absence of an activator protein (Zenzie-Gregory et al. 1993; B. Zenzie-Gregory and K. Emami, unpubl.), whereas basal transcription from the TATA promoter is readily detectable (O’Shea-Greenfield and Smale 1992; Zenzie-Gregory et al. 1992). Thus, it might not be surprising that a stable complex does not form on the Inr promoter in the absence of an activator. Perhaps a stable TFIID-containing complex forms on this promoter only when an activator is bound to an upstream site, or in the presence of TFIID interactions at DPEs (Burke and Kadonaga 1996). Alternatively, stable TFIID-containing complexes on an Inr promoter may require RNA polymerase II, TFIIB, and TFIIF (Weis and Reinberg 1997), CIF150 (Kaufmann et al. 1996; Kaufmann et al. 1998), or a specific Inr-binding protein such as TFII-I (Roy et al. 1991). Novel factors that stabilize TFIID binding to TATA-less, Inr-containing promoters may also exist.
Previously, we employed a functional complementation assay to identify CIF150, the human homolog of Drosophila TAF150 (Verrijzer et al. 1994, 1995), as an essential cofactor for Inr activity from a TATA–Inr promoter (Kaufmann et al. 1996). CIF150 does not appear to be present in the purified TFIID prepared by the method of Zhou et al. (1992), based on silver staining (Zhou et al. 1992; Kaufmann et al. 1996) and immunoblot analysis (Kaufmann et al. 1998), suggesting that all of the interactions analyzed in this study are independent of that protein. Despite the essential role of CIF150 for Inr activity, recent studies have shown that recombinant CIF150 stabilizes TFIID binding to a similar extent on the TATA–Inr and TATA promoters (Kaufmann et al. 1998). Thus, CIF150 does not appear to be responsible for Inr recognition itself and differs from TFIIA in that it does not selectively enhance TFIID binding to Inr-containing promoters. CIF150 must, therefore, carry out an essential role during Inr-mediated transcription that is largely independent of specific Inr recognition.
The Inr-dependent stabilization activity of TFIIA adds to the growing list of activities and functions for this intriguing protein (for review, see Kang et al. 1995; Ozer et al. 1996). TFIIA was first identified as a factor that enhanced specific transcription from the AdML promoter following fractionation of in vitro transcription extracts (Matsui et al. 1980). Subsequent cloning and characterization of this factor revealed that it greatly stabilizes the binding of TBP to a TATA box (Ranish et al. 1992; Ozer et al. 1994; Sun et al. 1994; Yokomori et al. 1994). Recent TFIIA mutant studies have led to the suggestion that it carries out functions during transcriptional activation that are independent of its TBP stabilization activity (Kang et al. 1995; Ma et al. 1996; Ozer et al. 1996). Finally, TFIIA plays an important role during the recruitment of TFIID to core promoters by activator proteins (see introductory section).
Although the basic TBP stabilization activity of TFIIA has been well characterized, the relevance of this activity during TFIID-mediated transcription appears to be uncertain, as the experiments shown here suggest that TFIIA has little effect on TFIID binding to a promoter containing only a TATA box. A more important activity of TFIIA may be its putative ability to induce a conformational change in the TFIID protein. The conformational change was proposed following the observation that TFIIA led to altered TFIID interactions downstream of a TATA box in DNase I footprinting and UV cross-linking assays (Oelgeschläger et al. 1996). It is noteworthy that the natural AdML promoter, containing a functional Inr element, was used for those studies.
An analysis of the two promoters for the Drosophila adh gene provided functional evidence that TFIIA plays an unusually important role during Inr-mediated transcription, as TFIIA selectively enhanced transcription from the promoter that contains a strong Inr element (Hansen and Tjian 1995). The Inr-dependent stabilization of TFIID binding by TFIIA provides a possible mechanism for the selective enhancement that was observed. With the synthetic promoters used here, TFIIA-depletion experiments have revealed that, in the presence of TFIID, TFIIA is essential for significant transcription from both the TATA–Inr and TATA promoters (A. Jain, unpubl.); this finding is consistent with the hypothesis that TFIIA is specifically involved in Inr activity, although it clearly is important for basal transcription both in the presence and absence of an Inr.
It is noteworthy that the AdE4 core promoter used for most of the previous TFIID–TFIIA recruitment studies (Lieberman and Berk 1994; Chi et al. 1995; Chi and Carey 1996) contains a sequence at its start site that, based on the Inr consensus sequence (Javahery et al. 1994; Lo and Smale 1996), is likely to have Inr activity. Thus, the inducible downstream contacts observed with that promoter may depend on the presence of the Inr element. Alternatively, inducible, Inr-independent interactions between TFIID and the AdE4 downstream region may occur, similar to those observed with the AdML promoter. The experiments shown here suggest that, with the synthetic promoter containing only a TATA box, activator did not stimulate TFIID–TFIIA binding to the start-site region or downstream region. Despite the absence of detectable downstream interactions, the TATA promoter can be strongly activated by GAL4–VP16, suggesting that downstream interactions by TFIID are not essential for transcription activation, even though the conformational change induced by activator and TFIIA may be essential.
The relevance of the TFIID interactions observed downstream of the natural AdML Inr element remains uncertain. AdML downstream sequences that appear to make specific contacts with TFIID had little or no effect on promoter strength in vitro. This negative result is consistent with a previous deletion analysis of the AdML promoter, which failed to identify a function for the AdML downstream sequences in an in vitro transcription assay (Hu and Manley 1981). In contrast, this region was important for efficient mRNA production in an in vivo assay (Lewis and Manley 1985). Perhaps, the sequences within the untranslated region are important in vivo for efficient elongation of transcription or for mRNA stability, activities that may be independent of the TFIID interactions. Alternatively, the downstream contacts by TFIID may play a role only when the DNA is associated with nucleosomes. To distinguish between these possibilities, an in vitro assay that relies on the downstream TFIID–DNA contacts will need to be established.
Materials and methods
Plasmids
The plasmids containing synthetic promoters with multiple GAL4-binding sites upstream of the AdML TATA box and/or the TdT Inr element were described previously (Emami et al. 1995). The GAL4–TATA–Inr(+3G) mutant was constructed by excising the mutant Inr from plasmid J3102 (Kaufmann and Smale 1994) by digestion with SacI and BamHI. The SacI–BamHI fragment was then inserted into the GAL4–TATA promoter cleaved with the same two enzymes.
The plasmids containing the wild-type and mutant AdML core promoters were derived from plasmid II (Smale et al. 1990), containing the AdML TATA region from −35 to −13 (relative to the transcription start site), inserted into the EcoRI and SacI sites of pSP72 (Promega). AdML core promoter sequences from −6 to +40 were amplified by PCR from a plasmid containing the AdML promoter and gene (obtained from Dr. Arnold Berk, University of California, Los Angeles). A SacI restriction site flanked the −6 sequence within one PCR primer and a HindIII restriction site flanked the +40 sequence within the other primer. The amplified product was inserted into plasmid II digested with SacI and HindIII. The resulting plasmid contains AdML sequences from −35 through +40, with a SacI restriction site in place of the sequences from −11 through −6. The AdML mutant promoter was prepared by the PCR strategy described above, but with two nucleotide substitutions (+1G and +3G) in the upstream PCR primer.
The plasmids with variable spacing between the TATA and Inr elements were derived from plasmids VII through XII (O’Shea-Greenfield and Smale 1992). The core promoters from these plasmids were excised with EcoRI and BamHI and inserted into the EcoRI and BamHI sites of GAL4–SP72 (Emami et al. 1995). All plasmids were purified by column chromatography (Qiagen, Inc.) and their sequences confirmed by DNA sequencing with a Sequenase kit (U.S. Biochemical).
Protein preparation
The hemagglutinin (HA) epitope-tagged TFIID complex was isolated from the LTRα3 cell line by immunoaffinity purification as described (Zhou et al. 1992). The recombinant TFIIA subunits were expressed in Escherichia coli and purified by the method of Ozer et al. (1994). GAL4–VP16 was a gift from the laboratories of Drs. Michael Carey (UCLA) and Arnold Berk (UCLA). Purified, full-length human TBP was prepared by the method of Zhou et al. (1992). It is worth noting that some preparations of TBP, including those obtained from a commercial source, yielded a weak hypersensitive site at +5, which was reminiscent of the TFIID contact at the Inr (data not shown). The origin of this hypersensitive site remains unknown, but it is unlikely to be related to the Inr-dependent hypersensitive site induced by the intact TFIID complex for the following reasons: It was observed only with very high concentrations of a subset of the TBP preparations tested; it was not reduced by the +3G mutation (data not shown); and it was not enhanced when GAL4–VP16 was included in the reactions (data not shown).
DNA-binding assays
Radiolabeled DNA probes for the DNase I footprinting reactions were prepared by PCR as described (Kaufmann and Smale 1994), by use of one unlabeled oligonucleotide PCR primer and a second primer that was 5′-end-labeled with [γ-32P]ATP and T4 polynucleotide kinase. For all probes, the radiolabeled downstream primer was the SP6 promoter primer (Promega, Inc.). For the promoters with GAL4-binding sites, for the promoters lacking activator binding sites, and for the AdML promoters, the unlabeled upstream primer was the T7 promoter primer (Promega, Inc.).
Binding reactions for the DNase I footprinting experiments were performed as described (Lieberman and Berk 1994), in a total volume of 13.5 μl, containing 6000 cpm of the 32P-labeled probe, 5 mm MgCl2, 2.2 μg of bovine serum albumin, 12.5 mm HEPES (pH 7.9), 12.5% glycerol, 0.2 mm EDTA, 70 mm KCl, 60 mm β-mercaptoethanol, and 30 μg/ml of poly[dG–dC] as nonspecific competitor DNA. After incubating the binding mixtures at 30°C for 60 min, the labeled probes were partially digested with DNase I, and the cleaved DNA molecules were analyzed by electrophoresis on an 8% denaturing polyacrylamide gel (see Lo et al. 1991).
Radiolabeled probes for Mg2+ agarose EMSAs were prepared as described above for the DNase I footprinting probes. Binding conditions were as described above. Mg2+ agarose EMSA gels were prepared and run as described (Zhou et al. 1992; Lieberman and Berk 1994). Signals were quantitated by PhosphorImager analysis (Molecular Dynamics, Inc.).
In vitro transcription assays
In vitro transcription reactions were performed with HeLa nuclear extracts (100 μg) and 300 ng of supercoiled plasmid DNA as described previously (Smale and Baltimore 1989). RNA products were analyzed by primer extension analysis by use of a 32P-labeled SP6 promoter primer (Promega; see Smale et al. 1990). cDNA products of the primer extension reactions were visualized by electrophoresis on an 8% denaturing polyacrylamide gel. Signals were quantitated by PhosphorImager analysis (Molecular Dynamics, Inc.).
Acknowledgments
We are extremely grateful to Michael Carey, Arnold Berk, and the members of their laboratories for many helpful discussions and for providing some of the GAL4–VP16 and TFIIA preparations used during the course of this study. We also thank Michael Carey for critical reading of the manuscript. S.T.S. is an Associate Investigator with the Howard Hughes Medical Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL steves@hhmi.ucla.edu; FAX (310) 206-8623.
References
- Bellorini M, Dantonel JC, Yoon JB, Roeder RG, Tora L, Mantovani R. The major histocompatibility complex class II Ea promoter requires TFIID binding to an initiator sequence. Mol Cell Biol. 1996;16:503–512. doi: 10.1128/mcb.16.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- Carey M, Leatherwood J, Ptashne M. A potent GAL4 derivative activates transcription at a distance in vitro. Science. 1990;247:710–712. doi: 10.1126/science.2405489. [DOI] [PubMed] [Google Scholar]
- Chi T, Carey M. Assembly of the isomerized TFIIA–TFIID–TATA ternary complex is necessary and sufficient for gene activation. Genes & Dev. 1996;10:2540–2550. doi: 10.1101/gad.10.20.2540. [DOI] [PubMed] [Google Scholar]
- Chi T, Lieberman P, Ellwood K, Carey M. A general mechanism for transcriptional synergy by eukaryotic activators. Nature. 1995;377:254–257. doi: 10.1038/377254a0. [DOI] [PubMed] [Google Scholar]
- Chiang C-M, Ge H, Wang Z, Hoffmann A, Roeder RG. Unique TATA-binding protein containing complexes and cofactors involved in transcription by RNA polymerases II and III. EMBO J. 1993;12:2749–2762. doi: 10.1002/j.1460-2075.1993.tb05936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami KH, Navarre WW, Smale ST. Core promoter specificities of the Sp1 and VP16 transcriptional activation domains. Mol Cell Biol. 1995;15:5906–5916. doi: 10.1128/mcb.15.11.5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Martinez E, Chiang CM, Roeder RG. Activator-dependent transcription by mammalian RNA polymerase II: In vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 1996;274:57–71. doi: 10.1016/s0076-6879(96)74008-9. [DOI] [PubMed] [Google Scholar]
- Goodrich JA, Cutler G, Tjian R. Contacts in context: Promoter specificity and macromolecular interactions in transcription. Cell. 1996;84:825–830. doi: 10.1016/s0092-8674(00)81061-2. [DOI] [PubMed] [Google Scholar]
- Hansen SK, Tjian R. TAFs and TFIIA mediate differential utilization of the tandem Adh promoters. Cell. 1995;82:565–575. doi: 10.1016/0092-8674(95)90029-2. [DOI] [PubMed] [Google Scholar]
- Hori R, Carey M. The role of activators in assembly of RNA polymerase II transcription complexes. Curr Opin Genet Dev. 1994;4:236–244. doi: 10.1016/s0959-437x(05)80050-4. [DOI] [PubMed] [Google Scholar]
- Horikoshi M, Carey MF, Kakidani H, Roeder RG. Mechanism of action of a yeast activator: Direct effect of GAL4 derivatives on mammalian TFIID-promoter interactions. Cell. 1988a;54:665–669. doi: 10.1016/s0092-8674(88)80011-4. [DOI] [PubMed] [Google Scholar]
- Horikoshi M, Hai T, Lin YS, Green MR, Roeder RG. Transcription factor ATF interacts with the TATA factor to facilitate establishment of a preinitiation complex. Cell. 1988b;54:1033–1042. doi: 10.1016/0092-8674(88)90118-3. [DOI] [PubMed] [Google Scholar]
- Hu SL, Manley JL. DNA sequence required for initiation of transcription in vitro from the major late promoter of adenovirus 2. Proc Natl Acad Sci. 1981;78:820–824. doi: 10.1073/pnas.78.2.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale ST. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser K, Meisterernst M. The human general co-factors. Trends Biochem Sci. 1996;21:342–345. [PubMed] [Google Scholar]
- Kang JJ, Auble DT, Ranish JA, Hahn S. Analysis of the yeast transcription factor TFIIA: Distinct functional regions and a polymerase II-specific role in basal and activated transcription. Mol Cell Biol. 1995;15:1234–1243. doi: 10.1128/mcb.15.3.1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann J, Smale ST. Direct recognition of initiator elements by a component of the transcription factor IID complex. Genes & Dev. 1994;8:821–829. doi: 10.1101/gad.8.7.821. [DOI] [PubMed] [Google Scholar]
- Kaufmann J, Verrijzer CP, Shao J, Smale ST. CIF, an essential cofactor for TFIID-dependent initiator function. Genes & Dev. 1996;10:873–886. doi: 10.1101/gad.10.7.873. [DOI] [PubMed] [Google Scholar]
- Kaufmann, J., K. Ahrens, R. Koop, S.T. Smale, and R. Müller. 1998. CIF150, a human cofactor for TFIID-dependent initiator function. Mol. Cell. Biol. 18: (in press). [DOI] [PMC free article] [PubMed]
- Kingston RE, Bunder CA, Imbalzano AN. Repression and activation by multiprotein complexes that alter chromatin structure. Genes & Dev. 1996;10:905–920. doi: 10.1101/gad.10.8.905. [DOI] [PubMed] [Google Scholar]
- Kobayashi N, Boyer TG, Berk AJ. A class of activation domains interacts directly with TFIIA and stimulates TFIIA-TFIID-promoter complex assembly. Mol Cell Biol. 1995;15:6465–6473. doi: 10.1128/mcb.15.11.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koleske AJ, Young RA. The RNA polymerase II holoenzyme and its implications. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- Lewis ED, Manley JL. Control of adenovirus late promoter expression in two human cell lines. Mol Cell Biol. 1985;5:2433–2442. doi: 10.1128/mcb.5.9.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman PM, Berk AJ. A mechanism for TAFs in transcriptional activation: Activation domain enhancement of TFIID–TFIIA–promoter DNA complex formation. Genes & Dev. 1994;8:995–1006. doi: 10.1101/gad.8.9.995. [DOI] [PubMed] [Google Scholar]
- Lo K, Smale ST. Generality of a functional initiator consensus sequence. Gene. 1996;182:13–22. doi: 10.1016/s0378-1119(96)00438-6. [DOI] [PubMed] [Google Scholar]
- Lo K, Landau NR, Smale ST. LyF-1, a transcriptional regulator that interacts with a novel class of promoters for lymphocyte-specfic genes. Mol Cell Biol. 1991;11:5229–5243. doi: 10.1128/mcb.11.10.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma D, Olave I, Merino A, Reinberg D. Separation of the transcriptional coactivator and antirepression functions of transcription factor IIA. Proc Natl Acad Sci. 1996;93:6583–6588. doi: 10.1073/pnas.93.13.6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez E, Chiang CM, Ge H, Roeder RG. TAFs in TFIID function through the initiator to direct basal transcription from a TATA-less class II promoter. EMBO J. 1994;13:3115–3126. doi: 10.1002/j.1460-2075.1994.tb06610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Segall J, Weil PA, Roeder RG. Multiple factors required for accurate initiation of transcription by purified RNA polymerase II. J Biol Chem. 1980;255:11992–11996. [PubMed] [Google Scholar]
- Nakajima N, Horikoshi M, Roeder RG. Factors involved in specific transcription by mammalian RNA polymerase II: Purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol Cell Biol. 1988;8:4028–4040. doi: 10.1128/mcb.8.10.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelgeschläger T, Chiang C-M, Roeder RG. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- O’Shea-Greenfield A, Smale ST. Roles of TATA and initiator elements in determining the start site location and direction of RNA polymerase II transcription. J Biol Chem. 1992;267:1391–1402. [PubMed] [Google Scholar]
- Ozer J, Moore PA, Bolden AH, Lee A, Rosen CA, Lieberman PM. Molecular cloning of the small (γ) subunit of human TFIIA reveals functions critical for activated transcription. Genes & Dev. 1994;8:2324–2335. doi: 10.1101/gad.8.19.2324. [DOI] [PubMed] [Google Scholar]
- Ozer J, Bolden AH, Lieberman PM. Transcription factor IIA mutations show activator-specific defects and reveal a IIA function distinct from stimulation of TBP-DNA binding. J Biol Chem. 1996;271:11182–11190. doi: 10.1074/jbc.271.19.11182. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Purnell BA, Gilmour DS. Contributions of sequences downstream of the TATA element to a protein–DNA complex containing the TATA-binding protein. Mol Cell Biol. 1993;13:2593–2603. doi: 10.1128/mcb.13.4.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell BA, Emanuel PA, Gilmour DS. TFIID sequence recognition of the initiator and sequence farther downstream in Drosophila class II genes. Genes & Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- Ranish JA, Lane WS, Hahn S. Isolation of two genes that encode subunits of the yeast transcription factor IIA. Science. 1992;255:1127–1129. doi: 10.1126/science.1546313. [DOI] [PubMed] [Google Scholar]
- Roberts SG, Choy B, Walker SS, Lin YS, Green MR. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr Biol. 1995;5:508–516. doi: 10.1016/s0960-9822(95)00103-5. [DOI] [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Roy AL, Meisterernst M, Pognonec P, Roeder RG. Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature. 1991;354:245–248. doi: 10.1038/354245a0. [DOI] [PubMed] [Google Scholar]
- Sauer F, Hansen SK, Tjian R. Multiple TAFIIs directing synergistic activation of transcription. Science. 1995;270:1783–1788. doi: 10.1126/science.270.5243.1783. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Kim J, Sharp PA. Activation of the TFIID–TFIIA complex with HMG-2. Genes & Dev. 1995;9:1354–1365. doi: 10.1101/gad.9.11.1354. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Kim J, Stewart L, Champoux JJ, Sharp PA. Topoisomerase I enhances TFIID–TFIIA complex assembly during activation of transcription. Genes & Dev. 1997;11:397–407. doi: 10.1101/gad.11.3.397. [DOI] [PubMed] [Google Scholar]
- Smale ST. Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Smale ST, Schmidt MC, Berk AJ, Baltimore D. Transcriptional activation by Sp1 as directed through TATA and initiator: Specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci. 1990;87:4509–4513. doi: 10.1073/pnas.87.12.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stargell LA, Struhl K. Mechanism of transcriptional activation in vivo: Two steps forward. Trends Genet. 1996;12:311–315. doi: 10.1016/0168-9525(96)10028-7. [DOI] [PubMed] [Google Scholar]
- Struhl K. Transcriptional enhancement by acidic activators. Biochim Biophys Acta. 1996;1288:015–017. doi: 10.1016/s0304-419x(96)00029-7. [DOI] [PubMed] [Google Scholar]
- Sun X, Ma D, Sheldon M, Yeung K, Reinberg D. Reconstitution of human TFIIA activity from recombinant polypeptides: A role in TFIID-mediated transcription. Genes & Dev. 1994;8:2336–2348. doi: 10.1101/gad.8.19.2336. [DOI] [PubMed] [Google Scholar]
- Tantin D, Chi T, Hori R, Pyo S, Carey M. Biochemical mechanisms of transcriptional activation by GAL4-VP16. Methods Enzymol. 1996;274:133–149. doi: 10.1016/s0076-6879(96)74013-2. [DOI] [PubMed] [Google Scholar]
- Usheva A, Shenk T. TATA-binding protein independent initiation: YY1, TFIIB and RNA polymerase II direct basal transcription on supercoiled template DNA. Cell. 1994;76:1115–1121. doi: 10.1016/0092-8674(94)90387-5. [DOI] [PubMed] [Google Scholar]
- Verrijzer CP, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- Verrijzer CP, Yokomori K, Chen J-L, Tjian R. Drosophila TAFII150: Similarity to yeast gene TSM-1 and specific binding to core promoter DNA. Science. 1994;264:933–941. doi: 10.1126/science.8178153. [DOI] [PubMed] [Google Scholar]
- Verrijzer CP, Chen J-L, Yokomori K, Tjian R. Binding of TAFs to core elements directs promoter selectivity by RNA polymerase II. Cell. 1995;81:1115–1125. doi: 10.1016/s0092-8674(05)80016-9. [DOI] [PubMed] [Google Scholar]
- Wang JC, Van Dyke MW. Initiator sequences direct downstream promoter binding by human transcription factor IID. Biochim Biophys Acta. 1993;1216:73–80. doi: 10.1016/0167-4781(93)90039-g. [DOI] [PubMed] [Google Scholar]
- Wang W, Gralla JD, Carey M. The acidic activator GAL4-AH can stimulate polymerase II transcription by promoting assembly of a closed complex requiring TFIID and TFIIA. Genes & Dev. 1992;6:1716–1727. doi: 10.1101/gad.6.9.1716. [DOI] [PubMed] [Google Scholar]
- Weis L, Reinberg D. Accurate positioning of RNA polymerase II on a natural TATA-less promoter is independent of TATA-binding protein-associated factors and initiator-binding proteins. Mol Cell Biol. 1997;17:2973–2984. doi: 10.1128/mcb.17.6.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Brou C, Wu J, Lutz Y, Moncollin V, Chambon P. The acidic transcriptional activator GAL-VP16 acts on preformed template-committed complexes. EMBO J. 1992;11:2229–2240. doi: 10.1002/j.1460-2075.1992.tb05282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokomori K, Zeidler MP, Chen J-L, Verrijzer CP, Mlodzik M, Tjian R. Drosophila TFIIA directs cooperative DNA binding with TBP and mediates transcriptional activation. Genes & Dev. 1994;8:2313–2323. doi: 10.1101/gad.8.19.2313. [DOI] [PubMed] [Google Scholar]
- Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- Zenzie-Gregory B, O’Shea-Greenfield A, Smale ST. Similar mechanisms for transcription initiation mediated through a TATA box or an initiator element. J Biol Chem. 1992;267:2823–2830. [PubMed] [Google Scholar]
- Zenzie-Gregory B, Khachi A, Garraway IP, Smale ST. Mechanism of initiator-mediated transcription: Evidence for a functional interaction between the TATA-binding protein and DNA in the absence of a specific recognition sequence. Mol Cell Biol. 1993;13:3841–3849. doi: 10.1128/mcb.13.7.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Liebermann PM, Boyer TG, Berk AJ. Holo-TFIID supports transcriptional stimulation by diverse activators and from TATA-less promoters. Genes & Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]