Abstract
Steps in the replication of HIV-1 occurring in the virus, but not the host are preferred targets of anti-retroviral therapy. Strand transfer is unique; the DNA strand being made by viral reverse transcriptase (RT) is moved from one RNA template position to another.1 Understanding the mechanism requires knowing whether the RT directly mediates the template exchange, or dissociates during the exchange, so that it occurs by polymer dynamics. Earlier work in vitro showed that the presence of an RT-trapping polymer would allow synthesis on the original or donor template, but completely block transfer and subsequent synthesis on the second or acceptor template. One interpretation is that the RT must dissociate during transfer, but an alternative is that sequestration of non-polymerizing RTs prevents polymerization-independent ribonuclease H (RNase H) cleavages of the donor template necessary for strand exchange. To resolve this ambiguity, we designed a primer-template system that allows strand transfer without RNase H activity. Using an RNase H negative mutant RT, we showed that a polymer trap still prevented strand transfer. This confirms that RT dissociates during strand transfer. The presence of HIV-1 nucleocapsid protein, which promotes strand exchange, had little effect on this outcome. Additional assays showed that both, the wild type RT and a multiple NRTI resistant HIV-1 RT, containing an extended fingers domain, which is characterized by its enhanced primer-template binding affinity, were both unable to transfer with the trapping polymer. This implies that common sequence variations among RTs are unlikely to alter the dissociation feature.
Keywords: HIV-1, reverse transcription, reverse transcriptase, RNase H, strand transfer, dissociation
Introduction
Human immunodeficiency virus-1 (HIV-1) is the causative agent for acquired immunodeficiency syndrome (AIDS), a disease that remains a major health concern. Like other retroviruses, HIV-1 replicates by reverse transcription, converting the single stranded viral RNA genome into double stranded DNA for incorporation into the host genome. The virus-encoded reverse transcriptase (RT) mediates this process using two enzymatic activities, polymerization and RNase H activity. Both activities were shown to be essential for completing DNA synthesis.
The steps of reverse transcription have been examined in detail. They include two essential strand transfer events occurring from the 5′ and 3′ ends of the viral genome, which are designated minus and plus strand transfer respectively4;5. The virus carries two complete RNA genomic strands. Depending on growth conditions, viral genome re-assortment and recombination is a common event during replication occurring three to thirty times per viral genome replication 6. As a consequence, recombination can result in the creation of functionally altered viruses with enhanced survival characteristics. These characteristics include a greater ability to evade host defenses and more effective resistance to therapeutic drugs. Strand transfer associated with viral recombination is thought to employ basically the same mechanism as replication transfer.
The mechanism of strand transfer has been characterized extensively in vitro 4. Typical reactions involve an initial RNA template designated the donor, which is primed with a labeled DNA. Extension of the primer by RT yields a distinct length donor extension product. The reaction also includes an acceptor RNA template, containing a sequence homology region with the donor template. This acceptor is longer at the 3′ end in order to detect strand transfer products and to differentiate it from the donor extension product. Primer transfer and extension on the acceptor template would result in a longer transfer product, that could be resolved using gel electrophoresis. Use of this assay demonstrated that strand transfer requires both the polymerization and RNase H activities of RT, and occurs by a strand-invasion mechanism. The RT employs polymerization-dependent RNase H activity that periodically nicks the template during synthesis. RTs that are not engaged in polymerization will bind the pre-formed nicks and perform polymerization-independent cleavages to create short gaps. The acceptor template invades these gaps to bind the DNA strand, and a branch migration process exchanges the donor for the acceptor template. Viral nucleocapsid protein, which has been shown to promote strand exchange, increases the efficiency of strand transfer in these assays.
The RT is a processive DNA polymerase, capable of adding hundreds of nucleotides without dissociating from the DNA primer. Processivity of synthesis was measured by pre-binding the RT to a primer terminus, then adding a polymer trap, and finally starting polymerization with dNTPs. The same approach was then also applied to determine whether the RT carried out strand transfer in a processive manner 18. The RT was pre-bound to the DNA primer on the donor template, followed by the addition of polymer trap and then dNTPs. In this case, the RT synthesized to the end of the donor template, but no synthesis on the acceptor template could be detected19. This result suggests that the RT dissociates before the primer terminus is transferred and then rebinds the primer after transfer to continue synthesis on the acceptor template.
However, since strand transfer involves RT-RNase H, there is an alternative explanation for these results. The addition of the polymer trap immediately sequesters and inactivates all of the RT molecules in the reaction except those bound to the DNA primer termini. This leaves no RTs to make the polymerization-independent cleavages necessary for acceptor invasion. The absence of these cleavages could be the actual factor responsible for inactivation of transfer by the polymer trap.
In order to determine whether the RT actually dissociates during transfer, we set out to devise a transfer reaction that did not require RT-RNase H. Fortunately, our previous work involved development of a substrate system that could support strand transfer mediated by an RT mutant that lacks RNase H functions15. Here, we report the use of this template and the RNase H negative RT to assess whether HIV-1 RT dissociates during strand transfer.
Results
Development of an Experimental System that Supports Strand Transfer without RNase H
Strand transfer in retroviruses has been proposed to occur by an invasion mechanism. The mechanism involves local cutting of the donor RNA by RT-RNase H after successful polymerization of the DNA primer. This cutting creates a short gap in the template exposing a single stranded region of the DNA. The acceptor template can then invade or base pair with the single stranded DNA region. The region of pairing can then spread by branch migration toward the DNA primer terminus. The branch migration spread is aided by cuts made in the donor template between the invasion site and the DNA primer terminus. We previously reported construction and use of a primed donor substrate that would support strand transfer without the need of RNase H activity 20. The substrate had two essential structural characteristics. First, the DNA primer extended over the 3′ end of the donor template to form a single stranded region that served as a “pre-created” invasion site. Second, the annealed region between the donor template and the DNA primer, after the primer was extended to the 5′ end of the donor, was still sufficiently short that strand exchange with the acceptor RNA after invasion could readily occur without the need of RNase H cleavages. This substrate is diagrammed in Figure 1b. Significantly, we demonstrate that this substrate could support strand transfer with HIV-1 E478Q RT. This mutant RT has an amino acid substitution in the RNase H active site. HIV-1 E478Q RT, displays normal polymerization activity, but no RNase H activity.
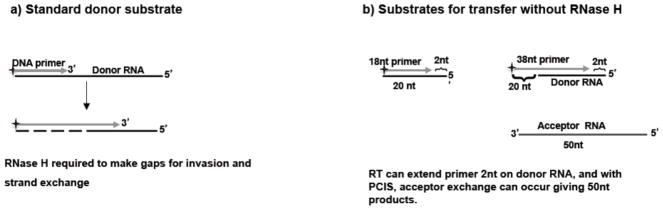
Figure 1. Substrate design for strand transfer without RNase H.
Schematic of substrates used in our experiments for measuring transfer without RNase H. Donor RNA template was either annealed to 5′ radiolabeled DNA primer (gray line with arrow showing direction of primer extension) with or without pre-created invasion site, PCIS. Shown in (a), is the standard donor RNA substrate (no PCIS) (black line) that requires RNase H to make gaps for invasion and strand exchange, while (b) shows substrates for transfer without RNase H. Substrates are shown with and without a PCIS region (20 nucleotides), where acceptor can exchange without cleavage. The PCIS primer used was 38 nucleotides long. Full extension on donor RNA yields a 40 nucleotide product while full-length extension on the acceptor yields a 50 nucleotide product.
Equipped with this substrate, we could address the objectives of the current study. First, is to test whether strand transfer can occur without RNase H. Second, is to test whether reverse transcriptase dissociation during strand transfer is essential.
Trapping polymer blocks strand transfer using wild-type RT
To explore the question of RT dissociation, we began by testing the suitability of our substrates for the polymer trap, strand transfer reactions. Initial measurements were made with the wild type RT, since it was important to know whether the substrate would support strand transfer without a trap, but completely fail at transfer when the trap was present. Such a result would show that we could achieve the same experimental outcome with the new substrate as we had with a standard strand transfer substrate that requires RNase H cleavages.
The first experiment was performed in the absence of NC, (Figure 2a). Several reactions served as controls for the proper expected behavior of the substrates for the strand transfer reactions. Lane 1 shows the un-extended labeled primer. Lane 2 shows that in the presence of dNTPs and the primed donor template, the RT extended the primer to the end of the donor template. Lane 6 shows extension of a primer, when it was annealed to the acceptor template. This shows that the RT is fully capable of primer elongation on the acceptor strand. Lane 4 shows the outcome of a strand transfer reaction, which included the primed donor template and an unprimed acceptor template. The fully extended 40 nucleotide donor extension product and 50 nucleotide transfer product are both visible. This indicates that RT extended primers to the end of the donor, and a significant portion of these primers transferred to the acceptor, and was extended to the full length. This set of controls show that the substrate system supports strand transfer.
Figure 2. Trapping polymer blocks transfer in the presence of wild type RT.
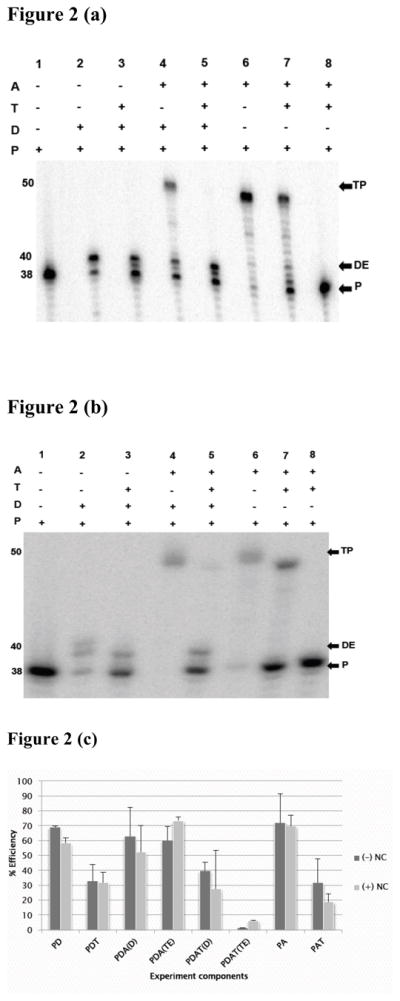
The primer-donor was pre-incubated for three min at 37°C with RT in the presence of Mg2+. Reaction was initiated by adding starter reagents, which could be dNTPs alone, dNTPs with trap, dNTPs and acceptor, or dNTPs plus acceptor and trap. Extension on the primed acceptor was also measured with and without trap. Incubation was for 15 min and the reactions stopped by adding a termination buffer (see Materials and Methods). The final product was separated on a 10% denaturing gel. Panels (a) and (b) are representative gel images of experiments with wild type RT in the absence, (−) NC, and presence, (+) NC, of nucleocapsid protein, respectively. Lanes labeled 1–8 on top of the figure represent an independent reaction. The (+) and (−) signs indicate the reagents included or omitted in each reaction. The abbreviations used denote: primer (P), acceptor (A), donor (D), trapping polymer (T), and strand transfer (TP) and donor extension (DE) products. The numbers on the side indicate the length of primer, donor extension and transfer products in nucleotides.
Panel (c) is a bar graph showing analysis of donor extension and strand transfer products obtained with wild type RT. The Y-axis represents % efficiency of product formation; the X-axis indicates the experimental component analyzed. The labels used here are: PD, representing measured full length primer extension efficiency on the donor template without trap; PDT, measured efficiency of primer extension on the donor in the presence of trap; PDA (D), measured efficiency of primer extension on the donor in the presence of unprimed acceptor; PDA (TE), measured transfer efficiency without trap; PDAT (D), measured efficiency of primer extension on the donor in the presence of unprimed acceptor and trap; PDAT (TE), measured transfer efficiency in the presence of trap; PA, measured extension efficiency on a primed acceptor, and PAT, measured extension efficiency on primed acceptor with trap. Black bars designate (−) NC and gray bars designate (+) NC, respectively.
The remaining lanes represent the trapped experiment. Lane 8 shows the important trap control. In this reaction, three hundred-fold excess of the oligo(dT)poly(rA) trapping polymer was added to a reaction containing a primed acceptor template. Then the RT and dNTPs were added. The virtual absence of any extension products proves that the trap is effective at fully sequestering RT molecules that are not bound to the labeled substrate. This control verifies that RTs dissociating from the donor template during a trapped transfer reaction will not be able to reinitiate on primers that have transferred to the acceptor template. Lane 3 shows a reaction, in which the RT was pre-bound to the primed donor template in the presence of magnesium, and then trap and dNTPs were added. Observed extension on the donor template proves that the trap is largely ineffective at sequestering RTs that are engaged in primer extension. Similarly, lane 7 shows that when RT was pre-bound to the primed acceptor, followed by addition of trap and dNTPs, the RT could fully extend the primer on the acceptor. The final experiment, in which strand transfer is assessed in the presence of trap, is shown in lane 5. Here, the RT was pre-bound to the primed donor template in the presence of magnesium. Then trap, acceptor template and dNTPs were added. Results show that the RT was able to extend the primer on the donor template, but virtually no synthesis was observed on the acceptor. Our quantitation of band densities indicated that the presence of trap reduced transfer efficiency from 54% to 1.2%. This result is consistent with previous observations with other heteropolymeric strand transfer substrates19. It shows that the trap almost completely prevents transfer, but does not reveal which of the two proposed mechanisms of transfer inhibition are employed.
NC does not significantly alter the blocking effect of polymer trap with wild type RT
We performed the same set of experiments in the presence of NC (Figure 2b). NC is known to have nucleic acid chaperone activity, and to affect template switching in reconstituted reactions22, suggesting that it could facilitate template exchange in the RT active site. NC was added at a level sufficient to coat 100% (1 molecule NC per 7–8 nucleotides) of the template in reactions without trap. Reactions with trap contained much more RNA and DNA and so would have required a much higher level of NC for complete coating. We tested for the levels of NC coating that was effective in coating the primers. Addition of NC at a level sufficient to fully coat all polymer in the reaction inhibited primer extension. This was an indication that the NC distributed preferentially to the heteropolymeric experimental substrate compared to the polymer trap. In consideration of this observation, we adjusted the NC concentration in trapped reactions to 10% coating level, which produced little inhibition.
NC facilitated strand transfer considerably in this system raising it to 73% in the absence of trap. Transfer was much less efficient with trap, with a measured value of 5.8 %. The transfer reaction with trap still shows that the trap is highly, though not completely, effective at preventing transfer; this result indicates that NC will allow a small percentage of the RT to stay bound during transfer.
Pre-created invasion sites (PCIS) increase the rate of strand transfer using E478Q mutant RT
Pre-created invasion sites have been shown to enhance strand transfer. We anticipated that transfer mediated by E478Q RT, which does not produce cleavages in RNA templates for invasion sites, would likely need a PCIS in the substrate for strand transfer to occur. To test this expectation, we compared transfer efficiencies on substrates with (Figure 3b) or without a PCIS region (Figure 3a). Reactions were initiated with E478Q RT, sampled at various times and analyzed by 10% PAGE. Figure 3(c) shows an analysis of transfer efficiency obtained with these substrates. With the substrate lacking a PCIS, transfer efficiency only reached 5% in the 15 minute sampling period. In the presence of a PCIS, transfer efficiency rose rapidly and consistently, reaching 50% by 3 minute and 85% by the end of the 15 minutes sampling period. These results highlight the importance of RNase H-created invasion sites to the basic transfer mechanism. Most significantly for our current studies, they show that our substrate is suitable for analyses of the mechanism of strand transfer using the E478Q RT.
Figure 3. Strand transfer with E478Q mutant RT using PCIS substrates.
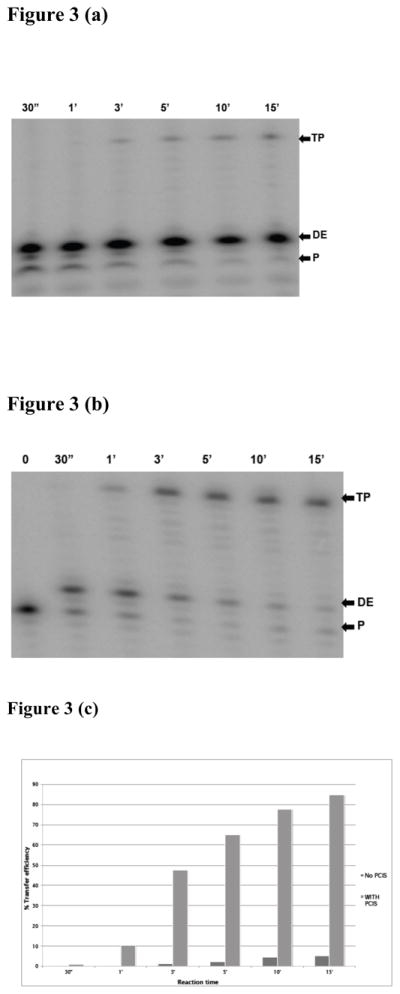
(a) A 10% denaturing gel image of time course reactions performed with E478Q mutant RT using the substrate without a pre-created invasion site (PCIS) (see Figure 1a) and (b) gel image of reactions with a PCIS substrate (Figure 1b). A master reaction was prepared containing primer-donor, acceptor, magnesium and E478Q mutant RT. After pre-incubation for three minutes at 37°C, the reaction was started by adding dNTPs. Aliquots were then taken and mixed with stop buffer after reaction times of 30 sec, 1, 3, 5, 10 and 15 min. TP denotes transfer product, DE, donor extension product and P, unextended primer. Panel (c) represents analysis of strand transfer products: shown on the Y-axis, % transfer efficiency, while on the X-axis are products analyzed on each time point using the substrate without (−) PCIS or with (+) PCIS.
Trapping polymer blocks strand transfer with E478Q mutant RT
Using our specially designed substrate that allows efficient strand transfer with E478Q mutant RT, we set out to test whether the RT dissociates during the transfer process. This group of experiments had the same design and controls as the analyses described with the wild type RT. Figure 4(a) and (b) present results without NC, and with NC, respectively. Both panels have control experiments without trapping polymer including extension on the donor template (lane 2), extension on the acceptor template (lane 6), and the transfer reaction (lane 4). Results demonstrate that the substrate supports effective transfer with the E478Q RT. The control experiment in lane 8 demonstrates the effectiveness of the polymer trap. Extension on the donor (lane 6) and acceptor (lane 7) were both efficient in spite of the presence of trap. The key strand transfer results with trap are shown in lane 5. In the presence and absence of NC, there was efficient synthesis on the donor template, but almost no transfer products.
Figure 4. Trap blocks strand transfer in the presence of E478Q mutant RT.
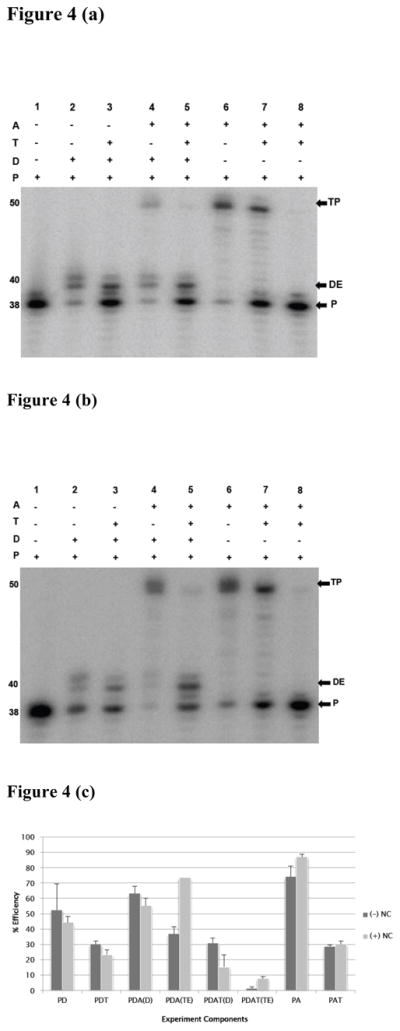
(a) A representative 10 % denaturing gel of strand transfer reactions in the absence (a) and presence (b) of NC. Substrates used are as described in Figure 1b. Lanes 1–8 represent independent reactions with reagents added as shown above the gels by the (+) or (−) signs. TP represents transfer product, DE, donor extension product while P is unextended labeled primer. The numbers on the side indicate the expected lengths of these products in nucleotides. Panel (c) summarizes the effects of the blocking polymer on donor extension and transfer efficiency. Black bars represent products without (−) NC and gray bars are products with (+) NC. Products analyzed here; PD, representing measured donor extension efficiency without trap; PDT, measured donor extension efficiency in the presence of trap; PDA(D), measured donor extension efficiency in the presence of unprimed acceptor; PDA(TE), measured transfer efficiency without trap; PDAT(D), measured donor extension efficiency in the presence of unprimed acceptor and trap; PDAT(TE), measured transfer efficiency in the presence of trap; PA, measured extension efficiency on a primed acceptor, and PAT, measured extension efficiency on primed acceptor with trap.
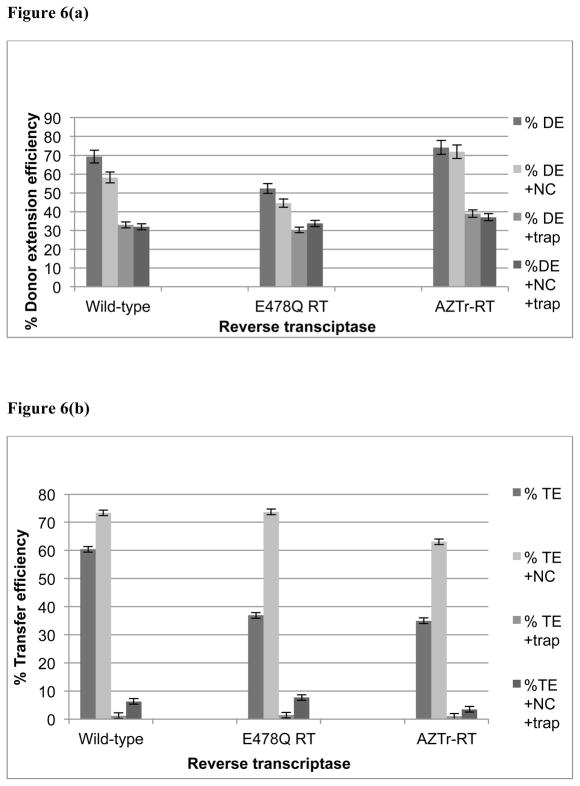
We then analyzed donor and transfer product efficiencies shown here in Figure 4c. In experiments without NC the donor extension efficiency with trap was 30.9 %, while transfer product efficiency was only 1.4%. The presence of NC improved transfer efficiency in the absence of trap from 36.8 to 73.7% indicating that NC is an effective facilitator of strand transfer with this combination of substrate and E478Q mutant RT. This enhanced transfer with NC was also noted in our experiments with wild-type RT, and is greater than had been observed previously with longer substrates. Possibly the ability of NC to facilitate strand exchange is enhanced on a short substrate. In the presence of both trap and NC transfer efficiency increased to 7.7%. Evidently, a small fraction of the RTs remain bound during transfer, and this fraction is greater in the presence of NC. This observation led us to test a mutant RT with a higher primer-template affinity for RT dissociation.
A mutant RT with higher primer-template affinity still dissociates during transfer
There are many natural variations in HIV-1 RT sequence, and numerous drug resistant mutant forms of RT have been identified and characterized. We questioned whether a minor variation in amino acid sequence could alter the properties of the RT so that it would remain bound to the T/P substrates during the transfer reaction. We anticipated that a mutation that improved polymer substrate binding during DNA synthesis was most likely to allow the RT to transfer without dissociation. Azidothymidine resistant reverse transcriptase (AZTr-RTs) can excise AZT monophosphate from the blocked 3′ end of the DNA primer strand and allow the RT to continue extension on the primer26. Several specific mutations have been associated with the development of resistance to AZT27. Certain mutations in the fingers domain of the RT increase the primer-template interaction affinity and the processivity of primer elongation. We obtained an AZTr-RT mutant isolated from a patient after multiple NRTI treatment that contains a Serine-Glycine (SG) insertion between amino acid 69 and 70 in RT. In addition to the dipeptide insertion, this mutant contains a T215Y thymidine analog mutation (TAM) and several other amino acid substitutions, which are not TAM. This mutant was ideal, having both the substitutions and insertions in the RT polymerase linked to improved binding and polymerization. Moreover, our measurements show that this mutant RT has an approximately seven-fold higher binding affinity to primer-template than the wild type RT (details under review elsewhere). Figures 5(a) and (b) display representative results obtained with AZTr-RT in the presence or absence of NC. The control experiments generally indicated similar behavior to the wild type RT. AZTr-RT synthesized well on the donor and acceptor templates, and also transferred efficiently. The trap was fully effective at sequestering the RT in the absence or presence of NC. The trap allowed synthesis on the donor, but virtually eliminated transfer.
Figure 5. Trap blocks strand transfer with AZT resistant-RT (AZTr-RT).
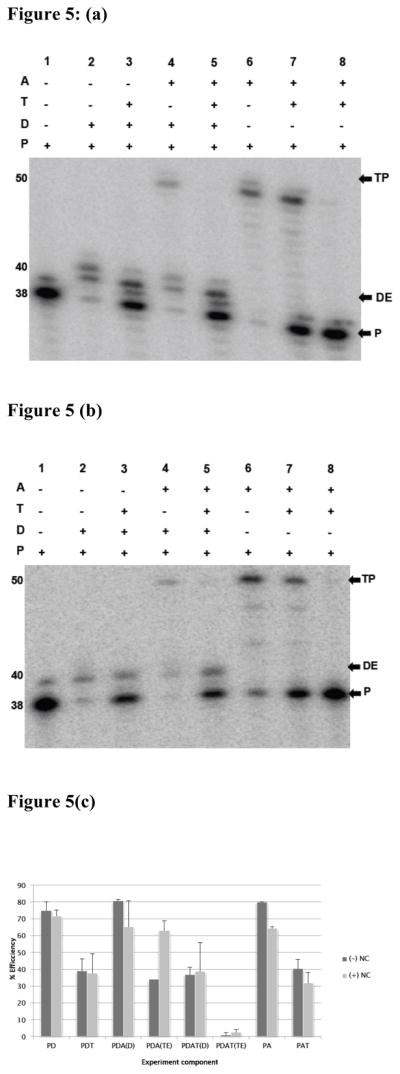
Experiments examining the behavior of a mutant reverse transcriptase with higher primer-template affinity. Panel (a) displays a gel image showing products of transfer reactions performed using AZTr-RT in the absence of NC, while panel (b) shows products of reactions performed in the presence of NC. The conditions of transfer reactions were as described under Materials and Methods. The numbers 1–8 shown above the gel images represent different experiment and control reactions. The (+) and (−) shown on top indicates the specific additives for each reaction. The labels P, DE and TP denote labeled unextended primer, donor extension and transfer product, respectively. The numbers 38, 40 and 50 represent the size of primer, donor extension and transfer product, respectively in nucleotides.
Panel (c) is a bar graph summarizing donor and transfer efficiency of independent reactions calculated and averaged from at least three reactions. Error bars represent standard error of the mean. Other abbreviations used are as described under Figure 4.
Figure 5(c) presents the analysis of products from these experiments. In the absence of NC transfer efficiency was 34% without trap and 1% with trap. In experiments with NC, transfer efficiency was 63.1% and 3.5% without and with trap, respectively. Clearly, the mutation did not alter the binding and dissociation of the RT during transfer. The improvement in transfer seen previously with NC was consistent.
Overall, observations with wild type RT, E478Q RT, and AZTr-RT, consistently show that the RT almost always dissociates during transfer. Moreover, NC increases the small fraction of transfer events in which the RT accompanies the primer terminus in its movement to the acceptor template.
Discussion
Considerable experimentation has been directed toward understanding the mechanism of HIV-1 strand transfer, a possible therapeutic target. The HIV-1 RT is a key component of the strand transfer reaction, although its specific roles are not fully understood. RT possesses several distinct activities, including DNA- and RNA-template-dependent DNA synthesis, RNase H cleavage, and strand displacement synthesis, all proposed to contribute to successful transfer. Analyses in vitro have led to the proposal of the invasion mechanism of transfer, described earlier. Since this transfer mechanism relies on the catalytic activities of the RT, it was reasonable to expect that the RT also direct the switching of templates from donor to acceptor.
In fact, the earliest analysis of strand transfer mechanism supported this expectation16. In this system oligo-dT was extended over poly-rA. The reaction was performed in the presence of an RT trapping molecule. Results showed that a single RT molecule could mediate a series of transfers without dissociation producing a transfer product much longer than the average template length. This could only occur if the RT did not dissociate during the primer transfer. Moreover, since each primer had to have transferred many times in the presence of the trap, it appeared that RT dissociation during transfer either never occurred or occurred very rarely.
Consistent with this interpretation, HIV-1 RT was found to be capable of simultaneous binding to the primer, donor and acceptor strands of a transfer intermediate33. Evidence for a three-strand complex with the RT implies that the active site of the RT is designed to mediate transfer while the templates are exchanging. Indeed, the formation of such complexes has been observed in different organisms as part of reaction intermediates.
Later, trapping experiments with a heteropolymeric set of substrates produced an entirely different result. In this case, the primer could extend on the donor in the presence of trap, but evidence of transfer to the acceptor and subsequent extension was entirely absent. This result was interpreted to mean that either the RT is obligated by requirements of the mechanism to dissociate during transfer, or, even though it could remain bound during transfer, it simply does not. Why were these opposite results obtained? One possibility lies in the structural dynamics of a homopolymeric T/A primer-template. Because the sequence is the same at every position, the primer can shift position on the template with no change in binding free energy. Such changes of position are also favored by the weak hydrogen bonding of AT base pairs, which should result in low transition energy for movement of the primer on the template. This situation could allow the primer to slide on the template into a single stranded 3′ overhang. The overhanging strand could then simply anneal to the next template, and then move back into the RT active site for more synthesis. Whereas strand sliding is not possible on the heteropolymeric template, it must employ a fundamentally different mechanism. Since natural transfer during infection involve heteropolymeric sequences, that mechanism is most likely the one used in vivo.
The detection of three strand complexes with the RT is consistent with RT-mediated transfer, but does not prove that it occurs. One can envision that although a three-strand complex can form, the actual completion of template exchange throughout the active site cleft of the RT may be blocked at some point.
Had measurements with the heteropolymeric substrates indicated that the RT does not dissociate during transfer, the result would have been unambiguous. However, as discussed above, the actual result showing that trapping the RT eliminates transfer had two equally probable interpretations. The first was that the RT almost always dissociates during the strand exchange. The second was that trapping all non-synthesizing RTs prevents polymerization-independent RNase H activity needed to facilitate transfer. We distinguished these possibilities by devising a substrate that allowed transfer in the absence of RNase H. Results showed that the trap still prevented transfer. We interpret this to mean that the RT dissociates during transfer. We emphasize that the result does not provide information on the importance of polymerization-independent RNase H for transfer, since the substrate was purposely designed so that that mode of cleavage was not required.
Does efficient transfer require polymerization-independent RNase H? This question may be even more difficult to address unambiguously than the issue of RT dissociation. We have attempted to perform strand transfer on a heteropolymeric template using progressively lower amounts of RT, to determine whether the transfer efficiency decreased. However, the low amount of synthesis precluded making an accurate measurement. Moreover, considering that the RT dissociates during transfer, the transfer efficiency may appear low at reduced RT levels because after the transfer there is a long delay before an RT comes to re-initiate synthesis on the acceptor template.
Is dissociation of the RT a fundamental requirement made necessary by the structure of the RT active site? Clearly it is not required with the homopolymer substrate. We considered that some natural variants of the RT might also transfer on heteropolymer templates without dissociation. To maximize our chances of seeing such a phenomenon, we tested the AZTr mutant RT, that displays an seven-fold greater substrate binding affinity than the wild type, having an (SG) insertion mutation between amino acids 69 and 70, and TAM. This finding that this mutant RT also dissociated during transfer suggests that common variants all dissociate, and that dissociation is the norm and not the exception.
Interestingly, when the E478Q RT was tested with NC protein, a small fraction of the extended primers transferred successfully in the presence of trap. This result suggests that the RT could have evolved to remain bound to the DNA primer terminus during the transfer reaction if there had been natural selective pressure to do so. The implication of this result is that the final strand exchange that leads to terminus transfer does not need protein-mediated assistance. In fact, the exchange may be slowed in its movement through the active site cleft in the RT. Such a kinetic disadvantage, might have naturally selected for RT variants that dissociate during transfer.
The invasion mechanism of transfer has been proposed to have three steps: acceptor invasion at a short gap in the donor template created by the RT-RNase H activity, propagation of the double strand formed between the primer and the acceptor RNA, and final primer terminus transfer. Based on the results we have obtained here, we propose that the entire process proceeds most efficiently without the direct help of the RT. The initial invasion would occur in a region of the primer-donor that has already cleared the back end of the RT. Propagation of the primer-acceptor hybrid would proceed to the active site cleft and possibly induce RT dissociation. DNA primer terminus transfer would then proceed to completion.
Materials and Methods
Enzymes and Substrates
DNA and RNA oligonucleotides were obtained from Integrated DNA Technologies (Coralville, IA). Polymerase-trapping polymer, poly (rA) x (dT)15 was from Roche Applied Sciences. The 32P-ATP (6000Ci/mmol) isotope was purchased from Perkin-Elmer Life Sciences. The wild type HIV-1 reverse transcriptase was purified in our laboratory as previously described. HIV-1 E478Q RT (specific activity = 40,000 U/mg) was generously provided by Dr. Stuart F. J. Le Grice (RT Biochemistry Section, HIV Drug Resistance Program, National Cancer Institute, Frederick, MD, USA). AZTr RT (specific activity 20,000 U/mg) was also purified in our laboratory isolated from a patient after multiple NRTI treatments that contains a Serine-Glycine (SG) insertion between amino acid 69 and 70 in RT. This AZTr-RT also contains a T215Y, thymidine analog mutation (TAM), a common AZT resistance mutation. NC protein was provided by Dr. Robert J. Gorelick, National Cancer institute, National Institutes of Health. Other reagents used in these experiments were of analytical grade level sourced from different suppliers.
DNA and RNA Templates
Donor, acceptor RNA and DNA primers were purchased from Integrated DNA Technologies. The primer sequences used were: PCIS 86 (no invasion site): (5′ – TGG TAA ACA TTC TTG AGT – 3′) PCIS 87 (with invasion site): (5′ – CCG GTT CTA TAA CGG TAT GAT GGT AAA CAT TCT TGA GT – 3′).
Acceptor RNA, PCIS 91 (3′– GGC CAA GAU AUU GCC AUA CUA CCA UUU GUA AGA ACU CAC GAG CUA GAC UA –5′), Donor RNA, PCIS 94 (3′ A CCA UUU GUA AGA ACU CAC G -5′).
Substrate preparation
DNA primers were 5′ labeled with γ-32ATP and T4 polynucleotide kinase (Invitrogen). Labeled primers were cleared of unincorporated radionucleotides with Micro Bio-spin columns (BioRad).
Strand transfer assay
Transfer reactions were performed as previously described with some modifications. Radio labeled primer (0.64nM) was mixed with donor template (4nM) or acceptor RNA (8nM) in 50mM Tris, pH 8.0, 50mM KCl, 1mM DTT, and 1mM ethylenediamine tetra acetic acid (EDTA), pH 8.0. The solution was heated at 95°C for 5 minutes and left to cool at room temperature. Eight μl of the pre-annealed donor-primer (0.64nM primer, 4nM donor RNA) was pre-incubated in Mg2+ (1.5mM), either wild type RT, E478Q RT, or AZTr RT, to a final concentration of 17nM enzyme at 37°C for 3 minutes. The start-mix contained dNTPs alone, dNTPs and trap, dNTPs and acceptor, dNTPs, acceptor and a trap to a final concentration of 100μM dNTPs, 4μM trap, 0.64nM primer, 8.0nM acceptor and 4.0nM donor. The reaction was stopped after 15 minutes by taking 6.25μl and mixing with 2x stop dye solution, which contains {10mM EDTA (pH 8.0) to chelate out magnesium ions from RT, 90 % formamide (v/v), and 0.1% xylene cyanole and bromophenol blue}. The trap control reaction was performed by pre-incubating RT/Mg+ with trap at 37°C for 3 minutes, then adding them to pre-annealed primer-acceptor at the same concentrations as before, and incubating for an additional 3 minutes at 37°C before adding the start mix containing dNTPs (100μM final concentration) and incubating for 15 minutes at 37°C.
For reactions performed in the presence of NC protein, an appropriate amount of NC was added to the mixture before addition of the start-mix and incubated at 37°C for15 minutes. The amount of NC used in these reactions was 100% coating for reactions without trap, and 10 % coating for reactions with the trap assuming that one NC molecule covers 7nts.
Detection and Analysis
The reaction products were separated on a 10% denaturing PAGE, dried and exposed overnight. The gels were imaged with STORM 820 or 860 Phosphor Imager and quantified with ImageQuant software version 1.2, all from GE Healthcare (Piscataway, NJ). Transfer efficiency was determined with the equation TE = [TP/(DE + TP)] ×100% while donor extension efficiency was determined using the equation DE = [DE/(DE+P)] ×100% in which TP is measured intensity of strand-transfer products, DE is measured intensity of full-length donor-template extension products and P is the measured intensity of primer not extended.
Figure 6. Donor extension and strand transfer behavior using different RT enzymes.
Panel (a) shows percentage donor extension efficiency obtained with different RT enzymes with or without NC protein. The Y-axis is percentage efficiency while the X-axis represents the type of RT enzyme used for each set of products. The bars represent donor extension product without trap (DE), donor extension with NC (DE+NC), donor extension product with trap (DE+trap), and donor extension with NC and trap (DE+NC+trap). Panel (b) shows transfer efficiency using wild type RT, E478Q RT and AZTr-RT. The bars represent, transfer efficiency without trap (TE), transfer efficiency, with NC (TE-NC), transfer efficiency, with trap, (TE-trap), and transfer efficiency, with NC and trap, (TE+NC+trap).
Acknowledgments
This work was supported by the National Institutes of Health grant GM049573. NC protein preparation was supported by the NIH contract HHSN261200800001E.
We gratefully thank Joseph Liberman for his contributions in the preliminary studies of this research. Special thanks to Dr Stuart F.J. Le Grice of NIH for providing E478Q RT. We thank Dr. Carrie Dykes, Dr. Dorota Piekna-Przybylska, Wen Shen and Daniel Levine for their helpful input and discussions.
Abbreviation used
- RT
reverse transcriptase
- RNase
ribonuclease
- PCIS
pre-created invasion site
- NC
nucleocapsid protein
- NRTI
nucleoside reverse transcriptase inhibitors
- AZTr-RT
azidothymidine resistant reverse transcriptase
- DE
donor extension
- TP
transfer product
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
John M. Muchiri, Email: John_muchiri@urmc.rochester.edu.
Sean T. Rigby, Email: sean.rigby@gmail.com.
Laura A. Nguyen, Email: Laura_nguyen@urmc.rochester.edu.
Baek Kim, Email: Baek_kim@urmc.rochester.edu.
Robert A. Bambara, Email: Robert_bambara@urmc.rochester.edu.
References
- 1.Dion AS, Williams CJ, Moore DH. RNase H and RNA-directed DNA polymerase: associated enzymatic activities of murine mammary tumor virus. J Virol. 1977;22:187–93. doi: 10.1128/jvi.22.1.187-193.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tan CK, Zhang J, Li ZY, Tarpley WG, Downey KM, So AG. Functional characterization of RNA-dependent DNA polymerase and RNase H activities of a recombinant HIV reverse transcriptase. Biochemistry. 1991;30:2651–5. doi: 10.1021/bi00224a013. [DOI] [PubMed] [Google Scholar]
- 3.Smith CM, Smith JS, Roth MJ. RNase H requirements for the second strand transfer reaction of human immunodeficiency virus type 1 reverse transcription. J Virol. 1999;73:6573–81. doi: 10.1128/jvi.73.8.6573-6581.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basu VP, Song M, Gao L, Rigby ST, Hanson MN, Bambara RA. Strand transfer events during HIV-1 reverse transcription. Virus Res. 2008;134:19–38. doi: 10.1016/j.virusres.2007.12.017. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Balakrishnan M, Roques BP, Fay PJ, Bambara RA. Mechanism of minus strand strong stop transfer in HIV-1 reverse transcription. J Biol Chem. 2003;278:8006–17. doi: 10.1074/jbc.M210959200. [DOI] [PubMed] [Google Scholar]
- 6.Henry KR, Weber J, Quinones-Mateu ME, Arts EJ. The impact of viral and host elements on HIV fitness and disease progression. Curr HIV/AIDS Rep. 2007;4:36–41. doi: 10.1007/s11904-007-0006-9. [DOI] [PubMed] [Google Scholar]
- 7.Burke DS. Recombination in HIV: an important viral evolutionary strategy. Emerg Infect Dis. 1997;3:253–9. doi: 10.3201/eid0303.970301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saksena NK, Wang B, Ge YC, Xiang SH, Dwyer DE, Cunningham AL. Coinfection and genetic recombination between HIV-1 strains: possible biological implications in Australia and South East Asia. Ann Acad Med Singapore. 1997;26:121–7. [PubMed] [Google Scholar]
- 9.DeStefano JJ, Roberts B, Shriner D. The mechanism of retroviral recombination: the role of sequences proximal to the point of strand transfer. Arch Virol. 1997;142:1797–812. doi: 10.1007/s007050050198. [DOI] [PubMed] [Google Scholar]
- 10.Fang G, Weiser B, Kuiken C, Philpott SM, Rowland-Jones S, Plummer F, Kimani J, Shi B, Kaul R, Bwayo J, Anzala O, Burger H. Recombination following superinfection by HIV-1. AIDS. 2004;18:153–9. doi: 10.1097/00002030-200401230-00003. [DOI] [PubMed] [Google Scholar]
- 11.Ramirez BC, Simon-Loriere E, Galetto R, Negroni M. Implications of recombination for HIV diversity. Virus Res. 2008;134:64–73. doi: 10.1016/j.virusres.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 12.Bretscher MT, Althaus CL, Muller V, Bonhoeffer S. Recombination in HIV and the evolution of drug resistance: for better or for worse? Bioessays. 2004;26:180–8. doi: 10.1002/bies.10386. [DOI] [PubMed] [Google Scholar]
- 13.Johnson PE, Turner RB, Wu ZR, Hairston L, Guo J, Levin JG, Summers MF. A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription. Biochemistry. 2000;39:9084–91. doi: 10.1021/bi000841i. [DOI] [PubMed] [Google Scholar]
- 14.Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010:7. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rigby ST, Van Nostrand KP, Rose AE, Gorelick RJ, Mathews DH, Bambara RA. Factors that determine the efficiency of HIV-1 strand transfer initiated at a specific site. J Mol Biol. 2009;394:694–707. doi: 10.1016/j.jmb.2009.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber HE, McCoy JM, Seehra JS, Richardson CC. Human immunodeficiency virus 1 reverse transcriptase. Template binding, processivity, strand displacement synthesis, and template switching. J Biol Chem. 1989;264:4669–78. [PubMed] [Google Scholar]
- 17.Bavand MR, Wagner R, Richmond TJ. HIV-1 reverse transcriptase: polymerization properties of the p51 homodimer compared to the p66/p51 heterodimer. Biochemistry. 1993;32:10543–52. doi: 10.1021/bi00091a003. [DOI] [PubMed] [Google Scholar]
- 18.DeStefano JJ. Human immunodeficiency virus nucleocapsid protein stimulates strand transfer from internal regions of heteropolymeric RNA templates. Arch Virol. 1995;140:1775–89. doi: 10.1007/BF01384341. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Rodriguez L, Tsuchihashi Z, Fuentes GM, Bambara RA, Fay PJ. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J Biol Chem. 1995;270:15005–11. doi: 10.1074/jbc.270.25.15005. [DOI] [PubMed] [Google Scholar]
- 20.Rigby ST, Rose AE, Hanson MN, Bambara RA. Mechanism analysis indicates that recombination events in HIV-1 initiate and complete over short distances, explaining why recombination frequencies are similar in different sections of the genome. J Mol Biol. 2009;388:30–47. doi: 10.1016/j.jmb.2009.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hanson MN, Balakrishnan M, Roques BP, Bambara RA. Evidence that creation of invasion sites determines the rate of strand transfer mediated by HIV-1 reverse transcriptase. J Mol Biol. 2006;363:878–90. doi: 10.1016/j.jmb.2006.08.068. [DOI] [PubMed] [Google Scholar]
- 22.Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA biology. 2010;7:754–74. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin JG, Guo J, Rouzina I, Musier-Forsyth K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism. Progress in nucleic acid research and molecular biology. 2005;80:217–86. doi: 10.1016/S0079-6603(05)80006-6. [DOI] [PubMed] [Google Scholar]
- 24.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends in biochemical sciences. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 25.Negroni M, Buc H. Retroviral recombination: what drives the switch? Nature reviews. Molecular cell biology. 2001;2:151–5. doi: 10.1038/35052098. [DOI] [PubMed] [Google Scholar]
- 26.Arion D, Kaushik N, McCormick S, Borkow G, Parniak MA. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–17. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 27.Larder BA, Kemp SD. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–8. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 28.Ly JK, Margot NA, MacArthur HL, Hung M, Miller MD, White KL. The balance between NRTI discrimination and excision drives the susceptibility of HIV-1 RT mutants K65R, M184V and K65r+M184V. Antivir Chem Chemother. 2007;18:307–16. doi: 10.1177/095632020701800603. [DOI] [PubMed] [Google Scholar]
- 29.Kim B, Ayran JC, Sagar SG, Adman ET, Fuller SM, Tran NH, Horrigan J. New human immunodeficiency virus, type 1 reverse transcriptase (HIV-1 RT) mutants with increased fidelity of DNA synthesis. Accuracy, template binding, and processivity. J Biol Chem. 1999;274:27666–73. doi: 10.1074/jbc.274.39.27666. [DOI] [PubMed] [Google Scholar]
- 30.Meyer PR, Matsuura SE, Mian AM, So AG, Scott WA. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 31.Pop MP. In vitro analysis of the HIV-1 second strand-transfer reaction. Biochim Biophys Acta. 1996;1307:193–204. doi: 10.1016/0167-4781(96)00043-7. [DOI] [PubMed] [Google Scholar]
- 32.Fuentes GM, Palaniappan C, Fay PJ, Bambara RA. Strand displacement synthesis in the central polypurine tract region of HIV-1 promotes DNA to DNA strand transfer recombination. J Biol Chem. 1996;271:29605–11. doi: 10.1074/jbc.271.47.29605. [DOI] [PubMed] [Google Scholar]
- 33.Canard B, Sarfati R, Richardson CC. Binding of RNA template to a complex of HIV-1 reverse transcriptase/primer/template. Proc Natl Acad Sci U S A. 1997;94:11279–84. doi: 10.1073/pnas.94.21.11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rose RJ, Welsh TS, Waksman G, Ashcroft AE, Radford SE, Paci E. Donor-strand exchange in chaperone-assisted pilus assembly revealed in atomic detail by molecular dynamics. J Mol Biol. 2008;375:908–19. doi: 10.1016/j.jmb.2007.10.077. [DOI] [PubMed] [Google Scholar]
- 35.Remaut H, Rose RJ, Hannan TJ, Hultgren SJ, Radford SE, Ashcroft AE, Waksman G. Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted beta strand displacement mechanism. Mol Cell. 2006;22:831–42. doi: 10.1016/j.molcel.2006.05.033. [DOI] [PubMed] [Google Scholar]
- 36.Mizrahi V, Lazarus GM, Miles LM, Meyers CA, Debouck C. Recombinant HIV-1 reverse transcriptase: purification, primary structure, and polymerase/ribonuclease H activities. Arch Biochem Biophys. 1989;273:347–58. doi: 10.1016/0003-9861(89)90493-1. [DOI] [PubMed] [Google Scholar]
- 37.Stahlhut M, Li Y, Condra JH, Fu J, Gotlib L, Graham DJ, Olsen DB. Purification and characterization of HIV-1 reverse transcriptase having a 1:1 ratio of p66 and p51 subunits. Protein Expr Purif. 1994;5:614–21. doi: 10.1006/prep.1994.1084. [DOI] [PubMed] [Google Scholar]
- 38.Stahlhut MW, Olsen DB. Expression and purification of retroviral HIV-1 reverse transcriptase. Methods Enzymol. 1996;275:122–33. doi: 10.1016/s0076-6879(96)75010-3. [DOI] [PubMed] [Google Scholar]