Abstract
We analyzed the function of the downstream promoter element (DPE), a distinct 7-nucleotide core promoter element that is ∼30 nucleotides downstream of the transcription start site of many TATA-box-deficient (TATA-less) promoters in Drosophila. There is a strict requirement for spacing between the Inr and DPE motifs, as an increase or decrease of 3 nucleotides in the distance between the Inr and DPE causes a seven- to eightfold reduction in transcription as well as a significant reduction in the binding of purified TFIID. These results suggest a specific and somewhat rigid interaction of TFIID with the Inr and DPE sequences. Photo-cross-linking analysis of purified TFIID with a TATA-less DPE-containing promoter revealed specific cross-linking of dTAFII60 and dTAFII40 to the DPE, with a higher efficiency of cross-linking to dTAFII60 than to dTAFII40. These data, combined with the previously well-characterized interactions between the two TAFs and their homology to histones H4 and H3, suggest that a dTAFII60–dTAFII40 heterotetramer binds to the DPE. Human and Drosophila transcription factors exhibit essentially the same requirements for DPE sequence and for Inr–DPE spacing. In addition, the TATA-less promoter of the human interferon regulatory factor-1 (IRF-1) gene contains a DPE that is important for transcriptional activity both in vitro and in cultured cells. Hence, these studies provide evidence for a direct role of TAFs in basal transcription of TATA-less DPE-containing genes and collectively indicate that the DPE is, in many respects, a downstream counterpart to the TATA box that is present in Drosophila to humans.
Keywords: RNA polymerase II, in vitro transcription, Inr element, TATA box, TFIID, TBP, TAFs
Transcription is centrally involved in an array of biological processes, which include growth, development, and response to external stimuli. In eukaryotes, protein-coding genes are transcribed by the RNA polymerase II transcriptional machinery, which comprises RNA polymerase II and other factors that are required for basal and regulated transcription. Transcription by RNA polymerase II is directed by cis-acting DNA sequences that typically consist of a core promoter along with regulatory elements, such as enhancers, that contain binding sites for sequence-specific transcriptional activator and/or repressor proteins. Thus, the study of both the trans-acting protein factors and the cis-acting DNA elements is necessary to gain a better understanding of the fundamental mechanisms by which genes are transcribed (for recent reviews, see Björkland and Kim 1996; Burley and Roeder 1996; Orphanides et al. 1996; Roeder 1996; Verrijzer and Tjian 1996; Ptashne and Gann 1997; Sauer and Tjian 1997; Smale 1997; Tansey and Herr 1997).
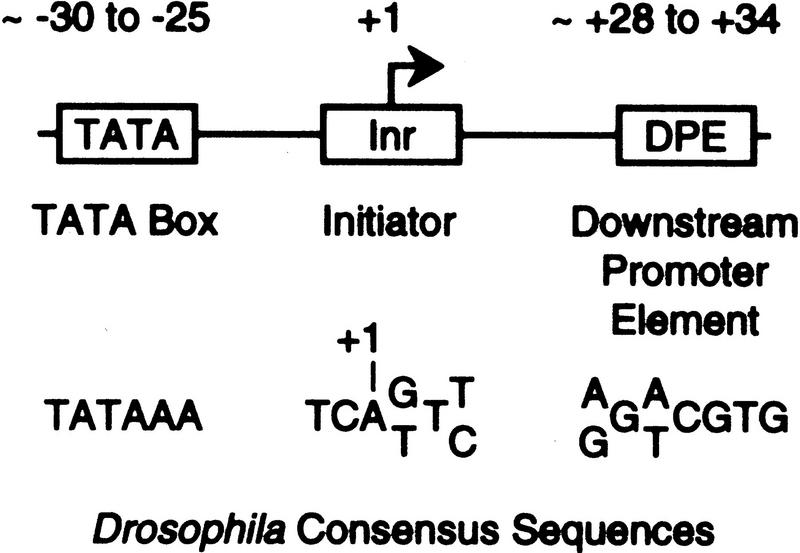
The key DNA element that is essential for transcription by RNA polymerase II is the core promoter—the DNA sequences, which encompass the transcription start site (within about −40 to +40 relative to the +1 start site) and are sufficient to direct the accurate initiation of transcription. Two important core promoter motifs are the TATA box and the initiator (Inr) (Fig. 1). The TATA box is an A/T-rich sequence that is located ∼25–30 nucleotides upstream of the RNA start site of many, but not all, promoters. It is recognized by the TATA box-binding polypeptide (TBP), which is a component of the multisubunit TFIID complex. The Inr encompasses the RNA start site, and like the TATA box, it is also present in many, but not all, core promoters (Smale and Baltimore 1989; Smale 1994, 1997). Inr elements have been characterized in various TATA-less and TATA-containing promoters, and the Inr consensus sequence is Py-Py-A+1-N-T/A-Py-Py (where A+1 is the transcription start site) in mammalian genes (Smale and Baltimore 1989; Bucher 1990; Javahery et al. 1994) and T-C-A+1-G/T-T-T/C in Drosophila genes (Hultmark et al. 1986; Purnell et al. 1994; Arkhipova 1995).
Figure 1.
The TATA box, Inr, and DPE are core promoter elements. The consensus sequences and locations of the TATA box, Inr, and DPE motifs are indicated. The TATA box and DPE appear to be functionally redundant, and promoters generally do not contain both elements.
Many promoters contain functionally important sequences that are downstream of the transcription start site. Such downstream promoter sequences have been found in TATA-containing promoters (see, e.g., Lewis and Manley 1985; Nakatani et al. 1990; Lee et al. 1992; Emanuel and Gilmour 1993; Purnell and Gilmour 1993), as well as in TATA-less promoters (see, e.g., Biggin and Tjian 1988; Perkins et al. 1988; Soeller et al. 1988; Smale and Baltimore 1989; Jarrell and Meselson 1991; Contursi et al. 1995; Minchiotti et al. 1997). It appears that many of these downstream promoter sequences are involved in basal transcription, but it is also important to consider that some downstream promoter sequences might be binding sites for sequence-specific transcriptional activators.
In the analysis of the interaction of purified TFIID with TATA-less promoters, a conserved downstream core promoter element was found to be required for the sequence-specific binding of TFIID to a subset of TATA-less promoters (Burke and Kadonaga 1996). This motif was termed the downstream promoter element (DPE). The DPE is a distinct 7-nucleotide element that is located at about +30 (typically, from +28 to +34) relative to the transcription start site (Fig. 1). It is present in many, but not all, promoters and is bound by TFIID but not by TBP. In addition, nearly all of the promoters that have been found to contain DPE-like sequences are TATA-less promoters. Modification of the DPE by clustered point mutagenesis was found to cause a decrease in the binding of purified TFIID to the promoter as well as an 8- to 100-fold reduction in basal transcriptional activity in vitro. Mutational analysis of the Inr and DPE motifs revealed that the DPE acts in conjunction with the Inr to provide a binding site for TFIID in the absence of a TATA box. Interestingly, the loss of transcriptional activity upon disruption of the TATA box in a TATA-containing promoter could be recovered by the insertion of a DPE at a downstream position (+28 to +34) in the defective promoter. Thus, the DPE appears to be a functionally important, conserved downstream core promoter element.
We are, however, still in the early stages of understanding the DPE. In this work we have investigated several fundamental questions regarding the role of the DPE in the transcription process. These experiments provide evidence for the function of TBP-associated factors (TAFs) in DPE-driven basal transcription and reveal that the DPE has many properties that are analogous to those of the TATA box.
Results and Discussion
The spacing between the Inr and DPE motifs is important for promoter function
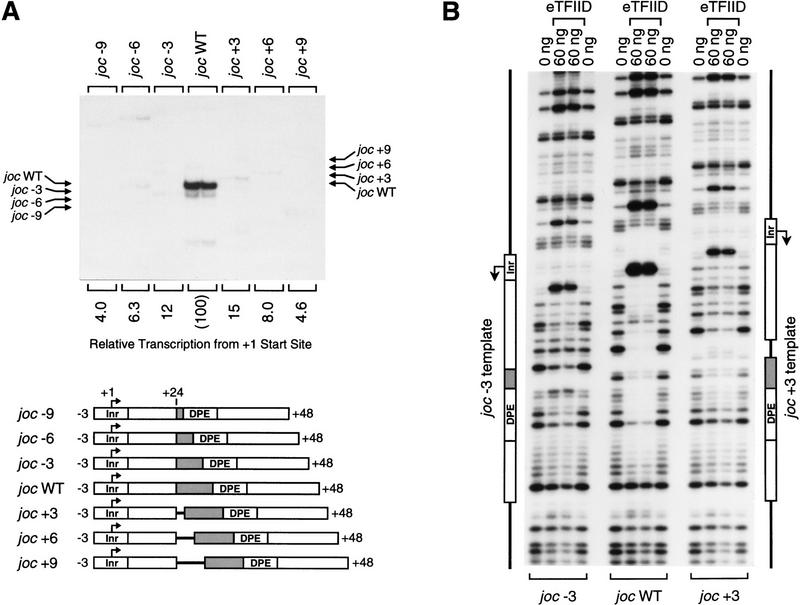
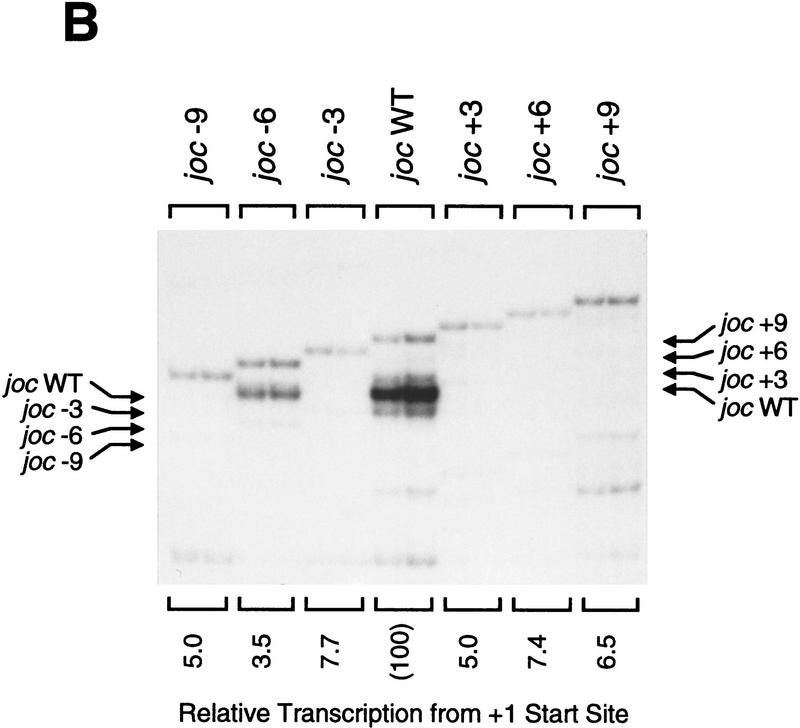
To gain a better understanding of the fundamental parameters that govern the function of the DPE, we tested whether the spacing between the DPE and the Inr is important for promoter activity. Analysis of a Drosophila promoter database revealed that DPE consensus sequences (or partial, DPE-like quadruplet motifs; Arkhipova 1995) are typically found ∼30 nucleotides downstream of the transcription start site (Burke and Kadonaga 1996). We therefore constructed a series of minimal Drosophila jockey (joc; Mizrokhi et al. 1988) mutant promoters in which the distance between the Inr and the DPE was either increased or decreased by 3, 6, or 9 nucleotides (Fig. 2A). In vitro transcription analysis of these promoters revealed that a 3-nucleotide increase or decrease in the Inr to DPE spacing results in a seven- to eight-fold reduction in transcription, whereas changes of 6 or 9 nucleotides in the interval between the Inr and DPE cause a greater reduction (12- to 25-fold) in promoter activity. Thus, transcriptional activity is significantly reduced upon alteration of the spacing between the Inr and DPE with a naturally occurring core promoter.
Figure 2.
The position of the DPE relative to the Inr is important for transcriptional activity and TFIID binding. (A) In vitro transcription analysis of wild-type and mutant Drosophila joc promoters. A series of mutant joc core promoters (comprising sequences from −3 to +48 relative to the RNA start site) was constructed in which the spacing between the DPE and the Inr was either increased or decreased by 3-nucleotide increments, as depicted. These templates were transcribed in vitro with the Drosophila SK nuclear extract, and the resulting transcripts were subjected to primer extension analysis. The positions of the reverse transcription products from each template are indicated by arrows. Transcriptional activity from the +1 start site for each template is reported as relative to that of the wild-type joc promoter (joc WT). (B) DNase I footprinting analysis of the binding of TFIID to the joc promoter. The mutant versions of the promoter contained either a 3-nucleotide deletion (joc −3) or a 3-nucleotide insertion (joc +3) between the Inr and DPE elements, as in A. The amount of purified Drosophila eTFIID in each reaction is indicated. The positions of the Inr and DPE elements in the mutant promoters are depicted at each side of the autoradiograph.
Because the DPE is a recognition site for the binding of TFIID to the joc promoter, we carried out DNase I footprinting experiments to examine whether alteration of the Inr to DPE spacing affects DNA binding by purified TFIID. As shown in Figure 2B, a 3-nucleotide deletion (joc −3) or insertion (joc +3) between the Inr and the DPE causes a substantial reduction in the TFIID footprint relative to that seen with the wild-type joc promoter (joc WT). The spacing mutant promoters exhibited a decrease in DNase I protection at several sites in the region encompassing the Inr to DPE, as well as a decrease in the DNase I hypersensitive sites that flank the Inr. Therefore, these results indicate that the native spacing between the Inr and the DPE is important for TFIID binding and for transcriptional activity of TATA-less, DPE-containing promoters. This strict spacing requirement suggests that the interaction of TFIID with the Inr and DPE elements is somewhat inflexible. It seems likely, however, that the optimal number of nucleotides between the Inr and DPE will not be identical for all DPE-containing promoters, because differences in the structure of the intervening DNA sequences in different promoters could affect the binding of TFIID to the Inr and DPE motifs.
DPE activity is not restricted to the complete 7-nucleotide consensus sequence
The DPE-containing promoters that we have studied thus far [Drosophila joc and Drosophila Antennapedia P2 (Antp P2) promoters] contain a 7-nucleotide sequence that conforms to the DPE consensus (Fig. 1) and was derived from sequences of 11 Drosophila promoters (Burke and Kadonaga 1996). There are, however, other promoters that contain a partial DPE sequence, G-A/T-C-G (which is a subset of the 7-nucleotide DPE consensus), at ∼30 nucleotides downstream of the transcription start site where a DPE motif would be located (see, e.g., Burke and Kadonaga 1996). Because these DPE-related sequences may, in some instances, be genuine DPE motifs, we examined two naturally occurring promoters that contain these partial DPE-like sequences.
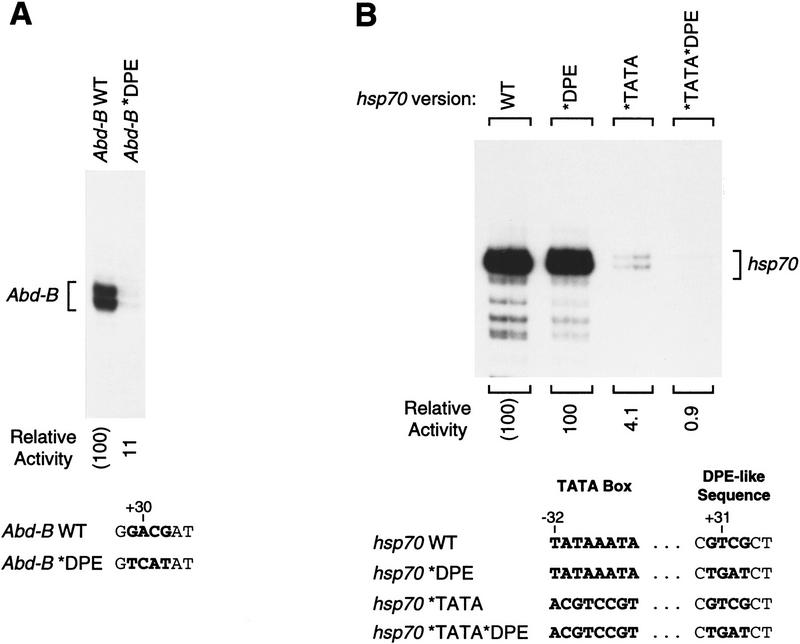
First, we tested the TATA-less Drosophila Abdominal-B (Abd-B) promoter (DeLorenzi et al. 1988). As shown in Figure 3A, we analyzed wild-type and mutant versions of the Abd-B core promoter and found that disruption of the partial DPE sequence causes a nine-fold reduction in transcriptional activity. Hence, it appears that DPE function is not restricted to the full 7-nucleotide DPE consensus sequence.
Figure 3.
Analysis of the partial DPE sequence, GA/TCG, in a TATA-less and a TATA-containing promoter. (A) A DPE-related motif in the TATA-less Drosophila Abd-B core promoter is important for transcriptional activity. Wild-type and DPE mutant versions of the Abd-B core promoter (comprising sequences from −17 to +41 relative to the major upstream start site) were constructed, as depicted at the bottom. These templates were transcribed in vitro with the Drosophila SK nuclear extract, and the resulting transcripts were subjected to primer extension analysis. Transcriptional activity is reported as relative to that of the wild-type Abd-B minimal promoter. (B) The DPE-like sequence in the TATA-containing Drosophila hsp70 promoter does not compensate for the loss of the TATA box. Wild-type and mutant versions of the TATA-containing hsp70 core promoter (comprising sequences from −35 to +40 relative to the RNA start site) were constructed, as shown. Transcription reactions were performed with the indicated template DNAs and Drosophila SK nuclear extract. Transcriptional activity is reported as relative to that of the wild-type hsp70 minimal promoter.
We then examined a TATA-containing promoter, Drosophila hsp70, for potential function of a 4-nucleotide DPE-like sequence. It had been shown previously that TFIID interacts with downstream sequences of an hsp70 promoter at approximately +18 and +28 relative to the transcription start site (Purnell and Gilmour 1993; Purnell et al. 1994). Inspection of this downstream region of hsp70 reveals a GTCG sequence at a position (+30 to +33) where a DPE would be located. Hence, we and others (Roeder 1996) had postulated that this hsp70 promoter may be an atypical DPE-containing promoter with an upstream TATA box element. To test this hypothesis, we analyzed the properties of mutant versions of this hsp70 promoter in which the TATA box and partial DPE-like sequence had been modified individually and in combination (Fig. 3B). These experiments revealed that mutation of the TATA box causes a significant (25-fold) reduction in transcription (Fig. 3B, cf. WT with *TATA), while in contrast, mutation of the DPE-like sequence does not affect promoter activity (Fig. 3B, cf. WT with *DPE). Thus, the DPE-like sequence in this hsp70 promoter is not required for basal transcriptional activity and, in contrast to the Antp P2 DPE (Burke and Kadonaga 1996), is not able to compensate for the loss of a TATA box. If, however, both the TATA box and the DPE-like sequence are mutated, then the amount of transcription is severalfold less than that seen upon mutation of the TATA box alone (Fig. 3B, cf. *TATA with *TATA*DPE). Hence, the DPE-like motif in this hsp70 promoter may contain weak DPE activity in the absence of the TATA box. These results indicate that the partial DPE-like sequence in this TATA-containing hsp70 promoter possesses little or no DPE activity.
We do not yet know the precise consensus sequence for the DPE. However, based on these results as well as on the significantly higher occurrence of consensus DPE or partial DPE-related sequences in the +30 downstream region in TATA-less promoters relative to TATA-containing promoters (Arkhipova 1995; Burke and Kadonaga 1996), it seems likely that DPE motifs mainly function in TATA-less promoters.
Alteration of the DPE sequence does not affect transcript stability
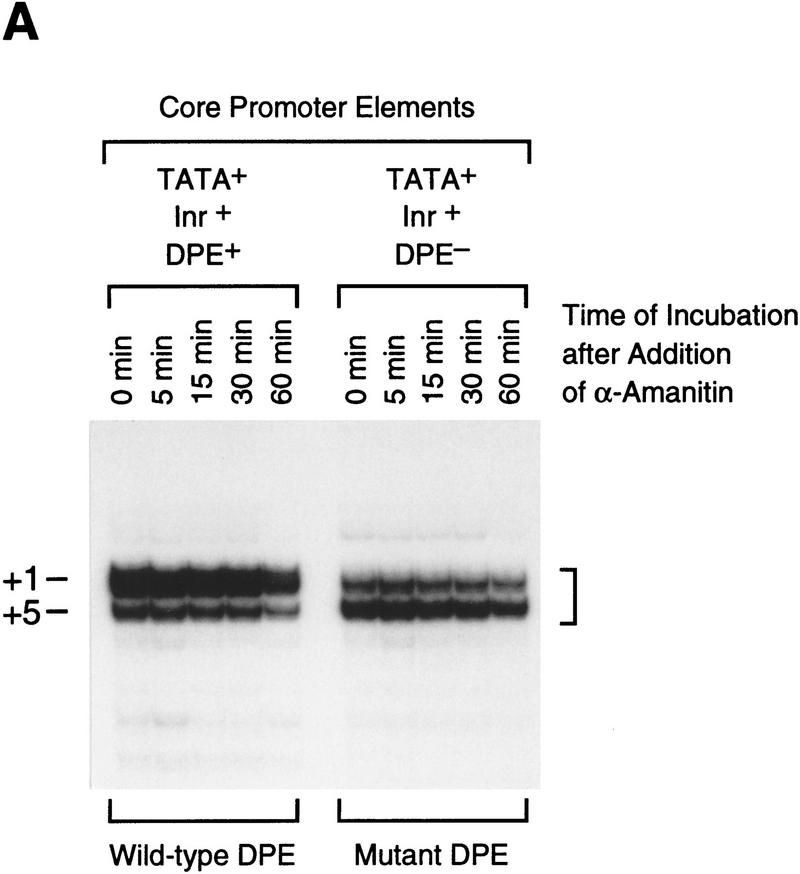
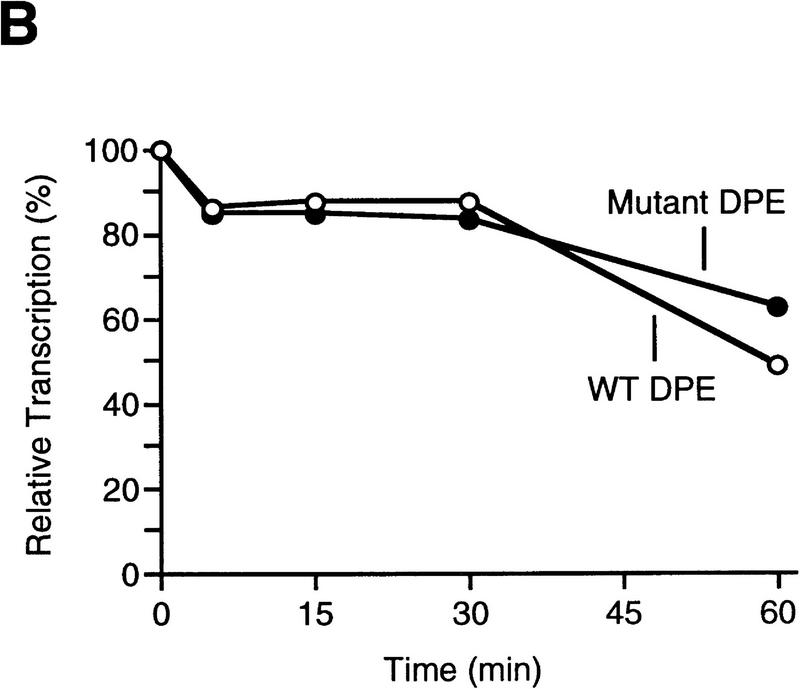
Because the DPE is downstream of the transcription start site, mutation of the DPE sequence also results in alteration of the RNA transcript. It is therefore possible that transcripts containing sequences derived from the mutant DPE might be less stable than the wild-type transcripts and that this instability might contribute to the reduction in transcriptional activity that is observed upon mutation of the DPE. To test for this possibility, we performed in vitro transcription reactions with two related hybrid promoter templates consisting of the Drosophila hunchback P2 (hb P2) core promoter (which possesses TATA and Inr elements) fused to downstream sequences of the Antp P2 (Perkins et al. 1988) promoter that contain either the wild-type DPE or a nonfunctional mutant DPE. As shown previously (Burke and Kadonaga 1996), both of these templates, hb P2–Antp P2 (TATA+, Inr+, DPE+) and hb P2–Antp P2*DPE (TATA+, Inr+, DPE−), are transcriptionally active because they each contain TATA box and Inr elements. Transcription reactions were performed with each of these templates, and α-amanitin was added to inhibit transcription. The reaction mixtures were incubated for an additional period of up to 60 min, and the resulting RNA was detected by primer extension analysis. As shown in Figure 4, both the wild-type and the mutant DPE transcripts were of comparable stability upon incubation up to 60 min in the reaction medium. It thus appears that the decrease in transcriptional activity that is seen upon mutation of the DPE sequence in TATA-less promoters is due to reduced efficiency of the transcription process rather than to reduced transcript stability.
Figure 4.
Mutation of the DPE sequence does not affect transcript stability. (A) In vitro transcription reactions were performed with the hb P2–Antp P2 (TATA+Inr+DPE+) and hb P2–Antp P2*DPE (TATA+Inr+DPE−) hybrid minimal promoters (Burke and Kadonaga 1996). After 30-min reaction time, α-amanitin was added (to 4 μg/ml final concentration) to inhibit transcription, and the reaction mixtures were further incubated for the indicated times to determine the stability of the transcripts with the wild-type DPE relative to those with the mutant DPE. The reverse transcription products are indicated by the bracket (right). Note that the preferential use of the +1 start site in the DPE-containing promoter is probably due to the optimal positioning of the DPE to the +1 start site relative to the +5 site, as discussed in Burke and Kadonaga (1996). (B) Graph of data shown in A.
Drosophila TAFII60 is specifically cross-linked to the DPE
Previously, we had found that purified TFIID, but not TBP, is able to bind to TATA-less, DPE-containing promoters (Burke and Kadonaga 1996). It therefore appeared likely that binding of TFIID to the DPE involved one or more of the TAF subunits. To determine which polypeptide (or polypeptides) of TFIID was in close proximity to the DPE (and, hence, likely to interact directly with the DPE), we carried out photo-cross-linking experiments with core promoter probes containing the thymidine analog 5-[N-(p-azidobenzoyl)-3-amino allyl]–deoxyuridine 5′-monophosphate (N3RdUMP; Bartholomew et al. 1990, 1991, 1995). N3RdUMP has a photoreactive aryl azide moiety on a 9- to 10-Å tether that probes the space in the vicinity of the DNA major groove (Bartholomew et al. 1990), and it has been used extensively in the study of proteins that interact with DNA. For instance, in the context of these studies, it is pertinent to note that N3RdUMP was used in the analysis of the binding of human TFIID to the TATA-containing, DPE-less adenovirus major late promoter (Oelgeschläger et al. 1996). Thus, by the incorporation of N3RdUMP and an adjacent radioactive label at specific positions in the joc promoter, we were able to map close interactions of individual TFIID subunits with the DPE (Fig. 5).
Figure 5.
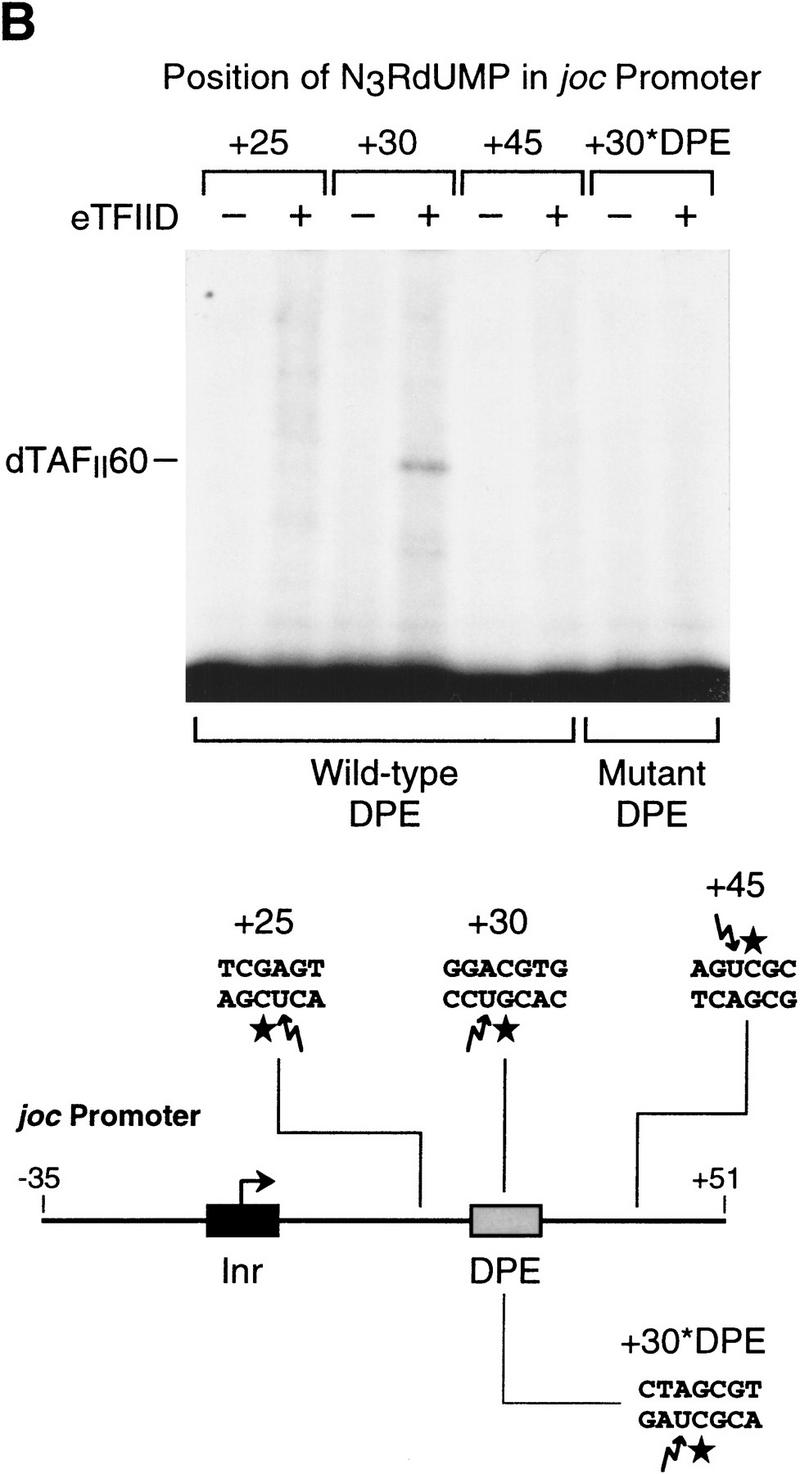
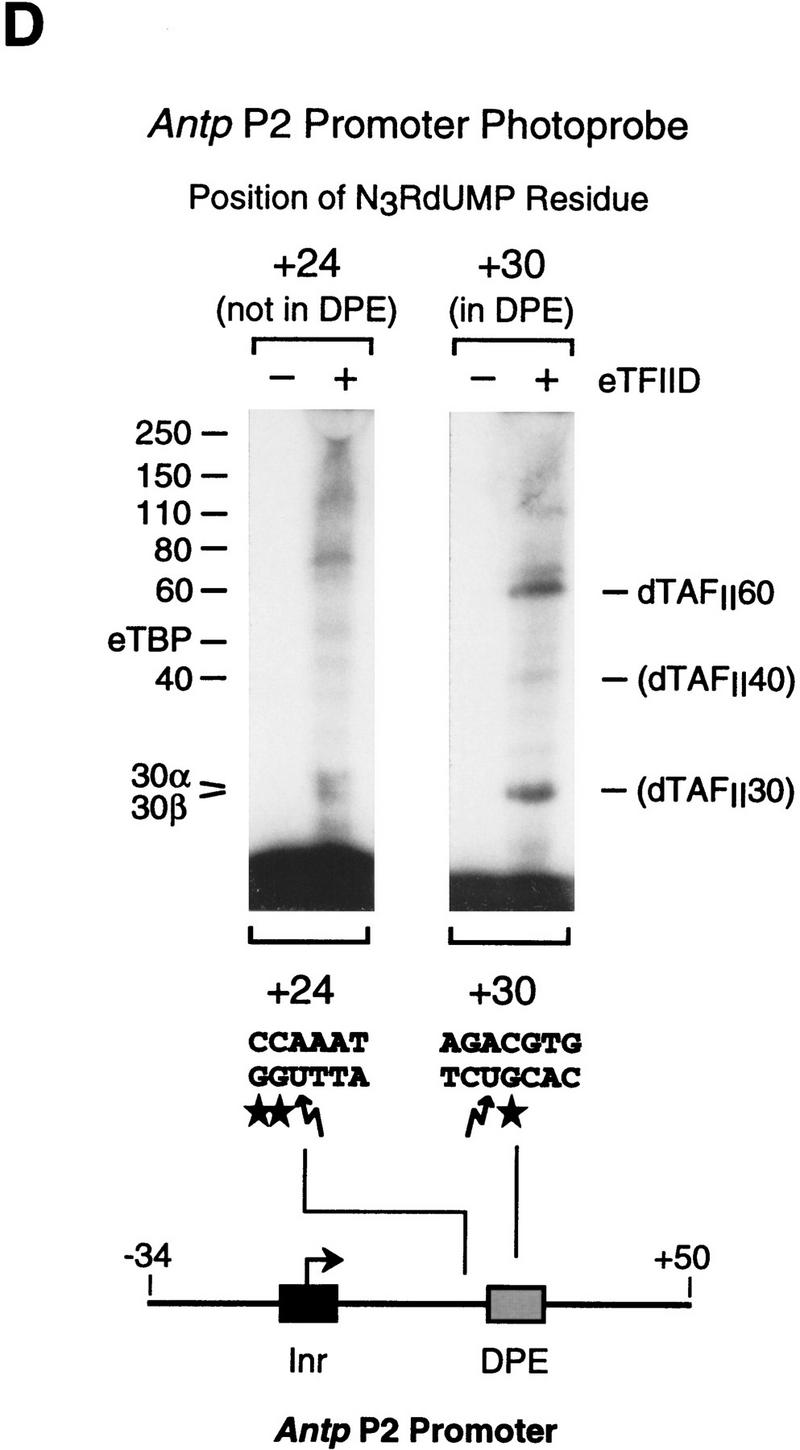
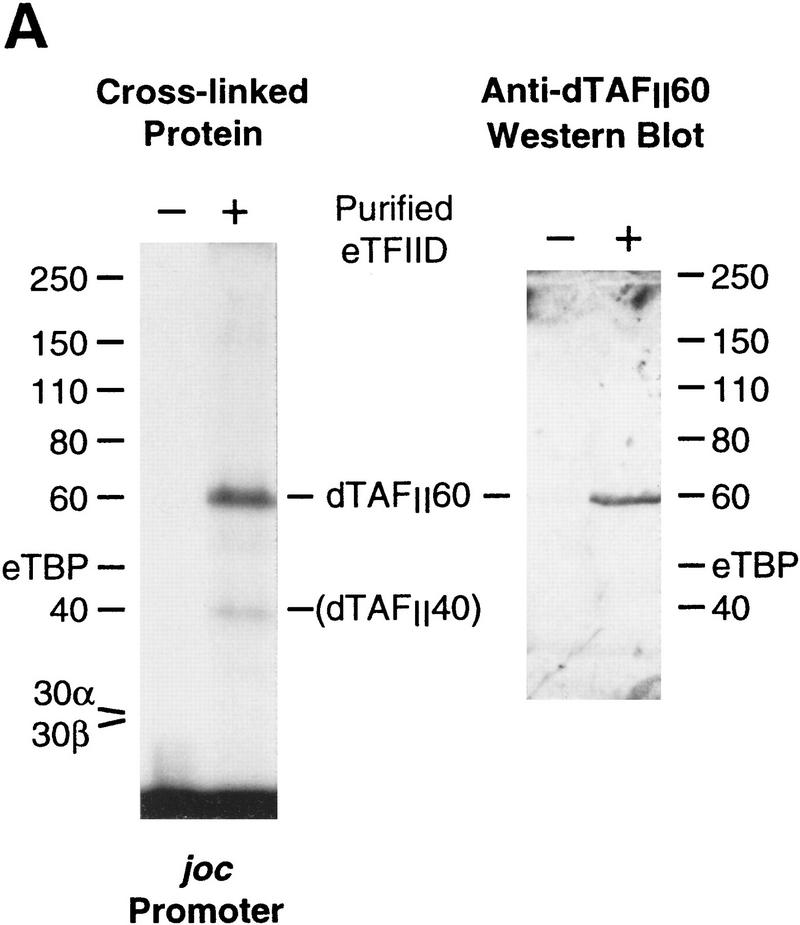
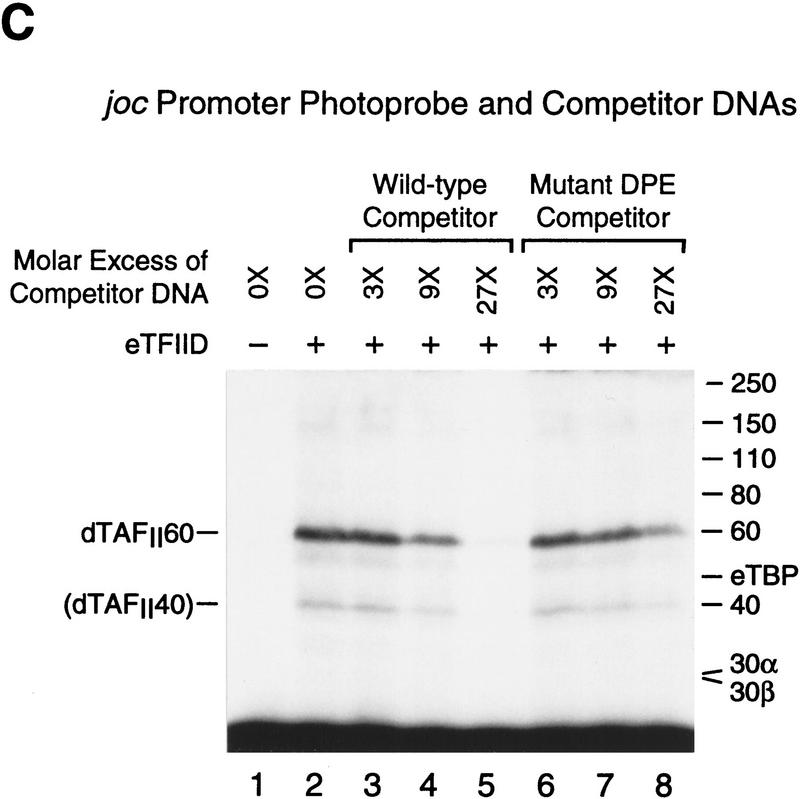
The histone H4-related dTAFII60 is cross-linked to the DPE. (A) dTAFII60 is cross-linked to the joc DPE. Photo-cross-linking experiments were performed with purified TFIID and the joc core promoter. The joc promoter photoprobe was a DNA fragment comprising sequences from −35 to +51 relative to the start site, which contained N3RdUMP in the DPE as well as an adjacent radiolabeled nucleotide (see the +30 probe at the bottom of B). TFIID was incubated with the DNA photoprobe, and the samples were subjected to UV irradiation. The complexes were then digested with nucleases to remove excess DNA, and the resulting radiolabeled proteins were resolved by electrophoresis on a 10% polyacrylamide–SDS gel and detected by autoradiography (left). The migration of each of the polypeptides that comprise TFIID is indicated. Western blot analysis of the cross-linked protein further confirmed that the major radiolabeled band corresponds to dTAFII60 (right). Weaker, yet clearly apparent cross-linking to a species that comigrates with dTAFII40 is also indicated. (B) Sequence-specific photo-cross-linking of the dTAFII60 subunit of TFIID to the wild-type DPE, but not to a mutant DPE. Four different joc promoter photoprobes, which are indicated at the bottom, were each incubated with TFIID and subjected to cross-linking. The positions of N3RdUMP residues (arrows) and radiolabeled nucleotides (stars) are indicated. In the mutant DPE (+30*DPE) probe, the wild-type DPE motif GGACGTG was changed to CTAGCGT. (C) The interaction of dTAFII60 with the DPE photoaffinity probe can be specifically inhibited by competition with a DPE-containing promoter fragment. TFIID was photo-cross-linked to the +30 DPE probe (see B) in the absence or the presence of the indicated amounts (reported as the molar ratio of competitor DNA to probe DNA) of wild-type DPE or mutant DPE promoter DNA fragments. The wild-type DPE competitor DNA fragment was identical to the photoprobe, except that it was not radiolabeled and contained a thymidine residue instead of N3RdUMP. The corresponding mutant DPE competitor DNA contained GGACGTG to CTAGCGT substitutions at the DPE. (D) Photo-cross-linking of TFIID to the DPE in the Antp P2 core promoter. The positions of N3RdUMP residues (arrows) and radiolabeled nucleotides (stars) are indicated.
Analysis of purified Drosophila TFIID with the joc promoter containing a N3RdUMP residue in the DPE (at +30 relative to the start site) revealed strong cross-linking of an ∼60-kD polypeptide and weaker cross-linking of an ∼40-kD polypeptide (Fig. 5A). Comparison of the migration of these polypeptides with that of TFIID subunits indicated that these two cross-linked polypeptides co-migrate with the core histone-like TAFs, dTAFII60 and dTAFII40 (also known as dTAFII62 and dTAFII42; see, e.g., Burley and Roeder 1996), which possess regions of sequence similarity to histones H4 and H3, respectively. The coincident migration of the ∼60-kD cross-linked polypeptide with dTAFII60 was also confirmed by Western blot analysis of cross-linked TFIID (Fig. 5A).
To determine whether the cross-linking of dTAFII60 was specific for the DPE motif, we prepared a series of photoprobes in which N3RdUMP was incorporated at different locations in the downstream promoter region (Fig. 5B, bottom). As shown in Figure 5B (top), cross-linking of dTAFII60 was observed when N3RdUMP was in the wild-type DPE (+30) but not when the photoreactive nucleotide was located either upstream (+25) or downstream (+45) of the DPE. In addition, when the consensus DPE sequence in the photoprobe was mutated (Fig. 5B, +30*DPE), dTAFII60 was not cross-linked to N3RdUMP at the +30 position. These results indicate that dTAFII60 is in close proximity to the DPE motif and that the interactions leading to dTAFII60 cross-linking are specific for the DPE motif, as seen in the binding of TFIID to DPE-containing promoters.
Next, we tested the specificity of dTAFII60 interactions with the DPE by carrying out cross-linking experiments in the presence of unlabeled competitor DNA (Fig. 5C). These experiments revealed that cross-linking of dTAFII60 was specifically inhibited by a competitor DNA fragment with a wild-type DPE motif but not by a competitor DNA fragment with a mutated DPE. For instance, the presence of a 27-fold molar excess of wild-type competitor inhibited cross-linking of dTAFII60 nearly completely (Fig. 5C, lane 5), whereas the inclusion of the same amount of mutant competitor led to only a modest decrease in cross-linking efficiency (Fig. 5C, lane 8). It is also notable that the apparent cross-linking to dTAFII40 was inhibited more potently by the wild-type DPE competitor DNA than by the mutant DPE competitor DNA.
To gain broader insight into TFIID components that are in the vicinity of the DPE, we carried out photo-cross-linking experiments with the Antp P2 core promoter (Fig. 5D). These experiments revealed specific cross-linking of dTAFII60 and dTAFII40 to the Antp P2 DPE, as seen with the joc DPE. Interestingly, cross-linking of a 30-kD species, which might be the histone H2B-like dTAFII30α (also known as p28/22 or dTAF28/22; Yokomori et al. 1993; Kokubo et al. 1994; Hoffmann et al. 1996), was also observed with the Antp P2 DPE. These findings provide additional evidence that dTAFII60 and dTAFII40 interact with the DPE and also suggest that a 30-kD TAF (perhaps the H2B-like dTAFII30α) is in the vicinity of the DPE.
Hence, these data, combined with the knowledge that dTAFII60 and dTAFII40 (which are structurally related to histones H4 and H3) can interact with one another and form an α2β2 heterotetramer (Goodrich et al. 1993; Weinzierl et al. 1993; Kokubo et al. 1994; Nakatani et al. 1996; Xie et al. 1996), suggest that a dTAFII60–dTAFII40 heterotetramer interacts with the DPE motif as a component of the TFIID complex. It is intriguing that an H3–H4 tetramer-like structure in TFIID appears to interact with the DPE in a sequence-specific manner.
Human transcription factors require the DPE for basal transcription from the Drosophila joc promoter
To investigate whether the DPE is conserved from Drosophila to humans, we initially tested whether human basal transcription factors exhibit a requirement for a DPE in a Drosophila promoter. We therefore examined the ability of a HeLa (human) cell nuclear extract to transcribe wild-type and mutant versions of the TATA-less, DPE-containing joc promoter. First, we analyzed a series of clustered point mutant versions of the joc promoter and found that mutation of the DPE motif, but not flanking sequences, results in a 7- to 25-fold reduction in transcription by HeLa factors (Fig. 6A). The relative specificity of the HeLa factors to transcribe each of these mutant promoters was nearly identical to that observed with Drosophila transcription factors (Burke and Kadonaga 1996). Then, we tested the ability of the HeLa factors to transcribe the joc spacing mutants shown in Figure 2A. In these experiments we found that transcription with the HeLa factors was strongly reduced upon alteration of the spacing between the Inr and DPE motifs (Fig. 6B), as seen with the Drosophila transcription factors (Fig. 2A). Hence, human and Drosophila transcription factors exhibit essentially the same requirements for DPE sequence and for Inr–DPE spacing, which suggests that DPE function is conserved from Drosophila to humans.
Figure 6.
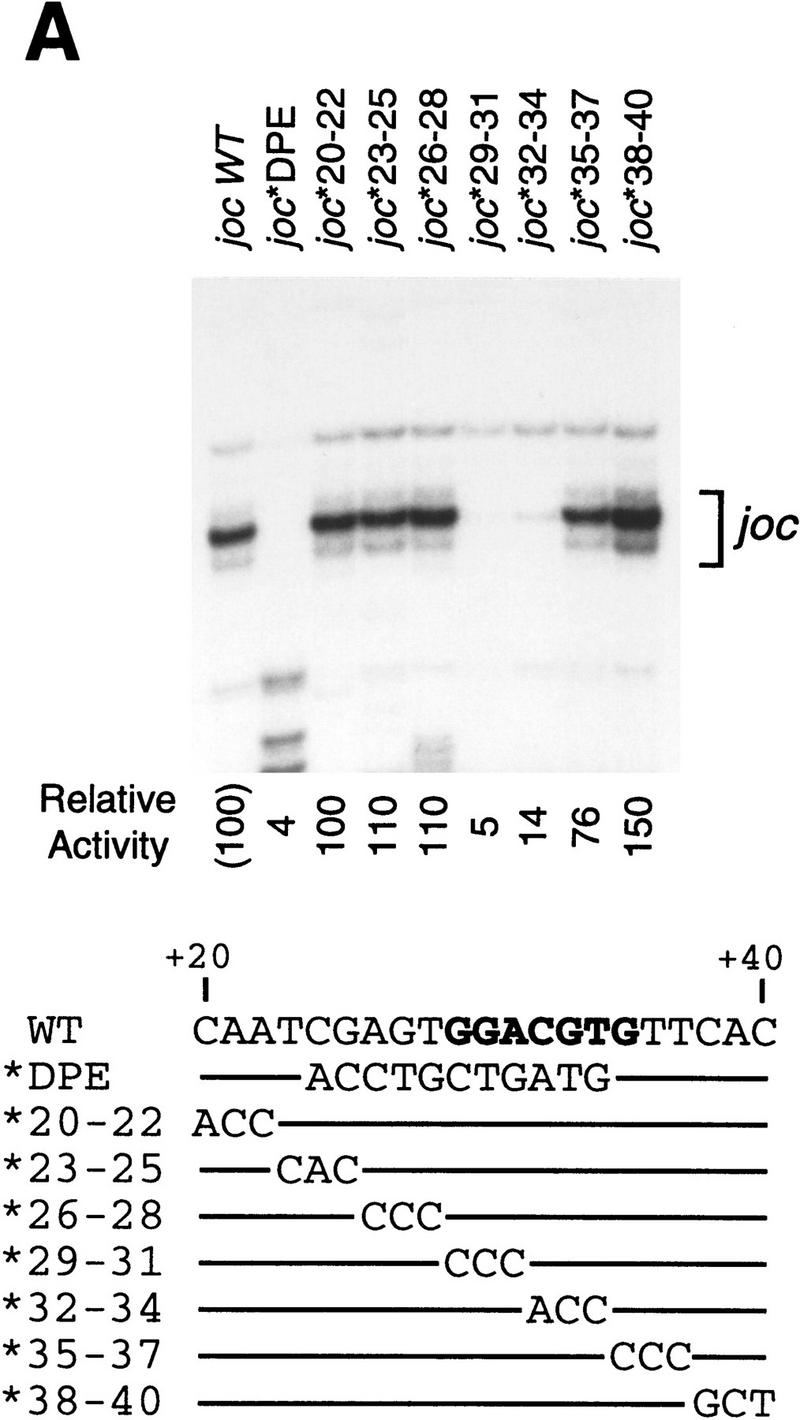
Human transcription factors can mediate basal transcription via a Drosophila DPE. (A) The conserved DPE consensus sequence is required for basal transcription by HeLa factors. A systematic set of clustered triple point mutant versions of the joc minimal promoter (from −3 to +48 relative to the major RNA start site; Burke and Kadonaga 1996) was subjected to in vitro transcription analysis with a HeLa nuclear extract. The joc promoter sequence from +20 to +40 relative to the transcription start site is shown with the corresponding nucleotide substitutions below. Lines indicate unchanged sequences. The DPE consensus sequence is shown in boldface type. The transcriptional activity is reported relative to that of the wild-type joc minimal (−3 to +48) promoter. (B) The spacing between the Inr and the DPE elements is important for basal transcription by HeLa factors. A series of mutant joc promoters in which the spacing between the Inr and DPE is varied by 3-nucleotide increments, as depicted in Fig. 2A, was subjected to in vitro transcription analysis with a HeLa nuclear extract. The reverse transcription products are indicated by arrows. Transcriptional activity from the +1 start site of each promoter is reported relative to that of the wild-type joc minimal promoter.
Interestingly, with the joc −6 mutant promoter, the DPE had directed inappropriate transcription from sequences in the pUC plasmid backbone that were located ∼6 nucleotides upstream of the normal transcription start site in the Inr. Moreover, this spurious transcription from −6 with the joc −6 promoter was seen with the HeLa factors (Fig. 6B) but not with the Drosophila factors (Fig. 2A). This difference may reflect the more degenerate Inr consensus sequence in mammals than in Drosophila, as the HeLa factors may possess less stringent specificity for Inr sequences than the Drosophila factors. Thus, these results indicate the strong propensity of the DPE motif to direct basal transcription and further reinforce the rigidity of the requirement for optimal Inr–DPE spacing.
We also tested whether purified human TFIID (provided courtesy of I. Olave and D. Reinberg, University of Medicine and Dentistry of New Jersey), which appears to be deficient in a human homolog of dTAFII150, binds to the TATA-less DPE-containing joc promoter. DNase I footprinting experiments revealed that human TFIID binds to the wild-type joc promoter but not to a mutant joc*DPE promoter with an altered DPE (data not shown). These results are consistent with the requirement for the DPE for transcription by the human basal factors (Fig. 6) and also suggest that dTAFII150 is not necessary for binding of TFIID to TATA-less DPE-containing promoters.
A mammalian DPE that is functionally important both in vitro and in vivo
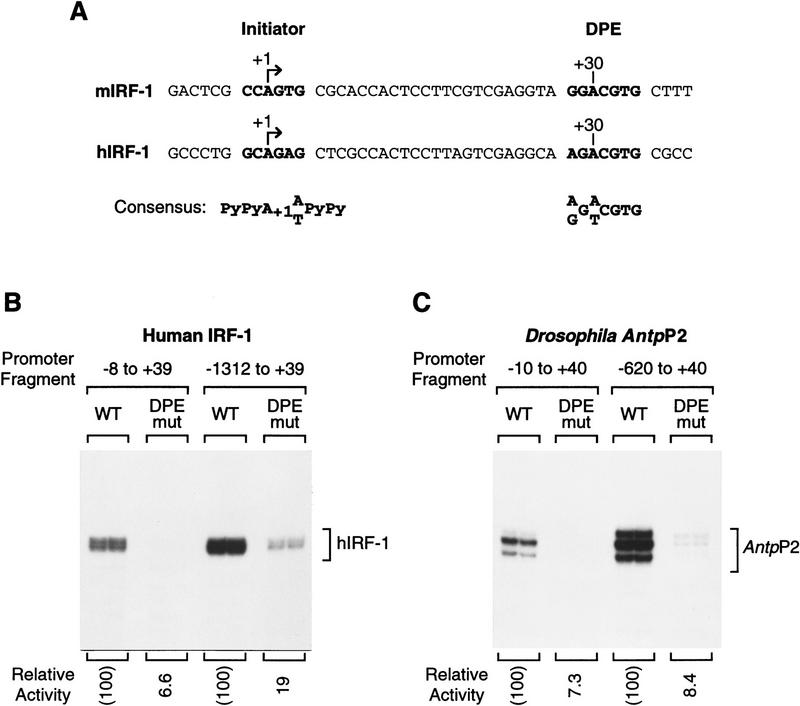
The ability of human transcription factors to recognize the DPE suggests that there is a homologous downstream element in mammals. We therefore examined the sequences of some mammalian promoters and found that the promoters of the genes encoding human (h) and mouse (m) interferon regulatory factor-1 (IRF-1; Miyamoto et al. 1988; Sims et al. 1993) is TATA-less and contains a sequence that conforms to the 7-nucleotide DPE consensus at the appropriate +30 downstream position (Fig. 7A). To study the functional importance of this DPE-like sequence, we constructed wild-type and DPE–mutant versions of the hIRF-1 promoter either as a minimal promoter (from −8 to +39) or as a larger promoter fragment (from −1312 to +39). The minimal promoter enables the precise examination of core promoter function, whereas the larger promoter fragment reveals the role of the DPE in a broader context of the gene. As shown in Figure 7B, disruption of the hIRF-1 DPE-like sequence causes a 15- and 5-fold reduction in transcriptional activity with the minimal and large promoters, respectively, when transcribed in vitro with a human HeLa cell nuclear extract. Similar results were seen in the analysis of minimal and larger promoter fragments of the Antp P2 promoter (Fig. 7C). In addition, mutation of the DPE in the minimal mIRF-1 promoter (from GGACGTG to TTCATGT) was found to reduce promoter activity by ∼17-fold (data not shown). Therefore, the DPE-like sequence in the IRF-1 gene appears to be functionally homologous to the Drosophila DPE motif.
Figure 7.
The mammalian IRF-1 gene has a TATA-less, DPE-containing core promoter. (A) The promoters of the IRF-1 genes from mice (m) and humans (h) contain a consensus DPE sequence that is located downstream of the Inr element with spacing that is identical to that seen in many Drosophila DPE-containing promoters. The mammalian Inr and Drosophila DPE consensus sequences are indicated. (B) The hIRF-1 DPE sequence is important for basal transcription in the context of either a minimal core promoter or a larger promoter fragment. The human IRF-1 promoter constructions, which contained the indicated sequences relative to the RNA start site, were transcribed with a HeLa nuclear extract. In the mutant promoter, the DPE sequence was altered from AGACGTG to CTCATGT. The amount of transcription from each mutant promoter is reported as the percentage of transcription from the corresponding wild-type promoter. (C) The Drosophila Antp P2 DPE sequence is important for basal transcription in the context of either a minimal core promoter or a larger promoter fragment. The Antp P2 promoter constructions, which contained the indicated sequences relative to the RNA start site, were transcribed with a Drosophila SK nuclear ex-tract. In the mutant promoter, the DPE sequence was altered from AGACGTG to CTCATGT. The amount of transcription from each mutant promoter is reported as the percentage of transcription from the corresponding wild-type promoter.
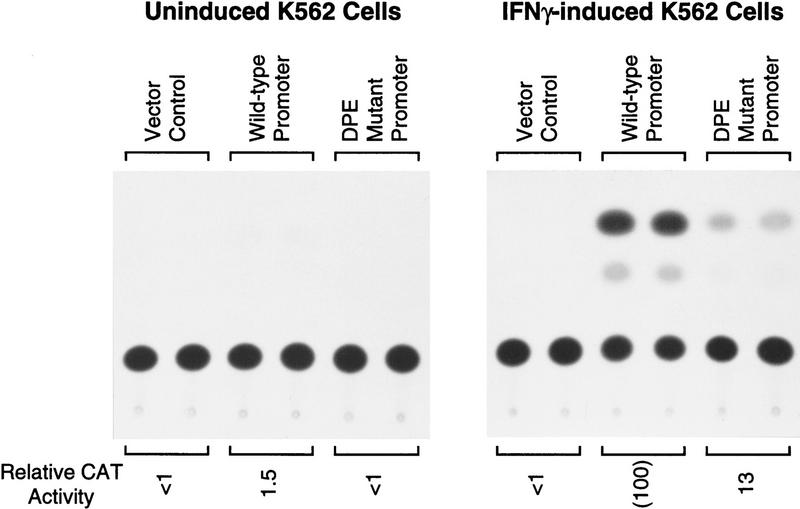
Next, we tested the activity of the human IRF-1 DPE in cultured cells (Fig. 8). Plasmids were constructed with the wild-type or DPE–mutant versions of the hIRF-1 promoter (from −1312 to +39), fused to the chloramphenicol acetyltransferase (CAT) reporter gene, and then transfected by electroporation into K562 erythroleukemia cells. In K562 cells, the hIRF-1 gene is inducible with γ interferon (IFN-γ; Sims et al. 1993). Low levels of basal activity were observed in uninduced cells, and transcriptional activity was found to increase by ∼67-fold upon addition of IFN-γ (Fig. 8). Under these conditions, mutation of the DPE motif resulted in an approximately eight-fold reduction in promoter activity, which is comparable to that seen in vitro (Fig. 7B). These experiments therefore provide evidence that the IRF-1 promoter is an example of a human TATA-less, DPE-containing promoter, and that the IRF-1 DPE is required for efficient transcription both in vitro and in vivo.
Figure 8.
The DPE sequence in the human IRF-1 promoter is important for IFN-γ-induced transcription in cultured cells. The hIRF-1 promoter (containing sequences from −1312 to +39 relative to the transcription start site) with a CAT reporter gene was transfected into K562 cells, which were then grown in the presence or the absence of IFN-γ (1500 U/ml), as indicated. The promoterless vector was transfected in parallel as a control. CAT activity is reported relative to that seen with the wild-type promoter in the presence of IFN-γ.
Thus, these data suggest that the DPE is a core promoter element that is used in both Drosophila and mammals. This conclusion is in accord with the general observation that basal transcription by RNA polymerase II is conserved throughout eukaryotes and that transcriptional mechanisms in Drosophila and humans appear to be very closely related. For instance, Drosophila promoters can be transcribed with human factors, and vice versa. In addition, each of the Drosophila basal transcription factors can be functionally substituted with its human counterpart, and vice versa (see, e.g., Wampler et al. 1990; Sauer and Tjian 1997).
It may be notable that Inr sequences exhibit a much more distinct consensus sequence in Drosophila than in mammals, in which the Inr consensus appears to be more degenerate. By analogy, it is possible that the consensus DPE sequence might be more distinctly apparent in Drosophila than in mammals. Hence, although the IRF-1 promoter DPE sequence conforms to the tentative Drosophila DPE consensus, it seems likely that there are many mammalian promoters that do not fit this consensus but nevertheless do possess fully functional DPE motifs.
The DPE—a downstream counterpart to the TATA box?
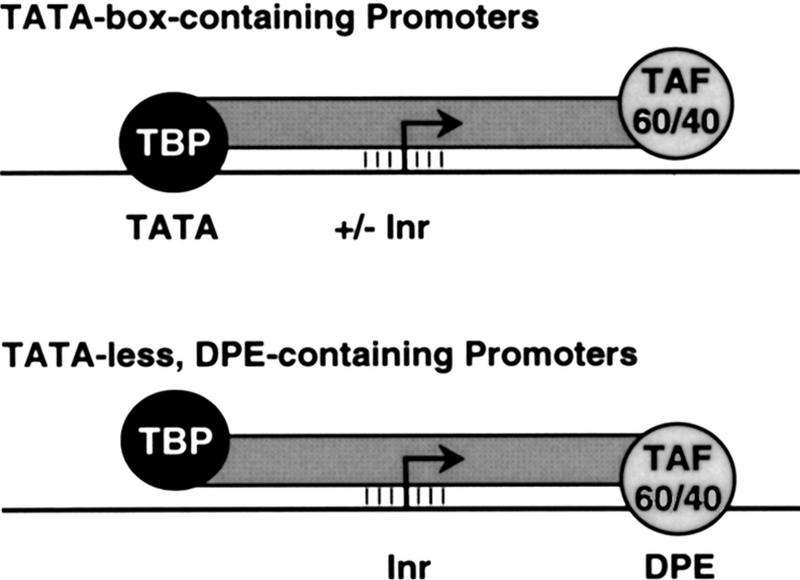
Comparison of the properties of the TATA box and the DPE reveals the following. First, both are core promoter elements that are recognized and bound by TFIID. The TATA box is ∼30 nucleotides upstream of the start site and is bound by TBP, whereas the DPE is ∼30 nucleotides downstream of the start site and appears to be bound by dTAFII60–TAFII40, probably as a heterotetramer that resembles the histone H3–H4 tetramer. However, purified TBP polypeptide alone is able to bind to the TATA box, whereas, in contrast, TFIID binds to DPE-containing promoters via cooperative interactions between the Inr and DPE motifs (Burke and Kadonaga 1996). The strict requirement for spacing between the Inr and DPE motifs further suggests a specific and somewhat rigid interaction of TFIID with the Inr and DPE sequences. Thus, the interaction of TFIID with the DPE is likely to be different from the well-characterized binding of TBP to the TATA box. Nevertheless, it is likely that interactions of TFIID with either the TATA box or the DPE are important early events in transcription of genes that contain these motifs.
Second, the TATA box and DPE are functionally redundant. For instance, the loss of a TATA box can be compensated by the addition of a DPE (Burke and Kadonaga 1996). The DPE, like the TATA box, is present in some but not all core promoters, but all known or putative DPE-containing promoters are TATA-less, which is consistent with the absence of a requirement for both elements in a core promoter.
Third, the DPE motif appears to be conserved from Drosophila to humans and thus likely has a fundamental and important role in basal transcription. These findings collectively suggest, as depicted in Figure 9, that the DPE is a downstream counterpart to the TATA box.
Figure 9.
A model for the interaction of TFIID with TATA box-containing vs. TATA-less, DPE-containing promoters. Depicted are two postulated TFIID–promoter interactions, not excluding other types of functionally important interactions between TFIID and core promoters. The lines between TFIID and the Inr indicate interactions between components of TFIID, such as TAFII150 and/or TAFII250, and the Inr.
DPE or TATA box?
Why would a promoter have a DPE instead of a TATA box? It is likely that there are many different answers to this question, which range from a basic biological necessity to random chance. Examination of Drosophila promoters that contain either the 7-nucleotide DPE consensus or the G-A/T-C-G partial DPE sequence at the +30 downstream position reveals an apparently random assortment of genes, with the exception of several long interspersed nuclear elements (LINEs), which include joc, Doc, F, G, and I elements (see, e.g., Burke and Kadonaga 1996; Minchiotti et al. 1997). LINEs are mobile elements that do not contain long terminal repeats (LTRs) and require an internal promoter for transposition. Hence, LINE promoters exemplify the use of the DPE in a situation where a downstream core promoter is needed for biological function.
We also postulate that the DPE might be involved in transcriptional regulation, such as in the communication between enhancers and promoters. It is possible, for instance, for enhancers to regulate nearby genes differentially based on the structure of the core promoter (see, e.g., Li and Noll 1994; Merli et al. 1996). Variation in the structure and function of core promoters, such as presence of DPE or TATA motifs, can vastly increase the range of potential mechanisms for transcriptional regulation.
Summary and perspectives
The DPE is a distinct core promoter element that appears to be conserved from Drosophila to humans. It is, in many respects, a downstream analog of the TATA box. In Drosophila, all known or putative DPE-containing promoters are TATA-less, but only a fraction (perhaps ∼20%) of TATA-less promoters appear to contain a DPE. It thus seems likely that there are additional core promoter motifs that remain to be identified. Analysis of interactions of purified TFIID with the DPE revealed strong cross-linking of dTAFII60 as well as weak cross-linking of dTAFII40. These data, combined with the well-characterized interactions between the two TAFs, suggest that a dTAFII60–dTAFII40 heterotetramer binds to the DPE. Hence, these studies provide evidence for a direct role of TAFs in basal transcription of TATA-less, DPE-containing genes. It is additionally intriguing to consider that sequence-specific binding of the DPE by the dTAFII60–dTAFII40 complex may resemble the interaction of DNA with the histone H3–H4 tetramer. Thus, in summary, the analysis of the function of the DPE in basal transcription has revealed both similarities and differences in the fundamental mechanisms of transcription from TATA-containing, DPE-less promoters versus TATA-less, DPE-containing promoters. It will be interesting, in the future, to investigate whether the distinctions between TATA-containing and DPE-containing promoters are used as a mode of transcriptional regulation at the level of the core promoter.
Materials and methods
UV cross-linking
N3RdUTP (5-[N-(p-azidobenzoyl)-3-amino allyl]-deoxyuridine 5′-triphosphate; also called AB–dUTP) was generously provided by Drs. George Kassavetis and E. Peter Geiduschek (University of California, San Diego, La Jolla). The N3RdUTP was synthesized and used as described previously (Bartholomew et al. 1990, 1995), except that the DNA probes were prepared from synthetic oligonucleotides instead of single-stranded M13 DNA. The specific DNA sequences and reaction conditions are available upon request.
Transient transfection assays
K562 human erythroleukemia cells were maintained at 37°C (5% CO2) in Dulbecco’s modified Eagle medium (DMEM) plus phenol red, supplemented with 10% fetal calf serum, 2 mm l-glutamine, 100 U/ml of penicillin, and 100 μg/ml of streptomycin. The cells, which were grown in suspension, were split by dilution daily for 3 days prior to transfection by electroporation. Cells were harvested and washed with 0.5 culture volume of ice-cold PBS (137 mm NaCl, 2.7 mm KCl, 3 mm Na2HPO4, 1.5 mm KH2PO4). Cells were resuspended in ice-cold PBS, counted with a hemocytometer, and diluted to ∼2 × 107 cells/ml in PBS. Aliquots of cell suspension (0.5 ml) were transferred into prechilled electroporation cuvettes (Bio-Rad Gene Pulser Cuvettes, 0.4 cm). Each sample received 20 μg of CAT reporter plasmid plus 10 μg of pCMVβ plasmid in a total volume of 10 μl, and samples were mixed and incubated on ice for 10 min. Cuvettes were placed in the electroporation apparatus (Bio-Rad Gene Pulser) and shocked once at a voltage of 0.2 kV and a capacitance of 960 μF (no resistor), and then returned to ice for 15 min. Transfected cells were then transferred to 10-cm plates containing 10 ml of culture medium (as above), in the presence or the absence of 1500 U/ml interferon-γ (Promega), as indicated, and the cuvettes were rinsed with medium to remove all transfected cells. Transfected cells were then incubated at 37°C (5% CO2). Twenty hours after electroporation, the transfected cells were harvested, washed with ice-cold PBS, and resuspended in 250 mm Tris-HCl at pH 7.6 (200 μl). Extracts were prepared by multiple freeze–thaw cycles, as described previously (Kraus et al. 1994). CAT assays, which were normalized for the β-galactosidase activity of each extract, were performed as described previously (Kraus et al. 1994). Quantitation of the CAT assay results was performed with a PhosphorImager (Molecular Dynamics).
In vitro transcription analysis
Transcription reactions were carried out as described previously (Wampler et al. 1990; Tyree et al. 1993; Burke and Kadonaga 1996) by using supercoiled DNA templates (500 ng) with either SK (Soeller–Kornberg) extract from Drosophila embryos (Soeller et al. 1988; Burke and Kadonaga 1996) or HeLa nuclear extract from HeLa (human) cells (Dignam et al. 1983) in a total volume of 25 μl. The resulting transcripts were subjected to primer extension analysis, as described previously (Kadonaga 1990; Wampler et al. 1990), with the M13 reverse sequencing primer, which is complementary to sequences downstream of the transcription start sites in pUC plasmid vector. The numerical designations of the promoters are given relative to the transcription start site (+1). The specific promoter sequences in each of the plasmids and the cloning strategies are available upon request. Quantitation of the in vitro-synthesized RNA was carried out with a PhosphorImager (Molecular Dynamics). All experiments were performed a minimum of two times (but typically at least three times) to ensure reproducibility of the data.
DNase I footprint analysis
DNase I footprint probes were prepared by PCR with primers flanking the promoter region, as described previously (Burke and Kadonaga 1996). PCR primers used for amplification of minimal promoter DNA templates were the M13 universal (upstream) and M13 reverse (downstream) sequencing primers. The PCR amplification products were gel-purified on 5% nondenaturing polyacrylamide gels to remove free primer and spurious reaction by-products. Binding reactions for DNase I footprinting experiments were performed for 30 min at 25°C in a total volume of 50 μl, and contained 12.5 mm HEPES (K+) at pH 7.6, 50 mm KCl, 0.05 mm EDTA, 5% glycerol, 0.05% NP-40, 0.5 mm dithiothreitol, 2% polyvinyl alcohol, and 32P-labeled probe (6000 cpm). Nonspecific competitor DNA was not included in the reactions. The labeled probes were then partially digested with DNase I, and the reaction products were purified and analyzed by electrophoresis on a 6% denaturing polyacrylamide gel.
Acknowledgments
We thank Jessica Tyler, Catherine George, Lee Kraus, Beth Blackwood, Jenny Butler, and Mike Pazin for critical reading of the manuscript. We are grateful to Drs. George Kassavetis and E. Peter Geiduschek for providing advice and N3RdUTP for the photo-cross-linking experiments; Aarron Willingham for preparation of the Abd-B promoter constructions; Ivan Olave and Danny Reinberg for the gift of purified human TFIID; Drs. Y. Cha Henderson and A. Deisseroth for the hIRF-1 promoter clone; Drs. Cathy Thut and Robert Tjian for the dTAFII60 cDNA; and Drs. W. Lee Kraus and Beth Blackwood for advice regarding the transient transfection assays. This work was supported by a grant from the National Institutes of Health (GM 41249) to J.T.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jkadonaga@ucsd.edu; FAX (619) 534-0555.
References
- Arkhipova IR. Promoter elements in Drosophila melanogaster revealed by sequence analysis. Genetics. 1995;139:1359–1369. doi: 10.1093/genetics/139.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B, Kassavetis GA, Braun BR, Geiduschek EP. The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J. 1990;9:2197–2205. doi: 10.1002/j.1460-2075.1990.tb07389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B, Kassavetis GA, Geiduschek EP. Two components of Saccharomyces cerevisiae transcription factor IIIB (TFIIIB) are stereospecifically located upstream of a tRNA gene and interact with the second-largest subunit of TFIIIC. Mol Cell Biol. 1991;11:5181–5189. doi: 10.1128/mcb.11.10.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B, Tinker RL, Kassavetis GA, Geiduschek EP. Photochemical cross-linking assay for DNA tracking by replication proteins. Methods Enzymol. 1995;262:476–494. doi: 10.1016/0076-6879(95)62039-7. [DOI] [PubMed] [Google Scholar]
- Biggin MD, Tjian R. Transcription factors that activate the Ultrabithorax promoter in developmentally staged extracts. Cell. 1988;53:699–711. doi: 10.1016/0092-8674(88)90088-8. [DOI] [PubMed] [Google Scholar]
- Björklund S, Kim Y-J. Mediator of transcriptional regulation. Trends Biochem Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- Burke TW, Kadonaga JT. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- Contursi C, Minchiotti G, Di Nocera PP. Identification of sequences which regulate the expression of Drosophila melanogaster Doc elements. J Biol Chem. 1995;270:26570–26576. doi: 10.1074/jbc.270.44.26570. [DOI] [PubMed] [Google Scholar]
- DeLorenzi M, Ali N, Saari G, Henry C, Wilcox M, Bienz M. Evidence that the Abdominal-B r element function is conferred by a trans-regulatory homeoprotein. EMBO J. 1988;7:3223–3231. doi: 10.1002/j.1460-2075.1988.tb03189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel PA, Gilmour DS. Transcription factor TFIID recognizes DNA sequences downstream of the TATA element in the hsp70 heat shock gene. Proc Natl Acad Sci. 1993;90:8449–8453. doi: 10.1073/pnas.90.18.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Hoey T, Thut CJ, Admon A, Tjian R. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell. 1993;75:519–530. doi: 10.1016/0092-8674(93)90386-5. [DOI] [PubMed] [Google Scholar]
- Hoffman A, Chiang C-M, Oelgeschläger T, Xie X, Burley SK, Nakatani Y, Roeder RG. A histone octamer-like structure within TFIID. Nature. 1996;380:356–359. doi: 10.1038/380356a0. [DOI] [PubMed] [Google Scholar]
- Hultmark D, Klemenz R, Gehring W. Translational and transcriptional control elements in the untranslated leader of heat-shock gene hsp26. Cell. 1986;44:429–438. doi: 10.1016/0092-8674(86)90464-2. [DOI] [PubMed] [Google Scholar]
- Jarrell KA, Meselson M. Drosophila retrotransposon promoter includes an essential sequence at the initiation site and requires a downstream sequence for full activity. Proc Natl Acad Sci. 1991;88:102–104. doi: 10.1073/pnas.88.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale S. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadonaga JT. Assembly and disassembly of the Drosophila RNA polymerase II complex during transcription. J Biol Chem. 1990;265:2624–2631. [PubMed] [Google Scholar]
- Kokubo T, Gong D-W, Wootton JC, Horikoshi M, Roeder RG, Nakatani Y. Molecular cloning of Drosophila TFIID subunits. Nature. 1994;367:484–487. doi: 10.1038/367484a0. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Montano MM, Katzenellenbogen BS. Identification of multiple, widely spaced estrogen-responsive regions in the rat progesterone receptor gene. Mol Endocrinol. 1994;8:952–969. doi: 10.1210/mend.8.8.7997237. [DOI] [PubMed] [Google Scholar]
- Lee H, Kraus KW, Wolfner MF, Lis JT. DNA sequence requirements for generating paused polymerase at the start of hsp70. Genes & Dev. 1992;6:284–295. doi: 10.1101/gad.6.2.284. [DOI] [PubMed] [Google Scholar]
- Lewis ED, Manley JL. Control of adenovirus late promoter expression in two human cell lines. Mol Cell Biol. 1985;5:2433–2442. doi: 10.1128/mcb.5.9.2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Noll M. Compatibility between enhancers and promoters determines the transcriptional specificity of gooseberry and gooseberry neuro in the Drosophila embryo. EMBO J. 1994;13:400–406. doi: 10.1002/j.1460-2075.1994.tb06274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merli C, Bergstrom DE, Cygan JA, Blackman RK. Promoter specificity mediates the independent regulation of neighboring genes. Genes & Dev. 1996;10:1260–1270. doi: 10.1101/gad.10.10.1260. [DOI] [PubMed] [Google Scholar]
- Minchiotti G, Contursi C, Di Nocera PP. Multiple downstream promoter modules regulate the transcription of the Drosophila melanogaster I, Doc and F elements. J Mol Biol. 1997;267:37–46. doi: 10.1006/jmbi.1996.0860. [DOI] [PubMed] [Google Scholar]
- Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-β gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- Mizrokhi LJ, Georgieva SG, Ilyin YV. jockey, a mobile Drosophila element similar to mammalian LINEs, is transcribed from the internal promoter by RNA polymerase II. Cell. 1988;54:685–691. doi: 10.1016/s0092-8674(88)80013-8. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Horikoshi M, Brenner M, Yamamoto T, Bresnard F, Roeder RG, Freese E. A downstream initiation element required for efficient TATA box binding and in vitro function of TFIID. Nature. 1990;348:86–88. doi: 10.1038/348086a0. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Bagby S, Ikura M. The histone folds in transcription factor TFIID. J Biol Chem. 1996;271:6575–6578. doi: 10.1074/jbc.271.12.6575. [DOI] [PubMed] [Google Scholar]
- Oelgeschläger T, Chiang C-M, Roeder RG. Topology and reorganization of a human TFIID-promoter complex. Nature. 1996;382:735–738. doi: 10.1038/382735a0. [DOI] [PubMed] [Google Scholar]
- Orphanides G, Lagrange T, Reinberg D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Perkins K, Dailey G, Tjian R. In vitro analysis of the Antennapedia P2 promoter: Identification of a new Drosophila transcription factor. Genes & Dev. 1988;2:1615–1626. doi: 10.1101/gad.2.12a.1615. [DOI] [PubMed] [Google Scholar]
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- Purnell BA, Gilmour DS. Contribution of sequences downstream of the TATA element to a protein-DNA complex containing the TATA-binding protein. Mol Cell Biol. 1993;13:2593–2603. doi: 10.1128/mcb.13.4.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell BA, Emanuel PA, Gilmour DS. TFIID sequence recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes & Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- Roeder RG. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- Sauer F, Tjian R. Mechanisms of transcriptional activation: Differences and similarities between yeast, Drosophila, and man. Curr Opin Genet & Dev. 1997;7:176–181. doi: 10.1016/s0959-437x(97)80126-8. [DOI] [PubMed] [Google Scholar]
- Sims SH, Cha Y, Romine MF, Gao P-Q, Gottlieb K, Deisseroth AB. A novel interferon-inducible domain: Structural and functional analysis of the human Interferon Regulatory Factor-1 gene. Mol Cell Biol. 1993;13:690–702. doi: 10.1128/mcb.13.1.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smale ST. Core promoter architecture for eukaryotic protein-coding genes. In: Conaway RC, Conaway JW, editors. Transcription: Mechanisms and regulation. New York, NY: Raven Press, Ltd.; 1994. pp. 63–80. [Google Scholar]
- ————— Transcription initiation from TATA-less promoters within eukaryotic protein-coding genes. Biochim Biophys Acta. 1997;1351:73–88. doi: 10.1016/s0167-4781(96)00206-0. [DOI] [PubMed] [Google Scholar]
- Smale ST, Baltimore D. The “initiator” as a transcription control element. Cell. 1989;57:103–113. doi: 10.1016/0092-8674(89)90176-1. [DOI] [PubMed] [Google Scholar]
- Soeller WS, Poole SJ, Kornberg T. In vitro transcription of the Drosophila engrailed gene. Genes & Dev. 1988;2:68–81. doi: 10.1101/gad.2.1.68. [DOI] [PubMed] [Google Scholar]
- Tansey WP, Herr W. TAFs: Guilt by association? Cell. 1997;88:729–732. doi: 10.1016/s0092-8674(00)81916-9. [DOI] [PubMed] [Google Scholar]
- Tyree SM, George CP, Lira DeVito LM, Wampler SL, Dahmus ME, Zawel L, Kadonaga JT. Identification of a minimal set of proteins that is sufficient for accurate initiation of transcription by RNA polymerase II. Genes & Dev. 1993;7:1254–1265. doi: 10.1101/gad.7.7a.1254. [DOI] [PubMed] [Google Scholar]
- Verrijzer CP, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–342. [PubMed] [Google Scholar]
- Wampler SL, Tyree CM, Kadonaga JT. Fractionation of the general RNA polymerase II transcription factors from Drosophila embryos. J Biol Chem. 1990;265:21223–21231. [PubMed] [Google Scholar]
- Weinzierl ROJ, Ruppert S, Dynlacht BD, Tanese N, Tjian R. Cloning and expression of Drosophila TAFII60 and human TAFII70 reveal conserved interactions with other subunits of TFIID. EMBO J. 1993;12:5303–5309. doi: 10.1002/j.1460-2075.1993.tb06226.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X, Kokubo T, Cohen SL, Mirza UA, Hoffmann A, Chait BT, Roeder RG, Nakatani Y, Burley SK. Structural similarity between TAFs and the heterotetrameric core of the histone octamer. Nature. 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- Yokomori K, Chen J-L, Admon A, Zhou S, Tjian R. Molecular cloning and characterization of dTAFII30α and dTAFII30β: Two small subunits of Drosophila TFIID. Genes & Dev. 1993;7:2587–2597. doi: 10.1101/gad.7.12b.2587. [DOI] [PubMed] [Google Scholar]