Abstract
We have previously shown that copper supplementation extends the replicative life span of Saccharomyces cerevisiae when grown under conditions forcing cells to respire. We now show that copper’s effect on life span is through Fet3p, a copper containing enzyme responsible for high affinity transport of iron into yeast cells. Life span extensions can also be obtained by supplementing the growth medium with 1mM ferric chloride. Extension by high iron levels is still dependent on the presence of Fet3p. Life span extension by iron or copper requires growth on media containing glycerol as the sole carbon source, which forces yeast to respire. Yeast grown on glucose containing media supplemented with iron show no extension of life span. The iron associated with cells grown in media supplemented with copper or iron is 1.4–1.8 times that of cells grown without copper or iron supplementation. As with copper supplementation, iron supplementation partially rescues the life span of superoxide dismutase mutants. Cells grown with copper supplementation display decreased production of superoxide as measured by dihydroethidium staining.
Keywords: Aging, mitochondria, FET3, respiration, copper, iron
1. Introduction
Both theory and experimental evidence support the premise that mitochondria are central to the aging process (Balaban et al., 2005; Wallace, 2005). Energy production in mitochondria is dependent on the efficient transfer of electrons between the complexes of the electron transport chain. Efficient transfer of electrons between complexes depends on the presence of iron and copper as cofactors (iron in complex I, III, IV, coenzyme Q and cytochrome C, and copper in complex IV). Iron deficiency can lead to the loss of heme synthesis, a specific decrease in of complex IV levels, and an increase in oxidative damage (Atamna et al., 2002).
To utilize the budding yeast, Saccharomyces cerevisiae, as a tool to investigate the role of mitochondria in cellular aging we have altered the standard replicative life span assay, replacing glucose with glycerol as the carbon source, forcing the yeast cells to respire. We have used this system to analyze the effect of iron supplementation on yeast longevity. Though iron is essential, in excess it can be toxic to cells, hence iron uptake into the cell is stringently regulated. Iron is abundant in the environment, but mostly as ferric hydroxide which is not very soluble (Yun et al., 2000). Mechanisms that Saccharomyces cerevisiae use to obtain and regulate iron are well described (Philpott & Protchenko, 2008). Yeast have several mechanisms for iron uptake, all of which are under the control of Aft1p, which is the major iron dependent transcription factor in yeast (Yamaguchi-Iwai et al., 1996). Ferric iron in growth media is first reduced by one of two plasma membrane metalloreductases FRE1p or FRE2p (Dancis et al., 1990; Georgatsou & Alexandraki, 1994). Uptake through the plasma membrane occurs through either a high affinity system that utilizes a multicopper oxidase, Fet3p, and permease Ftr1p (Stearman et al., 1996), or a low affinity system that utilizes the permease, Fet4p (Dix et al., 1997). Another mechanism yeast can use to accumulate iron is through the capture of siderophores. Siderophores are secreted molecules that can specifically bind ferric iron with high affinity (Wandersman & Delepelaire, 2004). The iron that is bound to these siderophores can be captured by specific cellular transport systems in yeast. Yeast are not capable of secreting their own siderophores but are able to obtain siderophore bound iron though the use of the Arn and Fit families of polypeptides which recognize and bind different siderophores (Philpott et al., 2002).
High affinity uptake of iron by yeast requires copper as a cofactor for Fet3p. We have previously shown that copper supplementation increases replicative life span of yeast grown on media requiring respiratory metabolism (Kirchman & Botta, 2007). We now show that copper’s influence on life span is through its involvement in iron import by Fet3p/Ftr1p.
2. Materials and Methods
2.1 Strains and growth conditions
S. cerevisiae strains used: BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 uraΔ0); fet3 (BY4742 fet3::KanMX4); sod1 (BY4742 sod1::KanMX4); sod2 (BY4742 sod2::KanMX4). Cells were grown in YPG medium (1% yeast extract, 2% peptone, 3% glycerol, nanopure water). All BY strains were obtained from Open Bio Systems. Plates used for analyses contained 2% agar. For life span or iron analyses, when indicated, ferric chloride was added to a final concentration of 1mM and copper sulfate was added to a final concentration of 62μM. When required, media was acidified by the addition of 1 M HCl to pH 6.2.
2.2 Life span analysis
Life span analyses were conducted as described previously (Kirchman et al., 1999). In short, 1μl of logarithmically growing cells from YPG liquid media were placed onto YPG plates. The unbudded cells were further separated from the cellular population with the use of a micromanipulator. These cells were allowed to grow and produce buds. These buds were removed and used as the cohort for life span analysis. The removal of an individual bud from a cell was counted as a generation. The cells were grown at 30ºC during the day and at 12.5ºC overnight. It has been shown that cells grown at low temperatures are not affected in their overall life span (Muller et al., 1980). All life spans data presented are representative examples of life span analyses, which were all repeated a minimum of 3 times. Statistical analyses of individual life span experiments were performed using the nonparametric Mann-Whitney test.
2.3 Iron assay
BY4742 was grown in 250 ml YPG at 30ºC shaking at 220 RPM. For growth under iron supplementation conditions, iron was added to a concentration of 200μM for 30 minutes before increasing the concentration to 1mM for growth. Cultures were started at an OD600 of less than 0.5 and cells were harvested in mid to late log phase when the OD600 reached between 4.5 and 5.2 by centrifugation at 1000×g. Cell pellets were washed three times with nanopure water and then the slurries were transferred to pre-weighed porcelain crucibles. Cell pellets were dried for 30 minutes at 80ºC in a vacuum oven and then weighed to determine the dry pellet weight. Samples were ashed in a muffle furnace at 550ºC for >16 hours and then weighted to determine the ash weight. To assay iron, the following procedure (adapted from Atkins, 1975) was used. Three hundred μl of nanopure water were used to wet the ash, which was then dissolved by adding 600μl of 6M HCl and heating. Samples were then brought to 1.2ml by the addition of nanopure water and the pH was adjusted to approximately 3.5 by the addition of between 350 and 400 μl 10 M NaOH. Samples were divided into 3 tubes and the iron was reduced by the addition of 100 μl of 10 mg/ml hydroquinone. As a blank 150 μl of water was added to one of the 3 samples. To the other 2 samples was added 150 μl of o-phenanthroline, prepared by dissolving 1.25 g in 50 mL of ethanol and adding 450 mL of water. After more than one hour absorbance was measured at 508 nM. Iron concentration per microgram of ash was normalized to the iron concentration of ash from cells grown in unsupplemented YPG. Statistical analyses of the differences in iron content were conducted using a one-tailed paired t-test. Standard curves were prepared from a stock of 8 mM Fe(NH4)2(SO4)2 ·6H2O pH 3.5.
2.4 Flow cytometric analysis
Log phase cells were collected by centrifugation at 1000 × g and resuspended in yeast nitrogen base medium containing 1/10th volume YPG and 5 μg/ml dihydroethidium (Invitrogen). Cells were stained for 10 minutes at 30ºC shaking at 220 RPM, collected by centrifugation at 1000 × g, and resuspended in cold yeast nitrogen base medium containing 1/10th volume YPG. Flow cytometry was performed on a Gallios flow cytometer (Beckman Coulter). Fluorescence of cells was analyzed using an excitation of 488 and emission of 575/30. Data were analyzed using FlowJo software (TreeStar, Inc., Ashland, OR, USA). Statistical analyses of the differences in DHE staining were conducted using a one-tailed paired t-test.
3. Results
3.1 Life span extension by copper requires FET3
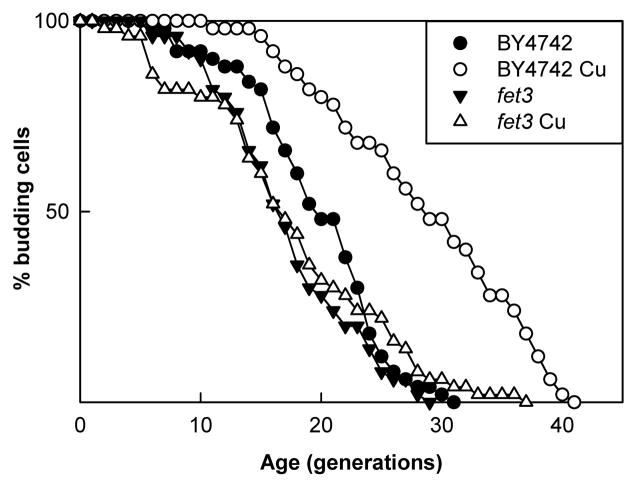
We previously determined that copper’s influence on yeast longevity was not through the copper containing enzyme Cu/Zn SOD (Kirchman & Botta, 2007). This lead us to hypothesize that copper was acting indirectly to influence longevity. Fet3p is a plasma membrane ferroxidase that requires four copper ions for activity and is required for high affinity iron import. In order to explore the relationship between this system and increased life span, we examined the life span of a fet3 mutant grown with and without copper supplementation. Copper supplementation extends life span between 45 and 60% in multiple repeats of the experiment, but this extension is lost in a fet3 strain. Although the fet3 mutant displayed slower growth both in liquid and solid media, as has been described for this mutant on respiratory media (Yuan et al., 1995), it did not have a shortened life span compared to the BY4742 control. Fig. 1, a representative experiment, clearly shows the life span extension due to copper supplementation and its loss in a fet3 strain.
Fig. 1.
Extension of replicative life span by copper is lost in a fet3 deletion strain. Supplementation of YPG medium with copper provided BY4742 with an increase in life span (p<0.001) from a mean of 19.7 without copper (●) to a mean of 28.7 with copper (○). A fet3 mutant strain’s life span is not extended (p=0.8) on media supplemented with copper, averaging 17.3 without copper supplementation (▼) and 17.6 with copper supplementation (△).
3.2 Supplementation with iron also extends life span
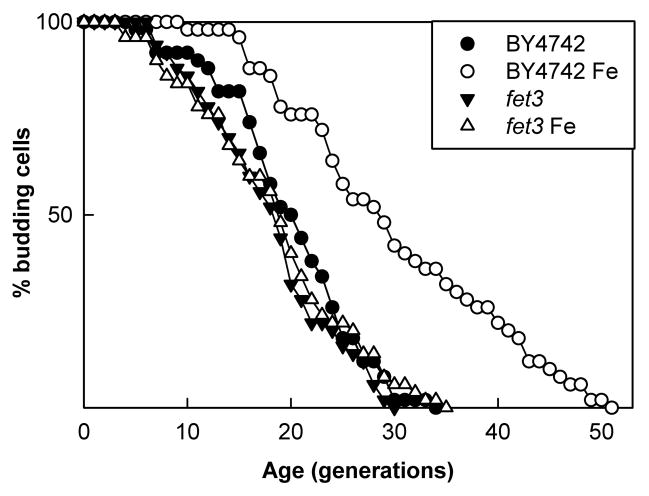
If the extension of life span by copper supplementation is due to an increase in iron importation by Fet3p, then a similar result might also be obtained if Fet3p were induced to higher activity by another means. While increased iron does result in approximately 3 fold lower levels of Fet3p (Felice et al., 2005) we hypothesized that, if iron concentration is the rate limiting step, then the higher extracellular concentration might drive import into the cell through the remaining Fet3p. Ferric chloride was added to YPG media to a final concentration of 1mM, which is greater than a 50 fold increase over what we found in unsupplemented YPG media. While much of this iron is expected to precipitate, much is also expected to complex with organic material in the medium (Theis and Singer, 1974) and life span analyses suggest that the increased concentration does result in increased iron available to the cells. Life span analyses show that iron supplementation does reproducibly extend the life span of the control strain BY4742 between 35 and 50%, but the fet3 strain does not show a significant increase in life span (a representative experiment is shown in Fig. 2). The addition of FeCl3 lowers the pH of the media from 6.8 to 6.2. We assessed the effect of acidification on life span and found no extension due to decreased pH (not shown). Thus, the extension is entirely dependent on FET3.
Fig. 2.
Iron supplementation extends replicative life span and requires FET3. Supplementation of YPG medium with 1mM iron provided BY4742 with an increase in life span (p<0.001) from a mean of 20.1 without iron (●) to a mean of 30.1 with iron (○). A fet3 mutant strain’s life span is not extended on media supplemented with iron (p=0.73), averaging 18.2 without iron supplementation (▼) and 18.6 with iron supplementation (△).
3.3 Copper or iron extend life span through the same pathway
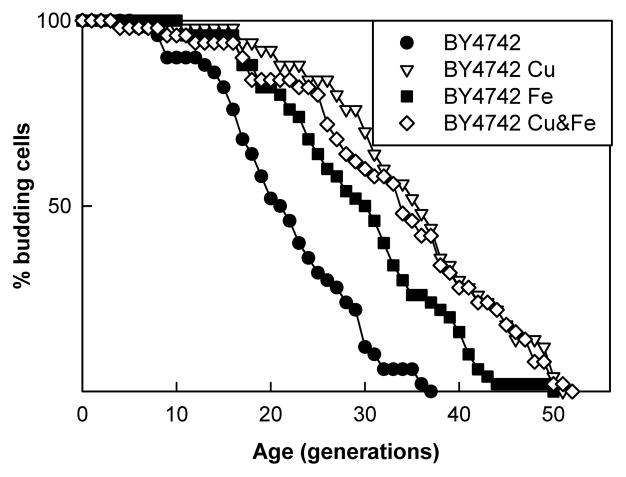
Supplementation of solid YPG media with 62μM copper or 1mM iron each increased life span compared to controls with no metal supplementation. Each of these effects require FET3, implying that copper’s influence on life span is solely due to increased iron import. To confirm that copper and iron operate in the same pathway, through FET3, life span analyses were performed on media supplemented with each metal individually and both metals simultaneously. Media with either metal or both metals increases longevity, but copper and iron combined did not have additive effects on life span extension beyond either metal alone. An example of one experiment is shown in Fig. 3.
Fig. 3.
Copper and iron supplements do not additively extend replicative life span. Life spans of BY4742 grown on YPG media supplemented with copper, iron, or both copper and iron. Average life spans were: no supplements (●), 21.8; copper (▽), 35.3; iron (■), 29.7; copper and iron (◇), 33.4. Metal supplementation increased life span (p<0.001 for all 3 conditions) but no additive effect was observed (p>0.05 for either copper or iron vs. copper and iron).
3.4 Life span extension by iron requires respiratory growth
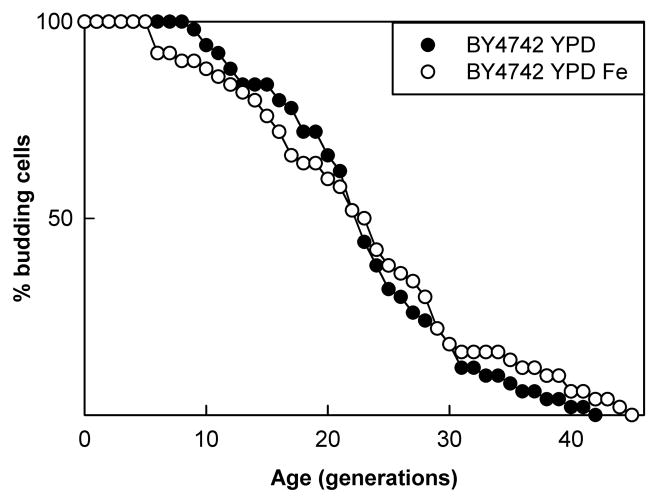
We have previously shown that the extension of life span by copper supplementation only occurs on media requiring respiratory growth (Kirchman & Botta, 2007). Since copper supplementation extends life span by providing increased iron import through Fet3p, life span extension by iron supplementation should also be limited to respiratory growth. This turned out to be the case, as BY4742 grown on YPD media, containing glucose instead of glycerol, showed no increase in life span when grown on YPD supplemented with iron. An example of one such experiment is shown in Fig. 4.
Fig. 4.
Iron supplementation does not increase replicative life span under conditions allowing fermentation. The mean life span of BY4742 on YPD media (●) was 21.4 and on YPD supplemented with 1 mM iron (○) was 22.2 (p=0.64).
3.5 Supplementation with iron or copper results in increased cell associated iron
To confirm that iron supplementation resulted in an increase in iron associated with cells, we measured the iron present in cells grown with and without supplementation. Cell pellets were ashed and a colorimetric assay was used to determine the iron content of the ash. The results of the assay are shown in Fig. 5. The iron found in the pellet of cells grown with iron supplementation was 1.8 fold (average of 2 trials) more than was found in cells grown in unsupplemented YPG. Because we were concerned that the iron could simply be associated with the surface of the cells due to the high concentration in the media, we also tested the iron content in cell grown in media supplemented with copper. If copper supplementation does increase the import of iron, then iron levels should also be higher in copper supplemented cells. We found that the iron concentration was increased 1.4 fold (average of 3 trials) in the pellet of cells grown with copper supplementation (Fig. 5).
Fig. 5.
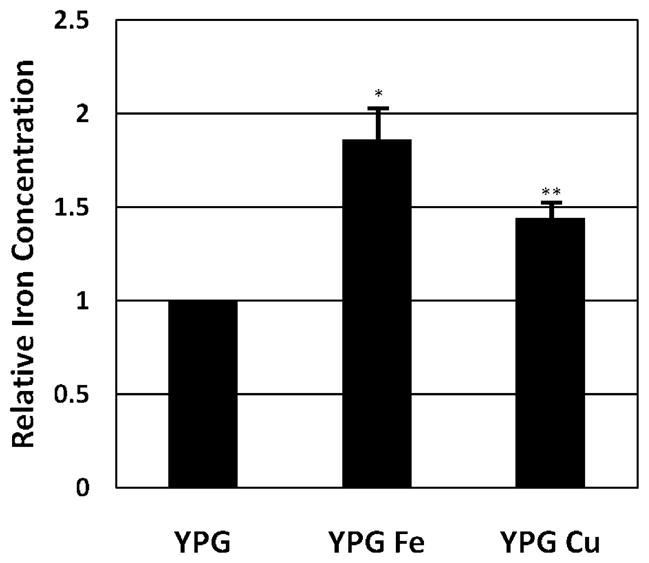
Cellular iron levels are increased in cells grown in media supplemented with iron or copper. The iron concentration of cells grown with iron supplementation (YPG Fe) averaged a 1.8 fold increase in two trials (* p<0.05). The iron concentration of cells grown with copper supplementation (YPG Cu) averaged a 1.4 fold increase in three trials (** p<0.01). Error bars represent one standard deviation.
3.6 Iron Partially Rescues the life span of SOD Mutants
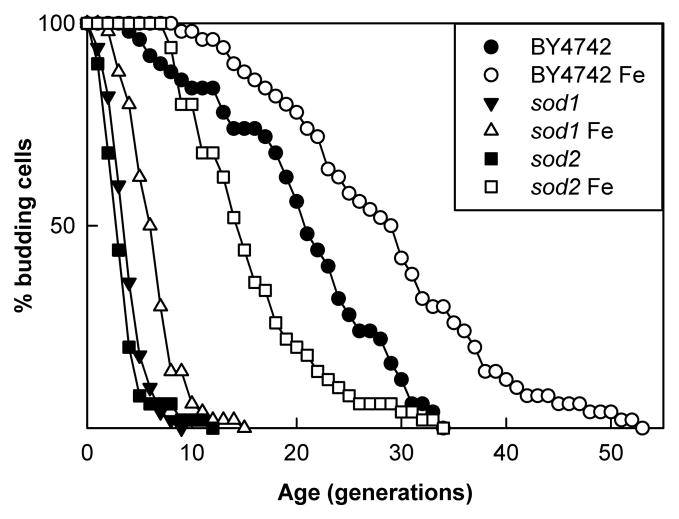
Iron deficiency can lead to oxidative stress (Ames et al., 2005). To examine the possibility that the increased iron reduces oxidative stress to extend life span we analyzed the effect of iron supplementation on the life spans of SOD1 (Cu/Zn SOD) and SOD2 (MnSOD) mutants. Previous research has shown that copper supplementation partially restores life span in these mutants (Kirchman & Botta, 2007). If copper’s effect was in fact due to increased iron uptake by the cells, then iron supplementation should have similar results. Both SOD mutants were assayed on YPG media with and without iron supplementation. Supplementation with iron reproducibly increases the life span of the sod1 strain, and almost completely restores the life span of the sod2 strain to the level of the unsupplemented BY4742 control (Fig. 6). These results do not exactly mimic the result of our previous findings with copper, since iron supplementation does not rescue the sod1 mutant as well as copper supplementation. However, the extension of life span in both the sod1 and sod2 strains with either iron or copper is significant and suggests that the increased iron in cells grown with iron or copper supplementation decreases oxidative stress.
Fig. 6.
Iron supplementation increases the replicative life span of superoxide dismutase mutants. BY4742 had increase in life span (p<0.001) from a mean of 20.7 without iron (●) to a mean of 28.6 with iron (○). SOD1 mutants had an increase in mean life span (p<0.001) from 4.1 without iron (△) to 6.5 with iron (▼), and SOD2 mutants had an increase in mean life span (p<0.001) from 3.5 (■) without iron to 15.8 when supplemented with iron (□).
3.7 Cells grown with copper supplementation show lower levels of superoxide production
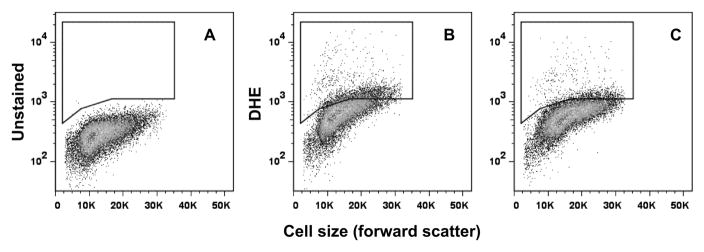
To determine if the higher cellular iron concentrations resulted in lower oxidative stress, cells were stained with dihydroethidium (DHE) and analyzed for fluorescence by flow cytometry. When DHE reacts with superoxide, ethidium is formed and can be detected by fluorescent emission. Cells grow with or without copper supplementation were stained and analyzed as shown in figure 7. A gate was generated that excluded unstained cells. Cells stained with DHE were then assayed for fluorescence and the percentage of cells with fluorescent intensity above the unstained control was determined. Twenty five thousand cells grown with or without copper supplementation were analyzed for the BY4742, sod1, and sod2 strains. The percentage of fluorescent cells above the threshold decreased significantly when cells were grown with copper supplementation in each of the three strains, indicating that superoxide production is decreased in cells grown with copper supplementation (Figure 8).
Fig. 7.
Superoxide levels in cells grown with copper supplementation. Logarithmically growing cells were incubated with dihydroethidium (DHE) and analyzed by flow cytometry. A total of 25,000 cells are included in each analysis. (A) Unstained BY4742 grown in YPG medium. Only 0.012% fall within the gated area. (B) BY4742 grown in YPG medium and stained with DHE. The gated area contains 11.4% of the cells. (C) BY4742 grown in copper supplemented YPG medium. The gated area contains 4.5% of the cells.
Fig. 8.
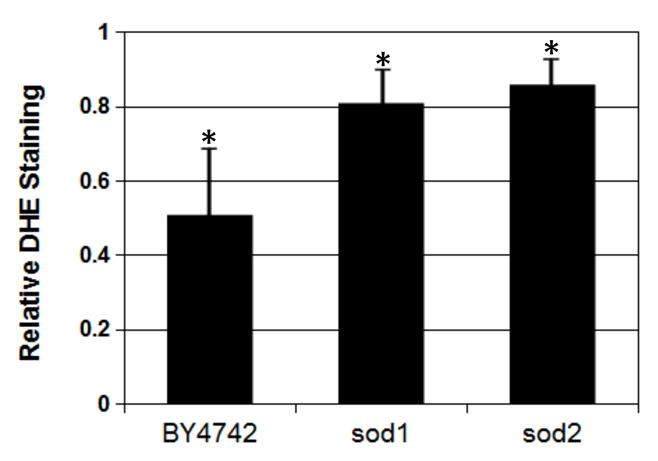
Growth in copper supplemented YPG reduces the production of superoxide detected by DHE staining and flow cytometry. Bars represent the percentage of DHE stained cells in strains grown with copper supplementation relative to the same strain grown in unsupplemented medium. The results are the average of 3 trials. The average percentage of fluorescent cells was reduced by 49% in BY4742 (*p<0.05), by 19% in the sod1 deletion strain (*p<0.05), and by 14% in the sod2 deletion strain (*p<0.05). Error bars represent 1 standard deviation.
4. Discussion
The copper containing ferroxidase, Fet3p, has been shown to increase iron concentrations in yeast (Stearman et al., 1996). The life span of fet3 mutants is not increased by either copper or iron supplementation, showing both that the extension by copper is due to the increase in iron supplied to the cell through Fet3p, and that life span extension by iron is dependent on Fet3p for entry into the cell. Even at high iron concentrations the iron responsible for life span extension is entering cells through the high affinity transporter Fet3p. While iron is still required, the low affinity transporter, Fet4p, is sufficient to provide iron for survival. Fet4p is a low affinity transporter and the amount of iron it imports does not change much over a range of iron concentrations (Dix et al., 1997). In fact, Fet4p transport of iron is less than optimal even for growth since the fet3 strain displays a growth defect on non-fermentable carbon sources (Yuan et al., 1995 and our unpublished observation).
The extension of life span in cells supplied with iron or copper is possibly due to a reduction in oxidative stress, which is supported by our findings that strains with deletions of either SOD1 or SOD2 have greatly reduced life spans that are partially rescued by iron supplementation (Fig. 6), and that copper supplemented cells have reduced production of superoxide (Fig.8). We did not directly measure superoxide production in cells supplemented with iron because the precipitate caused by the high iron concentration titrates out the DHE, resulting in poor staining. To remove this precipitate for the iron assay required extensive washing, which would likely result in metabolically inactive cells. Since copper supplementation increases cellular iron the decrease in superoxide production is likely due to the increased iron in cells.
Initially the idea that increasing iron concentration extends life span seems counter-intuitive due to the possibility of superoxide production by the often referenced Fenton reaction. Disorders that result in high levels of iron, such as hemochromatosis (Toyokuni, 2009), or Friedreich’s ataxia (Armstrong, et al., 2010) show increased oxidative stress due to the Fenton reaction, in which ferrous iron (Fe2+) catalytically generates hydoroxyl radicals (Wardman and Candeias, 1996). However, evidence for the existence of free ferrous iron in non-diseased eukaryotic cells or organisms is rare due to a lack of methods to detect it (Pate, et al., 2006; Toyokuni, 2009). Anemia on the other hand is a confirmed problem in elderly humans (Vanasse and Berliner, 2010). Therefore, the more realistic danger in normal aging may be a lack of iron that results in the inhibition of an efficient electron flow.
A variety of situations that inhibit electron transfer through the electron transport chain, such as disease (Esposito et al., 1999; Calabrese et al., 2005; Napoli et al., 2006) or chemical inhibitors (St-Pierre et al., 2002) are known to increase mitochondrial production of reactive oxygen species (ROS). Yeast cells in which electron transport is inhibited either chemically, or through mutation or loss of mitochondrial DNA display an increase in nuclear mutations due to increased ROS production (Rasmussen et al., 2003). Iron deficiency, in the form of anemia in humans, has been shown to reduce heme content in mitochondrial and lead to the increase in ROS (Atamna et al., 2002; Ames et al., 2005). This may be the finding most similar to the results presented here. The yeast strains were grown on rich media containing yeast extract. Even without supplementation the iron concentration is far from limiting for growth, as colonies have no problem growing in these conditions and supplemented liquid cultures grow at the same rate as unsupplemented (not shown). However, the life span assay looks not for survival of the colony, but for survival of the individual. In order to produce exponential growth each cell only needs to bud twice. For maximal life span the iron requirements are higher than they are for basic survival, which is likely true for a variety of nutrients in all organisms given the opportunity to reach maximum longevity (Ames et al., 2005). The reduced ability of cells to obtain and/or incorporate iron into the components of the electron transport chain could, in part, account for increased oxidative stress and damage found in tissue with increasing age (Moghaddas et al., 2003; Choksi et al., 2008). Lam et al. (2011) have recently shown that oxidative stress begins to accumulate early in replicatively aging yeast. Even slightly increased levels of ROS at earlier ages due to less efficient electron transport could result in a shortening of replicative life span.
It has been known for some time that increased respiration leads to increased longevity in yeast. Lin et al. (2002) showed that the shift in metabolism toward respiration is responsible for the increased replicative life span of yeast grown under conditions that mimic caloric restriction in yeast (0.5% glucose vs. the normal 2% glucose). Although copper or iron supplementation does not increase the life span of glucose grown yeast, we have preliminary evidence that copper or iron supplementation does increase the replicative life span of yeast grown on raffinose, although no to the same extent as on glycerol (not shown). This is likely due to the fact that on raffinose mitochondrial biogenesis is not repressed to the same extent as it is on glucose, resulting in increased respiratory metabolism (Szekely & Montgomery, 1984). Further support comes from Barros et al. (2004) who found that that increasing respiration by lowering glucose concentration or adding an uncoupling agent increases yeast chronological life span and lowers the production of ROS.
Veatch et al. (2009) have shown that older yeast cells (>20 generations) produce buds that have lost heterozygosity (LOH). They propose that decreased iron-sulfur cluster (ISC) biogenesis results from the loss of mitochondrial DNA and alters the iron regulon in cells, leading to LOH. In our system, with haploid cells grown on a non-fermentable carbon source, the loss of either nuclear or mitochondrial chromosomes will result in cell death. Their system uses diploid cells grown on a fermentable carbon source, allowing viability and detection of both cells that have lost mitochondrial DNA or cells that have lost chromosomes. What initially causes the loss of mitochondrial DNA is an open question. Iron-sulfur clusters are known to be targets of ROS damage (Sideri et al., 2009) and, when damaged can release free iron, leading to DNA damage (Keyer and Imlay, 1996). A protein containing an iron-sulfer cluster, which is also known to be involved in mitochondrial DNA maintenance, is mitochondrial aconitase (Chen et al., 2005). This role is independent of its catalytic activity in the Krebs cycle. Mitochondrial aconitase is part of a mitochondrial nucleoid that protects mitochondrial DNA from oxidative damage (Chen et al., 2007) and is regulated by the retrograde response (Liu & Butow, 2006), which coordinates metabolism and aging (Jazwinski, 2005). Interestingly, mitochondrial aconitase has recently been shown to be asymmetrically inherited in yeast, with the oxidatively-damaged, inactive molecules remaining in the mother cell (Klinger, et al., 2010). If increased iron counteracts the initial decrease in iron-sulfur cluster or heme production, thereby reducing oxidative stress, it would lead to the extension of life span seen in our system by decreasing the rate of damage to mitochondrial aconitase and mitochondrial DNA. These mechanisms clearly fit into the free radical theory of aging (Harman, 1973) and the various “vicious cycle” theories that follow (Alexeyev, et al., 2004).
Iron regulation and utilization is clearly a complex process. The Saccharomyces cerevisiae genome database (www.yeastgenome.org) lists 75 genes involved with iron. We are currently looking for the point at which iron supplementation influences life span. While it appears from our data that iron supplementation may act in the mitochondrion to reduce oxidative stress, the effect may be complex. The analysis is made more difficult by the fact that many of the genes involved cause respiratory deficiency and are therefore not assayable in our system. While this does present a problem, we continue to believe that analyses on media requiring yeast to respire will lead to new insights in cellular longevity, which is demonstrated by the fact that neither copper nor iron supplementation have any effect on life span when yeast cells are allowed to ferment.
Highlights.
Increased yeast life span by copper is dependent on the ferroxidase Fet3p
Iron supplementation also results in a Fet3p dependent increase in life span
Respiration is necessary for the increase in life span by iron supplementation
Cells grown with copper or iron supplementation contain more iron
Iron supplementation partially rescues the life span of SOD mutants
Acknowledgments
We thank Eugene Smith for help with the iron assay and Herb Weissbach for critically reading the manuscript. This work was funded in part by an NIH academic research enhancement award (1 R15 AG021956-01) to PAK.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexeyev MF, Ledoux SP, Wilson GL. Mitochondrial DNA and aging. Clin Sci (Lond) 2004;107:355–64. doi: 10.1042/CS20040148. [DOI] [PubMed] [Google Scholar]
- Ames BN, Atamna H, Killilea DW. Mineral and vitamin deficiencies can accelerate the mitochondrial decay of aging. Mol Aspects Med. 2005;26:363–78. doi: 10.1016/j.mam.2005.07.007. [DOI] [PubMed] [Google Scholar]
- Armstrong JS, Khdour O, Hecht SM. Does oxidative stress contribute to the pathology of Friedreich's ataxia? A radical question. FASEB J. 2010;24:2152–63. doi: 10.1096/fj.09-143222. [DOI] [PubMed] [Google Scholar]
- Atamna H, Killilea DW, Killilea AN, Ames BN. Heme deficiency may be a factor in the mitochondrial and neuronal decay of aging. Proc Natl Acad Sci U S A. 2002;99:14807–12. doi: 10.1073/pnas.192585799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins RC. Colorimetric determination of iron in vitamin supplement tablets. A general chemistry experiment. J Chem Educ. 1975;52:550. doi: 10.1021/ed052p550. [DOI] [PubMed] [Google Scholar]
- Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–95. doi: 10.1016/j.cell.2005.02.001. [DOI] [PubMed] [Google Scholar]
- Barros MH, Bandy B, Tahara EB, Kowaltowski AJ. Higher respiratory activity decreases mitochondrial reactive oxygen release and increases life span in Saccharomyces cerevisiae. J Biol Chem. 2004;279:49883–8. doi: 10.1074/jbc.M408918200. [DOI] [PubMed] [Google Scholar]
- Calabrese V, Lodi R, Tonon C, D'Agata V, Sapienza M, Scapagnini G, Mangiameli A, Pennisi G, Stella AM, Butterfield DA. Oxidative stress, mitochondrial dysfunction and cellular stress response in Friedreich's ataxia. J Neurol Sci. 2005;233:145–62. doi: 10.1016/j.jns.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Wang X, Butow RA. Yeast aconitase binds and provides metabolically coupled protection to mitochondrial DNA. Proc Natl Acad Sci U S A. 2007;104:13738–43. doi: 10.1073/pnas.0703078104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Wang X, Kaufman BA, Butow RA. Aconitase couples metabolic regulation to mitochondrial DNA maintenance. Science. 2005;307:714–7. doi: 10.1126/science.1106391. [DOI] [PubMed] [Google Scholar]
- Choksi KB, Nuss JE, Deford JH, Papaconstantinou J. Age-related alterations in oxidatively damaged proteins of mouse skeletal muscle mitochondrial electron transport chain complexes. Free Radic Biol Med. 2008;45:826–38. doi: 10.1016/j.freeradbiomed.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dancis A, Klausner RD, Hinnebusch AG, Barriocanal JG. Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:2294–301. doi: 10.1128/mcb.10.5.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix D, Bridgham J, Broderius M, Eide D. Characterization of the FET4 protein of yeast. Evidence for a direct role in the transport of iron. J Biol Chem. 1997;272:11770–7. doi: 10.1074/jbc.272.18.11770. [DOI] [PubMed] [Google Scholar]
- Esposito LA, Melov S, Panov A, Cottrell BA, Wallace DC. Mitochondrial disease in mouse results in increased oxidative stress. Proc Natl Acad Sci U S A. 1999;96:4820–5. doi: 10.1073/pnas.96.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felice MR, De Domenico I, Li L, Ward DM, Bartok B, Musci G, Kaplan J. Post-transcriptional regulation of the yeast high affinity iron transport system. J Biol Chem. 2005;280:22181–90. doi: 10.1074/jbc.M414663200. [DOI] [PubMed] [Google Scholar]
- Georgatsou E, Alexandraki D. Two distinctly regulated genes are required for ferric reduction, the first step of iron uptake in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3065–73. doi: 10.1128/mcb.14.5.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman D. Free radical theory of aging. Triangle. 1973;12:153–8. [PubMed] [Google Scholar]
- Jazwinski SM. Rtg2 protein: at the nexus of yeast longevity and aging. FEMS Yeast Res. 2005;5:1253–9. doi: 10.1016/j.femsyr.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Keyer K, Imlay JA. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci U S A. 1996;93:13635–40. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman PA, Botta G. Copper supplementation increases yeast life span under conditions requiring respiratory metabolism. Mech Ageing Dev. 2007;128:187–95. doi: 10.1016/j.mad.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae. Genetics. 1999;152:179–90. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinger H, Rinnerthaler M, Lam YT, Laun P, Heeren G, Klocker A, Simon-Nobbe B, Dickinson JR, Dawes IW, Breitenbach M. Quantitation of (a)symmetric inheritance of functional and of oxidatively damaged mitochondrial aconitase in the cell division of old yeast mother cells. Exp Gerontol. 2010;45:533–42. doi: 10.1016/j.exger.2010.03.016. [DOI] [PubMed] [Google Scholar]
- Lam YT, Aung-Htut MT, Lim YL, Yang H, Dawes IW. Changes in reactive oxygen species begin early during replicative aging of Saccharomyces cerevisiae cells. Free Radic Biol Med. 2011;50:963–70. doi: 10.1016/j.freeradbiomed.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–8. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Liu Z, Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet. 2006;40:159–85. doi: 10.1146/annurev.genet.40.110405.090613. [DOI] [PubMed] [Google Scholar]
- Moghaddas S, Hoppel CL, Lesnefsky EJ. Aging defect at the QO site of complex III augments oxyradical production in rat heart interfibrillar mitochondria. Arch Biochem Biophys. 2003;414:59–66. doi: 10.1016/s0003-9861(03)00166-8. [DOI] [PubMed] [Google Scholar]
- Muller I, Zimmermann M, Becker D, Flomer M. Calendar life span versus budding life span of Saccharomyces cerevisiae. Mech Ageing Dev. 1980;12:47–52. doi: 10.1016/0047-6374(80)90028-7. [DOI] [PubMed] [Google Scholar]
- Napoli E, Taroni F, Cortopassi GA. Frataxin, iron-sulfur clusters, heme, ROS, and aging. Antioxid Redox Signal. 2006;8:506–16. doi: 10.1089/ars.2006.8.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pate KT, Rangel NA, Fraser B, Clement MH, Srinivasan C. Measuring “free” iron levels in Caenorhabditis elegans using low-temperature Fe(III) electron paramagnetic resonance spectroscopy. Anal Biochem. 2006;358:199–207. doi: 10.1016/j.ab.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott CC, Protchenko O. Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell. 2008;7:20–7. doi: 10.1128/EC.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philpott CC, Protchenko O, Kim YW, Boretsky Y, Shakoury-Elizeh M. The response to iron deprivation in Saccharomyces cerevisiae: expression of siderophore-based systems of iron uptake. Biochem Soc Trans. 2002;30:698–702. doi: 10.1042/bst0300698. [DOI] [PubMed] [Google Scholar]
- Rasmussen AK, Chatterjee A, Rasmussen LJ, Singh KK. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:3909–17. doi: 10.1093/nar/gkg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sideri TC, Willetts SA, Avery SV. Methionine sulphoxide reductases protect iron-sulphur clusters from oxidative inactivation in yeast. Microbiology. 2009;155:612–23. doi: 10.1099/mic.0.022665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Buckingham JA, Roebuck SJ, Brand MD. Topology of superoxide production from different sites in the mitochondrial electron transport chain. J Biol Chem. 2002;277:44784–90. doi: 10.1074/jbc.M207217200. [DOI] [PubMed] [Google Scholar]
- Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, Dancis A. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science. 1996;271:1552–7. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- Szekely E, Montgomery DL. Glucose represses transcription of Saccharomyces cerevisiae nuclear genes that encode mitochondrial components. Mol Cell Biol. 1984;4:939–46. doi: 10.1128/mcb.4.5.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyokuni S. Role of iron in carcinogenesis: cancer as a ferrotoxic disease. Cancer Sci. 2009;100:9–16. doi: 10.1111/j.1349-7006.2008.01001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanasse GJ, Berliner N. Anemia in elderly patients: an emerging problem for the 21st century. Hematology Am Soc Hematol Educ Program. 2010;2010:271–5. doi: 10.1182/asheducation-2010.1.271. [DOI] [PubMed] [Google Scholar]
- Veatch JR, McMurray MA, Nelson ZW, Gottschling DE. Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell. 2009;137:1247–58. doi: 10.1016/j.cell.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandersman C, Delepelaire P. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol. 2004;58:611–47. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- Wardman P, Candeias LP. Fenton chemistry: an introduction. Radiat Res. 1996;145:523–31. [PubMed] [Google Scholar]
- Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 1996;15:3377–84. [PMC free article] [PubMed] [Google Scholar]
- Yuan DS, Stearman R, Dancis A, Dunn T, Beeler T, Klausner RD. The Menkes/Wilson disease gene homologue in yeast provides copper to a ceruloplasmin-like oxidase required for iron uptake. Proc Natl Acad Sci U S A. 1995;92:2632–6. doi: 10.1073/pnas.92.7.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun CW, Ferea T, Rashford J, Ardon O, Brown PO, Botstein D, Kaplan J, Philpott CC. Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae. Evidence for two pathways of iron uptake. J Biol Chem. 2000;275:10709–15. doi: 10.1074/jbc.275.14.10709. [DOI] [PubMed] [Google Scholar]