Abstract
We have previously shown that in osteoblasts Sox2 expression can be induced by Fgfs, and can inhibit Wnt signaling and differentiation. Furthermore, in mice in which Sox2 is conditionally deleted in the osteoblastic lineage, bones are osteopenic, and Sox2 inactivation in cultured osteoblasts leads to a loss of proliferative ability with a senescent phenotype. To help understand the role of Sox2 in osteoblast development we have specifically expressed Sox2 in bone from a Col1α1 promoter, which extended Sox2 expression into more mature osteoblasts. In long bones, trabecular cartilage remodeling was delayed and the transition from endochondral to cortical bone was disrupted, resulting in porous and undermineralized cortical bone. Collagen deposition was disorganized, and patterns of osteoclast activity were altered. Calvarial bones were thinner and parietal bones failed to develop the diploic space. Microarray analysis showed significant up- or downregulation of a variety of genes coding for non-collagenous extracellular matrix proteins, with a number of genes typical of mature osteoblasts being downregulated. Our results position Sox2 as a negative regulator of osteoblast maturation in vivo.
Keywords: Sox2, osteoblasts, bone, differentiation, transgenic
Introduction
Bone formation is dependent on the regulated commitment, differentiation and maturation of osteoblasts from undifferentiated mesenchymal precursor cells. The major transcription factors in the acquisition of osteoblast fate and function are Runx2, Osterix, and Atf4, which control the expression of genes crucial to bone formation. However, these proteins act within a wider network of transcription factors necessary for osteoblasts to build functional skeletal elements, including homeobox proteins (Msx1, Msx2, Dlx5, Dlx6) and helix-loop-helix proteins (Twist-1, -2). In turn, the expression and activity of transcription factors are regulated by extracellular signaling systems such as those controlled by the Bmp, Tgfβ, Fgf and Wnt families of growth factors [1, 2]. The understanding of what transcription factors are induced by such signaling proteins, and to what effect, is incomplete.
Sox2 is a member of the Sry-related HMG-box family of transcription factors [3]. It is expressed in embryonic stem (ES) cells and the inner cell mass of the blastocyst, and is required for epiblast and extraembryonic ectoderm survival [4]. Sox2 is also expressed in embryonic and adult neural stem cells and is required for proper neurogenesis [5–7]. SOX2 mutations cause anophthalmia in humans [8]. We have previously shown that Sox2 is induced in response to FGF stimulation in both cultured primary osteoblasts and a variety of osteoblast cell lines, is expressed in the osteogenic fronts of the post-natal murine calvaria, and that overexpression of Sox2 can inhibit osteoblast differentiation. In culture, Fgf signaling inhibits osteoblast differentiation and antagonizes the effect of Wnt signaling [9, 10], and Sox2 plays a role in this process by impairing β-catenin transcriptional activity. We have studied the effects of a conditional knock-out of Sox2 in the osteoblast lineage and found that Sox2 inactivation not only resulted in an osteopenic phenotype, but that Sox2 was essential for the self-renewal of osteoblasts in culture, such that its inactivation resulted in the cessation of proliferation and the onset of a senescent phenotype [11].
To further investigate the role of Sox2 in osteoblast function we have generated a transgenic line of mice expressing Sox2 from the osteoblast-specific Col1α1 promoter. We show that Sox2 expression adversely affects mature osteoblast function, resulting in decreased bone quality and mineralization, indicating that Sox2 negatively regulates osteoblast maturation in vivo.
Materials and methods
Generation of Col1α1-Sox2 mice
The Sox2-expressing plasmid pJSox2 was generated from pJ251 (kindly supplied by B. Crombrugghe), which contains the 2.3 kb Col1α1 promoter followed by a LacZ gene [12], by deleting the LacZ gene with BamH1 digestion and replacing it with a 1270 bp BglII fragment from pCMV-Sox2 [13] containing the murine Sox2 coding region flanked by 135 bp and 180 bp of 5’ and 3’ sequence, respectively. Restriction mapping and DNA sequencing across the areas of ligation confirmed the integrity of the final construct. Transgenic animals were derived by pronuclear injection of the eggs of FVB/N donors and five viable FVB/N strains containing the Sox2 transgene were obtained.
PCR
Genotyping was performed by PCR of genomic DNA with control primers targeted to the Sox2 open reading frame (Sox2QF3: 5’CTGCAGTACAACTCCATGAC3’; Sox2QR2: GGAGTGGGAGGAAGAGGTAA3’) and primers specific to the Sox2/mouse protamine region of the transgene (TGSPF: 5’CCATCCCATCCAAATTAACGC3’; TGSPRM: 5’GAGATGCTCTTGAAGTCTGGTA3’). The transgene copy number for each line was estimated by quantitative real-time PCR of genomic DNA with both the Sox2QF3/Sox2QR2 primer combination and with primers specific to the mouse protamine gene (MPF: 5’GAACAATGCCACCTGTCAATAA3’; MPR: 5’CATTTGACCAGTCATGTTCCCTAA3’. In each case levels were normalized to that of primers specific for the 3’ untranslated region of Sox2 not present in the transgene (DIR2: 5’TATGGTTTGTAATATTTCTGTAAATTG3’; REV2: 5’AAATGTAGCTGTTATAAGGATGG3’). Sox2 RNA expression levels in newborn (P1) calvaria were estimated by quantitative real-time PCR of cDNA using the Sox2QF3/R2 combination normalized to the level of β-actin RNA in each sample. The following primer pairs were used for microarray validations: Ameloblastin (5’GAGCTGATAGCACCAGATGA3’, 5’ATTGGTTTGCTCCATAAGACA3’); Claudin-10 (5’TGGAATGAAATGTACCAAAGTCG3’, 5’CCCAATGATGCAGAGAGAAG3’); Osteocalcin (5’TGTGAGCTTAACCCTGC3’, 5’CTGTGACATCCATACTTGC3’); Mepe (5’TGCCCTCTCACAGTCTTAGTA3’, 5’TCACCATGACTCTCACTAGAAC3’); Sclerostin (5’TAAGGTCGTTGGAGGAAACT3’, 5’GCTTCTCAGCATATGTATAACACT3’). RNA and cDNA was prepared as previously described [14]. Quantitative real-time PCR of cDNA was performed with the LightCycler FastStart DNA master SYBR green 1 kit (Roche) on a LightCycler system (Roche).
Western Blotting
Protein was prepared from E17.5 calvaria in RIPA buffer using a Polytron tissue homogenizer (Kinematika, Switzerland), and 20 µg was resolved on a 10% SDS-PAGE gel before transfer to nitrocellulose and detection with a rabbit antibody against Sox2 (AB5603, Chemicon).
RNA in situ hybridization (ISH)
Frozen sections of E16.5 murine calvaria were prepared and hybridized with DIG-labeled RNA riboprobes as described previously [14]. The Sox2 probe consists of the cDNA from amino acids 121–319 cloned into pBS KS (Stratagene). Probes for rat Osteopontin [15] and Osteocalcin [16] have been described. The Ameloblastin probe was transcribed from an 1119 bp fragment obtained by PCR (5’GAGCTGATAGCACCAGATGA3’; 5’GTGTCACATTTCCTGGGCATA3’) and cloned into pCRII-TOPO (Invitrogen).
Histological staining
Histological staining was performed on dissected femurs or calvaria fixed overnight in 4% PFA, demineralized in 10% EDTA/1xPBS for 10 days, dehydrated, paraffin-embedded, and sectioned at a thickness of 10 µm. Staining for hemotoxylin and eosin (HE) and Alcian blue was performed using standard procedures. Staining for tartrate-resistant acid phosphatase (TRAP) to identify osteoclasts or residual extracellular TRAP activity was performed using the Leukocyte Acid Phosphatase (TRAP) Kit (387A-1, Sigma-Aldrich) according to the manufacturer’s instructions.
Microarray analysis
RNA from individual E17.5 calvaria (frontal, parietal, and interparietal bones stripped of extraneous membranes) was prepared with Trizol (Roche), DNase–treated (Qiagen), then purified with the RNeasy MinElute Cleanup Kit (Qiagen). Fragmented biotinylated cRNA probes for microarray analysis were prepared from 2.5 µg of RNA using the Genechip Expression 3’-Amplification One-Cycle cDNA Synthesis Kit and IVT labeling Kit (Affymetrix), and 15 µg was hybridized to the mouse genome 430 2.0 array, and scanned by the GeneArray Scanner (Affymetrix) at the Columbia University Microarray Facility (New York, NY). Results were analyzed with Genespring 7.0 and 11.0 software (Agilent), and expression changes were averaged between paired samples.
Dual Energy X-ray Absorptiometry (DXA)
The bone mineral density (BMD) of femurs prepared by maceration in KOH was quantified using a Lunar PIXImus (GE).
Faxitron and SEM
Radiographs were acquired with a Hewlett Packard Faxitron 43805N X-Ray System set to 2.5 mA tube current, 25 kVp, and an exposure of 12 seconds. Electron microscopy was performed with a Zeiss EVO 50 variable pressure scanning electron microscope operated at 50 Pa variable pressure and beam parameters of 600 pA and 15 kV accelerating voltage. Topographic images were acquired with a variable pressure secondary electron detector (VPSE-SEM) and density-dependent images were acquired with a solid-state 4-quadrant backscattered detector (BSE-SEM). All VPSE-SEM- and BSE-SEM-imaged bones were prepared by maceration with KOH. For BSE-SEM-imaged bone cross sections, bones were embedded in polymethylmethacrylate (PMMA).
Growth analysis and bone measurements
Neonatal mice were weighed on the stated day prior to sacrifice. For growth curves, animals were weighed weekly between 3 and 50 weeks. Weaning was invariably at four weeks. Femurs prepared by maceration in KOH were measured using Measuregraph 123 (Rose Technologies). Parietal thickness was determined in Photoshop on para-sagittal calvarial sections at three points within the parietal bone of each section (rostral, central, and caudal), on n = 3 (WT; TG) at 5 weeks, and n = 3 WT and 4 TG at 52 weeks, and combining these measurements to give a single average thickness.
Statistical Analysis
Where given, body weights and bone parameters are presented as the mean ± standard deviation (s.d.), and were analyzed using the unpaired, two-tailed Student's t test. Differences with a P value ≤ 0.05 were considered significant.
Results
Col1α1-Sox2 mice have a decreased growth rate
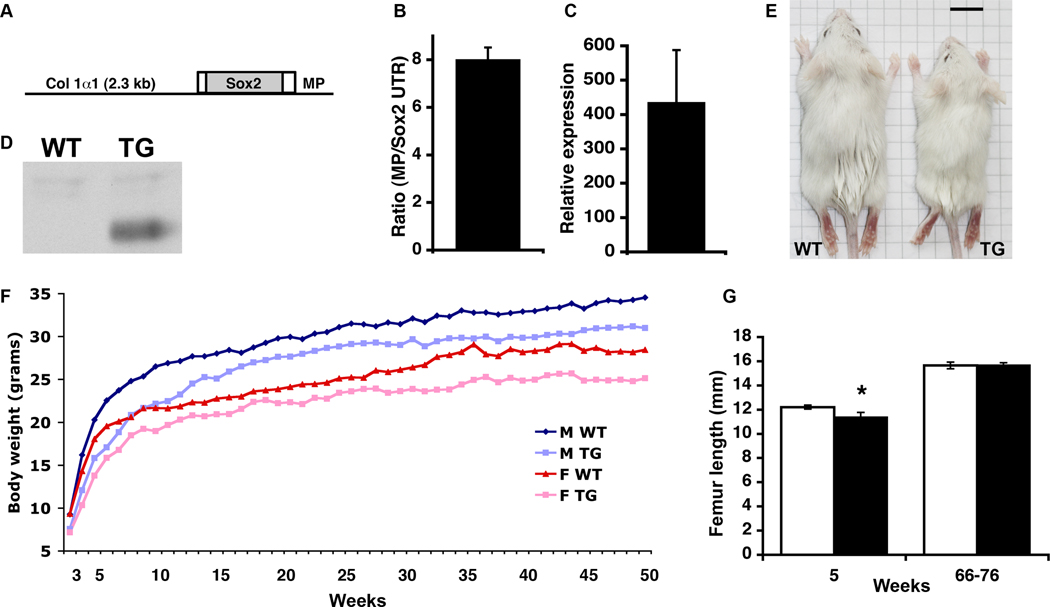
To investigate the role of Sox2 in bone formation we created transgenic mouse lines that expressed murine Sox2 from the 2.3 kb Col1α1 promoter, which is active in osteoblasts and odontoblasts [12] (Fig. 1A). Five lines transmitting the transgene were established, and all were viable and fertile. The transgene copy number of each line was determined by quantitative real-time PCR of genomic DNA. The two lines with the highest copy number were Line 18 (Fig. 1B) and Line 9 (not shown), with approximately 14 and 40 copies of the transgene, respectively. The levels of Sox2 RNA expression in these two transgenic lines were compared to wild-type (WT) levels by quantitative RTPCR of RNA from newborn (P1) calvaria. Line 18 expressed the highest levels of Sox2 (Fig. 1C and not shown), and the data presented in this report are derived from this line. Overexpression of Sox2 protein was confirmed by Western blotting of protein from E17.5 calvaria (Fig. 1D). While the strongest endogenous Sox2 expression is limited to the calvarial osteogenic fronts [9], by in situ hybridization and immunohistochemistry we confirmed that transgenic Sox2 expression was in more mature osteoblasts (Fig. 5A and not shown). The large fold increase of Sox2 expression in transgenic mice of both lines therefore reflects the ectopic expression of Col1α1-Sox2 across a broader range of osteoblasts compared to the WT. The other three lines had one copy of the transgene, WT levels of Sox2 RNA expression, and are not discussed further.
Fig. 1.
Sox2 transgenic construct, expression, and growth of mice. (A) The Sox2 transgene consists of the mouse Col 1α1 2.3 kb promoter, the mouse Sox2 ORF with short 5’ and 3’ UTRs, and the mouse protamine 1 3’ UTR/polyA region (MP), and the pJ251 backbone. (B) Ratio of mouse protamine 3’ UTR gDNA between transgenic (TG) and wild-type (WT; value = 1) mice, normalized to non-transgene Sox2 3’ UTR gDNA, from which transgene copy number was estimated. (C) Relative expression level of calvarial Sox2 RNA in Line 18 between TG and WT newborn littermates (WT value = 1). (D) Western blot for Sox2 in protein from E17.5 WT and TG calvaria. (E) Female WT and TG mice at 4 weeks of age. Scale bar = 1 cm. (F) Growth curves of male (M) and female (F) WT and TG mice weighed weekly between 3 and 50 weeks of age. The minimum number of mice used for any point is n = 7. Error bars are omitted for clarity. (G) Femur lengths of male WT (white bars) and TG (black bars) mice. At 5 weeks, n = 4 WT, 5 TG, P = 0.006; at 66–76 weeks, n = 5 WT, 3 TG, P = 0.914. Similar results were found for female femurs (not shown).
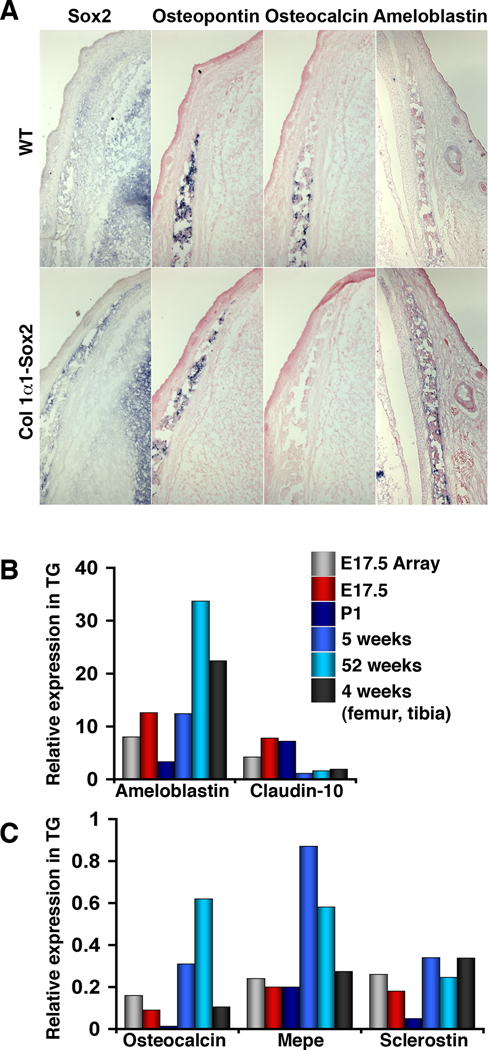
Fig. 5.
Validation of gene expression changes. (A) RNA ISH shows unchanged Osteopontin expression, but downregulated Osteocalcin and upregulated Ameloblastin expression, in areas of Col1α1-Sox2 expression. Sections are adjacent transverse sections of E16.5 frontal bone, except for Ameloblastin. (B, C) For the genes indicated, quantitative RTPCR was used to compare expression in TG calvaria to WT littermates (expression value = 1) at E17.5 (using littermates of those mice used for the microarray; red), P1 (dark blue), 5 weeks (medium blue), and 52 weeks (light blue) and in the cortical bone of femur and tibia at 4 weeks (dark grey). The microarray value of each gene (light grey) is included for comparison.
Line 18 transgenic mice were noticeably smaller compared to WT around the time of weaning (Fig. 1E), so the growth rates of animals in Line 18 were monitored over a fifty-week period. No size or weight difference was seen between neonatal WT and transgenic pups. (At post-natal day (P) 2.5, WT = 2.18 +/− 0.13 grams, n = 10; TG = 2.10 +/− 0.39 grams, n = 8; P = 0.55). By three weeks of age, transgenic mice of either sex were significantly smaller than the WT, and large differences persisted for up to sixteen weeks for males and twelve weeks for females (Fig. 1F). After this, transgenic mice still averaged a statistically significant lower weight than WT mice throughout the fifty-week period (approximately 10% and 12% less than WT for transgenic males and females, respectively). This size difference was reflected in the femur length in younger mice, which was shorter in transgenic animals at five weeks, although no difference was seen by around one year of age (Fig. 1G). A second cohort of WT and transgenic mice bred from a single mating pair but on a mixed background gave similar growth results (not shown).
Bone quality is reduced in Col1α1-Sox2 mice
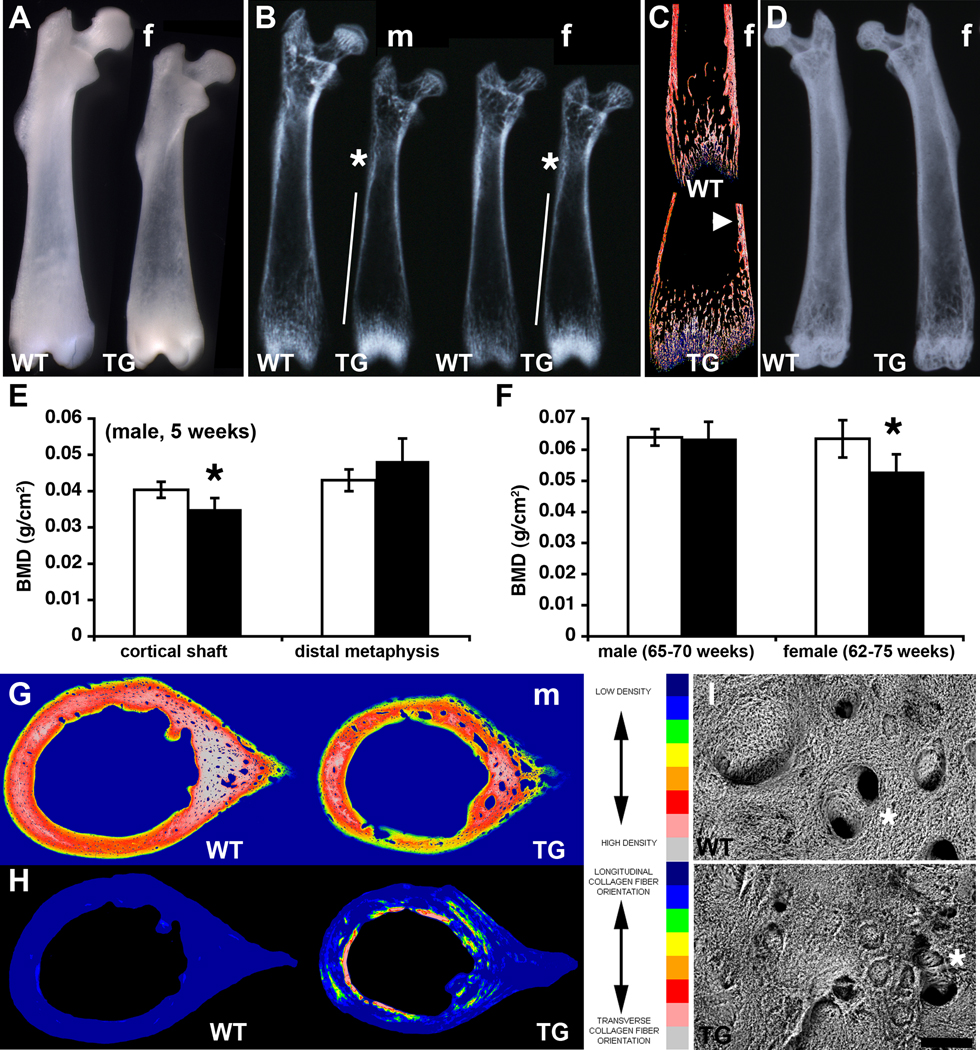
Alcian blue/alizarin red staining for cartilage/mineralized bone did not reveal any obvious difference between WT and transgenic skeletons at birth (not shown). At five weeks of age, when the size difference between WT and transgenic animals was pronounced, bones throughout the transgenic skeleton were visibly less dense than WT. For example, the intact femoral diaphysis of transgenic mice was clearly more translucent than the WT, although interestingly the distal metaphysis was less translucent (Fig. 2A). X-ray analysis revealed extensive areas of the transgenic cortical bone in the shaft that were thinner or less dense, while the distal metaphysis was denser than the WT, in both sexes (Fig. 2B). Backscattered electron imaging in the scanning electron microscope (BSE-SEM) of sectioned distal femurs confirmed the presence of thinner cortical bone with inclusions of mineralized trabecular cartilage remnants (Fig. 2C). A build-up of trabeculae in the transgenic growth plates was also evident, explaining their greater density (Fig. 2C). Although the size difference between WT and transgenic femurs was lost in older bones, the lower radiodensity of transgenic bones persisted in females, and included the distal femur. Thinner and more porous cortical bone was clearly evident (Fig. 2D). The differences in bone mineral density (BMD) were quantified by DXA. At 5 weeks the male transgenic cortical shaft was 15% less dense than the WT (Fig. 2E), while in old females the difference between entire femurs was almost 20% (Fig. 2F). Mid-shaft cortical bone quality was also directly examined by BSE-SEM imaging. Areas of transgenic bone matrix were less dense than the WT, included significantly more mineralized cartilage fragments, and was highly porous (Fig. 2G). The orientation of collagen deposition was markedly perturbed in the transgenic femur, with regions of collagen fibers running in the transverse orientation instead of the normal longitudinal orientation (Fig. 2H). Variable pressure secondary electron imaging in the scanning electron microscope (VPSE-SEM) of bone-forming surfaces of the femoral midshaft revealed disorganized collagen fiber bundles, surface irregularities, and occluded or misshapen capillary channels in transgenic mice (Fig. 2I).
Fig. 2.
Osteopenia and altered collagen deposition in TG femurs. (A) The femoral diaphysis of transgenic 5-week old mice is more translucent than WT, while the distal femur is denser. (f, female) (B) Radiography of 5-week old male (m) and female (f) femurs confirms areas of less dense (*) and thinner (vertical line) cortex in the TG but a denser distal femur compared to WT. Female femurs are from the same animals as (A). (C) BSE-SEM of distal female femurs in (B) confirms thinner cortex with cartilage inclusions (arrowhead) and denser trabecular cartilage remnants in TG mice. (D) Radiography of 54-week old female femurs shows that TG are less dense than WT. (E) The difference in bone mineral density (BMD) between male WT (white bars) and TG (black bars) cortical shaft and distal metaphysis at 5 weeks was quantified by DXA (*P = 0.045, n = 4 WT, 6 TG). (F) The difference in BMD between WT (white bars) and TG (black bars) male (65–70-weeks old) and female (62–75-weeks old) femurs was quantified by DXA (*P = 0.002, n = 9 WT, 8 TG). (G) Mid-shaft cortical bone density of 5-week old male femurs analyzed by BSE-SEM shows thinner, less dense, and more porous TG cortices. (H) Collagen fiber orientation in the mid-shaft of 5-week old male femurs in (G) analyzed by SEM shows aberrant circumferential orientation in TG mice. (I) VPSE-SEM of the surface of the 5 week-old femurs in (A) reveals a chaotic collagen deposition and less ordered or occluded vascular channels (*) in TG mice. Scale bar = 50 µm.
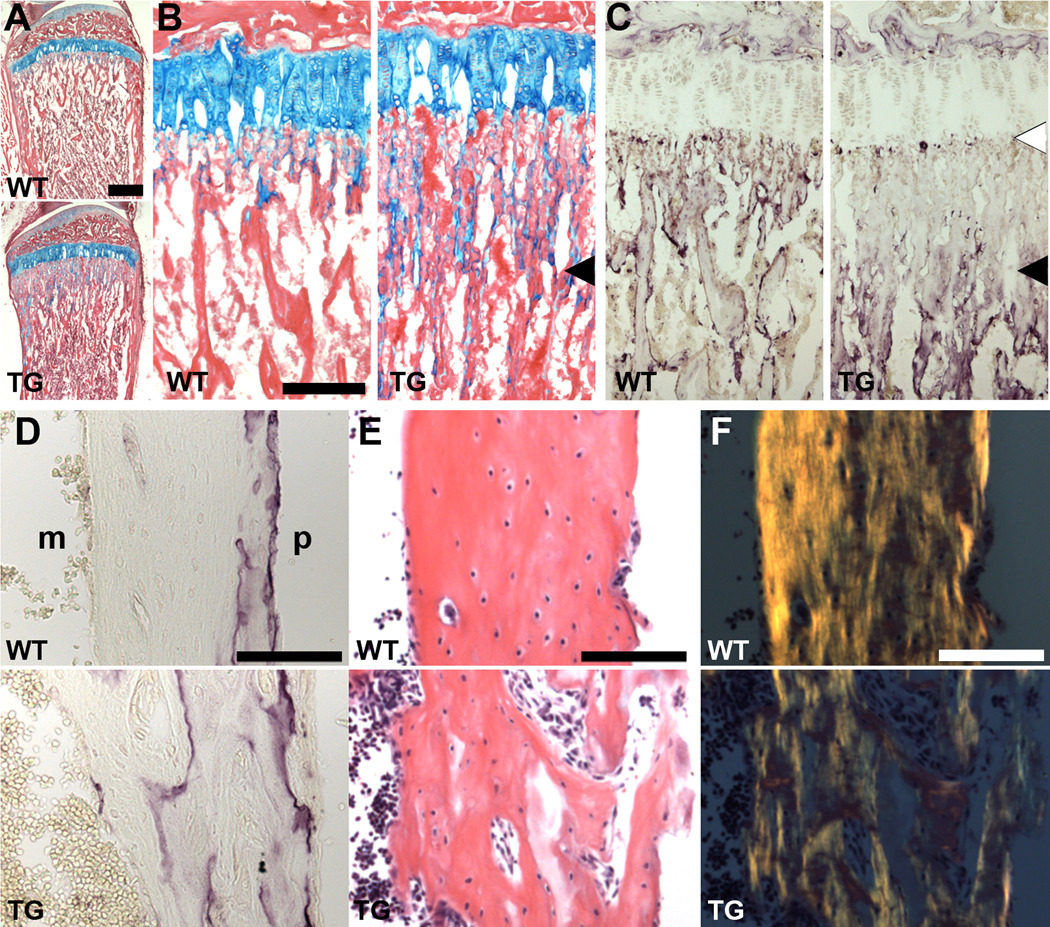
We further examined the growth plates and cortices of demineralized tibias and femurs histologically. In tibias of 5-week old transgenic mice, the proximal growth plate had a comparable length and composition to WT, but the mineralized trabecular cartilage remnants, with attached osteoblasts secreting osteoid, extended further into the diaphysis (Fig. 3A,B), consistent with what was seen in the distal femur by BSE-SEM imaging (Fig. 2C). Cartilage staining of newborn growth plates did not show any obvious differences (not shown). Staining for tartrate-resistant acid phosphatase (TRAP) activity of osteoclasts, which remove this cartilage and remodel the trabecular bone, showed that while osteoclasts appeared on the more distant trabeculae, they had not migrated into the region closer to the growth plate (Fig. 3C), which corresponds to the limit of extended trabecular cartilage (compare TG panels in 3B and C). Interestingly, TRAP-positive cells were still present at the chondro-osseous border (Fig. 3C). Residual extracellular TRAP activity in the absence of osteoclasts was also seen within cortical bone in sections of transgenic femurs (Fig. 3D; 4-week old), indicating that trabecular bone was not effectively remodeled during incorporation into the growing cortical bone shaft. Accordingly, this cortical bone was disordered and highly porous, with irregular endosteal and periosteal surfaces (Fig. 3E). The altered collagen fiber orientation noted at five weeks (Fig. 2H) is also clearly seen in sections of transgenic femoral cortical bone (Fig. 3F). The presence of osteocytes (Fig. 3E) indicates that terminal differentiation of osteoblasts is not totally blocked by Sox2 expression, although the integrity of their canalicular network and functionality was not assessed. In endochondral bone formation, therefore, Sox2 expression in mature osteoblasts disrupts the processes of trabecular cartilage remodeling, and incorporation of endochondral metaphyseal bone into cortical shaft bone. Osteoclasts form but are hindered in the remodeling of trabecular cartilage.
Fig. 3.
Histological analysis of growing tibias and femurs. (A–C) Proximal tibial growth plates of 5-week old mice (A, B) Alcian blue/eosin staining shows that while cartilage growth plates are comparable between WT and TG mice, the network of trabecular cartilage coated in osteoid extends further into the TG diaphysis (arrowhead). (C) TRAP staining shows that osteoclasts form in the TG but are largely absent from the area of extended trabecular cartilage (black arrowhead). TRAP-positive cells still appear at the chondro-osseus border (white arrowhead). (D–F) Femoral cortical bone of 4-week old mice. (D) Extracellular TRAP activity left by osteoclasts remains in the poorly remodeled TG cortex. m, marrow space; p, periosteal surface. (E) Trabeculae incorporated into growing TG cortical bone is not compacted, but osteocytes still differentiate. (F) Polarized light imaging of sections in (E) reveals the altered collagen orientation in TG cortical bone. Scale bars = 500 µm (A), 200 µm (B, C) and 100 µm (D–F).
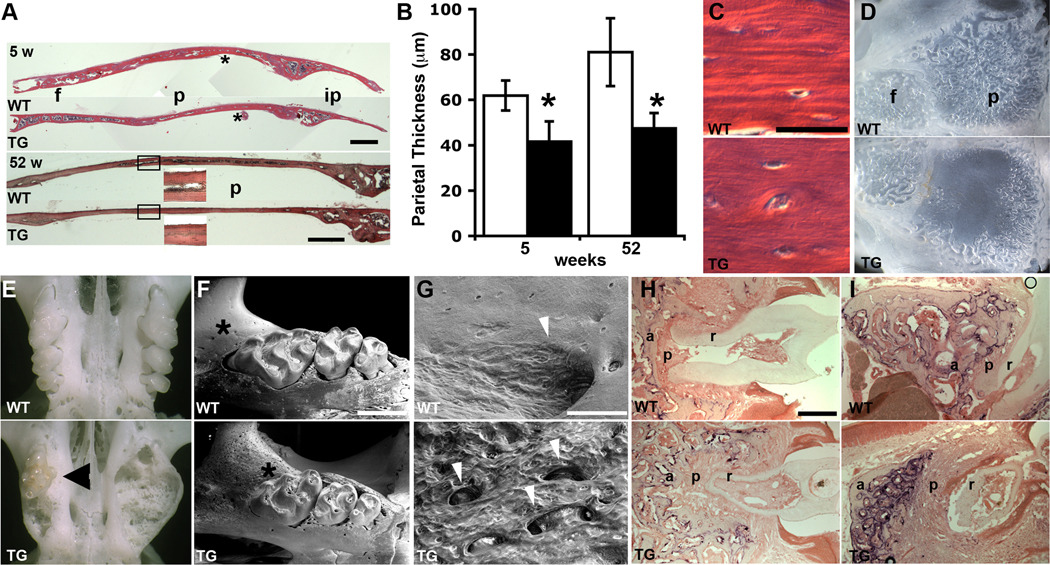
Calvarial bones form directly by intramembranous ossification, as opposed to endochondral ossification of cartilage templates in long bones. Sectioning of calvaria at five and 52 weeks showed that the transgenic frontal, parietal and interparietal bones were thinner compared to WT (Fig. 4A,B), while the lamellar organization of bone deposition was less distinct (Fig. 4C). Osteocytes were present at a similar density to WT calvaria (Fig. 4C). In adult mice of both sexes the diploic space separating the inner and outer tables of the parietal bone failed to form in the central portion of parietal bones (Fig. 4A,D). Transgenic mice of both sexes also had a remodeling defect of the maxillary alveolar bone (Fig. 4E) that was seen in prepared skeletons as early as five weeks of age (Table 1), which often led to the loss of some molar teeth in older mice as seen during skeletal preparation, and grew in frequency and severity with age (Table 1; compare Figs. 4F,E and H,I). Molars were mildly altered in form with a thinner dentin layer (Fig. 4F,H,I), and frequently became discolored compared to WT (Fig. 4E). VPSE-SEM imaging of the jaw in the early stages of this condition reveals an erosion surface reflecting greatly enhanced osteoclast activity coupled with increased angiogenesis (Fig. 4F,G). To further investigate the remodeling defect, alveolar bone and molars were sectioned at 5 and 52 weeks and stained for TRAP activity. At 5 weeks, transgenic alveolar bone is more trabecular with a higher osteoclast presence, and there is a thicker periodontal ligament surrounding the molar roots (Fig. 4H). At 52 weeks these defects are more exaggerated, and the highly resorbed alveolar bone is thinner and separated from the molars by an even greater volume of periodontal ligament (Fig. 4I). Interestingly, this condition was also seen in most old (15–20 months) WT mice examined (Table 1). It did not seem to impair feeding, as transgenic mice were able to accrue weight throughout the time monitored. Very occasionally this phenotype was also seen in the mandible of transgenic mice. Some transgenic mice also had problems with fragile lower incisors and failed to thrive on standard rodent chow after weaning at three weeks, unless provided with powdered food. These mice were not included in the growth curve.
Fig. 4.
Craniofacial bone alterations. (A) TG calvarial bones are thinner than WT at 5 and 52 weeks, and the diploic space fails to develop (boxed insets). f, frontal; p, parietal; ip, interparietal. Each 5-week old calvaria is a montage of three photos. * indicates the area enlarged in (C). Scale bars = 500 µm. (B) Quantification of parietal thickness. (*P = 5.0E-05, 5 weeks; 1.3E-06, 52 weeks). (C) The lamellar deposition of bone is less distinct in TG calvaria. Osteocytes still differentiate. Scale bar = 25 µm. (D) The thinner TG parietal bones lack the venous channels of the diploic space. Calvaria are of 16-month old male mice. (E) The maxillary alveolar bone of TG mice becomes grossly distorted, and molars (arrowhead in TG) are discolored. Maxillae at 55 weeks. (F) At 9 weeks, tooth morphology is mildly altered and the alveolar bone surface (*) (G) shows a broad erosion field indicating greatly increased osteoclast activity and vascularization (arrowheads). Scale bar = 1 mm (F) and 100 µm (G). (H, I) TRAP staining of maxillae at 5 (H) and 52 (I) weeks shows excessive osteoclast activity and alveolar bone remodeling, with increased periodontal ligament surrounding molars, in TG mice. a, alveolar bone; p, periodontal ligament; r, molar root. Scale bar = 50 µm.
Table 1.
Frequency of Maxillary Alveolar Bone Phenotype
| Age | WT | TG |
|---|---|---|
| 5–9 weeks | 0/8 | 6/11 |
| 50–55 weeks | 1/11 | 11/11 |
| 60–80 weeks | 14/16 | 19/19 |
Osteoblasts control osteoclast differentiation through the ratio of expression of Osteoprotegerin and Rankl [17]. Given the regional variations in osteoclast activity, we used quantitative real-time RTPCR to measure the expression of these regulators in osteoblasts of calvaria and long bones, but found no significant differences in their expression or ratios in young or old bones of WT and TG mice (not shown). This suggests that Col1α1-Sox2 osteoblasts indirectly alter osteoclast activity through changes in the extracellular matrix, rather than directly influencing their differentiation.
Sox2 reduces the expression of markers of mature osteoblasts
As endogenous Sox2 expression appeared highest in less mature osteoblasts, such as in the calvarial osteogenic fronts [9], we focused our initial molecular analysis of gene expression changes on newborn or younger ages, when this population is still significant. To identify the molecular changes underlying the impaired bone development, we initially used quantitative real-time RTPCR to compare the expression levels in newborn WT and transgenic calvaria of genes characteristic of osteoblasts. Of these, only Osteocalcin (Bglap2) was affected, being strongly downregulated in transgenic calvaria (Fig. 5A,C and discussed below). We therefore used microarray analysis to comprehensively screen for changes in gene expression, using E17.5 calvaria. This age was chosen as a minimum age at which the transgene was strongly expressed, individual calvaria were large enough to yield enough RNA for analysis and contain the full range of osteoblast development, and to minimize the contribution of other tissues such as vascular endothelia, haematopoietic cells, and osteoclasts. Two calvaria of each genotype, from both Line 18 and Line 9, were used for analysis. Results are presented in Tables 2 and 3, which show the averaged fold-change in expression for genes that were flagged as “Present” and expressed above a low minimum (100 units) in at least four of the eight samples, and whose expression was changed at least two-fold in Line 18 transgenic samples compared to WT, and was changed in the same direction, though to a lesser extent, in Line 9. While changes in the expression of a much larger number of genes was seen in the microarray, these genes were usually expressed at low levels and changes were below two-fold compared to WT.
Table 2.
Genes downregulated in Sox2 transgenic calvaria at E17.5 (Line 18)
| Gene symbol | Fold change | Genbank | Gene name |
|---|---|---|---|
| Bglap2 | 6.19 | NM_007541 | Osteocalcin |
| Ranbp3l | 6.00 | BB496379 | Ran binding protein 3-like |
| Mepe | 4.09 | NM_053172 | Matrix extracellular phosphoglycoprotein with ASARM motif (bone) |
| ChaC1 | 4.04 | BC025169 | ChaC1, cation transport regulator-like 1 |
| Sost | 3.80 | BB212560 | Sclerostin |
| A430110A21Rik | 2.63 | AK020789 | RIKEN cDNA A430110A21 gene |
| Itgb3 | 2.63 | AV352983 | Integrin beta 3 |
| Vit | 2.52 | AF454755 | Vitrin |
| Hpgd | 2.46 | AV026552 | Hydroxyprostaglandin dehydrogenase 15 (NAD) |
| Mmp13 | 2.39 | NM_008607 | Matrix metalloproteinase -13 |
| Mt3 | 2.36 | NM_013603 | Metallothionein-3 |
| Mmp9 | 2.20 | NM_013599 | Matrix metalloproteinase -9 |
| Slc37a2 | 2.19 | BC022752 | Solute carrier family 37 (glycerol-3-phosphate transporter), member 2 |
| Nhedc2 | 2.18 | AV251613 | Na+/H+ exchanger domain containing 2 |
| Lipc | 2.15 | NM_008280 | Lipase, hepatic |
| Snx10 | 2.14 | AK010399 | Sorting nexin 10 |
| Cd68 | 2.14 | BC021637 | Cluster of Differentiation 68 antigen |
| Gpnmb | 2.08 | NM_053110 | Osteoactivin |
| Asns | 2.08 | AV212753 | Asparagine synthetase |
| Gpr137b-ps | 2.06 | AK009736 | G protein-coupled receptor 137B, pseudogene |
| Cp | 2.03 | BB009037 | Ceruloplasmin |
Table 3.
Genes upregulated in Sox2 transgenic mouse calvaria at E17.5 (Line 18)
| Gene symbol | Fold change | Genbank | Gene name |
|---|---|---|---|
| Ambn | 8.06 | NM_009664 | Ameloblastin |
| Cldn10 | 4.20 | BC021770 | Claudin 10 |
| Angpt1 | 3.56 | BB453314 | Angiopoietin 1 |
| Fetub | 2.90 | NM_021564 | Fetuin beta |
| Ncf1 | 2.76 | AI844633 | Neutrophil cytosolic factor 1 |
| unknown | 2.57 | AI785329 | unknown |
| Rgs2 | 2.47 | AF215668 | Regulator of G-protein signaling 2 |
| Tox2 | 2.44 | AI851523 | TOX high mobility group box family member 2 |
| Car3 | 2.43 | NM_007606 | Carbonic anhydrase 3 |
| Pcdha1 | 2.38 | NM_054072 | Protocadherin alpha 1 |
| Scd1 | 2.25 | NM_009127 | Stearoyl-Coenzyme A desaturase |
| Gpcpd1 | 2.07 | BB550273 | Glycerophosphocholine phosphodiesterase GDE1 homolog |
| Slc24a6 | 2.05 | NM_133221 | Solute carrier family 24 (sodium/potassium/calcium exchanger), member 6 |
| Tmem20 | 2.04 | BM121082 | Transmembrane protein 20 |
| Gfra1 | 2.04 | BE534815 | Glial cell line derived neurotrophic factor family receptor alpha 1 |
As we found in our initial RTPCR screening, many genes typically expressed in osteoblasts were expressed at similar levels between WT and transgenic calvaria- these include genes such as Runx2, Osterix, Atf4, Osteopontin (Spp1), Collagen1α1, and Alkaline phosphatase (not shown). A number of genes downregulated in transgenic calvaria (Table 2) are well characterized in osteoblast development, including Bglap2, Matrix extracellular glycoprotein (Mepe), Sclerostin (Sost), and Matrix metalloproteinase-13 (Mmp13). Of the genes upregulated in transgenic calvaria (Table 3), only Ameloblastin (Ambln) is well characterized, but is associated with tooth morphogenesis. A number of genes associated with osteoclasts and macrophages showed changes in expression. Ncf1 and Car3 were upregulated (Table 3), while Mmp9, Slc37a2, Nhedc2, Lipc, and Cd68 were downregulated (Table 2). TRAP was also downregulated, but by 1.9-fold (not shown). These contrasting changes suggest a change in the metabolic or differentiation state of osteoclasts at this location and time. The changes in expression for a small number of genes were validated by RNA in situ hybridization (ISH) and quantitative real-time RTPCR (Fig. 5). By ISH we confirmed that Bglap2 was strongly downregulated in both the calvaria (Fig. 5A) and long bone at E16.5 (not shown), and Ambln expression was upregulated in calvaria (Fig. 5A). As expected, Spp1 expression was unchanged in osteoblasts (Fig. 5A). Validation of expression changes by RTPCR was extended over a range of calvarial ages, and in long bones at 4 weeks, and was invariably consistent with that seen on the microarray (Fig. 5B,C).
Discussion
We have generated transgenic mice expressing Sox2 in bone from the Col1α1 2.3 kb promoter fragment. As we had previously identified Sox2 expression in the less mature osteoblasts of the neo-natal osteogenic fronts, and the Col1α1 2.3 kb promoter is expressed in more mature osteoblasts, this line represents an ectopic extension of strong Sox2 expression into more mature osteoblasts.
Sox2 overexpression in the osteoblast lineage had clearly adverse effects on both endochondral and intramembranous bone development. Patterning of the skeleton was unaffected, as may be expected by the restriction of ectopic Sox2 expression to more differentiated osteoblasts, and osteoblasts were able to achieve terminal differentiation as osteocytes, but the function of mature osteoblasts was strikingly perturbed. In the long bones of growing mice there was a delay between bone deposition on mineralized trabecular cartilage and their subsequent remodeling by osteoclasts. Consequently, bony trabeculae incorporating into the growing cortical shaft during endosteal bone formation were not remodeled effectively into compact cortical bone, creating porous cortices with high levels of mineralized cartilage inclusions and irregular endosteal and periosteal surfaces. Overall, the cortices were thinner and less dense. This delayed cartilage remodeling could account for the early decreased growth rate, which resolved with age. The thinner cortices and lower bone density of long bones persist at least in females throughout life, while the thinner calvaria and underdeveloped diploic space persisted in adults of both sexes. More fundamentally, the organization and non-collagenous protein content of the extracellular matrix (ECM) produced by transgenic osteoblasts was markedly altered. Collagen fiber deposition was disordered and the lamellar organization of the cortical bone in both long bones and calvaria was disrupted. Despite these changes in bone quality, under laboratory conditions transgenic mice suffered no observed bone fractures or breakages. Another notable consequence of Sox2 expression was excessive remodeling and angiogenesis of the alveolar bone around the maxillary molars in both sexes. Intriguingly, this was similar to the condition of much older WT mice, but with a much earlier onset and severity.
Microarray analysis of gene expression in WT and transgenic calvaria at E17.5 showed that the greatest changes involved genes for non-collagenous ECM proteins. RTPCR demonstrated that most of these changes persisted in the calvaria throughout life and in long bones till at least 4 weeks, the oldest age analyzed, and by ISH we could confirm that Bglap2 was also repressed in embryonic calvaria and long bones. Hence, the major transcriptional consequences of Sox2 overexpression are consistent in osteoblasts in both intramembranous and endochondral bones, and over time.
We therefore compared the phenotype of Col1α1-Sox2 mice to phenotypes reported in the literature for overexpression or deletion of genes up- or downregulated in our microarray screen. Considering some of the genes most heavily downregulated - Bglap2, Mepe, and Sost, all of which are expressed by mature osteoblasts and osteocytes and normally restrict ECM production by osteoblasts to restrain bone formation - the bone phenotype of the Col1α1-Sox2 mice differs from those seen in the individual gene knockouts. Bglap2 and Sost null mice have increased cortical bone volume, but heterozygotes are unaffected [18, 19], and it is possible that enough Bglap2 and Sost are produced in Col1α1-Sox2 mice to provide sufficient function. Mice either heterozygous or homozygous for a null allele of Mepe show increased trabecular bone formation that is more pronounced with age [20], but this is not seen in the Col1α1-Sox2 mice. However, at least in calvaria, Mepe expression was near WT levels at five weeks of age in transgenic mice, so Mepe may be sufficiently expressed by early adulthood. On the other hand, the deletion of Mmp13 in osteoblasts [21] gives a phenotype very similar to that seen in Col1α1-Sox2 mice, including persistence of trabecular cartilage and a temporary increase in trabecular bone, with a delay in osteoclast progression towards the chondro-osseous border, during the active growth period in early adulthood. Mmp13 cleaves collagen and promotes the access and activity of osteoclasts on mineralized cartilage, and is specifically upregulated in newly recruited osteoblasts on mineralized cartilage matrix [21–23], so this site of rapid bone formation may be particularly sensitive to a decrease in Mmp13 expression. Interestingly, Bglap2 promotes the recruitment of osteoclasts onto bone [24]. A role of endogenous Sox2 expression may therefore be to delay the development of an ECM environment appropriate for osteoclastic bone remodeling.
Most genes upregulated by Sox2 overexpression have not been extensively studied with regard to their roles in osteoblast biology. However, Ambln, the gene whose expression was most strongly altered by Sox2 expression, has been characterized in dental development. It is essential for normal tooth enamel formation, and promotes the adhesion and differentiation, and inhibits the proliferation, of the ameloblasts that express this extracellular calcium-binding phosphoprotein [25]. While its function outside of tooth development is unknown, it is expressed in newly-formed woven bone in craniofacial and regenerating bones and is downregulated in mature bone [26, 27]. Ambln expression was highly upregulated in Col1α1-Sox2 mice throughout their life, and is likely to create an extracellular environment disruptive to normal osteoblast and osteoclast function. Angiopoietin-1 (Angpt1), which is expressed by osteoblasts and promotes endothelial cell migration and vascular stabilization, has been over-expressed in bone with the Col1α1 promoter, where it caused increased trabecular BMD and volume in 8-week old femurs and tibias [28]. This is consistent with our observations in younger mice, but a collagen remodeling or growth defect was not reported in this case. Claudin-10 is involved in tight junction formation, and therefore its upregulation may alter osteoblast adhesion [29]. Fetuin B plays an unknown role in bone biology, but like Fetuin A it binds calcium phosphate, and Fetuin A-null animals have increased BMD [30–32]; increased Fetuin B could therefore contribute to a decrease in BMD in Col1α1-Sox2 mice. In summary, the proteins made by those genes most strongly upregulated by Sox2 expression are likely to significantly alter the function of mature osteoblasts and the nature of the ECM that they produce.
The Col1α1-Sox2 phenotype can also be usefully compared to that of Col1α1-Sox8 [33] and Col1α1-dominant negative (dn) Runx2 mice [34]. Sox8 is expressed in osteoblasts, and Col1α1-Sox8 mice have a phenotype superficially to that seen in Col1α1-Sox2 mice - osteopenia, with smaller bone size and poorly organized cortical bone, although bone fractures frequent in Col1α1-Sox8 mice were not seen in Col1α1-Sox2 mice. While it was proposed that Sox8 exerted its effect by reducing Runx2 expression, with a resultant decrease in the expression of Runx2 target genes such as Col1α1, Bone sialoprotein, and Bglap2 [33], we did not see a decrease in Runx2 expression in Col1α1-Sox2 mice, and many Runx2 target genes were unaffected. More recently, however, a dominant negative Runx2 construct was expressed in bone with the mouse Col1α1 2.3 kb promoter to yield a very different phenotype than might be expected from Runx2 inhibition and very different from both the Sox8 and Sox2 overexpression phenotypes [34]. In these mice cortical bone was only mildly thinner and less dense, while trabecular bone was expanded and took on the quality of cortical bone; transgenic mice did not suffer from bone fractures. It was concluded that higher levels of Runx2 promoted the production of immature bone, while decreased levels in WT mature osteoblasts, or in the transgenic osteoblasts in which the 2.3 kb Col1α1 promoter is active, promoted the production of more organized cortical bone. Furthermore, Runx2 activity did not seem necessary for steady state expression of major bone matrix genes such as Col1α1, Spp1, or Bglap2 [34]. Therefore, as extended Sox factor expression impairs cortical bone organization, we suggest that while the Col1α1-Sox2 phenotype may have mechanistic causes in common with the Col1α1-Sox8 mice, it is not caused by a general inhibition of Runx2 activity.
In summary, persistent expression of Sox2 produced a qualitative change in osteoblast metabolic and secretory activity, resulting in a structurally and biochemically altered ECM. The most relevant molecular changes appear to be downregulation of Mmp13, impairing endochondral bone development, and upregulation of Ambln, with its potential effects on osteoblast adhesion, differentiation, and proliferation. It should be noted that most of the strongly downregulated genes are markers of osteoblast differentiation, consistent with the notion that Sox2 expression inhibits osteoblast maturation and function [9]. Its endogenous expression in less mature osteoblasts may therefore act to retard osteoblast differentiation, as it does when expressed in osteoblasts in vitro. The phenotype of the Col1α1-Sox2 is superficially similar to that of the osteoblast-specific conditional Sox2 knockout that we have recently described [11]. Both of these mice have early reduced growth, osteopenia, and reduced BMD. The occurrence of overlapping bone phenotypes has been seen with overexpression and deletion of other factors such as Fgf2 or Sox8 [33, 35, 36], and highlights the importance of restricting regulators of cell function to specific time windows. At the moment we believe the most likely interpretation of our results is that Sox2 inactivation impairs osteoprogenitor self-renewal and expansion, while Sox2 overexpression inhibits osteoblast maturation and function, both resulting in defective bone formation. Osteoblast differentiation can be inhibited by Fgf signaling and promoted by Wnt signaling. As Sox2 is expressed in the osteogenic fronts coincident with Fgfrs, is induced by Fgf treatment, and can inhibit Wnt signaling by inhibiting β-catenin activity [9, 10], one mode of action of Sox2 could be to inhibit Wnt signaling to impede the differentiation of osteoprogenitors to mature osteoblasts.
Research Highlights.
-
-
Prolonged Sox2 expression in osteoblasts in vivo disrupts normal bone morphology.
-
-
Gene expression is significantly altered in Sox2 transgenic osteoblasts.
-
-
Sox2 expression antagonizes osteoblast maturation and function.
Acknowledgments
This work was supported by grant AR051358 from the NIAMS. Microinjection to generate transgenic mice was performed by the NYUMC Transgenic Facility. Technical assistance was provided by the NYU Langone Research Histology Core, and Bin Hu of the Department of Biomaterials and Biomimetics, New York University College of Dentistry. We thank Dr Bruce Cronstein and Tuere Wilder (NYU School of Medicine) for use of the PIXImus machine.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Karsenty G, Kronenberg HM, Settembre C. Genetic control of bone formation. Annu Rev Cell Dev Biol. 2009;25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 2.Marie PJ. Transcription factors controlling osteoblastogenesis. Arch Biochem Biophys. 2008;473:98–105. doi: 10.1016/j.abb.2008.02.030. [DOI] [PubMed] [Google Scholar]
- 3.Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 4.Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferri AL, Cavallaro M, Braida D, Di Cristofano A, Canta A, Vezzani A, Ottolenghi S, Pandolfi PP, Sala M, DeBiasi S, Nicolis SK. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 6.Ellis P, Fagan BM, Magness ST, Hutton S, Taranova O, Hayashi S, McMahon A, Rao M, Pevny L. SOX2, a persistent marker for multipotential neural stem cells derived from embryonic stem cells, the embryo or the adult. Dev Neurosci. 2004;26:148–165. doi: 10.1159/000082134. [DOI] [PubMed] [Google Scholar]
- 7.Pevny LH, Nicolis SK. Sox2 roles in neural stem cells. Int J Biochem Cell Biol. 2010;42:421–424. doi: 10.1016/j.biocel.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 8.Fantes J, Ragge NK, Lynch SA, McGill NI, Collin JR, Howard-Peebles PN, Hayward C, Vivian AJ, Williamson K, van Heyningen V, FitzPatrick DR. Mutations in SOX2 cause anophthalmia. Nat Genet. 2003;33:461–463. doi: 10.1038/ng1120. [DOI] [PubMed] [Google Scholar]
- 9.Mansukhani A, Ambrosetti D, Holmes G, Cornivelli L, Basilico C. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J Cell Biol. 2005;168:1065–1076. doi: 10.1083/jcb.200409182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambrosetti D, Holmes G, Mansukhani A, Basilico C. Fibroblast growth factor signaling uses multiple mechanisms to inhibit Wnt-induced transcription in osteoblasts. Mol Cell Biol. 2008;28:4759–4771. doi: 10.1128/MCB.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basu-Roy U, Ambrosetti D, Favaro R, Nicolis SK, Mansukhani A, Basilico C. The transcription factor Sox2 is required for osteoblast self-renewal. Cell Death Differ. 2010;17:1345–1353. doi: 10.1038/cdd.2010.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossert J, Eberspaecher H, de Crombrugghe B. Separate cis-acting DNA elements of the mouse pro-alpha 1(I) collagen promoter direct expression of reporter genes to different type I collagen-producing cells in transgenic mice. J Cell Biol. 1995;129:1421–1432. doi: 10.1083/jcb.129.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 14.Holmes G, Rothschild G, Roy UB, Deng CX, Mansukhani A, Basilico C. Early onset of craniosynostosis in an Apert mouse model reveals critical features of this pathology. Dev Biol. 2009;328:273–284. doi: 10.1016/j.ydbio.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oldberg A, Franzen A, Heinegard D. Cloning and sequence analysis of rat bone sialoprotein (osteopontin) cDNA reveals an Arg-Gly-Asp cell-binding sequence. Proc Natl Acad Sci U S A. 1986;83:8819–8823. doi: 10.1073/pnas.83.23.8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deckelbaum RA, Majithia A, Booker T, Henderson JE, Loomis CA. The homeoprotein engrailed 1 has pleiotropic functions in calvarial intramembranous bone formation and remodeling. Development. 2006;133:63–74. doi: 10.1242/dev.02171. [DOI] [PubMed] [Google Scholar]
- 17.Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Ominsky MS, Niu QT, Sun N, Daugherty B, D'Agostin D, Kurahara C, Gao Y, Cao J, Gong J, Asuncion F, Barrero M, Warmington K, Dwyer D, Stolina M, Morony S, Sarosi I, Kostenuik PJ, Lacey DL, Simonet WS, Ke HZ, Paszty C. Targeted deletion of the sclerostin gene in mice results in increased bone formation and bone strength. J Bone Miner Res. 2008;23:860–869. doi: 10.1359/jbmr.080216. [DOI] [PubMed] [Google Scholar]
- 19.Ducy P, Desbois C, Boyce B, Pinero G, Story B, Dunstan C, Smith E, Bonadio J, Goldstein S, Gundberg C, Bradley A, Karsenty G. Increased bone formation in osteocalcin-deficient mice. Nature. 1996;382:448–452. doi: 10.1038/382448a0. [DOI] [PubMed] [Google Scholar]
- 20.Gowen LC, Petersen DN, Mansolf AL, Qi H, Stock JL, Tkalcevic GT, Simmons HA, Crawford DT, Chidsey-Frink KL, Ke HZ, McNeish JD, Brown TA. Targeted disruption of the osteoblast/osteocyte factor 45 gene (OF45) results in increased bone formation and bone mass. J Biol Chem. 2003;278:1998–2007. doi: 10.1074/jbc.M203250200. [DOI] [PubMed] [Google Scholar]
- 21.Stickens D, Behonick DJ, Ortega N, Heyer B, Hartenstein B, Yu Y, Fosang AJ, Schorpp-Kistner M, Angel P, Werb Z. Altered endochondral bone development in matrix metalloproteinase 13-deficient mice. Development. 2004;131:5883–5895. doi: 10.1242/dev.01461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gack S, Vallon R, Schmidt J, Grigoriadis A, Tuckermann J, Schenkel J, Weiher H, Wagner EF, Angel P. Expression of interstitial collagenase during skeletal development of the mouse is restricted to osteoblast-like cells and hypertrophic chondrocytes. Cell Growth Differ. 1995;6:759–767. [PubMed] [Google Scholar]
- 23.Johansson N, Saarialho-Kere U, Airola K, Herva R, Nissinen L, Westermarck J, Vuorio E, Heino J, Kahari VM. Collagenase-3 (MMP-13) is expressed by hypertrophic chondrocytes, periosteal cells, and osteoblasts during human fetal bone development. Dev Dyn. 1997;208:387–397. doi: 10.1002/(SICI)1097-0177(199703)208:3<387::AID-AJA9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 24.Glowacki J, Lian JB. Impaired recruitment and differentiation of osteoclast progenitors by osteocalcin-deplete bone implants. Cell Differ. 1987;21:247–254. doi: 10.1016/0045-6039(87)90479-9. [DOI] [PubMed] [Google Scholar]
- 25.Fukumoto S, Kiba T, Hall B, Iehara N, Nakamura T, Longenecker G, Krebsbach PH, Nanci A, Kulkarni AB, Yamada Y. Ameloblastin is a cell adhesion molecule required for maintaining the differentiation state of ameloblasts. J Cell Biol. 2004;167:973–983. doi: 10.1083/jcb.200409077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spahr A, Lyngstadaas SP, Slaby I, Pezeshki G. Ameloblastin expression during craniofacial bone formation in rats. Eur J Oral Sci. 2006;114:504–511. doi: 10.1111/j.1600-0722.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 27.Tamburstuen MV, Reseland JE, Spahr A, Brookes SJ, Kvalheim G, Slaby I, Snead ML, Lyngstadaas SP. Ameloblastin expression and putative autoregulation in mesenchymal cells suggest a role in early bone formation and repair. Bone. 2011;48:406–413. doi: 10.1016/j.bone.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki T, Miyamoto T, Fujita N, Ninomiya K, Iwasaki R, Toyama Y, Suda T. Osteoblast-specific Angiopoietin 1 overexpression increases bone mass. Biochem Biophys Res Commun. 2007;362:1019–1025. doi: 10.1016/j.bbrc.2007.08.099. [DOI] [PubMed] [Google Scholar]
- 29.Gunzel D, Stuiver M, Kausalya PJ, Haisch L, Krug SM, Rosenthal R, Meij IC, Hunziker W, Fromm M, Muller D. Claudin-10 exists in six alternatively spliced isoforms that exhibit distinct localization and function. J Cell Sci. 2009;122:1507–1517. doi: 10.1242/jcs.040113. [DOI] [PubMed] [Google Scholar]
- 30.Price PA, Lim JE. The inhibition of calcium phosphate precipitation by fetuin is accompanied by the formation of a fetuin-mineral complex. J Biol Chem. 2003;278:22144–22152. doi: 10.1074/jbc.M300744200. [DOI] [PubMed] [Google Scholar]
- 31.Denecke B, Graber S, Schafer C, Heiss A, Woltje M, Jahnen-Dechent W. Tissue distribution and activity testing suggest a similar but not identical function of fetuin-B and fetuin-A. Biochem J. 2003;376:135–145. doi: 10.1042/BJ20030676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Szweras M, Liu D, Partridge EA, Pawling J, Sukhu B, Clokie C, Jahnen-Dechent W, Tenenbaum HC, Swallow CJ, Grynpas MD, Dennis JW. alpha 2-HS glycoprotein/fetuin, a transforming growth factor-beta/bone morphogenetic protein antagonist, regulates postnatal bone growth and remodeling. J Biol Chem. 2002;277:19991–19997. doi: 10.1074/jbc.M112234200. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt K, Schinke T, Haberland M, Priemel M, Schilling AF, Mueldner C, Rueger JM, Sock E, Wegner M, Amling M. The high mobility group transcription factor Sox8 is a negative regulator of osteoblast differentiation. J Cell Biol. 2005;168:899–910. doi: 10.1083/jcb.200408013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maruyama Z, Yoshida CA, Furuichi T, Amizuka N, Ito M, Fukuyama R, Miyazaki T, Kitaura H, Nakamura K, Fujita T, Kanatani N, Moriishi T, Yamana K, Liu W, Kawaguchi H, Komori T. Runx2 determines bone maturity and turnover rate in postnatal bone development and is involved in bone loss in estrogen deficiency. Dev Dyn. 2007;236:1876–1890. doi: 10.1002/dvdy.21187. [DOI] [PubMed] [Google Scholar]
- 35.Sobue T, Naganawa T, Xiao L, Okada Y, Tanaka Y, Ito M, Okimoto N, Nakamura T, Coffin JD, Hurley MM. Over-expression of fibroblast growth factor-2 causes defective bone mineralization and osteopenia in transgenic mice. J Cell Biochem. 2005;95:83–94. doi: 10.1002/jcb.20389. [DOI] [PubMed] [Google Scholar]
- 36.Montero A, Okada Y, Tomita M, Ito M, Tsurukami H, Nakamura T, Doetschman T, Coffin JD, Hurley MM. Disruption of the fibroblast growth factor-2 gene results in decreased bone mass and bone formation. J Clin Invest. 2000;105:1085–1093. doi: 10.1172/JCI8641. [DOI] [PMC free article] [PubMed] [Google Scholar]