Abstract
Although androgen is considered an anabolic hormone, the consequences of androgen receptor (AR) overexpression in skeletally-targeted AR-transgenic lines highlight the detrimental effect of enhanced androgen sensitivity on cortical bone quality. A compartment-specific anabolic response is observed only in male but not female AR3.6-transgenic (tg) mice, with increased periosteal bone formation and calvarial thickening. To identify anabolic signaling cascades that have the potential to increase bone formation, qPCR array analysis was employed to define expression differences between AR3.6-tg and wild-type (WT) periosteal tissue. Notably, categories that were significantly different between the two genotypes included axonal guidance, CNS development and negative regulation of Wnt signaling with a node centered on stem cell pathways. Further, fine mapping of AR3.6-tg calvaria revealed that anabolic thickening in vivo is not uniform across the calvaria, occurring only in frontal but not parietal bones. Multipotent fraction 1 progenitor populations from both genotypes were cultured separately as frontal bone neural crest stem-like cells (fNCSC) and parietal bone mesenchymal stem-like cells (pMSC). Both osteoblastic and adipogenic differentiation in these progenitor populations was influenced by embryonic lineage and by genotype. Adipogenesis was enhanced in WT fNCSC compared to pMSC, but transgenic cultures showed strong suppression of lipid accumulation only in fNCSC cells. Osteoblastogenesis was significantly increased in transgenic fNCSC cultures compared to WT, with elevated alkaline phosphatase (ALP) activity and induction of mineralization and nodule formation assessed by alizarin red and von Kossa staining. Osteocalcin (OC) and ALP mRNA levels were also increased in fNCSC cultures from AR3.6-tg vs. WT, but in pMSC cultures ALP mRNA levels, mineralization and nodule formation were decreased in AR3.6-tg cells. Expression differences identified by array in long bone periosteal tissue from AR3.6-tg vs. WT were recapitulated in the fNCSC samples while pMSCs profiles reflected cortical expression. These observations reveal the opposing effects of androgen signaling on lineage commitment and osteoblast differentiation that is enhanced in cells derived from a neural crest origin but inhibited in cells derived from a mesodermal origin, consistent with in vivo compartment-specific responses to androgen. Combined, these results highlight the complex action of androgen in the body that is dependent on the embryonic lineage and developmental origin of the cell. Further, these data these data suggest that the periosteum surrounding long bone is derived from neural crest.
Keywords: Androgen, periosteum, neural crest, testosterone, mesenchymal stem cell
Introduction
Androgen action in the skeleton remains poorly understood [1-3]. Although androgen is considered an anabolic hormone, the consequences of androgen action in the skeleton are complex [4]. Androgen receptor (AR) is expressed in bone cells [5], and androgen plays an important role during the development of a sexually dimorphic skeleton [6], with a well recognized effect to specifically enhance periosteal apposition during puberty in males [7]. In contrast, androgen inhibits bone formation in the endocortical compartment [8, 9] and long-term non-aromatizable dihydrotestosterone therapy reduces bone density in the spine [10]. These observations are consistent with the generally disappointing results observed with androgen treatment in men [11, 12], with only the most hypogonadal benefiting [13].
The molecular and cellular response(s) responsible for the opposite effect of androgen signaling on bone formation in these distinct bone compartments are not understood. To identify specific cellular targets for androgen responses in bone and to better characterize the direct effects of androgen signaling in the skeleton, we created and characterized two distinct AR-transgenic families with targeted AR overexpression. Two different promoters were employed to direct overexpression in bone tissue derived from fragments of the type I collagen promoter, that results in overlapping and distinct expression profiles. Creation and characterization of the two lines has been described [8, 9]; they include the AR3.6-transgenic family (driven by the 3.6kb promoter fragment) with AR overexpression in stromal cells and throughout the osteoblast lineage including mature osteoblasts, and the AR2.3-transgenic mice (driven by a 2.3kb promoter fragment) with overexpression limited to the mature osteoblast/osteocyte. These genetic models provide for the characterization of enhanced androgen signaling in distinct skeletal compartments in vivo, through a comparison of the phenotypes observed in both lines. In common between the two families, we observed a phenotype of reduced bone quality and increased fracture risk, and inhibition of bone formation was observed at the endocortical envelope. Based on these observations, we have proposed that androgen inhibition of medullary bone formation at the endosteal surface in males may subserve an important physiological adaptive function, being key for appropriate spatial distribution and maintenance of the total amount/weight of bone in the cortical envelope [9]. Pathways that are implicated in the inhibitory actions of androgen on cortical bone, including reduced BMP signaling, were identified in qPCR array analysis. Androgen inhibition of proliferation and differentiated function was cell autonomous in primary osteoblast cultures [14].
Although the two AR transgenic models shared a similar bone phenotype, the most striking contrast between the two AR-transgenic models is observed at periosteal surfaces in AR3.6-transgenic males. In this model, anabolic activity is noted with increased cortical bone formation in the periosteum and dramatic intramembranous calvarial thickening [8, 9]. This finding was expected, given col3.6 transgene targeting to the periosteum and, conversely, the lack of expression at the same compartment with col2.3 transgene expression. The specificity of the periosteal anabolic effect in AR3.6-transgenic males is consistent with previous reports documenting the importance of androgen signaling in periosteal expansion [8]. However, the biological basis of this differential and opposite response to androgen in these two bone compartments is not understood. Periosteal apposition occurs through intramembranous bone formation, distinct from the endochondral process seen in endocortical bone. Experimental osteotomies have demonstrated the importance of the periosteum rather than bone marrow cells during repair [15]. Although the cellular source of the surface periosteum surrounding the diaphysis in long bone has not been clearly identified in fate mapping studies, it has been suggested that except for the facial skeleton, the rest of the skeleton is derived from mesoderm [16].
Androgen signaling also influences body composition [17]. With declining testosterone levels and during aging, changes in bone quality, body composition, physical and cognitive function occur that are of clinical importance [18]. Androgen treatment [13] and anabolic steroid use/abuse [19] can alter and/or ameliorate these changes. Clinically, increased total body and visceral fat are associated with low testosterone concentrations [20]. Declining testosterone concentrations have also been associated with an increase in the risk for type 2 diabetes and metabolic syndrome [21]. In addition, androgen deficiency induced by gonadotropin-releasing hormone agonist administration in healthy men results in increased adipose tissue mass [22]. In contrast, testosterone replacement in eugonadal men results in a reduction in adipose tissue mass and alteration of regional distribution [23]. Finally, sexual dimorphism is observed in distinct fat depots; females have increased subcutaneous fat while males have more visceral fat [24]. Thus, clinical evidence suggests that androgens also regulate body fat accumulation in humans. Interestingly, androgen signaling in male AR3.6-tg mice also results in a profound body composition phenotype, with increased muscle but decreased fat mass [25].
In the present study, we sought to characterize mechanisms underlying the positive effects of androgen action in the skeleton. To identify the basis or consequences of androgen-mediated enhancement of bone formation in vivo, alterations in molecular signatures in periosteal bone were characterized by qPCR array analysis with a focus on pathways previously associated with bone formation. Differences between male AR-overexpressing mice and their WT counterparts in the expression patterns of osteoblast-associated genes was characterized employing periosteal bone samples. For biological confirmation of the signaling pathways identified in vivo, and to characterize potential cell autonomous effects of androgen treatment, the functional consequences of androgen signaling in osteoblastic and adipogenic cells was evaluated in normal primary cultures isolated from WT and from AR-transgenic animals. Progenitor cells were isolated from neural crest-derived bone vs. mesenchymal-derived bone in order to characterize the consequences of androgen signaling in cultures of precursor populations of distinct embryonic origin. Expression profiles from these cultures were then compared to bone tissue derived from periosteum vs. endocortical compartments.
Materials and Methods
Animal procedures
The generation of AR transgenic mice employing the 3.6 kb α11 collagen promoter fragment to drive expression has been described previously [8]. AR3.6-transgenic animals (hereafter referred to as AR3.6-tg mice) were bred to WT B6D2F1 mice (Jackson Labs, Bar Harbor, ME) employing both genders. The mice were maintained under a 12 h light-dark cycle, had free access to tap water and were fed a standard rodent chow containing 4.5% fat and 23% protein (LabDiet 5001, PMI Nutrition Int., St. Louis, MO) ad libitum. All animal studies were performed according to institutional, local, state, federal and NIH guidelines for the use of animals in research under an Institutional Animal Use and Care Committee (IACUC)-approved protocol.
Real-time quantitative reverse transcription-polymerase chain reaction (qPCR) array and bioinformatic analyses
The AR3.6-tg line was used as a source to identify androgen-regulated transcripts in periosteal bone associated with increased bone formation. Transgenic mice offer the advantage of skeletal targeting and do not show alterations in circulating sex steroid concentrations that could indirectly alter bone metabolism through effects on other organ systems. Males were employed because the female cohort does not demonstrate a significant bone phenotype, likely a consequence of low endogenous androgen concentrations in females relative to males. Mice representing both independently-derived AR3.6-tg families (104 and 106) at 3-4 months of age were employed for array analyses. To isolate periosteal bone RNA, femora and tibiae were dissected from five to six mice of each genotype and the periosteum was harvested after five sequential enzymatic digestions at 37°C each for 30 min using a mixture of collagenase type II and trypsin (0.1% and 0.125%) in Hanks’ Balanced Salt Solution (HBSS) (Invitrogen Corp., Carlsbad, CA). Fractions 2 to 4 were pooled for RNA isolation. RNA was extracted with and contaminating DNA was removed as previously described [14]. RNA integrity was confirmed on a 1% agarose gel stained with SYBR Gold Nucleic Acid Gel Stain (Molecular Probes, Invitrogen Corp.). Gene expression differences between WT and AR-overexpressing tissues were assessed using commercially available, pre-validated pathway-specific gene expression mouse qPCR arrays (StellARray, Bar Harbor Biotechnology, Inc., Trenton, ME) for each AR3.6-tg family. The analysis was replicated to determine statistical differences and represents collapsed data across both AR3.6-tg families for comparison between genotypes. A two-step RT-PCR reaction employed 500 ng total RNA per 20 μl reaction with a mix of random and oligo-dT primers, using qScript cDNA Supermix (Quanta Biosciences, Gaithersburg MD). Four 96-well arrays chosen to create a 384-well plate with amplicons targeting bone homeostasis for transcripts involved in i) Osteoporosis, ii) Targets of Wnt/β-catenin Signaling containing Tcf binding sites, iii) TGF-β Signaling and iv) NFκB Signaling. A list of the 384 genes is available online at http://array.lonza.com. Expression changes were assayed simultaneously by qPCR. The qPCR reactions and subsequent quantitative analysis were performed by Bar Harbor Biotechnology, Inc. Expression difference was determined after normalization without predefined normalizers using a Global Pattern Recognition (GPR) algorithm that determines invariant genes for normalization in every experiment. Significance was determined in an iterative process using a two-tailed heteroscedastic t-test, with a p < 0.05 considered significant using a weighted geometric mean. The cutoff for expression was at 37.5 cycles. This approach uses a statistical gene ranking system without fold change bias. Overrepresented functional groups were identified using WebGestalt Gene Set Analysis Toolkit software at http://bioinfo.vanderbilt.edu/webgestalt, with the 384 genes on the custom array used as the background gene list for GO pathway analysis. Regulated biological networks were identified using pathway analysis (IPA v7.0; Ingenuity Systems, http://www.ingenuity.com), based on our specific user dataset as the reference set for comparison. As a third independent bioinformatic analysis, Pathway Architect software (GeneSpring GX10, Agilent Technologies, http://www.chem.agilent.com) was used to identify associations among the differentially expressed genes and biological processes for follow-up confirmation.
Primary calvarial cultures
Normal primary progenitor cultures were derived from the surface of calvarial bones harvested from 3 to 6 day old neonatal WT and AR3.6- transgenic mice by sequential collagenase-P digestions. Briefly, individual calvariae were pooled after genotyping, the frontal and parietal bones were dissected free of the suture lines and then subjected to 15-minute digestions in a mixture containing 0.05% trypsin and 0.1% collagenase-P at 37°C. Fraction 1 frontal bone neural crest stem cells (fNCSC) vs parietal bone mesenchymal cells (pMSC) were isolated, pelleted, and resuspended in αMEM media supplemented with 10% lot-selected fetal bovine serum (Hyclone, Logan, UT). Proliferation rates are similar between fNCSC and pMSC cultures. Standard markers for neural crest [Nagoshi, 2008 #5946] are expressed in fNCSC cells, including p75, Sox10, nestin and snail in increasing abundance (data not shown); however there are no standard markers for mesenchymal cells for pMSC cultures [Billon, 2008 #5872]. For the osteogenic and adipogenic assay first passage osteoblasts were seeded at 8000 cells/cm2. Beginning at confluence (day 7, hereafter referred to as day 0), cultures were then induced toward the osteoblast lineage with osteoinductive media containing αMEM supplemented with 10% FBS containing 50 μg/ml ascorbic acid and 5 mM β-glycerophosphate. For adipocyte differentiation, cells were isolated as above and at confluence were switched to adipogenic media containing 0.5 mM isobutylmethylxanthine (IBMX), 1 μM dexamethasone, and 10 μg/ml insulin and grown for 3 days. Medium was then replaced with the basal medium containing only 5 μg/ml insulin and cultures were then grown for 7-10 days with a media change every 3 days. All media, buffers, supplements and reagents for cell culture were obtained from GIBCO BRL-Life Technologies (Grand Island, NY) and Sigma Chemical Co. (St Louis, MO).
Confirmation of expression differences in primary cultures by qPCR analysis
Since screening of all differentially expressed genes identified in array analysis was not feasible, we selected four genes of interest for further confirmatory analysis in primary cultures derived from frontal vs. parietal periosteum from WT and AR3.6-transgenic littermates. Genes were selected to represent both up- and down-regulation, and span a range of statistical significance, fold expression difference, or biological interest and potential involvement in stem cell signaling. Osteoblast and adipocyte marker genes were also evaluated by qPCR analysis. RNA was harvested as described above. The qPCR reactions were carried out in 25 μl with 20 ng of total RNA in a reaction mix containing 1X QuantiTect SYBR Green RT-PCR Master Mix (Qiagen, Valencia CA) and 0.5 μM each primer. Real time qPCR was performed with the iCycler IQ Real Time PCR detection system (Bio-Rad Laboratories, Inc., Hercules, CA) using a one-step QuantiTect SYBR Green RT-PCR kit (Qiagen, Valencia, CA) on DNase-treated total RNA. Relative expression of the PCR product was determined using the comparative ΔΔCt method, after normalizing expression to total RNA measured with RiboGreen (Molecular Probes) as previously described [26]. Data are presented as fold change relative to control samples. Primers for calvarial culture RNA analysis were purchased pre-designed from Qiagen (Valencia, CA). Following PCR, reaction products were melted over the temperature range 55°C to 95°C in 0.5°C increments, 10 seconds per increment to ensure only the expected PCR product was amplified per reaction. The efficiency of amplification was determined for each primer set from serial dilutions, and did not vary significantly from 2. Transgenic-AR expression primers were forward 5′-AAGTGCCCAAGATCCTT-3′ and reverse 5′-ACAACAGATGGCTGGCAACT-3′ and specifically amplified Transgenic-AR and normalized to RiboGreen levels. Transgenic AR primers were designed by Oligo software using the sites in rat AR 3′ region and bovine GH within the col3.6-AR cDNA construct [8].
Histochemical analysis of calvaria
Male AR3.6-transgenic mice were employed for in vivo histological analysis at 2 months of age. Calvaria were isolated, fixed in 4% paraformaldehyde, decalcified in Immunocal (Decal Corp., Tallman, NY), and then processed for paraffin embedding as previously described [8]. Sagittal sections across frontal and parietal bones were cut (5–6 μm) and stained with hematoxylin and eosin (H&E) and von Giesson to identify newly synthesized collagen bundles as previously described [8].
ALP activity and mineral accumulation
Confluent primary cultures from frontal vs. parietal periosteum were grown in differentiation medium. ALP expression was determined by ALP histochemical staining (Sigma) at day 10 after osteoblastogenic differentiation. Analysis of mineralization was determined by von Kossa staining (day 10) and by alizarin red-S (AR-S) staining and extraction (day 14). Cultures were analyzed simultaneously for ALP histochemical staining and mineral staining by von Kossa. Cells were first subjected to ALP histochemical staining, then plates were rinsed briefly with water and covered with 5% AgNO3 for 1 h under bright light. Bone nodules were visualized by light microscopy and quantified from digital photographs using LAS v3.3.0 software (Leica Microsystems Inc., Bannockburn, IL). For AR-S staining, cells were rinsed with HBSS, fixed for 2 h in 70% ethanol at 4°C, rinsed with 1 mM HEPES and calcium deposition was then visualized after incubation with 4 mM AR-S pH 4.2. AR-S was extracted by destaining with 10 mM HCl in 70% ethanol, and mineral accumulation was quantified on a microplate reader at 520 nm. The cell layer was then solubilized in lysis buffer (10 mM formamide, 50 mM sodium acetate, 1% SDS, pH6) and protein content was assessed with BCA protein assay reagent (Pierce, Rockford, IL). Results were calculated as nmol alizarin red-S per μg protein and expressed as fold vs. undifferentiated control.
Adipogenesis analysis
To determine the extent of adipogenesis, cultures from both genotypes and both lineages were grown to confluence and differentiation was induced with either adipogenic media or osteogenic media. After 10 days, cells were fixed in 10% neutral buffered formalin, rinsed with 60% isopropanol, stained with 0.5 % Oil Red O in isopropanol, rinsed clear and photographed. Total adipocyte numbers containing at least 10% Oil Red O positive cells were determined on day 10 of differentiation.
Statistical analysis
Data were analyzed using Prism software v5.02 (GraphPad Software, Inc., San Diego, CA). Two-way ANOVA was used to evaluate the effects of genotype and embryonic lineage in progenitor cultures. When there was a significant interaction, significance of difference between groups was assessed by an unpaired two-tailed Student’s t-test. Confirmation of differential gene expression by qPCR identified in array analysis employed a one-tailed t-test. All data is expressed as mean ± SD and all experiments were performed two to four times. A p < 0.05 level was considered significant.
Results
Identification of pathways important in bone growth and osteoblastogenesis by characterization of periosteal bone gene expression
Androgen signaling differentially influences bone at the periosteal vs. enodcortical bone surface in long bone by stimulating or inhibiting bone formation respectively [8, 9]. Thus, bone tissue harvested from these distinct compartments can provide insight into specific pathways associated with inhibition vs. stimulation of bone formation. We have previously characterized signaling pathways that may mediate inhibition of bone formation in the endocortical bone compartment [14], which identified the transforming growth factor-beta (TGF-β) superfamily and bone morphogenetic protein (BMP) signaling as a primary androgen target. In the studies presented here, we sought to gain a better understanding of the transcriptional basis for anabolic stimulation of osteoblast function and bone formation as a consequence of androgen signaling in the periosteum.
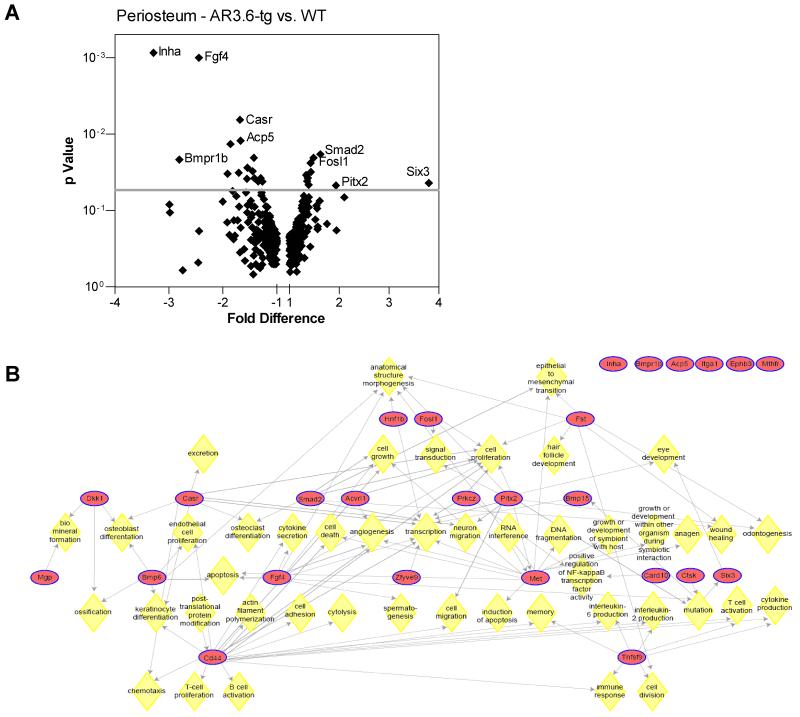
To identify important pathways that are physiologically relevant, we used an in vivo gene profiling approach with isolated periosteal bone tissue harvested from male transgenic mice with enhanced sensitivity to circulating androgens. AR3.6-tg mice were employed because the anabolic response is not present in AR2.3-tg males [8, 9], and gene expression differences were surveyed using qPCR arrays. Regulated transcripts were identified in periosteal tissue derived from WT vs. AR3.6-tg mice. Because little is known about androgen regulation of gene expression in bone, we constructed a qPCR array to contain pathways with established importance in bone development, formation and remodeling and tested the hypothesis that enhancement of periosteal formation by androgen signaling may be mediated by pathways that have previously been associated with bone formation. Expression analysis revealed fewer significantly regulated transcripts in periosteal bone compared with endocortical bone; of the 384 array genes examined by qPCR analysis, expression of a total of 26 genes (~7%) were significantly different between WT and transgenic bones vs. 78 genes (20%) in the endocortical comparisons [14]. Significantly upregulated sequences are shown in Table 1 and downregulated sequences are shown in Table 2. A volcano plot, employed to visualize differentially expressed genes, shows genes arranged along dimensions of biological impact (fold change) versus statistical significance (for reliability of change) in Fig. 1A. The horizontal dimension shows fold change between the two groups and the vertical axis represents the p-value for a t-test of differences between samples. Interestingly, there is no overlap with highly regulated sequences identified in endocortical bone [see 14]; only four genes reached significance in both comparisons (i.e., Casr, Inha, Six3 and Zfyve9) and all four were regulated in opposite directions between periosteal vs. endocortical bone.
Table 1.
Upregulation of expression in array qPCR analyses in male transgenic mice from periosteal bone samples
| Rank | Gene Symbol |
Gene ID | Change (fold/control) |
p-value |
|---|---|---|---|---|
| 27 | Six3 | 20473 | 7.0437 | 0.0436 |
| 29 | Pitx2 | 18741 | 1.9325 | 0.0471 |
| 9 | Smad2 | 17126 | 1.6197 | 0.0184 |
| 11 | Fosl1 | 14283 | 1.4813 | 0.0204 |
| 16 | Tnfsf9 | 21950 | 1.4284 | 0.0313 |
| 13 | Mthfr | 17769 | 1.4239 | 0.0239 |
| 28 | Met | 17295 | 1.3839 | 0.0461 |
| 20 | Acvrl1 | 11482 | 1.3677 | 0.0375 |
| 24 | Fst | 14313 | 1.3523 | 0.0411 |
| 19 | Itga1 | 109700 | 1.3341 | 0.0343 |
Results show genes with significant expression changes when comparing samples after global normalization. Rank number is listed in order of significant changes was significant at p < 0.05, calculated with respect to normalizers based on the GPR algorithm. Fold/control is shown as the expression change between transgenic and WT control. Analysis was from two samples in replicate representing both AR3.6-transgenic families.
Table 2.
Downregulation of expression in array qPCR analyses in male transgenic mice from periosteal bone samples
| Rank | Gene Symbol |
Gene ID | Change (fold/control) |
p-value |
|---|---|---|---|---|
| 1 | Inha | 16322 | −3.2852 | 8.63 E-4 |
| 12 | Bmpr1b | 12167 | −2.8082 | 0.0216 |
| 2 | Fgf4 | 14175 | −2.4430 | 0.0010 |
| 18 | Prkcz | 18762 | −1.9146 | 0.0330 |
| 7 | Ctsk | 13038 | −1.8610 | 0.0135 |
| 17 | Mgp | 17313 | −1.7063 | 0.0321 |
| 5 | Casr | 12374 | −1.6835 | 0.0065 |
| 6 | Acp5 | 11433 | −1.6740 | 0.0122 |
| 23 | Dkk1 | 13380 | −1.5534 | 0.0385 |
| 14 | Bmp15 | 12155 | −1.5470 | 0.0276 |
| 15 | Ephb3 | 13845 | −1.4499 | 0.0306 |
| 10 | Zfyve9 | 230597 | −1.4257 | 0.0204 |
| 22 | Bmp6 | 12161 | −1.4160 | 0.0378 |
| 25 | Tcf2 | 21410 | −1.3460 | 0.0417 |
| 21 | Card10 | 105844 | −1.3005 | 0.0376 |
| 26 | Cd44 | 12505 | −1.2698 | 0.0418 |
Analysis was as described in Table 1.
Figure 1. Gene expression differences in AR3.6-transgenic periosteal bone samples.
A. A volcano plot of significant expression differences identified in AR3.6-transgenic bone compared with WT periosteal bone. This identifies sequences that are differentially expressed as a consequence of enhanced androgen signaling in vivo. B. Pathway analyses of androgen-regulated gene expression in periosteal bone from AR-transgenic males. Significantly regulated genes expressed differentially between WT and AR3.6-transgenic periosteal bone samples were organized into functional groups to identify associations, based on biological processes using the PathwayArchitect module of GeneSpring GX 10. Biological processes are indicated by yellow diamonds. CD44 signaling is a central node for significantly regulated differences.
We used three independent computational approaches to identify the biological targets that are impacted by these expression differences: significantly enriched gene ontology (GO) categories were identified using WebGestalt software and biological processes were characterized using Ingenuity Pathway Analysis (IPA) and Pathway Architect software. Of the GO categories identified, the most significantly regulated were ‘retinal ganglion cell axon guidance’ (p=0.016), ‘negative regulation of Wnt receptor signaling pathway’ (p=0.016) and ‘central nervous system (CNS) development’ (p=0.020). Examination of expression differences highlights the number of sequences for genes involved in nervous system development including Hnf1b, Pitx2, Dkk1, Met, and Six3. To identify associations among the significantly regulated genes and characterize biological processes that were impacted by androgen signaling in the periosteum, we employed additional bioinformatic analyses to identify associations among the regulated genes. IPA analysis was used to characterize associated network functions, and identified ‘Tissue Development, Cellular Growth and Proliferation, Cellular Development’ with the highest score. The higher the score, the more interconnected the regulated molecules are within a given network. Finally, a third independent analysis was performed using Pathway Architect software. Examination of the results revealed several notable findings and indicate that stem cell differentiation is a target for androgen in the periosteal compartment (Fig. 1B). Consistent with this analysis, the most significantly regulated biological processes in the IPA analysis were developmental process/developmental process of tissue (p = 6.49 E-10) with genes that influence development including the genes Acp5, Acvrl1, Bmpr1b, Cd44, Ctsk, Ephb3, Fgf4, Fosl1, Hnf1b, Itga1, Met, Pitx2, Six3, Smad2; and developmental process of embryonic tissue (p = 9.96 E-05) with genes Cd44, Fgf4, Hnf1b, Pitx2, Six3, Smad2. In addition, Sox9 was highly expressed in periosteum and there was a trend for significance between WT and transgenic (p = 0.0555). Thus, bioinformatic analyses of the expression differences suggest that enhanced androgen action in the periosteum positively affects osteoblastogenesis by modulation of stem cells.
Fine mapping of calvarial structure in transgenic mice demonstrates neural crest stem cells contribute to androgen anabolic action in vivo to enhance osteogenesis
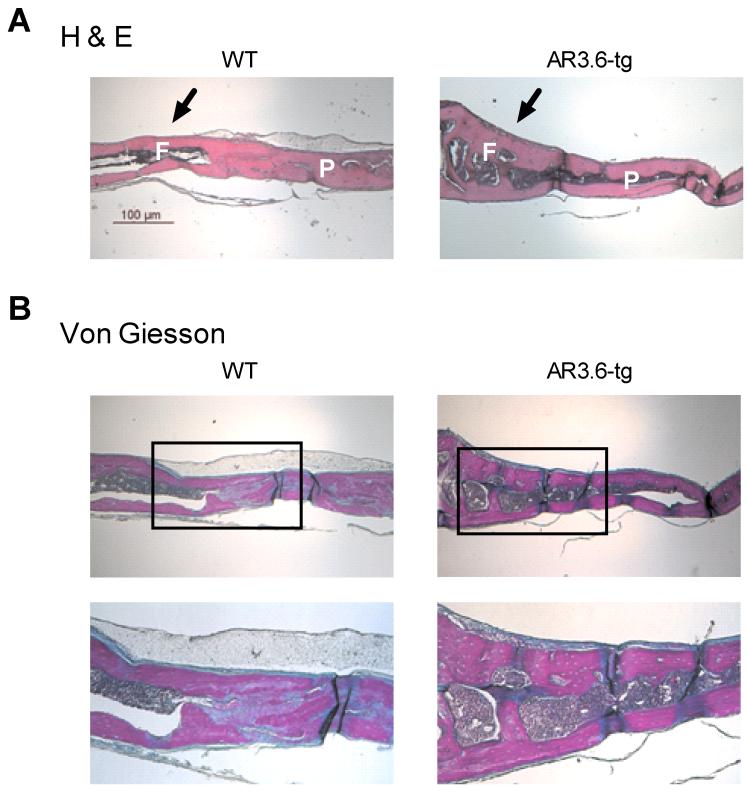
Combined, these results prompted us to examine more closely androgen-associated changes in periosteal apposition. We have previously shown that calvaria demonstrate periosteal thickening (as woven bone) in male AR3.6-tg but not in AR2.3-tg mice [8, 9]. In order to better characterize periosteal expansion in the calvaria, we fine-mapped the anabolic response over the surface of calvaria in 2 month old male WT vs. AR3.6-tg mice. Saggital sections were analyzed with H&E staining for morphology (Fig. 2A) and for new bone formation with von Giesson stains (Fig. 2B). As shown, there was no difference in WT animals between frontal and parietal bones. However in transgenic mice, thickening of the calvarial bone structure is observed. Thickening is not uniform across the surface with only the frontal bone demonstrates an anabolic thickening but not the parietal bone. Thus, enhanced androgen signaling in vivo appears to target distinct bones in the calvaria likely through alteration of stem cell pathways.
Figure 2. Histological analysis of site-specific anabolic action in calvaria.
Fine mapping of the calvarial response over the surface of the calvaria harvested from 2-month male WT and AR3.6-tg mice. Saggital sections across frontal and parietal bones stained with H-E (A) and von Gieson (B) indicate that in transgenic males only the frontal bone demonstrates an anabolic thickening (arrow), but not the parietal bones. Inset shows region of higher magnification image below. F indicates frontal bone, P indicates parietal bone. Total magnification 40X, inset 100X. Scale bar = 100 μm.
Ex vivo characterization of NCSC vs. MSC cultures derived from distinct calvarial bones
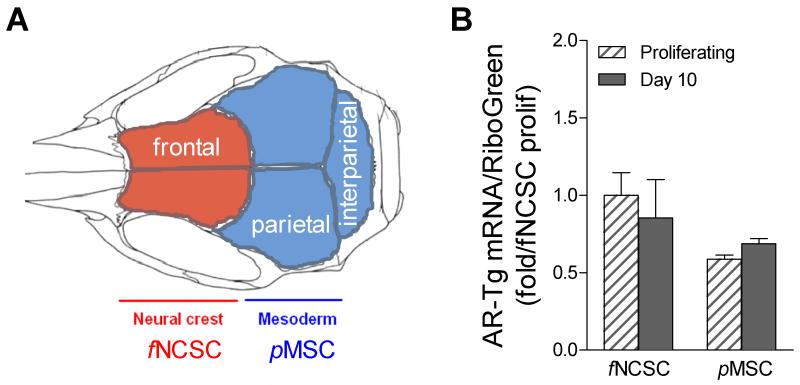
The calvaria provides an interesting model for analysis of bones derived from distinct embryological origins [for review, see 27]. As shown in Fig. 3A, the derivation of individual bones in the calvarial cap is distinct, with frontal bones derived from neural crest stem cells (NCSCs) while parietal bones are derived from a mesodermal mesenchymal origin (MSCs). Given the differential response to androgen observed in calvarial bone formation, we sought to evaluate the differentiation potential of progenitor cells derived from a neural crest (frontal) vs. mesodermal (parietal) origin. Precursor populations were isolated from the surface of dissected calvarial bones from both genotypes by collagenase digestion and characterized ex vivo. Cultures were established from frontal bones to provide progenitor neural crest stem-like cells (f while parietal bones were used to derive mesodermal mesenchymal stem-like cells (pMSC). There were no significant differences in proliferation potential between stem cell populations isolated from the two bone sites (data not shown). Because of the differential response in calvarial bone formation observed in vivo between the genotypes, we first determined transgene level in fNCSC vs. pMSC cells. Total RNA was harvested from proliferating and differentiating cultures from both lineages in cultures derived from neonatal transgenic mice. AR transgene levels were characterized by qPCR analysis (Fig. 3B). Cultures derived from both embryonic lineages show high AR-transgene expression. The level of transgene was similar between fNCSC and pMSC cultures and was not different in proliferating or in differentiating cultures at day 10.
Figure 3. Ex vivo model for analysis of progenitors derived from bone from neural crest vs. mesenchymal embryonic origins.
A. Calvaria bones are derived from distinct embryological origins, with frontal (colored red) from neural crest while parietal (colored blue) is derived from mesodermal-mesenchymal sources. Progenitors from the surface of each bone were harvested from the first collagenase digestion and grown for analysis of differentiation. B. AR transgene levels in fNCSC and pMSC cultures derived from AR3.6-transgenic bones were evaluated by qPCR. Data are expressed as fold vs. fNCSC control and shown as mean ± SD (n = 2-3).
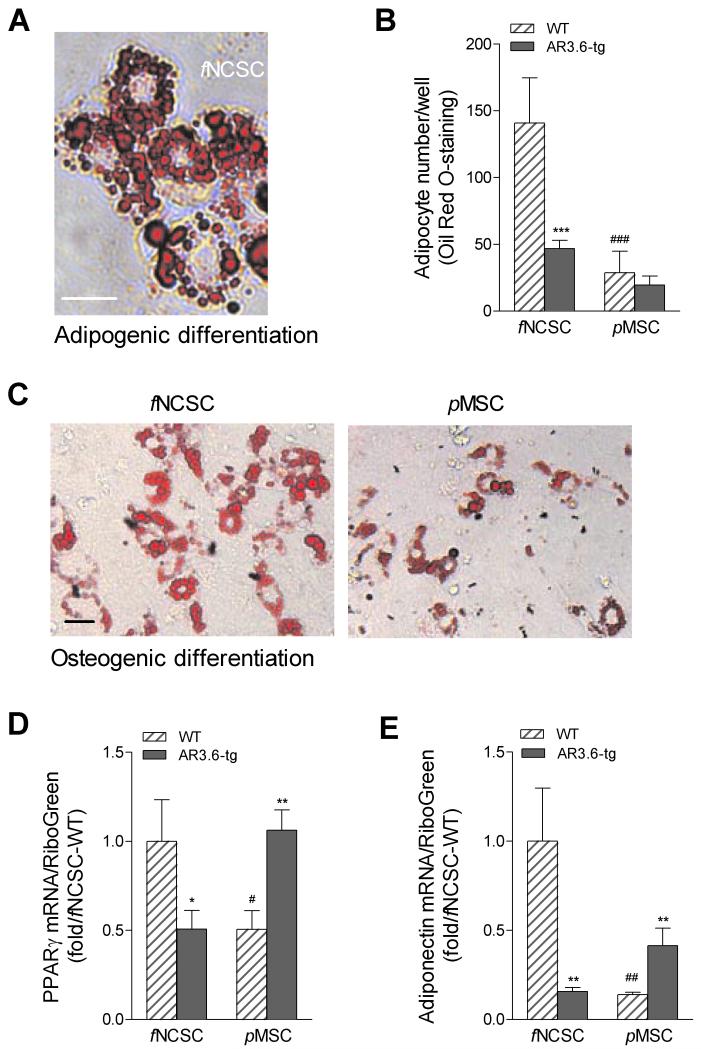
Both NCSC and MSC progenitors are multipotent and, depending on the signaling milieu of growth factors and hormones received, can give rise to osteoblasts and adipocytes. In the first sets of studies, we determined the adipogenic potential of fNSCS and pMSC cells. Primary cultures from fraction 1 frontal bone fNCSC vs. parietal bone pMSC were isolated by collagenase digestion from neonatal calvaria after genotyping. Cultures from both genotypes and both embryonic lineages were grown to confluence, switched to adipogenic medium and lipid accumulation was determined by Oil Red O staining on day 10 of adipogenic differentiation (Fig. 4A). A comparison in WT samples between embryonic origin shows that neural crest progenitor populations have enhanced adipogenic capacity with increased lipid accumulation compared to mesenchymal sources (Fig. 4B; p < 0.001). In addition, there was a difference in the ability of transgene to suppress adipogenic differentiation between the two progenitor populations. Transgenic cultures from fNCSC exhibited significant reduction in adipocyte number compared to WT controls (p < 0.001) but there was no significant difference in adipogenic capacity between the genotypes in pMSC cultures. Because adipogenesis also occurs during osteoblast differentiation [28], we evaluated lipid accumulation and gene expression differences in fNCSC and pMSC cultures after induction in osteogenic media. As shown in Fig. 4C, adipogenesis is observed after induction of osteoblastogenesis in WT progenitors from both embryonic lineages, with higher levels of adipocyte formation in fNCSC cultures. We also evaluated expression of the adipocyte markers PPARγ and adiponectin using RNA isolated from differentiating cultures from both genotypes and both lineages. WT cells from pMSC cultures showed a significant reduction of PPARγ and adiponectin expression vs. fNSCS WT cultures (Fig. 4D,E; p < 0.05 and p < 0.01 vs. fNCSC WT respectively). Evaluation of the effect of transgene in both cultures showed an opposite response in fNSCS cultures vs. pMSC cultures, with PPARγ and adiponectin mRNA levels decreased in fNCSC cultures from AR3.6-tg vs. WT control (p < 0.05 and p < 0.01). In contrast in pMSC cultures, PPARγ and adiponectin levels were increased in transgenic samples.
Figure 4. Adipogenesis in fNCSC vs. pMSC progenitors by Oil Red O staining.
A. Representative image of adipogenesis in differentiating cultures from fNCSC cultures after adipogenic induction as described in methods. Total magnification = 400X. Scale bar = 50 μm. B. Quantification of adipocyte numbers between WT and AR3.6-transgenic mice derived from fNCSC and pMSC cultures. Data are expressed as adipocyte number/well and shown as mean ± SD (n = 4). Two-way ANOVA for the effects of genotype and embryonic lineage showed a significant interaction, so t-test was used to determine significance. *** p < 0.001 vs. WT; ###, p < 0.001 vs. fNCSC WT. C. Adipocyte development in WT fNSCS vs. pMSC cultures after induction of osteoblastogenesis. Representative image demonstrates increased adipocyte differentiation even during induction of osteoblastogenesis in fNSCS cultures. Total magnification = 200X. Scale bar = 50 μm. Expression of adipocyte markers PPARγ (D) and adiponectin (E) was also evaluated by qPCR. Levels were determined using RNA isolated from differentiating cultures from both genotypes and both lineages as described in methods. Data are reported as mean ± SD (n = 3). Two-way ANOVA for the effects of genotype and embryonic lineage showed a significant interaction, so t-test was used to determine significance between samples. *, p < 0.05; **, p < 0.01 compared to WT and #, p < 0.05; ##, p < 0.01 vs. fNCSC WT.
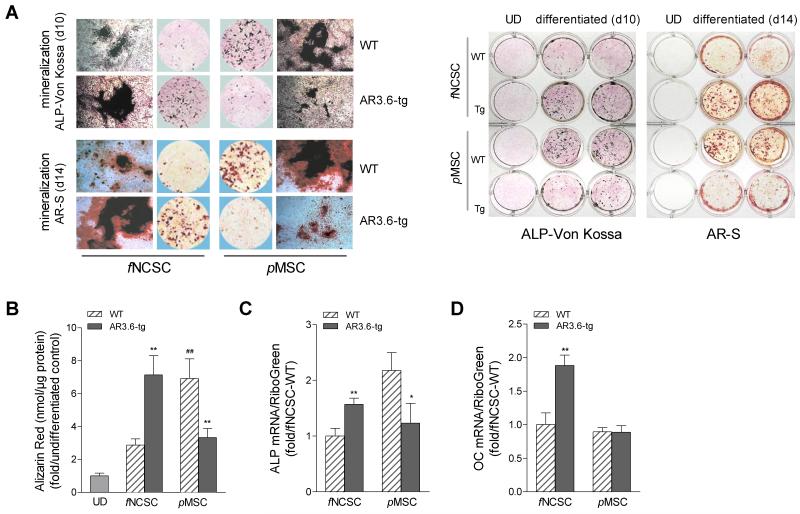
We next characterized osteogenic potential in neural crest vs. mesenchymal lineages and the effect of androgen on differentiation. Progenitors from both lineages and both genotypes were isolated by collagenase digestion from the surface of neonatal calvaria as above, and cultures were switched to osteoinductive media at confluence. Osteoblastogenesis assessed by alizarin red S (AR-S) accumulation and ALP and von Kossa staining as mineralization is the hallmark of the mature osteoblast. In WT cultures, pMSC cultures exhibited higher mineral accumulation and nodule formation was increased compared to fNCSC cultures (see Fig. 5A,B). Interestingly, this result with surface (fraction 1) cells was not observed when more mature cells were derived from deeper bone layers, since fraction 2-5 cultures showed enhanced osteoblastogenesis in frontal bone-derived cells compared to those from mesodermal bone (data not shown). The effect of transgene on progenitor differentiation was also evaluated at day 10 of osteoblastogenic differentiation. Elevated ALP and von Kossa staining was seen in AR3.6-tg cultures from fNCSC compared to controls while inhibition in cultures from pMSC (Fig. 5A). Quantitative analysis of mineral formation by alizarin red extraction at day 14 showed differences between WT cells from neural crest vs. mesenchymal origin with pMSC cultures showing higher mineral accumulation compared to fNCSC WT cultures (p < 0.01). Between the genotypes (Fig. 5B), a significant stimulation of osteoblastogenesis was noted in fNCSC cultures (p < 0.01) but significant inhibition in pMSC cells (p < 0.01) in transgenic cells. Changes in expression for osteoblast marker genes were assessed by qPCR analysis. Primary cultures were grown in osteoinductive media and total RNA was isolated from mature mineralizing cultures at day 10 of differentiation. Both ALP and osteocalcin gene expression (Fig. 5C,D) was increased in transgenic cultures from fNCSC vs. WT controls (p < 0.01 for both). In contrast, in pMSC cultures we observed decreased ALP gene expression in transgenic cells vs. WT (p < 0.05) and no significant difference in osteocalcin expression between the genotypes. Thus, AR3.6-tg cultures from fNCSC demonstrated induction of mineralization and mineral nodule formation, while inhibition was observed in AR3.6-tg cultures from pMSC compared to WT.
Figure 5. Androgen-mediated opposite effects of ALP activity and mineralization during differentiation in cultures from AR3.6-tg fNCSC vs. pMSC.
Cultures were induced toward the osteoblast lineage with osteoinductive media and osteoblastogenesis assessed by ALP staining, alizarin red S (AR-S) and von Kossa staining. A. Characterization of osteoblastogenesis using ALP, von Kossa and AR-S staining. Both fNCSC and pMSC cells were grown with osteoblastogenic medium at confluence. At day 10, cultures were stained and mineralized nodule formation was assessed by von Kossa over ALP staining. For mineral accumulation analysis, both fNCSC and pMSC cells were grown with osteoblastogenic medium for 14 days, and mineralization was assessed by alizarin red-S (AR-S) staining. Similar analysis at lower power in 12-well plates is shown in right panel. B. AR-S quantitation after extraction. AR-S accumulation was expressed as fold vs. undifferentiated control. Data are expressed as mean ± SD (n = 3). Two-way ANOVA for the effects of genotype and embryonic lineage showed a significant interaction, so t-test was used to determine significance. **, p < 0.01 vs. WT; ##, p < 0.01 vs. fNCSC WT. Osteoblast markers ALP (C) and OC (D) gene expression. Levels were evaluated by qPCR using RNA isolated from mature mineralizing cells from both genotypes and both lineages grown in osteoblastogenic media for 10 days. Data are reported as mean ± SD (n = 3). *, p < 0.05; **, p < 0.01 compared to WT. UD = undifferentiated.
Analysis of comparison between bone compartments and culture models derived from WT and transgenic bones: gene expression from qPCR array analysis in periosteal and endocortical bone samples vs. fNCSC and pMSC cultures
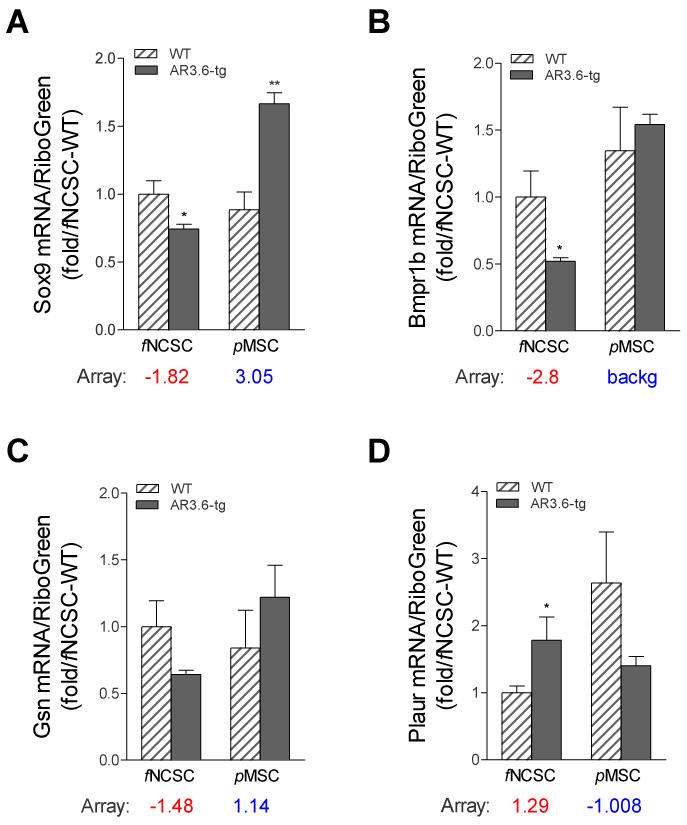
Combined, these results suggest that enhanced androgen signaling has opposite effects on lipid accumulation and on bone formation in progenitors depending on their embryonic origin. Given the similar responses observed in osteoblastogenesis between transgenic fNCSC cultures and periosteal bone formation, we determined whether gene expression patterns were also similar between the two samples. We have employed array analysis to describe changes in osteoblast-associated genes in two distinct bone compartments; endocortical bone samples from which both periosteum and marrow elements had been removed [14], and for periosteal bone samples that were stripped from long bone samples shown in Fig. 1. Expression profiles from both qPCR array studies were reviewed to identify candidates for evaluation. Genes were selected to represent both up- and down-regulation, and span a range of statistical significance, fold expression difference, and/or biological interest and potential involvement in stem cell signaling. Genes chosen included Sox9, Bmpr1b, Gsn, and Plaur, and expression levels were analyzed in both genotypes in fNCSC and pMSC cultures. Cells were cultured for 10 days after addition of osteogenic media, RNA harvested and qPCR was performed by qPCR (Fig. 6A-D). Array data comparing expression difference between the transgenic vs. WT bone sample is shown below each graph (red for periosteal bone samples and blue for endocortical bone samples). Significant differences were observed in fNCSC cultures between genotypes for Sox9, Bmpr1b, and Plaur (all p < 0.05). Sox9 was also significantly different in pMSC cultures but was decreased rather than increased in transgenic samples (p < 0.01). For all comparisons, the direction of regulation observed in expression profiles from AR3.6-tg vs WT long-bone periosteal tissue analyzed by array was recapitulated in fNCSC samples, while the pMSCs samples reflected similar expression patterns to that observed in cortical expression.
Figure 6. Comparison of gene expression from array analysis in periosteal tissue vs. fNCSC cells and cortical tissue vs. pMSC cultures.
Comparison between expression levels determined in bone samples by array results for long-bone periosteal and endocortical expression were compared to levels observed in fNCSC and pMSC cultures from both genotypes. fNCSC and pMSC cells were cultured for 10 days in osteogenic media and qPCR was performed for (A) Sox9, (B) Bmpr1b, (C) Gsn, and (D) Plaur expression. Array data is shown below each graph where red indicates qPCR array results from periosteal bone and blue indicates levels from endocortical bone samples. Each value represents the mean ± SD (n = 3). *, p < 0.05; **, p < 0.01 compared to WT control.
Discussion
A substantial body of evidence demonstrates that androgens increase periosteal bone formation, yet the mechanisms are incompletely characterized. Studies of transgenic mice with targeted AR overexpression reported here indicate that the effects of androgen are dictated by cell lineage and that the periosteal layer in long bone may have a neural crest origin. The mechanism(s) that underlie the distinct response to androgen treatment observed between bone compartments in vivo is not understood. Enhanced androgen sensitivity in vivo through targeted AR overexpression demonstrates this distinct response to androgen in cortical bone, with direct inhibition of bone formation on the endocortical surface but increased bone formation in the periosteum [8, 9]. Mesenchymal tissues such as bone and fat are derived from stem cells but the populations are diverse and not well characterized [29]. Our results demonstrate that androgen signaling in fNCSC results in enhanced osteoblastogenesis and bone matrix production, with elevated ALP activity, osteocalcin and ALP mRNA levels and induction of mineralization and nodule formation. In contrast, an opposite effect of androgen was observed in pMSC cultures, with decreased ALP mRNA levels, reduced mineralization and nodule formation in transgenic cells. In addition, gene expression profiles from these cultures recapitulate the response observed in periosteal vs. endocortical tissue. Opposite effects on adipocyte expression were also observed. Thus, the response to androgen in vivo likely depends on the embryonic origin of the osteoblastic cell, which will influence whether androgen treatment results in increased or decreased bone formation or, similarly, fat development.
A better understanding of periosteal bone formation is important since bone size correlates with bone strength, such that periosteal apposition greatly increases bone strength and is an important determinant of fracture resistance [30, 31]. Structurally, the periosteum is a connective tissue that is tightly attached to the bone surface. The periosteum is considered a unique tissue compartment in bone and site-specific differences within periosteal tissue in long bone have been reported, with both structural and cellular differences between metaphyseal and diaphyseal periosteum [32]. Thus, a promising target for androgen-mediated increases in bone width may be pursued with neural-crest derived cells as a target, since the origin of the cells influences the osteogenic response. The periosteum is also responsive to mechanical loading [33]. The involvement of androgen signaling in loading has been evaluated in a global AR null model, which surprisingly showed enhanced periosteal bone apposition in the null animal in response to loading compared to wild-type [34]. Because of global knockdown, the enhanced response to loading may be mediated in a variety of targets. As suggested by the authors, alteration in osteocyte function and expression is a primary candidate since the osteocyte functions as a central mediator of mechanotransduction [35, 36] and is inhibited by androgen signaling [9, 37].
The embryonic origin of the periosteum in long bone remains poorly characterized. Early work in an avian model suggested an osteogenic role for the periosteum and a potential relationship to neural crest in the avian head [38]. Another model for periosteal bone formation is described in studies characterizing regeneration of deer antlers, which are shed annually. Regeneration of the antler is derived from the specialized antlerogenic frontal pedicle periosteum [39, 40]. Although the pedicles do not develop until puberty and are androgen-dependent [41, 42], the specific role of androgen signaling to simulate bone formation is not well characterized [43]. Finally, fate mapping using labeled progenitors is a classic approach employed to directly characterize the contribution of cell linage and the embryonic origin of tissues [for example, see 27]. As noted, although not directly evaluated, it has been suggested that except for the facial skeleton, the rest of the skeleton is derived from mesoderm [16]. Results from our studies suggest that fine-mapping analysis of periosteal tissue in long bone should be a focus of future work to support the less refined anatomically-based approach employed here.
Adipogenic differentiation in these progenitor populations was also influenced by embryonic lineage and by genotype. In vivo, a body composition phenotype is observed in transgenic males, with increased lean mass but reduced fat mass [25]. Adipose tissue weight from transgenic males was reduced in both gonadal and perirenal white adipose tissue depots and adipocyte size was smaller than in wild-type littermates. Notably, we have shown elevated expression of PPARγ in proliferating pluripotent progenitor cultures from bone marrow stromal cells from transgenic mice [25], similar to the response observed in pMSC cultures but opposite to the response observed in fNCSC cells. High PPARγ expression is also observed in the subset of osteoblasts that switch fate to adipocytes in calvarial cultures [28]. In addition, significantly reduced Sox9 expression is observed in common between transgenic pMSC cultures and the calvarial osteoblasts that can switch fate to adipocytes. Although the developmental origin of adipocytes remains an area of debate, it is likely that there is a strong contribution from the neural crest at least in some fat depots [44]. Data presented here is consistent with the notion that androgen may stimulate adipogenesis in fat tissue derived from mesodermal origin, including bone marrow mesenchymal cells, but inhibit adipogenesis in fat derived from neural crest. Combined analysis in this AR targeted model suggests that the embryonic origin of progenitors determines response to androgen in both bone and fat tissue.
We pursued a novel approach to characterize the consequences of enhanced androgen signaling on the differentiation potential of progenitors derived from neural crest vs. mesenchymal lineages. Calvaria provide a unique resource for isolation of multipotent stem-like cells derived from distinct embryological origins, with frontal bones originating from ectodermal neural crest cells while parietal bones originate from mesoderm mesenchymal cells [27]. Progenitors were harvested from the surface of frontal vs. parietal bones by collagenase digestion from both genotypes for ex vivo analysis of relatively pure populations. Importantly, transgene expression was high and similar between fNCSC and pMSC cultures. Nevertheless, these distinct progenitor populations demonstrated functional differences, with more adipogenically primed cells but fewer osteoblastic cells in neural crest than mesenchymal progenitor populations. Notably, there is evidence that periosteal tissue harvested from neural crest-derived bone (from the mandible) vs. mesoderm-derived bone (tibia) is functionally different when implanted in bone grafts [16]. There are also expression differences in growth factors between frontal and parietal bones [45]. In contrast to our findings, accelerated osteoblast differentiation in neural crest-derived skeletal cells has been described in ex vivo analysis [16, 46]. A major methodological difference between these reports and the analysis presented in here is that our progenitors are harvested from the first collagenase digestion and represent surface cells that may be less restricted in development compared with later digestions of more mature cells [16, 46]. Consistent with this possibility, fractions from unstripped samples in our hands also show enhanced osteoblastogenesis from the deeper layers of frontal bone (data not shown). The functional differences observed in WT cultures between fNCSC and pMSC cells is consistent with a distinction between cranial (neural crest) and the appendicular (mesenchymal) osteoblasts observed in gene inactivation studies, where various genetic diseases that result from single gene disruption target cranial bones selectively while the appendicular skeleton is largely spared [for review, see 47].
Progenitors derived from neural crest are becoming recognized as a target of androgen signaling and sexually dimorphic responses [see 48]. For example, progenitor cells in the hair follicle are important in the development of androgenetic alopecia [49], which results from androgen suppression of that the conversion of hair follicle stem cells to progenitor cells. Cells from the bulge region of the hair follicle contain neural crest stem cells [50]. Notably, bioinformatic analysis of the effects of androgen signaling in the periosteum identified ‘hair follicle development’ as a target even in bone tissue (see Fig. 1B). The data reported here supports the concept that anabolic actions in the skeleton that are mediated by androgen treatment are likely to be restricted to tissues that are derived from a neural crest origin. Stem cell therapy is an emerging approach in regenerative medicine, and progenitor cells derived from periosteum have been employed to successfully repair bone defects [51, 52]. Utilizing progenitors derived exclusively from the surface of the frontal bone for example might provide an improved approach in males or when combined with androgen therapy.
Finally, qPCR array analysis was employed to identify expression differences in the periosteum between WT and transgenic males in order to identify pathways associated with increased bone formation. With reduced bone formation observed in endocortical bone, the ‘TGF-β/BMP/Activin signaling’ pathway was significantly regulated and was generally inhibited. In contrast, periosteal bone samples identified ‘axonal guidance’ and ‘CNS development’ pathways as activated, consistent with an influence of neural crest cells. Consistent with these distinct responses observed in cortical bone, analysis of AR3.6-tg calvaria revealed that anabolic thickening of the periosteum in vivo is not uniform across the calvaria, occurring only in frontal (neural crest) but not parietal (mesodermal) bones. In addition, ‘negative regulation of Wnt receptor signaling’ was an overrepresented pathway in the array analysis, of note since Wnt pathways have previously been implicated in regulating periosteal expansion [34, 53].
In summary, expression profiling identified stem cell pathways and organ morphogenesis as a predominant target of androgen in periosteum derived from male AR transgenic mice which have enhanced periosteal bone formation [8]. Fine-mapping of anabolic periosteal expansion in the calvaria demonstrated that the response was restricted to frontal bones derived from neural crest. Mesenchymal tissues such as bone and fat can be derived from stem cells from either neural crest or mesodermal origins. However, the contribution of neural crest to distinct mesenchymal tissues in the adult remains controversial. Ex vivo analysis was employed to characterize progenitor populations harvested from the periosteal surface of bones derived from distinct embryonic lineages in both WT and transgenic genotypes. Consistent with disparate responses in bone formation in distinct skeletal compartments observed in vivo in transgenic males, osteoblastogenesis was significantly increased in fNCSC cultures from transgenic compared to WT, but the opposite response was observed in pMSC cultures. Expression differences identified by array in long bone periosteal tissue from AR3.6-tg vs. WT were recapitulated in the fNCSC samples while pMSCs profiles reflected cortical expression. These observations reveal the opposing effects of androgen signaling on osteoblast differentiation that is enhanced in periosteal progenitors derived from a neural crest origin but inhibited in cells derived from a mesodermal origin. Adipogenesis was also differentially influenced by androgen signaling in fNCSCs vs. pMSCs. This data is consistent with the notion that sexually-dimorphic patterning associated with testosterone action in vivo in specific compartments is also influenced by the developmental origin of the cell in males vs. females. Combined, these results highlight the complex action of androgen in the body, which is dependent on the embryonic lineage and developmental origin of the cell, and provide a context in which to identify potential therapeutic targets for treatment of obesity and osteoporosis. Given the osteogenic response and similarity in gene expression patterns between the two models, these data also suggest that in the adult, the periosteum surrounding the diaphysis in long bone may have a neural crest origin.
Research Highlights.
Anabolic pathways in periosteal bone were identified in AR transgenic males by arrays Axonal guidance, CNS development and negative regulation of Wnt signaling identified Stem cell differentiation determined by neural crest or mesodermal embryonic lineage Lineage may determine sexually-dimorphic patterning associated with testosterone The periosteum surrounding long bone is likely derived from neural crest
Acknowledgments
The authors would like to thank Melissa Forquer (OHSU) for technical assistance and careful reading of the manuscript.
Role of the funding source This publication was made possible by grants from the United States Army Research Acquisition Activity Award No. W81XWH-05-1-0086 (KMW) and the National Institute of Diabetes, Digestive & Kidney Disease R01 DK067541 (KMW). KMW is a Research Career Scientist of the Department of Veterans Affairs and received support from the Veterans Affairs Merit Review Program. All work was performed in facilities provided by the Department of Veterans Affairs. The study sponsor(s) did not play a role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication. The information contained in this publication does not necessarily reflect the position or the policy of the government, and no official endorsement should be inferred.
Abbreviations
- ALP
alkaline phosphatase
- AR
androgen receptor
- fNCSC
frontal bone neural crest stem-like cell
- GPR
global pattern recognition
- pMSC
parietal bone mesenchymal stem-like cell
- OC
osteocalcin
- qPCR
real-time quantitative reverse transcription-polymerase chain reaction
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Vanderschueren D, Gaytant J, Boonen S, Venken K. Androgens and bone. Curr Opin Endocrinol Diabetes Obes. 2008;15:250–4. doi: 10.1097/MED.0b013e3282fe6ca9. [DOI] [PubMed] [Google Scholar]
- [2].Vignozzi L, Morelli A, Filippi S, Maggi M, Forti G. Male pubertal development: role of androgen therapy on bone mass and body composition. J Endocrinol Invest. 2011;33:27–32. [PubMed] [Google Scholar]
- [3].Frenkel B, Hong A, Baniwal S, Coetzee G, Ohlsson C, Khalid O, Gabet Y. Regulation of adult bone turnover by sex steroids. J Cell Physiol. 2010;224:305–310. doi: 10.1002/jcp.22159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wiren K. Androgens and bone growth: it’s location, location, location. Curr Opin Pharmacol. 2005;5:626–632. doi: 10.1016/j.coph.2005.06.003. [DOI] [PubMed] [Google Scholar]
- [5].Carnevale V, Romagnoli E, Cipriani C, Del Fiacco R, Piemonte S, Pepe J, Scillitani A, Minisola S. Sex hormones and bone health in males. Arch Biochem Biophys. 2010;503:110–117. doi: 10.1016/j.abb.2010.07.001. [DOI] [PubMed] [Google Scholar]
- [6].Callewaert F, Sinnesael M, Gielen E, Boonen S, Vanderschueren D. Skeletal sexual dimorphism: relative contribution of sex steroids, GH-IGF1, and mechanical loading. J Endocrinol. 2010;207:127–134. doi: 10.1677/JOE-10-0209. [DOI] [PubMed] [Google Scholar]
- [7].Callewaert F, Venken K, Kopchick J, Torcasio A, van Lenthe G, Boonen S, Vanderschueren D. Sexual dimorphism in cortical bone size and strength but not density is determined by independent and time-specific actions of sex steroids and IGF-1: evidence from pubertal mouse models. J Bone Miner Res. 2010;25:617–626. doi: 10.1359/jbmr.090828. [DOI] [PubMed] [Google Scholar]
- [8].Wiren KM, Zhang XW, Toombs AR, Kasparcova V, Gentile MA, Harada S, Jepsen KJ. Targeted overexpression of androgen receptor in osteoblasts: unexpected complex bone phenotype in growing animals. Endocrinology. 2004;145:3507–3522. doi: 10.1210/en.2003-1016. [DOI] [PubMed] [Google Scholar]
- [9].Wiren KM, Semirale AA, Zhang XW, Woo A, Tommasini SM, Price C, Schaffler MB, Jepsen KJ. Targeting of androgen receptor in bone reveals a lack of androgen anabolic action and inhibition of osteogenesis. A model for compartment-specific androgen action in the skeleton. Bone. 2008;43:440–451. doi: 10.1016/j.bone.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Idan A, Griffiths K, Harwood D, Seibel M, Turner L, Conway A, Handelsman D. Long-term effects of dihydrotestosterone treatment on prostate growth in healthy, middle-aged men without prostate disease: a randomized, placebo-controlled trial. Ann Intern Med. 2010;153:621–632. doi: 10.7326/0003-4819-153-10-201011160-00004. [DOI] [PubMed] [Google Scholar]
- [11].Kenny A, Kleppinger A, Annis K, Rathier M, Browner B, Judge J, McGee D. Effects of transdermal testosterone on bone and muscle in older men with low bioavailable testosterone levels, low bone mass, and physical frailty. J Am Geriatr Soc. 2010;58:1134–1143. doi: 10.1111/j.1532-5415.2010.02865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Benito M, Vasilic B, Wehrli FW, Bunker B, Wald M, Gomberg B, Wright AC, Zemel B, Cucchiara A, Snyder PJ. Effect of testosterone replacement on trabecular architecture in hypogonadal men. J Bone Miner Res. 2005;20:1785–1791. doi: 10.1359/JBMR.050606. [DOI] [PubMed] [Google Scholar]
- [13].Wang C, Cunningham G, Dobs A, Iranmanesh A, Matsumoto A, Snyder P, Weber T, Berman N, Hull L, Swerdloff R. Long-term testosterone gel (AndroGel) treatment maintains beneficial effects on sexual function and mood, lean and fat mass, and bone mineral density in hypogonadal men. J Clin Endocrinol Metab. 2004;89:2085–2098. doi: 10.1210/jc.2003-032006. [DOI] [PubMed] [Google Scholar]
- [14].Wiren K, Semirale A, Hashimoto J, Zhang X. Signaling pathways implicated in androgen regulation of endocortical bone. Bone. 2010;46:710–723. doi: 10.1016/j.bone.2009.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Schindeler A, Liu R, Little D. The contribution of different cell lineages to bone repair: exploring a role for muscle stem cells. Differentiation. 2009;77:12–18. doi: 10.1016/j.diff.2008.09.007. [DOI] [PubMed] [Google Scholar]
- [16].Leucht P, Kim J, Amasha R, James A, Girod S, Helms J. Embryonic origin and Hox status determine progenitor cell fate during adult bone regeneration. Development. 2008;135:2845–2854. doi: 10.1242/dev.023788. [DOI] [PubMed] [Google Scholar]
- [17].Bhasin S. Regulation of body composition by androgens. J Endocrinol Invest. 2003;26:814–822. doi: 10.1007/BF03345230. [DOI] [PubMed] [Google Scholar]
- [18].Matsumoto A. Andropause: clinical implications of the decline in serum testosterone levels with aging in men. J Gerontol A Biol Sci Med Sci. 2002;57:M76–99. doi: 10.1093/gerona/57.2.m76. [DOI] [PubMed] [Google Scholar]
- [19].Hartgens F, Kuipers H. Effects of androgenic-anabolic steroids in athletes. Sports Med. 2004;34:513–554. doi: 10.2165/00007256-200434080-00003. [DOI] [PubMed] [Google Scholar]
- [20].Seidell J, Bjorntorp P, Sjostrom L, Kvist H, Sannerstedt R. Visceral fat accumulation in men is positively associated with insulin, glucose, and C-peptide levels, but negatively with testosterone levels. Metabolism. 1990;39:897–901. doi: 10.1016/0026-0495(90)90297-p. [DOI] [PubMed] [Google Scholar]
- [21].Bassil N, Alkaade S, Morley J. The benefits and risks of testosterone replacement therapy: a review. Ther Clin Risk Manag. 2009;5:427–448. doi: 10.2147/tcrm.s3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Mauras N, Hayes V, Welch S, Rini A, Helgeson K, Dokler M, Veldhuis J, Urban R. Testosterone deficiency in young men: marked alterations in whole body protein kinetics, strength, and adiposity. J Clin Endocrinol Metab. 1998;83:1886–1892. doi: 10.1210/jcem.83.6.4892. [DOI] [PubMed] [Google Scholar]
- [23].Woodhouse LJ, Gupta N, Bhasin M, Singh AB, Ross R, Phillips J, Bhasin S. Dose-dependent effects of testosterone on regional adipose tissue distribution in healthy young men. J Clin Endocrinol Metab. 2004;89:718–726. doi: 10.1210/jc.2003-031492. [DOI] [PubMed] [Google Scholar]
- [24].Nedungadi T, Clegg D. Sexual dimorphism in body fat distribution and risk for cardiovascular diseases. J Cardiovasc Transl Res. 2009;2:321–327. doi: 10.1007/s12265-009-9101-1. [DOI] [PubMed] [Google Scholar]
- [25].Semirale A, Zhang X, Wiren K. Body composition changes and inhibition of fat development in vivo implicates androgen in regulation of stem cell lineage allocation. J Cell Biochem. 2011 doi: 10.1002/jcb.23098. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hashimoto JG, Beadles-Bohling AS, Wiren KM. Comparison of RiboGreen and 18S rRNA quantitation for normalizing real-time RT-PCR expression analysis. Biotechniques. 2004;36:54–6. 58–60. doi: 10.2144/04361BM06. [DOI] [PubMed] [Google Scholar]
- [27].Gross J, Hanken J. Review of fate-mapping studies of osteogenic cranial neural crest in vertebrates. Dev Biol. 2008;317:389–400. doi: 10.1016/j.ydbio.2008.02.046. [DOI] [PubMed] [Google Scholar]
- [28].Yoshiko Y, Oizumi K, Hasegawa T, Minamizaki T, Tanne K, Maeda N, Aubin J. A subset of osteoblasts expressing high endogenous levels of PPARgamma switches fate to adipocytes in the rat calvaria cell culture model. PLoS One. 2010;5:e11782. doi: 10.1371/journal.pone.0011782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Nombela-Arrieta C, Ritz J, Silberstein L. The elusive nature and function of mesenchymal stem cells. Nat Rev Mol Cell Biol. 2011;12:126–131. doi: 10.1038/nrm3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Allen M, Hock J, Burr D. Periosteum: biology, regulation, and response to osteoporosis therapies. Bone. 2004;35:1003–1012. doi: 10.1016/j.bone.2004.07.014. [DOI] [PubMed] [Google Scholar]
- [31].Seeman E. Periosteal bone formation--a neglected determinant of bone strength. N Engl J Med. 2003;349:320–323. doi: 10.1056/NEJMp038101. [DOI] [PubMed] [Google Scholar]
- [32].Fan W, Crawford R, Xiao Y. Structural and cellular differences between metaphyseal and diaphyseal periosteum in different aged rats. Bone. 2008;42:81–89. doi: 10.1016/j.bone.2007.08.048. [DOI] [PubMed] [Google Scholar]
- [33].McBride S, Dolejs S, Brianza S, Knothe U, Tate M Knothe. Net Change in Periosteal Strain During Stance Shift Loading After Surgery Correlates to Rapid De Novo Bone Generation in Critically Sized Defects. Ann Biomed Eng. 2011 doi: 10.1007/s10439-010-0242-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Callewaert F, Bakker A, Schrooten J, Van Meerbeek B, Verhoeven G, Boonen S, Vanderschueren D. Androgen receptor disruption increases the osteogenic response to mechanical loading in male mice. J Bone Miner Res. 2010;25:124–131. doi: 10.1359/jbmr.091001. [DOI] [PubMed] [Google Scholar]
- [35].Bonewald L. Mechanosensation and Transduction in Osteocytes. Bonekey Osteovision. 2006;3:7–15. doi: 10.1138/20060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Santos A, Bakker A, Klein-Nulend J. The role of osteocytes in bone mechanotransduction. Osteoporos Int. 2009;20:1027–1031. doi: 10.1007/s00198-009-0858-5. [DOI] [PubMed] [Google Scholar]
- [37].Wiren KM, Toombs AR, Semirale AA, Zhang X. Osteoblast and osteocyte apoptosis associated with androgen action in bone: requirement of increased Bax/Bcl-2 ratio. Bone. 2006;38:637–651. doi: 10.1016/j.bone.2005.10.029. [DOI] [PubMed] [Google Scholar]
- [38].Thorogood P. In vitro studies on skeletogenic potential of membrane bone periosteal cells. J Embryol Exp Morphol. 1979;54:185–207. [PubMed] [Google Scholar]
- [39].Goss R, Powel R. Induction of deer antlers by transplanted periosteum. I. Graft size and shape. J Exp Zool. 1985;235:359–373. doi: 10.1002/jez.1402350307. [DOI] [PubMed] [Google Scholar]
- [40].Li C, Mackintosh C, Martin S, Clark D. Identification of key tissue type for antler regeneration through pedicle periosteum deletion. Cell Tissue Res. 2007;328:65–75. doi: 10.1007/s00441-006-0333-y. [DOI] [PubMed] [Google Scholar]
- [41].Li C, Littlejohn R, Corson I, Suttie J. Effects of testosterone on pedicle formation and its transformation to antler in castrated male, freemartin and normal female red deer (Cervus elaphus) Gen Comp Endocrinol. 2003;131:21–31. doi: 10.1016/s0016-6480(02)00625-1. [DOI] [PubMed] [Google Scholar]
- [42].Bartos L, Schams D, Bubenik G. Testosterone, but not IGF-1, LH, prolactin or cortisol, may serve as antler-stimulating hormone in red deer stags (Cervus elaphus) Bone. 2009;44:691–698. doi: 10.1016/j.bone.2008.12.004. [DOI] [PubMed] [Google Scholar]
- [43].Kuzmova E, Bartos L, Kotrba R, Bubenik G. Effect of different factors on proliferation of antler cells, cultured in vitro. PLoS One. 2011;6:e18053. doi: 10.1371/journal.pone.0018053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Billon N, Monteiro M, Dani C. Developmental origin of adipocytes: new insights into a pending question. Biol Cell. 2008;100:563–575. doi: 10.1042/BC20080011. [DOI] [PubMed] [Google Scholar]
- [45].Quarto N, Behr B, Li S, Longaker M. Differential FGF ligands and FGF receptors expression pattern in frontal and parietal calvarial bones. Cells Tissues Organs. 2009;190:158–169. doi: 10.1159/000202789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Quarto N, Wan D, Kwan M, Panetta N, Li S, Longaker M. Origin matters: differences in embryonic tissue origin and Wnt signaling determine the osteogenic potential and healing capacity of frontal and parietal calvarial bones. J Bone Miner Res. 2010;25:1680–1694. doi: 10.1359/jbmr.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Helms J, Amasha R, Leucht P. Bone voyage: an expedition into the molecular and cellular parameters affecting bone graft fate. Bone. 2007;41:479–485. doi: 10.1016/j.bone.2007.06.023. [DOI] [PubMed] [Google Scholar]
- [48].Ray R, Novotny N, Crisostomo P, Lahm T, Abarbanell A, Meldrum D. Sex steroids and stem cell function. Mol Med. 2008;14:493–501. doi: 10.2119/2008-00004.Ray. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Garza L, Yang C, Zhao T, Blatt H, Lee M, He H, Stanton D, Carrasco L, Spiegel J, Tobias J, Cotsarelis G. Bald scalp in men with androgenetic alopecia retains hair follicle stem cells but lacks CD200-rich and CD34-positive hair follicle progenitor cells. J Clin Invest. 2011;121:613–622. doi: 10.1172/JCI44478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Yu H, Kumar S, Kossenkov A, Showe L, Xu X. Stem cells with neural crest characteristics derived from the bulge region of cultured human hair follicles. J Invest Dermatol. 2010;130:1227–1236. doi: 10.1038/jid.2009.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Knothe U, Springfield D. A novel surgical procedure for bridging of massive bone defects. World J Surg Oncol. 2005;3:7. doi: 10.1186/1477-7819-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Fuchs B, Steinmann S, Bishop A. Free vascularized corticoperiosteal bone graft for the treatment of persistent nonunion of the clavicle. J Shoulder Elbow Surg. 2005;14:264–268. doi: 10.1016/j.jse.2004.06.007. [DOI] [PubMed] [Google Scholar]
- [53].Minear S, Leucht P, Miller S, Helms J. rBMP represses Wnt signaling and influences skeletal progenitor cell fate specification during bone repair. J Bone Miner Res. 2010;25:1196–1207. doi: 10.1002/jbmr.29. [DOI] [PMC free article] [PubMed] [Google Scholar]