Abstract
The genes pannier (pnr) and u-shaped (ush) are required for the regulation of achaete-scute during establishment of the bristle pattern in Drosophila. pnr encodes a protein belonging to the GATA family of transcription factors, whereas ush encodes a novel zinc finger protein. Genetic interactions between dominant pnr mutants bearing lesions situated in the amino-terminal zinc finger of the GATA domain and ush mutants have been described. We show here that both wild-type Pannier and the dominant mutant form activate transcription from the heterologous α globin promoter when transfected into chicken embryonic fibroblasts. Furthermore, Pnr and Ush are found to heterodimerize through the amino-terminal zinc finger of Pnr and when associated with Ush, the transcriptional activity of Pnr is lost. In contrast, the mutant pnr protein with lesions in this finger associates only poorly with Ush and activates transcription even when cotransfected with Ush. These interactions have been investigated in vivo by overexpression of the mutant and wild-type proteins. The results suggest an antagonistic effect of Ush on Pnr function and reveal a new mode of regulation of GATA factors during development.
Keywords: Drosophila, pannier, u-shaped, heterodimerization, GATA, DNA-binding domain, bristle pattern
The GATA factors comprise a family of transcription factors that interact specifically with the (A/T)GATA(A/G) consensus sequence through a highly conserved zinc finger DNA-binding domain (Wall et al. 1988; Evans and Felsenfeld 1989; Tsai et al. 1989; Orkin 1992). At present, six members of this family have been identified in birds that have homologs in mammals and amphibians (Laverierre et al. 1994). GATA-binding proteins have also been isolated from fungi, yeast, flies, and worms (Kudla et al. 1990; Cunningham and Cooper 1991; Spieth et al. 1991; Abel et al. 1993; Ramain et al. 1993; Winnick et al. 1993; Lin et al. 1995).
The founding member of this family, GATA-1, was identified originally as an erythroid cell DNA-binding factor that interacts with the promoters and enhancers of many erythroid-specific genes (Evans and Felsenfeld 1988). Targeted disruption of GATA-1 in mice revealed that this protein is needed for the maturation of terminally differentiated erythroblasts (Pevny et al. 1991). High levels of expression of globin genes require regulatory sequences, dispersed over a wide area, that appear to cooperate with each other to drive globin expression (Crossley and Orkin 1993; Higgs and Wood 1993; Andrews and Orkin 1994). This cooperation is thought to involve protein–protein interactions and indeed two proteins that associate with GATA-1 have been described: Sp1, which is expressed ubiquitously, and the Krüppel-like factor EKLF, which is specific to erythroid cells (Miller and Bieker 1993; Merika and Orkin 1995). These proteins heterodimerize through their respective DNA-binding domains and the carboxy-terminal zinc finger of GATA-1 is sufficient to mediate physical association (Merika and Orkin 1995). When associated with GATA-1, Sp1 and EKLF have a synergistic effect on transcription.
Previously, we described the molecular cloning of pannier (pnr), a gene from Drosophila whose product bears a putative DNA-binding domain with two zinc fingers that are homologous to those of vertebrate GATA-1 (Ramain et al. 1993). pnr is required for the spatial regulation of the achaete and scute genes during bristle patterning in Drosophila. achaete and scute (sc) encode proteins bearing a basic helix–loop–helix motif and are required to provide cells with neural potential (Villares and Cabrera 1987; Ghysen and Dambly-Chaudière 1988; Campuzano and Modolell 1992). They are expressed in restricted groups of cells, the proneural clusters, at the sites where the macrochaete (large bristle) precursors form (Romani et al. 1989; Cubas et al. 1991; Skeath and Carroll 1991; Cubas and Modolell 1992). Mutants of pnr display changes in the number and positions of bristles that are correlated with changes in ac–sc expression (Ramain et al. 1993; Heitzler et al. 1996).
We show here that Pannier binds to the GATA core sequence and we have investigated some of its transcriptional properties making use of its ability to activate the heterologous α-globin promoter when transfected into chicken embryonic fibroblasts. One class of dominant pnr alleles is associated with point mutations causing a single amino acid change in the amino-terminal zinc finger (Ramain et al. 1993). In spite of these lesions, the mutant proteins also activate transcription in the transient expression assay. In vivo, however, the mutants display an increase of ac–sc expression and additional bristles on the thorax, but decreased ac–sc expression and a loss of bristles at other sites (Ramain et al. 1993; Heitzler et al. 1996). This suggests that additional factors, differentially distributed within the epithelium, may regulate the activity of Pnr during development.
We have identified another gene, u-shaped (ush), mutants of which interact genetically with the dominant pnr mutants (Heitzler 1993; Cubadda et al., this issue). Mutants of ush display additional thoracic bristles. Lowering the dosage of ush enhances, whereas increasing the dosage suppresses, the phenotype of flies heterozygous for the alleles of pnr-bearing point mutations in the amino-terminal zinc finger. ush encodes a protein containing nine zinc fingers (five C2HC fingers and four C2H2 fingers) clustered in the amino and carboxyl termini of the protein (Cubadda et al., this issue). We show here that Ush and Pnr dimerize and that this interaction is mediated by the amino-terminal zinc finger of Pnr. Presumably because they bear lesions in this motif, association of the mutant forms of Pnr with Ush is severely reduced. When coexpressed with Ush in the transient expression assay, activity of the wild-type, but not the mutant, form of Pnr is strongly antagonized. The consequences of overexpression of Ush and the different forms of Pnr on an ac-lacZ reporter in transgenic flies are consistent with an antagonistic effect of Ush on the activity of Pnr in the regulation of ac–sc expression and bristle development. Thus, we have identified a new cofactor for a GATA homolog that mediates its effects through protein–protein interactions involving the amino-terminal zinc finger, unlike the previously identified Sp1 and EKLF that associate with the carboxy-terminal zinc finger.
Results
Pnr binds to the GATAAG consensus sequence and activates the α-globin promoter in chicken embryonic fibroblasts
To determine a preferential binding sequence for Pnr, a random pool of degenerate oligonucleotides was screened for binding affinity to an immobilized fusion protein between glutathionine S-transferase (GST) and the Pnr DNA-binding domain (GST–Pnr–DBD) on a glutathione–agarose column (see Materials and Methods). The bound oligonucleotides were subcloned into the pBluescript SK+ plasmid after four cycles of selection and PCR amplification. Of the 29 clones sequenced, 27 were found to contain a single GATA motif, and two exhibited a GATA repeat (Fig. 1A). Alignment of the different sequences allowed us to derive the consensus sequence 5′-GATAAG-3′ (Fig. 1B). When the enriched pool was used in an electrophoretic mobility-shift assay (EMSA), two specific complexes were detected (Fig. 1C). The slower migrating one could be either oligonucleotides bearing a repeat of the GATA sequence with a single GST–Pnr–DBD molecule bound to each motif (Fig. 1A) or to a dimer of the GST–Pnr–DBD protein bound to a single GATA sequence. We have shown that Pnr, like GATA-1, is able to homodimerize through its GATA–DBD (Crossley et al 1995; R. Woehl and P. Ramain, unpubl.).
Figure 1.
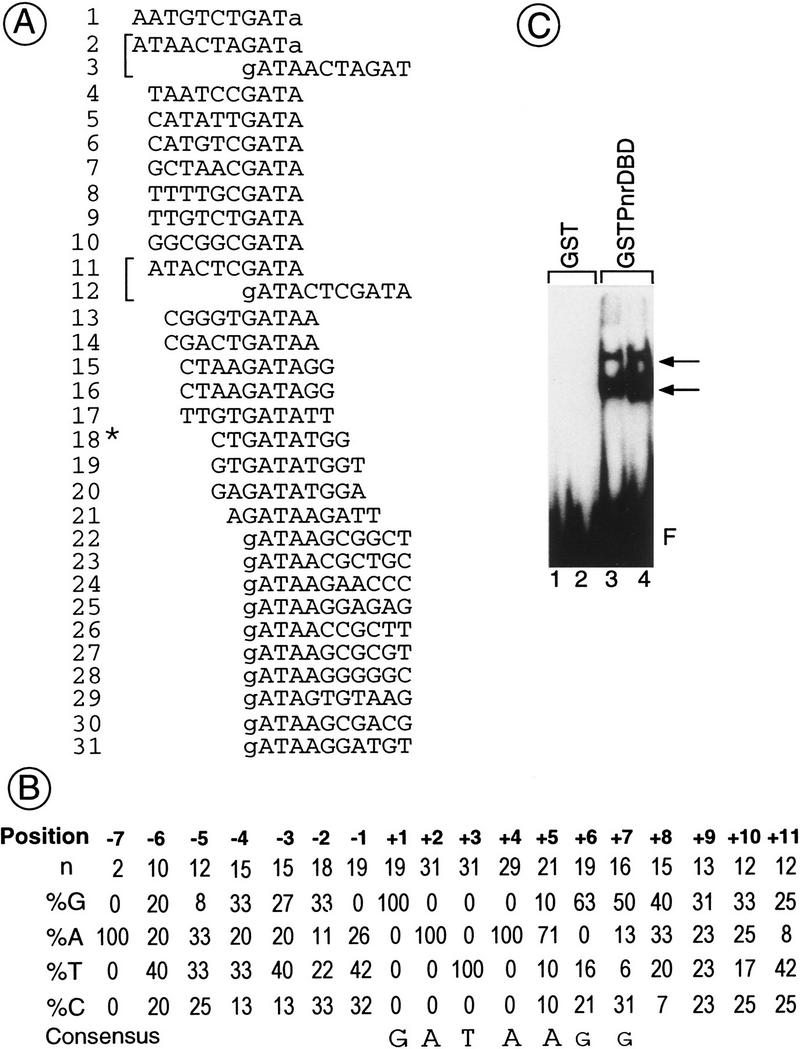
Definition of a consensus-binding site for Pnr. Degenerated oligonucleotides were loaded on a GST column where a fusion protein between the GST and the Pnr–DBD is bound (GST–Pnr–DBD). After elution, the selected oligonucleotides were PCR amplified and loaded again on the column (see Materials and Methods for details). After four cycles of selection and PCR amplification, the oligonucleotides were both subcloned for sequence analysis and used as a template in an electrophoretic mobility-shift assay (EMSA). (A) Alignment of 29 Pnr binding sequences recovered after four rounds of selection. The nucleotides included in the random region are shown in uppercase letters, whereas the bordering G and A nucleotides are in lowercase letters. The sequences are aligned with respect to the GATA motif. Brackets indicate oligonucleotides containing a repeat of the GATA sequence. The asterisk (*) denotes a template containing only nine nucleotides in the random region, which may correspond to either incomplete oligonucleotide synthesis or to PCR artifacts. (B) Consensus-binding site derived from the frequencies of the bases at the sites selected. (C) Autoradiography of an EMSA using as a template the pool of oligonucleotides selected and as a protein extract either purified GST (lanes 1,2: 20 and 200 ng, respectively) or GST–Pnr–DBD (lanes 3,4: 20 and 200 ng, respectively). (F) Unbound oligonucleotides. The arrows indicate two complexes of which the one migrating more slowly may correspond either to a template containing two GATA sequences (A) with one molecule bound to each motif or to a dimer bound to a template containing a single GATA sequence.
The consensus GATA sequence defined for Pnr binding is identical to that present in the α-globin promoter recognized by the chicken GATA-1 protein (cGATA-1). This promoter is active throughout development in erythroid cells and is unlikely to be regulated by stage- and tissue-specific factors. Furthermore, it can be activated by cGATA-1 after transfection in a chicken embryonic fibroblast (CEF) cell line (Evans and Felsenfeld 1991). To investigate transcriptional activity of Pnr we used a transient expression assay in CEF cells with a Pnr expression vector and a reporter in which the chloramphenicol acetyltransferase (CAT) sequences are under the control of the wild-type α-globin promoter. Figure 2B shows that expression of either cGATA-1 or Pnr stimulates activity of the α-globin reporter 35-fold. The effect of Pnr is mediated through a repeat of the GATA motif present in the promoter, as mutation of both GATA sequences abolishes activity (data not shown). Furthermore, experiments using mutated or multimerized GATA sequences from the globin gene upstream of an AdhCAT reporter gene (a minimal promoter from the Drosophila alcohol dehydrogenase gene) show that, like cGATA-1, Pnr binds as a monomer to the proximal GATA motif to stimulate transcription (data not shown). This is consistent with the consensus-binding sequence that we defined (Fig. 1C), since the distal GATA motif contains a G in position −1; therefore, it does not correspond to the target sequence for binding of Pnr.
Figure 2.
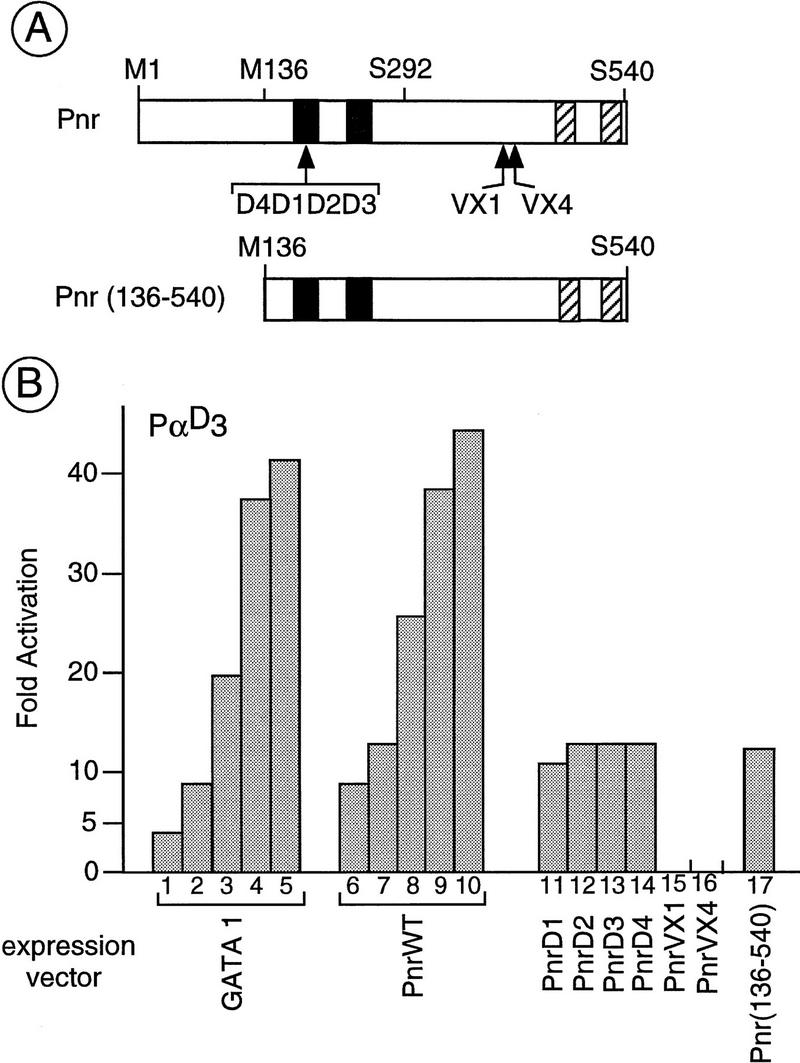
Transactivation of the α-globin promoter sequences by different forms of Pnr in chicken embryonic fibroblasts (CEF). (A) Structural features of the different pnr proteins used in the present study indicating the two zinc fingers (solid boxes) and two sequences organized as putative amphipathic α helices (hatched boxes). The point mutations associated with the pnrD alleles that result in a single amino acid exchange are located in the amino zinc finger, whereas the lesions associated with the pnrVX1/4 alleles and corresponding to a frameshift mutation in the open reading frame are localized in the amino-terminal of the two amphipathic helices. The amino acids are indicated with the single letter code and numbering refers to that given in Ramain et al. (1993). The methionine-136 (M136) is used as an internal start codon in the truncated protein Pnr (136–540) encoded by the pXJ(Sph–Not) expression vector (see Materials and Methods). In contrast, the chimeric proteins (see Fig. 4) contain either the amino acids methionine-1 to serine-292 (chimera Pnr–TEF-1) or the amino acids serine-292 to serine-540 (chimera Gal4–Pnr). (B) Transactivation of the α-globin promoter by the different forms of Pnr. The CEF cells were cotransfected with the α-globin reporter (6 μg of PαD3) and the expression vector for cGATA-1 (lanes 1–5; 25, 50, 100, 250, and 500 ng), wild-type Pnr (PnrWT; lanes 6–10; 25, 50, 100, 250, and 500 ng) and PnrD1, PnrD2, PnrD3, PnrD4, PnrVX1, PnrVX4, and Pnr (136–540), (lanes 11–17: 50 ng each). The level of activation is expressed relative to the reporter alone and as the average (s.d. ± 20%) of three independent experiments performed with two independent DNA preparations.
Previously we have described two mutant forms of the Pnr protein (Ramain et al. 1993). The dominant alleles pnrD1, pnrD2, pnrD3, and pnrD4 (collectively called pnrD) are associated with point mutations resulting in proteins with a single amino acid change in the amino-terminal zinc finger. They are associated with an overexpression of ac–sc and ectopic dorsocentral bristles on the thorax. In contrast, the alleles pnrVX1 and pnrVX4 (collectively called pnrVX1/4) are characterized by a frameshift deletion in the coding sequences deleting the two amphipathic α-helices present in the carboxyl terminus of the protein. They are associated with decreased ac–sc expression and a loss of dorsocentral bristles. The activity of these mutant proteins in the transient expression assay was tested.
The PnrVX1/4 proteins that contain a wild-type DNA-binding domain do not activate the α-globin promoter, suggesting the loss of an activation domain in the carboxyl part of the protein (Fig. 2B, lanes 15,16; see below). In contrast, we found that the PnrD forms of the protein, with a mutated DBD, activate the α-globin promoter as efficiently as the wild type (Fig. 2B, cf. lane 7 and lanes 11–14). This is consistent with in vitro observations showing that they interact with the GATA motif of the α-globin promoter in an EMSA (data not shown). In addition we also found that the amino part of Pnr can be removed without affecting activity (Fig. 2B, cf. lanes 7 and 17; see below).
Ush negatively regulates activation by Pnr but not by PnrD
The ush gene encodes a large zinc finger protein that also affects ac–sc expression and the number of bristles (Cubadda et al., this issue). The phenotype of pnrD, but not pnrVX1/4, heterozygotes is sensitive to the amount of Ush present. The number of ectopic bristles in pnrD/+ mutants increases in flies bearing only a single copy of ush+, but decreases when three copies are present (Cubadda et al., this issue). Activation of the α-globin promoter sequences in CEF cells by Pnr was used as an assay to study the effects of the Ush protein on the function of Pnr. When both Ush and wild-type Pnr are expressed simultaneously by cotransfection, activation of the promoter is abolished. Stimulation by Pnr is lost progressively in a concentration-dependent manner (Fig. 3A, lanes 1–4, 8–11). Similarly, activation by cGATA-1 is also lost after cotransfection with the Ush expression vector (Fig. 3A, lanes 12–14). Because Pnr and cGATA-1 have no homology outside their GATA–DBD, and as Ush alone has no effect on globin promoter activity (data not shown), these observations suggest that the function of Ush is mediated through the GATA–DBD.
Figure 3.
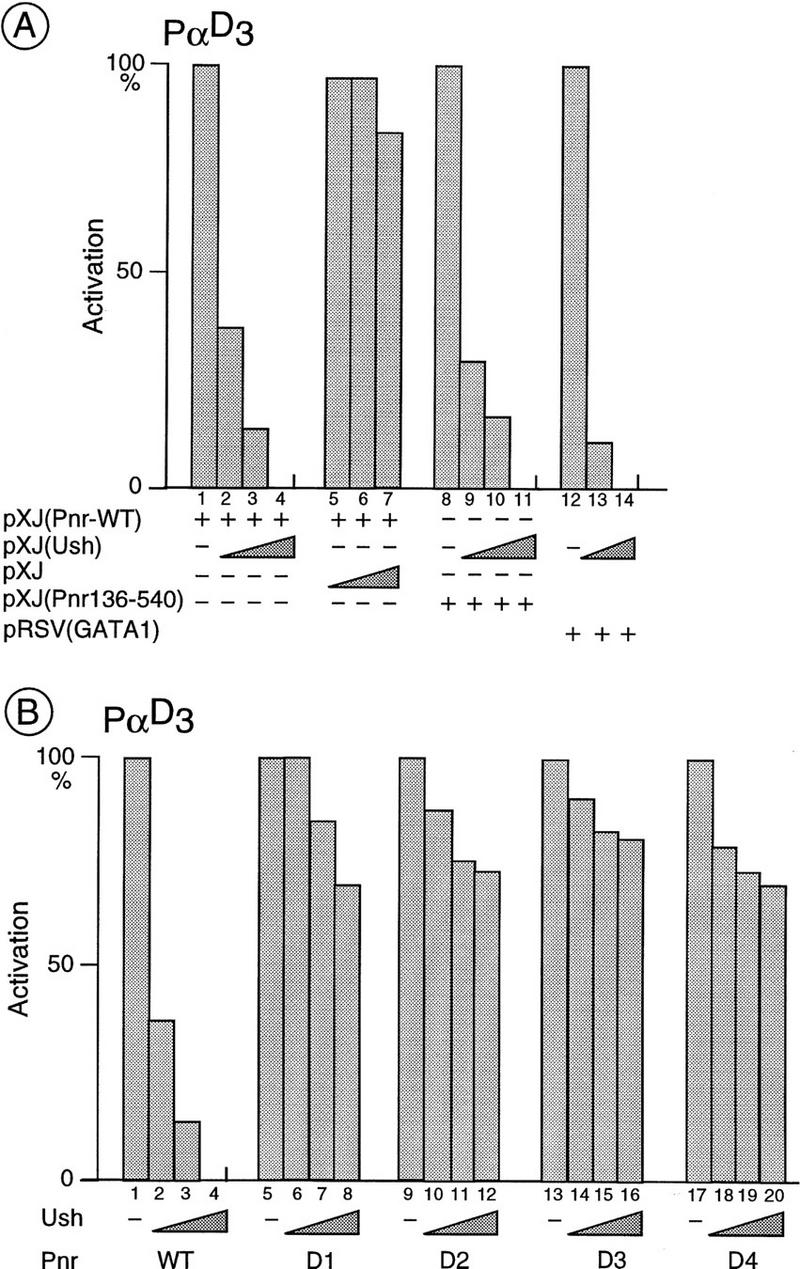
In vivo interactions between Ush and Pnr or cGATA-1 in CEFs. (A) Activation of the α-globin promoter sequences by cGATA-1 and wild-type Pnr is reduced strongly in the presence of Ush. CEF cells were cotransfected with the α-globin reporter (6 μg of PαD3) and an expression vector either for the full-length Pnr (lanes 1–7, 40 ng), or the truncated Pnr (136–540) (lanes 8–11, 40 ng), or cGATA-1 (lanes 12–14, 40 ng) and increasing amounts of an expression vector for Ush (lanes 2, 9, 13: 20 ng; lanes 3, 10, 14: 40 ng; lanes 4, 11: 80 ng). Cotransfection with the empty pXJ expression vector (lanes 5–7: 20, 40, and 80 ng, respectively) is used as a control. The CAT activities are expressed relative to the Pnr and cGATA-1 expression vectors alone, where the CAT activity is fixed arbitrarily at 100%. In each case, they represent the average (s.d. ± 20%) of three independent experiments performed with two independent DNA preparations. (B) Activation of the α-globin promoter sequences by the mutant protein PnrD is affected poorly by Ush. CEF cells are cotransfected with the α-globin reporter (6 μg of PαD3), an expression vector for either the wild-type Pnr or one of the mutated PnrD proteins (40 ng in each case; lanes 1–4: wild-type Pnr; lanes 5–8: PnrD1; lanes 9–12: PnrD2; lanes 13–16: PnrD3; lanes 17–20: PnrD4) and increasing amounts of a Ush expression vector (lanes 2,6,10,14,18: 20 ng; lanes 3,7,11,15,19: 40 ng; lanes 4,8,12,16,20: 80 ng). The activation represents the average (s.d. ± 20%) of three independent experiments performed with two independent DNA preparations and is expressed as a percentage of the full activation (100%) seen with Pnr alone.
To further investigate the interaction between Ush and the GATA–DBD-containing molecules, we used two chimeric proteins. In the first, the activation domain of transcriptional enhancer factor-1 (TEF-1), a Simian virus 40 enhancer-binding factor (Xiao et al. 1991; Hwang et al. 1993), is fused to the Pnr–GATA–DBD, and in the second, the carboxyl terminus of Pnr is fused to the Gal4–DBD (constructs Pnr–TEF-1 and Gal4–Pnr, respectively; Fig. 4). When expressed in CEF, the Pnr–TEF-1 fusion protein stimulates the globin promoter sequences. Cotransfection with a Ush expression vector, however, reduces this activation in a concentration-dependent manner (Fig. 4B, lanes 1–4). The Gal4–Pnr chimeric protein stimulates activity of the 17m5-TATA–CAT reporter, in which the CAT gene is under the control of five Gal4-binding sites, indicating the presence of an activating function in the carboxyl terminus. However, stimulation is not affected by cotransfection of the Ush expression vector (Fig. 4B, lanes 9–12). Therefore, Ush affects specifically stimulation by the proteins containing a GATA–DBD.
Figure 4.
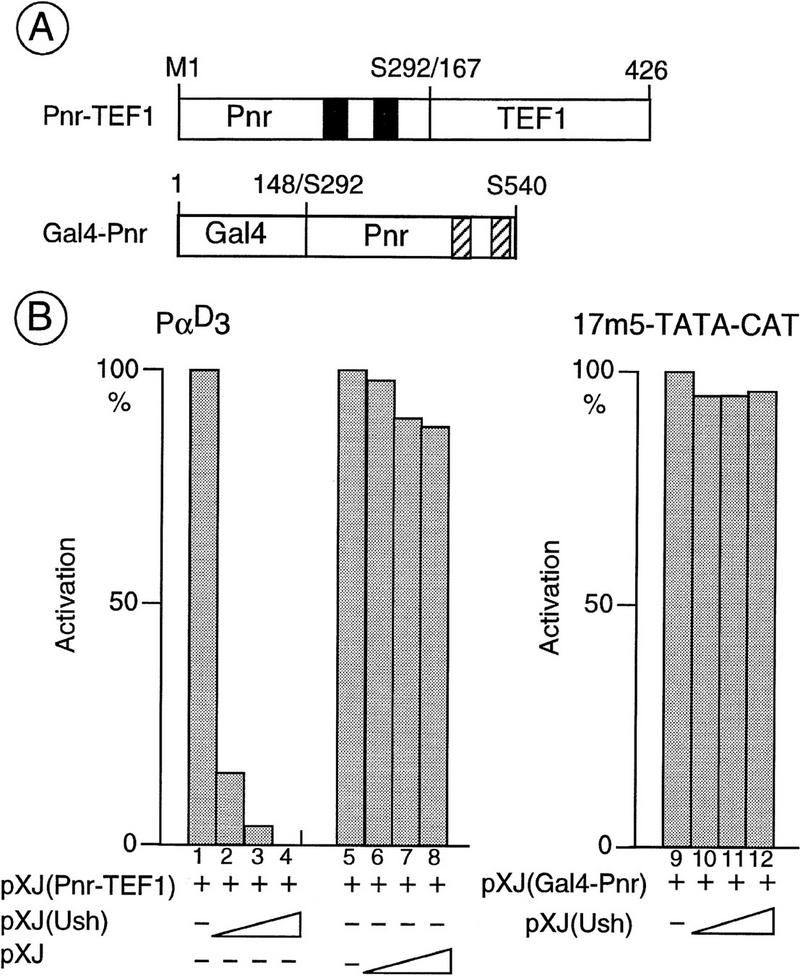
In vivo interactions between Ush and Pnr–DBD containing chimeric proteins. (A) Structural features of the chimeras used in the present study. The Pnr–TEF-1 chimera results from a fusion of the amino terminus of Pnr (M1–S292) including the DNA-binding zinc fingers (solid boxes), and the amino acids 167–426 of the SV40-enhancer binding protein TEF-1 that bears an activation function. The Gal4–Pnr chimera results from a fusion of the Gal4–DNA-binding domain (amino acids 1–148) and the carboxyl terminus of Pnr (S292–S540) containing the two α helices (hatched boxes). (B) Specific inhibition by Ush of the transcriptional activation induced by the Pnr–DBD-containing chimera. CEFs were cotransfected with either the α-globin reporter (6 μg of PαD3), or the upstream activating sequence (UAS) reporter (1 μg of 17m5–TATA–CAT), an expression vector encoding either the Pnr–TEF-1 (lanes 1–8: 40ng), or the Gal4–Pnr chimera (lanes 9–12: 40ng), and increasing amounts of a Ush expression vector (lanes 2,10: 20ng; lanes 3,11: 40ng; lanes 4,12: 80ng). Cotransfection of the empty pXJ expression vector (lanes 6–8: 20, 40, and 80 ng, respectively) is used as a control. The activation represents the average (s.d. ± 20%) of three independent experiments performed with two independent DNA preparations and is expressed as a percentage of the full activation (100%) seen with the chimera alone.
Despite bearing amino acid substitutions in the amino terminal zinc finger, the PnrD proteins stimulate the globin promoter as efficiently as wild-type Pnr (see Fig. 2B). Because it was shown earlier that the phenotype of the pnrD heterozygotes is sensitive to the amount of ush+ product, we looked for a possible interaction between Ush and these proteins. In contrast to the wild type, whose activity is reduced strongly by the presence of Ush, the PnrD proteins are poorly sensitive in this assay; stimulation by PnrD is not notably reduced when cotransfected with Ush (see Fig. 3B, cf. lanes 1–4 with lanes 5–8, 9–12, 13–16, and 17–20). Therefore, it is likely that the amino acids that have been mutated in PnrD are required for a molecular interaction between Pnr and Ush.
Pnr heterodimerizes with Ush through its GATA–DBD
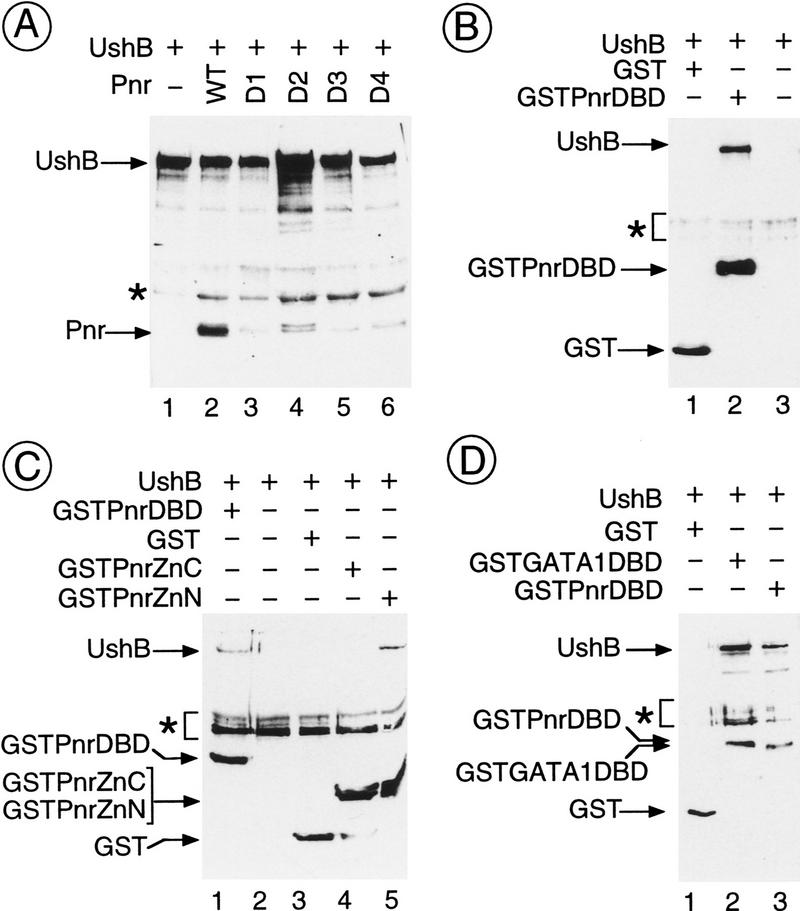
As Ush can antagonize transcriptional activation by Pnr, we then addressed the question as to whether there is a physical association between these two proteins. Protein extracts were made from Cos cells cotransfected with expression vectors for Pnr and a tagged Ush (see Materials and Methods). The two wild-type proteins coimmunoprecipitate and could be detected on Western blots with appropriate antibodies (Fig. 5A). In contrast, the PnrD proteins with mutated GATA–DBDs are only weakly associated with Ush in similar experiments (Fig. 5A, cf. lanes 3–6 with lane 2). This suggests that the association of the two proteins requires the amino-terminal zinc finger.
Figure 5.
Direct in vitro protein–protein interactions between Ush and Pnr. Cos cells were cotransfected with 5 μg of each expression vector, as denoted above each panel, and the corresponding protein extracts were either immunoprecipitated (A) with the B10 monoclonal antibody raised against the B epitope of the estrogen receptor inserted into the Ush open reading frame or incubated with glutathione–agarose (B–D). Selected proteins were analyzed by Western blot. Ush–B is detected with the B10 antibody, Pnr and PnrD with the specific monoclonal antibody 2B8, and the GST fusion proteins with the GST-specific monoclonal antibody 1D10. (A) In vitro interactions between Ush and wild-type Pnr as well as PnrD. Arrows indicate the Ush–B (lanes 1–6) and Pnr (lanes 2–6: wild-type Pnr, PnrD1, PnrD2, PnrD3 and PnrD4, respectively). The asterisk (*) denotes the location of the immunoglobulin heavy chain. (B) In vitro interactions between Ush and the isolated Pnr–DBD. Arrows indicate Ush–B (lane 2), GST (lane 1), and the fusion GST–Pnr–DBD (lane 2). The asterisk denotes artifactual bands. (C) In vitro interactions between Ush and Pnr isolated zinc fingers. Arrows indicate Ush–B (lanes 1,5), GST alone (lane 3), the fusion GST–Pnr–DBD (lane 1), and the fusion proteins GST–Pnr–ZnC (lane 4) and GST–Pnr–ZnN (lane 5). The asterisk denotes artifactual bands. (D) In vitro interactions between Ush and the cGATA–1–DBD. Arrows indicate Ush–B (lanes 2,3), GST alone (lane 1), and the fusion proteins GST–Pnr–DBD (lane 2) and GST-GATA-1–DBD (lane 3). The asterisk denotes artifactual bands.
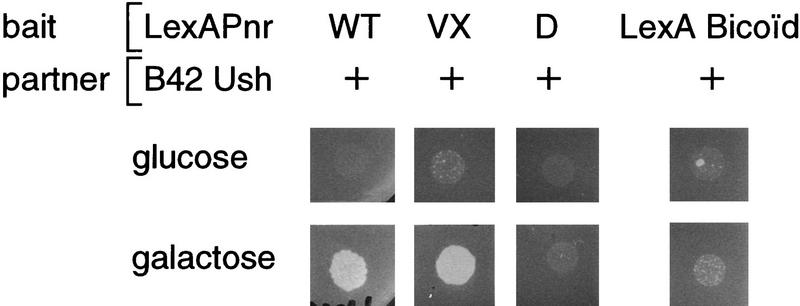
The interaction between Ush and Pnr was also tested in yeast. In these experiments physical association between a LexA–Pnr fusion protein and Ush fused to the B42 activation domain was detected by activation of a Leu2 reporter gene that contains upstream LexA-binding sites (Gyuris et al. 1993; Finley and Brent 1994; the wild type as well as the different mutated versions of Pnr do not activate transcription in yeast). As shown in Figure 6, Ush interacts with the LexA–Pnr+, LexA–PnrVX1, and LexA–PnrVX4 fusion proteins, which all carry a wild-type DBD. In contrast, this interaction is lost with fusion proteins constructed with the DBD carrying the point mutations characteristic of the pnrD class of dominant alleles (LexA–PnrD1 to LexA–PnrD4). The specificity of the interaction is further underlined by the use of the unrelated LexA–Bicoïd fusion protein (Fig. 6).
Figure 6.
In vivo interactions between Ush and Pnr in yeast. Yeast transformed by an expression vector encoding the bait LexA–Pnr (the different forms of the protein were wild-type Pnr, PnrVX1, or PnrD1) and an expression vector encoding the partner B42–Ush were plated on a medium lacking histidine, trytophane, and leucine, with either glucose, where the expression of B42–Ush is repressed, or with galactose, where expression of the partner is induced. The results were the same for LexA–PnrVX4, LexA–PnrD2, LexA–PnrD3, and LexA–PnrD4 (data not shown). The specificity of the interaction is further demonstrated by the use of the LexA–Bicoid unrelated fusion protein. Levels of LexA–Pnr fusion proteins in the different strains were monitored by Western blot analysis using the Pnr-specific monoclonal antibody 2B8 (data not shown).
To ascertain that Ush associates specifically with the DBD of Pnr, we performed GST fusion protein selection experiments with protein extracts from Cos cells cotransfected with vectors for the tagged Ush protein and a fusion between GST and the complete Pnr–DBD by itself (GST–Pnr–DBD; see Materials and Methods). As shown in Figure 5, Ush associates specifically with the GST–Pnr–DBD fusion protein but not with GST alone. Therefore, it interacts with the isolated Pnr–DBD. Furthermore, Ush also associates with a fusion protein between GST and the highly related DBD from the cGATA-1 transcription factor (GST–GATA-1–DBD; Fig. 5D, lane 3). Over a stretch of 115 amino acids in Pnr and 116 in cGATA-1, 85 are identical (74% identity).
To define further the domain of Pnr required for association with Ush, we expressed fusion proteins bearing either the amino or the carboxy-terminal zinc finger sequences fused to GST (fusion proteins GST–Pnr–ZnN and GST–Pnr–ZnC, respectively; Fig. 5C). Ush does not bind the GST–Pnr–ZnC fusion protein but does associate with the amino-terminal zinc finger fusion protein as efficiently as with the complete DBD (Fig. 5C, lanes 1,4,5). It is noteworthy that the PnrD proteins are all characterized by point mutations in the amino-terminal zinc finger and that the amino acids that are mutated are conserved perfectly within the GATA family of transcription factors.
Overexpression of wild-type and mutant pnr proteins in transgenic flies regulates ac–sc expression through the dorsocentral enhancer
We have shown that wild-type Pnr, as well as the pnrD proteins that bear lesions in the amino-terminal zinc finger, stimulate transcription from the heterologous α-globin promoter. In contrast the PnrVX1/4 proteins, which carry deletions of the amphipathic α-helices in the carboxyl region of Pnr, do not activate in this assay. Heterozygous flies mutant for pnrD differentiate extra dorsocentral bristles resulting from overexpression of ac–sc, whereas heterozygous flies mutant for pnrVX1/4 differentiate fewer dorsocentral bristles attributable to decreased ac–sc expression (Ramain et al. 1993). We have now analyzed the effects of overexpression of these three pnr proteins on the development of the dorsocentral bristles and on the activity of a lacZ reporter gene whose expression is under the control of specific enhancer sequences that drive ac–sc expression very strongly at the dorsocentral site (Gomez-Skarmeta et al. 1995). We made use of the GAL4/UAS system (Brand and Perrimon 1993), using as a driver the pnrMD237 strain that carries a GAL4-containing transposon inserted at the pnr locus. This gives an expression pattern indistinguishable from that of pnr and does not display a mutant bristle pattern on the thorax (Calleja et al. 1996; Heitzler et al. 1996).
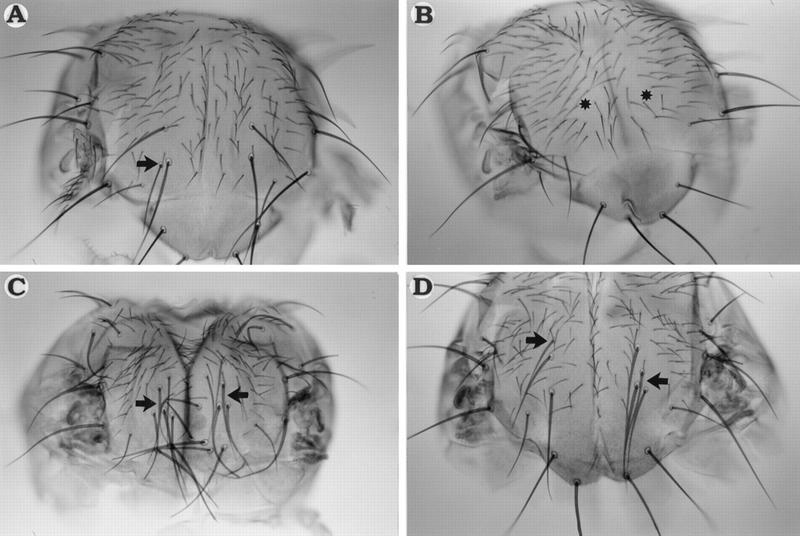
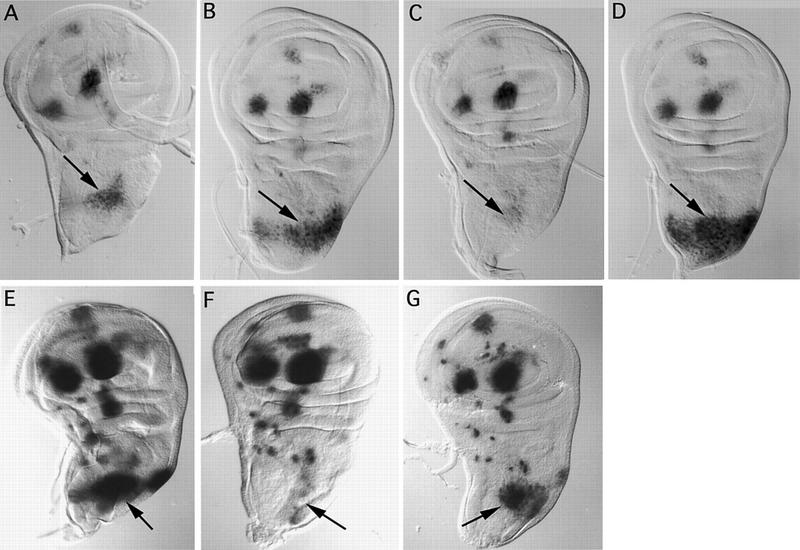
Overexpression of either the wild-type or the PnrD4 protein leads to an excess of dorsocentral bristles (Table 1; Fig. 7, A and C, respectively). The wild-type protein causes a modest effect of one or two additional bristles per hemithorax in 15%–25% of the transgenic flies. PnrD4, however, causes a dramatic increase of 4–10 additional bristles per hemithorax in all of the flies, and furthermore, viability of these animals is strongly reduced (Table 1). In contrast, overexpression of PnrVX4 leads to a loss of dorsocentral bristles (Table 1; Fig. 7B). These observations correlate perfectly with ac–lacZ reporter gene expression at the dorsocentral site (Gomez-Skarmeta et al. 1995; Fig. 8A). Staining is weakly reinforced by overexpression of Pnr (Fig. 8B), but becomes very intense after overexpression of PnrD4 and completely covers the dorsal-most region of the thoracic disc delimited by the prescutum, the postnotum, and the dorsocentral area (Fig. 8D). This is consistent with other observations showing that in pnrD4 mutants expression of this reporter is also increased (data not shown). This corresponds to the domain where pnr and ush are expressed simultaneously in the wild type. In contrast, overexpression of PnrVX4 leads to a strong reduction of lacZ staining in the dorsocentral area (Fig. 8C). Thus, there is a correlation between the effects of the three protein forms on ac–sc expression and bristle development and their transcriptional activity on the globin promoter in the transfection experiments.
Table 1.
Number of dorsocentral bristles present in flies of different genotypes following overexpression of Pnr, PnrD4, PnrVX4 or Ush
| Construct
|
Genetic background
|
Percent mutant heminota
|
Average no. dorsocentral bristles/heminota
|
No.
|
|---|---|---|---|---|
| None | pnrMD237/+ | 0 | 2.0 | 50 |
| UAS:PnrWTa | pnrMD237/+ | 19 | 2.2 ± 0.01 | 618 |
| UAS:PnrWT | ush−/+; pnrMD237/+ | 83 | 3.48 ± 0.07 | 178 |
| None | ush−/+; pnrMD237/+ | 0 | 2.0 | 86 |
| UAS: PnrVX4a | pnrMD237/+ | 99 | 0.29 ± 0.01 | 618 |
| UAS: PnrD4a | pnrMD237/+ | 100 | 7.35 ± 0.2 | 56b |
| UAS: Ush14AII | pnrMD237/+ | 100 | 0 | 8b |
| UAS: Ush14AII | pnrD1/pnrMD237 | 100 | 3.3 ± 0.6 | 22 |
| None | pnrD1/pnrMD237 | 100 | 7.6 ± 0.6 | 25 |
| None | pnrD1/+ | 100 | 2.7 ± 0.16 | 26 |
Each transgenic strain carried one transposon.
Averages of three independent insertion lines are given.
These animals showed strongly reduced viability.
Figure 7.
Effect of overexpression of the different pnr proteins on development of the dorsocentral bristles. Homozygous transgenic strains carrying a UAS–Pnr transposon were crossed with pnrMD237/TM6b; Tb flies and the Tb+ flies carrying the pnrMD237 driver were analyzed. Arrows denote additional dorsocentral bristles in A, C, and D, whereas asterisks (*) indicate the positions of the missing bristles in B. (A) Strain UAS–Pnr+. (B) Strain UAS–PnrVX4. (C) The transgenic strain UAS–PnrD4. (D) Strain UAS–Pnr+ (same as in (A) in the presence of a reduced amount of Ush [Df(3R)ushRev18, ush−/+].
Figure 8.
The effects of overexpression of the different forms of pnr proteins and ush on the lacZ reporter whose expression is driven by the dorsocentral-specific enhancer sequences (Gomez-Skarmeta et al. 1995). Arrows denote lacZ expression in the dorsocentral region of the thoracic disc. (A–D) Homozygous transgenic flies carrying a UAS–Pnr transposon were crossed with DC–enhancer–sc–lacZ; pnrMD237/TM6B Tb flies, and the resulting larvae of the appropriate genotype (see below) were dissected and the imaginal discs stained for β-galactosidase activity. In each case, the reaction was left for 3 hr at 22°C; longer reaction times led to background staining, which prevents comparison between the different pnr proteins. (E–G) Overexpression of ush leads to the reduction of ac–sc expression in pnrMD237/+ (F) but not pnrD/pnrMD237 mutants (G). In E–G the reaction was left for 6 hr at 37°C to emphasize the fact that overexpression of Ush leads to complete repression of the lacZ reporter.(A) Genotype (UAS-PnrWT/lacZ; TM6B, Tb/+); (B) genotype (UAS–PnrWT/lacZ; pnrMD237/+); (C) genotype (UAS–PnrVX4/lacZ; pnrMD237/+); (D) genotype (UAS–PnrD4/lacZ; pnrMD237/+); (E) genotype (UAS–Ush/lacZ; TM6B, Tb/+); (F) genotype (UAS–Ush/lacZ; pnrMD237/TM6B, Tb); (G) genotype (UAS–Ush/lacZ; pnrMD237/pnrD1).
Overexpression of the ush protein in transgenic flies reduces ac–sc expression in the wild-type but not in the pnrD mutants
Overexpression of Pnr in a genetic background associated with a reduced amount of the Ush product [Df(3R)ushRev18, ush−/+] leads to an increase in the frequency of phenotypically mutant flies, as well as an increase in the number of extra dorsocentral bristles (Table 1; Fig. 7D). Because Ush regulates Pnr negatively in the assay using the globin promoter, this suggests an antagonistic role of Ush on Pnr activity during bristle patterning. To verify this we looked at the effects of overexpression of Ush in flies expressing the mutant pnr protein PnrD, which displays in vitro a weaker association with Ush. Whereas in wild-type flies overexpression of Ush results in a loss of ac–sc reporter gene expression (Fig. 8F), in pnrD heterozygotes it has a milder effect and staining still remains fairly strong (Fig. 8G). This suggests that in the thoracic epithelium Ush antagonizes the effects of Pnr, leading to ac–sc expression and bristle development.
Discussion
Pnr binds to the consensus GATAAG sequence and activates transcription
The DBDs of members of the GATA family of transcription factors are highly related and here we demonstrate that Pnr is a transcription factor, it binds DNA, and can activate transcription of the GATA-1 target promoter of the α-globin gene in transfected CEFs. A binding site enrichment protocol has been used to define a consensus for the Pnr DNA-binding domain sequence and the optimal site appears to be a GATAAG motif. This is similar to the consensus proposed by Plumb et al. (1989) and close to the WGATAR sequence derived previously by an in vivo analysis (Yamamoto et al. 1990). Most GATA factors, like Pnr, carry two zinc fingers. In an extensive in vitro study, Whyatt et al. (1993) described two different classes of DNA-binding sites for GATA-1, the first requiring only the carboxy-terminal zinc finger to bind the motif GAT(A/T), the second requiring both zinc finger motifs to bind the (T/C)AAG sequence. Thus, it is possible that the consensus sequence that we have selected, GATAAG, may correspond to two overlapping recognition sequences where GATA is recognized by the carboxy-terminal zinc finger and TAAG by the complete DBD.
Our results suggest that in the case of Pnr, the amino-terminal zinc finger is not required for binding to the consensus sequence present in the globin promoter, or indeed activation, as the mutant PnrD proteins bearing lesions in the amino-terminal zinc finger are capable of transcriptional activation in a transient expression assay. It is noteworthy that, unlike the vertebrate proteins, the yeast GATA proteins bear only a single zinc finger, with a similar sequence to the carboxy-terminal zinc finger of vertebrate GATA factors. This suggests that, in the case of Pnr and the vertebrate proteins, the carboxy-terminal zinc finger is the crucial determinant for DNA binding. Indeed deletions within this region, or point mutations of the conserved cysteine residues, abolish binding of the protein. Furthermore, binding of the vertebrate proteins to the GAT(A/T) sequence correlates with transcriptional activation (deBoer et al. 1988; Mignotte et al. 1989a,b; Nicolis et al. 1991; Tsai et al. 1991; Simon et al. 1992; Philipsen et al. 1993) or with indirect transcriptional activity through the displacement of a repressor (Rahuel et al. 1992).
The amino-terminal zinc finger of GATA-1 does not bind DNA by itself, and may be involved in the discrimination between different GATA sites present in the control regions of erythrocyte genes (Yang and Evans 1992). There is recent evidence, however, that the amino-terminal zinc finger of GATA-2 and GATA-3 can bind a GATC sequence (Pedone et al. 1997). Binding is dependent on the presence of two basic regions on either side of the finger. One of these regions, the amino-terminal, is missing in GATA-1, which is unable to bind in the assay used by these investigators. This domain is also absent in Pnr.
Ush associates with the amino-terminal zinc finger of Pnr and regulates transcriptional activity negatively
The DNA-binding domain of GATA factors is also involved in dimerization processes. Thus, the GATA proteins homodimerize by means of either the amino- or carboxy-terminal zinc fingers (Crossley et al. 1995). GATA-1 has also been shown to heterodimerize with the transcription factors Sp1 and EKLF, which belong to the Krüppel family and carry C2H2 zinc finger motifs (Miller and Bieker 1993; Merika and Orkin 1995). This association requires the GATA carboxy-terminal zinc finger and the interaction leads to synergistic transcriptional activity. In contrast, our study illustrates a different mode of regulation of GATA function by means of heterodimerization through the amino-terminal zinc finger. We have shown that the function of Pnr is regulated by association with Ush, a large protein bearing both C2HC and C2H2 zinc fingers (Cubadda et al., this issue). The physical association requires a wild-type GATA–DBD. Indeed, when the DBD carries a single point mutation in the amino-terminal zinc finger, as in the PnrD proteins, association of the two proteins is reduced drastically. This suggests that the amino acids that have been lost either interact directly with Ush or are crucial determinants for the highly ordered structure of the zinc finger required for heterodimerization. All of the mutations involve amino acids that are conserved completely among the members of the GATA family, and furthermore pnrD1 and pnrD3 have substitutions of the conserved cysteine residues that probably maintain the structural integrity of the zinc finger (Ramain et al. 1993). Thus, heterodimerization involving the amino-terminal zinc finger of a GATA factor uncovers a novel mode of regulation of GATA function during development.
In the transient assay that we have used, Ush reduces the transcriptional activity of Pnr by heterodimerization. To determine whether the DNA-binding properties of Pnr are disrupted, we performed EMSA with protein extracts made from transfected Cos cells and we were unable to detect either ternary complex on the globin probe or reduced binding of Pnr in presence of Ush (data not shown). Further studies are required to resolve this point. Ush also interacts with cGATA-1, raising the possibility of the existence of vertebrate proteins homologous to Ush. Indeed, recently a zinc finger protein, FOG (friend of GATA-1), structurally similar but not homologous to Ush, has been recovered from a yeast two-hybrid screen using the vertebrate GATA-1 as bait (Tsang et al. 1997). Like Ush, this protein heterodimerizes with GATA-1 through the amino-terminal zinc finger. In contrast to Ush, however, FOG has a synergistic effect on the activation of transcription by GATA-1. These observations suggest the possible existence of a family of similar proteins that modulate the transcriptional activity of the different GATA factors.
Antagonistic activities of Pnr and Ush during bristle patterning
Mutants of pnr affect the level of ac–sc expression in the imaginal discs of the fly and alter the bristle pattern. The pnrD heterozygotes are associated with an over-expression of ac–sc in the dorsocentral area and the formation of additional bristles at this site (Ramain et al. 1993). On the other hand, pnrVX1/4 heterozygotes are associated with a loss of ac–sc expression and a loss of dorsocentral bristles. In an earlier study we interpreted the pnrD mutants as representing a loss of pnr function, thinking that the lesions in the amino-terminal zinc finger would abolish activity of the DBD. In this study, however, we have shown that the mutated PnrD proteins activate transcription as efficiently as the wild type in a transient expression assay involving the heterologous α-globin promoter. In contrast, the PnrVX1/4 proteins are unable to stimulate this promoter. Therefore, it is possible that in vivo Pnr acts as a transcriptional activator of ac and sc. Overexpression of Pnr or PnrD using the GAL4–UAS system also leads to the formation of additional bristles. We have shown that the effects of Pnr appear to be mediated by the specific enhancer sequences required for expression of ac–sc at the dorsocentral site (Gomez-Skarmeta et al. 1995). It remains to be seen whether Pnr acts directly on these sequences or indirectly through intermediate genes.
The direct association of the Pnr and Ush proteins that we describe here could be the molecular basis underlying the similar phenotypes of pnrD gain-of-function mutants and ush loss-of-function mutants, as well as the genetic interactions observed between mutant alleles of these two genes. Ush behaves genetically as a repressor of ac and sc; hypomorphic alleles accumulate higher levels of ac–sc and form additional bristles, whereas overexpression of Ush leads to a loss of bristles (Cubadda et al., this issue). PnrD heterodimerizes only poorly with Ush, which would mean that in vivo it would stimulate transcription more strongly and over a broader territory. Overexpression of PnrD4 causes a dramatic increase in the levels of ac–sc–lacZ, as does over-expression of wild-type Pnr in animals heterozygous for ush. The phenotype of heterozygous pnrD/+ flies is enhanced when the dosage of ush+ is reduced and suppressed when it is increased (Heitzler 1993; Cubadda et al., this issue). Other alleles do not display phenotypic changes in the presence of varying amounts of Ush.
One explanation for the specificity of this interaction could be attributable to the fact that the two genes are expressed in overlapping, but not precisely coincident, areas of the thorax; the pnr domain is slightly more extensive than that of ush (Ramain et al. 1993; Cubadda et al., this issue). Furthermore, the dorsocentral bristle precursors arise in a domain of high pnr expression but low ush expression (Cubadda et al., this issue). Down-regulation of Pnr by Ush is restricted presumably to the domain in which both genes are expressed. The PnrD protein would be more or less resistant to Ush, therefore, in pnrD flies Ac–Sc levels would increase and cause the appearance of ectopic bristles in the ush expression domain in addition to the ones that develop in the normal (Pnr+ Ush−) territory. On the other hand, the phenotype of alleles that cause a loss of pnr expression or function, such as pnrVX1/4, would be attributable to a loss of Pnr activity in the nonoverlapping domain of high pnr but low ush expression. Pnr in this area would not be regulated by Ush anyway.
Consistent with these observations, overexpression of Ush, which in wild-type flies leads to a loss of ac–sc expression, has only a mild effect on the levels of ac–sc–lacZ in pnrD mutant flies. Thus, Pnr and Ush appear to have antagonistic effects on the expression of ac–sc. Thus, it is possible that ac–sc levels are higher in areas of lower ush expression and that Pnr and Ush contribute to the precise positioning of dorsocentral and perhaps other bristles.
Materials and methods
All recombinant DNA work was performed according to standard procedures (Sambrook et al. 1989). Details concerning plasmid constructions, which were all verified by sequence analysis performed with an Applied Biosystems automated DNA sequencer, are available upon request. Nucleotides and amino acids are numbered with reference to our previous report (Ramain et al. 1993).
Plasmid constructions
A 2900-bp HindIII–EcoRI DNA fragment containing the complete wild-type open reading frame of Pnr and isolated from our original PNB40 clone (Ramain et al. 1993) was subcloned into pBluescript SK+ to give SK+Pnr+. The point mutations corresponding to the different PnrD proteins (D1 to D4), as well as the deletions corresponding to the PnrVX proteins (VX1 and VX4) were introduced by site-directed mutagenesis using the SK+Pnr+ single-strand DNA as a template and the following oligonucleotides: D1, 5′-AGGGACGCGAGTACGTCAATTGCGG-3′; D2, 5′-GGGATGGAACCGAACACTATCTGTG-3′; D3, 5′-GGACACTATCTGAGCAACGCCTGCG-3′; D4, 5′-GGCGAGGGACGCAAGTGCGTCAATT-3′; VXI, 5′-CCTTCACGGAGCTCTACACGCCCG-3′; VX4, 5′-CAGGAACGCAGCAGTTCCGGCGGA-3′.
For transfection experiments, the different HindIII–NotI fragments of the pBluescript SK+ derivatives were inserted into the pXJ vector (Xiao et al. 1991), whose expression is driven in transfected cells by the cytomegalovirus promoter sequences. Using M136 as an internal start codon, the truncated Pnr protein is produced with the expression vector pXJ containing the SphI(870)–NotI DNA fragment. A molecular mass of 45 kD, as expected, was found by Western blot analysis with the help of the monoclonal antibody 2B8, produced in mouse and raised against the peptide TTQQQHQQHGHSMTSSSGQA (amino acids 378–397). The expression vector carrying the Pnr–TEF-1 chimera was constructed by inserting the HindIII–BamHI DNA fragment containing the Pnr–DBD from SK+Pnr+ and the BamHI fragment from pXJ (Gal4–TEF-1; Xiao et al. 1991) containing the TEF-1 activation domain (amino acids 167–426) into pXJ. Similarly, the expression vector carrying the Gal4–Pnr chimera was constructed by exchanging the BamHI fragment from pXJ(Gal4–TEF-1) containing the TEF-1 activation domain with the BamHI fragment containing the carboxyl terminus of Pnr isolated from SK+Pnr+.
The expression vectors used in the yeast interaction assays were constructed in the plasmid pEG202, which encodes the bait fused to the LexA–DBD, and in the plasmid pJG4-5, where the putative partner is fused to the bacterial B42 activation domain that is efficient in yeast. A repaired NdeI–NotI fragment containing the complete Pnr open reading frame was isolated from a pBluescript SK+Pnr+ derivative where a NdeI restriction site was created on the ATG by site-directed mutagenesis using the oligonucleotide (5′-CGGCCATAAATCCATATGGGCATCTTACTG-3′; NdeI, underlined) and inserted into the repaired EcoRI–NotI cloning sites from pEG202 to give pEG202Pnr+. The pEG202PnrD1 to pEG20PnrD4 expression vectors were constructed by exchange of the wild-type SalI fragment from pEG202Pnr+ with the mutated SalI fragments from pXJPnrD1 to pXJPnrD4. The pEG202PnrVX1 and pEG202PnrVX4 expression vectors were constructed by exchange of the BamHI–NotI fragment from pEG202Pnr+ with the BamHI–NotI fragments from SK+PnrVX1/VX4. pJG4-5 was modified by insertion into the EcoRI and XhoI cloning sites of the complementary oligonucleotides (5′-AATTCGCTAGCTAAC-3′and 5′-TCGAGTTAGCTAGCG-3′) creating an XbaI cloning site (underlined). A 4-kb XbaI fragment isolated from SK+Ush and encompassing the complete open reading frame was then inserted into the XbaI site of the pJG4-5 derivative.
The selection of GST fusion proteins was performed with extracts made from Cos cells transfected with pBC derivatives (Stratagene, La Jolla, CA; Chatton et al. 1995) allowing the expression of GST fusion proteins in a cultured cell line. Thus, the complete Pnr–DBD as well as the isolated zinc fingers were generated by site-directed mutagenesis using as a template the SK+Pnr+ single-stranded DNA and the following oligonucleotides carrying an NheI restriction site (underlined): 5′-GGAAGGATTCGCTAGCGCGCATGCAC-3′ and 5′-CACTGGATCCGCTAGCGGCTCCACTT-3′ (Pnr–DBD); 5′-GGAAGGATTCGCTAGCGCGCATGCAC-3′ and 5′-TGTGAGTGCAGCTAGCACTGCCACCC-3′ (Pnr–ZnN); 5′-TGTGAGTGCAGCTAGC ACTGCCACCC-3′ and 5′-CACTGGATCCGCTAGCGGCTCCACTT-3′ (Pnr–ZnC).
The SK+ derivatives were then restricted by NheI and the DNA fragments encompassing the complete DBD as well as the isolated Pnr zinc fingers were inserted into the NheI cloning site of pBC to give pBC–Pnr–DBD, pBC–Pnr–ZnN, and pBC–Pnr–ZnC, allowing the production of GST fusion proteins containing the amino acids alanine 124–serine 292 (GST–Pnr–DBD), alanine 124–alanine 216 (GST–Pnr–ZnN), and threonine 217–serine 292 (GST–Pnr–ZnC), respectively. The pBC–GATA-1–DBD expression vector was generated by insertion of a 421-bp SmaI fragment containing the cGATA-1–DBD sequences (glycine 85–proline 224) and isolated from pRSV20.2 (Evans and Felsenfeld 1991) into the NdeI repaired cloning site of pBC.
PCR-assisted DNA-binding site selection
The NheI fragment containing the Pnr–DBD sequences was inserted into the pGEX-3X vector, allowing the production of GST fusion proteins in Escherichia coli. The resulting plasmid was introduced by transformation into BL21 (DE3) lysS competent cells. Five hundred milliliters of Luria broth plus 100 μg/ml of ampicillin were innoculated with a 10-ml saturated overnight culture grown at 30°C and incubated at the same temperature until it reached an OD of 0.6–0.7 at 600 nm. Isopropyl-β-thio-galactopyranoside (IPTG) was added to a final concentration of 1 mm and the culture was further incubated for 2 hr. Cells were harvested by centrifugation, washed in cold PBS, and lysed by sonication in 1× PBS with 1% Triton X-100. The bacteria debris was eliminated by centrifugation and the supernatant applied to a glutathione–agarose (Pharmacia) column equilibrated with 1× PBS, 1% Triton X-100. After extensive, successive washes with 1× PBS, 1× PBS, 1 m NaCl, 1% Triton X-100, and the binding buffer [50 mm Tris-HCl (pH 7.8), 50 mm KCl, 10% glycerol, 1 mm DTT, 100 μm ZnSO4, 100 μg/ml poly[d(I-C)], the labeled degenerated oligonucleotides were loaded on the column. The random sequence oligonucleotide: [5′-CTGGATCCTAGATGTCCCTG(N)10AGGCTCAAAGCTGAATTCCT-3′] was rendered double stranded with the Klenow polymerase by primed synthesis using the primer (PL192: 5′-AGGAATTCAGCTTTGAGCCT-3′) labeled with [γ-32P]ATP. After purification on an acrylamide gel, electroelution, and precipitation, the probe was resuspended in binding buffer. After extensive washes with the binding buffer plus 1% Triton X-100, the selected oligonucleotides were eluted stepwise with the binding buffer containing increasing concentrations of KCl, and the oligonucleotides eluted at a 500-mm KCl concentration were precipitated, recovered by centrifugation, and resuspended in water before PCR amplification with the PL192 and QK61(5′-CTGGATCCTAGATGTCCCTG-3′) primers labeled with [γ-32P]ATP. The amplified product, purified on an acrylamide gel, was applied again to the column and after four cycles of enrichment the selected oligonucleotides were restricted by BamHI and EcoRI and subcloned into pBluescript SK+ for further sequence analysis. They were also used as a template in an EMSA. The GST and GST–Pnr–DBD fusion proteins were purified with glutathione–agarose according to the recommendations of the manufacturer and the protein concentration was estimated by the Bradford assay and Coomassie staining after gel electrophoresis. For the EMSA, the purified proteins were incubated with the pool of selected oligonucleotides (50 × 103 cpm) in the binding buffer [20 mm HEPES (pH 7.9), 1 mm DTT, 20% glycerol, 100 mm KCl] during 20 min at room temperature and the samples were loaded on a 5% acrylamide gel (29:1 acrylamide/bis-acrylamide; 0.5× Tris Borate EDTA).
DNA transfections and CAT assays
CEF cells were used freshly prepared according to standard methods and transfected with the Ca3(PO4)2 precipitate technique. In addition to the expression vectors or gene reporters described in each figure, all transfections contained 100 ng of the β-galactosidase reporter CMV–β-gal as an internal standard and the amount of DNA was scaled up to 10 μg with pBluescript SK+ DNA as carrier. For analysis of the transactivation, the cells were recovered after scraping and centrifugation in cold PBS and lysed in 100 μl of 0.25 m Tris-HCl (pH 7.5) by three cycles of freeze–thawing. For each transfection, a 10 μl sample was analyzed for β-galactosidase activity and a volume containing a defined amount of activity was then assayed for CAT activity. The reactions were done twice for 1 hr (reporter pαD3) and twice for 20 min (reporter 17m5-TATA–CAT) at 37°C. They were then analyzed by standard thin-layer chromatography and after autoradiography, the conversion percentage was determined by a quantitative phosphoImager using a Fujix BAS 2000 apparatus. The results from three independent experiments performed with two independent DNA preparations were combined to determine the mean activities shown in the figures.
DNA transfections, immunoprecipitations, GST fusion protein selection, and Western blot analysis
Cos-7 cells, grown in 5% calf serum-supplemented Dulbecco’s medium (Sigma Chemical, St. Louis, MO), were transfected like the CEF cells with recombinant adjusted to 10 μg with pBluescript SK+ as carrier DNA. After 36 hr, the cells were harvested in cold PBS, pelleted, washed, and resuspended in lysis buffer [400 mm KCl, 20 mm Tris-HCl (pH 7.5), 20% glycerol, 5 mm DTT, 0.4 mm PMSF] containing 2.5 mg/ml of leupeptin, pepstatin, aprotinin, antipain, and chymostatin. After three cycles of freeze–thaw in liquid nitrogen, the resulting cell lysate was diluted four times with the lysis buffer without KCl to give a final concentration of 100 mm KCl and then cleared by centrifugation for 5 min at 13,000 rpm. The protein concentration was determined by the Bradford assay.
Two hundred fifty micrograms of protein extract, adjusted to 1 ml with the 100 mm KCl lysis buffer, was incubated for 2 hr in a coldroom with agitation either with glutathione–agarose or with protein G–Sepharose in the presence of the B10 monoclonal antibody produced in mouse and raised against the B epitope of the estrogen receptor. The Sepharose beads were then recovered by centrifugation and washed three times with 1 ml of RIPA buffer (1× PBS, 0.1% SDS, 0.5% sodium deoxycholate, 0.5% NP-40). The adsorbed proteins were dissociated by boiling for 5 min in 20 μl of Laemmli buffer and resolved by SDS–polyacrylamide gel electrophoresis. Proteins separated by electrophoresis were electrotransferred onto a nitrocellulose filter. Blocking, washing, and incubation of the membrane with antibodies were carried out in 1× PBS containing 5% skimmed dry milk and 0.5% Tween 20. As the proteins in our study have different molecular masses, the filter was cut and each part was probed with the appropriate antibody. We used the 2B8 monoclonal antibody (that recognizes an epitope in the carboxy-terminal part of Pnr) to detect Pnr and PnrD, the B10 monoclonal antibody for the B-tagged Ush, and the 1D10 monoclonal antibody, which recognizes the GST part of the fusion proteins. After washing (1× PBS, 0.5% Tween 20) and blocking (1× PBS, 5% skimmed dry milk, 0.5% Tween 20), the blot was further incubated with horseradish peroxidase-linked goat anti-mouse immunoglobulins (Jackson ImmunoResearch Laboratories, West Grove, PA). Specific immunocomplexes were visualized by the enhanced chemiluminescence (ECL) Western blotting detection system according to the recommendations of the manufacturer (Amersham International, Les Ulis, France).
Yeast interaction assays
Yeast interaction assays were performed essentially as described previously (Finley and Brent 1994). Briefly, the yeast strain EGY48 with an integrated leu2 reporter gene and upstream LexA operators was transformed with pEG202–Pnr+, pEG202–PnrD1/D4, or pEG202–PnrVX1/VX4, which allows expression of the full-length wild-type Pnr or one of its dominant versions, fused to the LexA–DBD. The different strains were then transformed with the pJG4–5–Ush plasmid, which allows galactose-dependent expression of the fusion protein containing in its amino-terminal moiety the bacterial B42 activation domain efficient also in yeast. After selection on a medium lacking histidine and tryptophane and selecting for the presence of the pEG202 and pJG4-5 plasmids, the transformed yeasts were plated on a medium lacking histidine, trytophane, and leucine, with either glucose, where the B42Ush fusion protein is repressed, or with galactose, where its expression is induced. Levels of LexA fusion proteins in the different strains were monitored by Western blots using the Pnr-specific monoclonal antibody 2B8.
Pnr and Ush overexpression and staining for β-galactosidase activity
The wild-type pnr cDNA as well as the dominant forms pnrD4 and pnrVX4 isolated from the pBluescript derivatives were subcloned in the appropriate restriction sites of the plasmid pUAST, which contains several binding sites for the GAL4 activator upstream to the basal hsp70 promoter sequences. The resulting pUAST–Pnr plasmids were used to transform embryos of a w1118 stock. Transgenic strains were established and crossed with the driver pnrMD237, an enhancer trap line where a GAL4-containing transposon is inserted at the pnr locus, giving an imaginal expression indistinguishable from that of the pnr gene. They were also crossed with DC–enhancer–sc–lacZ; pnrMD237/TM6B, Tb flies. The transgenic lines DC-3.2 harbor the 5.7-kb EcoRI fragment that contains the dorsocentral enhancer fragment fused to 3.7sc–lacZ (3.7 kb of the sc promotor region fused to the lacZ gene; for details see Gomez-Skarmeta et al. 1995). The transgenic line containing the UAS–Ush construct is described in Cubadda et al. (this issue). Larvae were dissected and discs stained for β-galactosidase activity according to standard methods.
Acknowledgments
We thank Gary Felsenfeld for the gift of the GATA expression vectors and the globin reporter, Juan Modolell for his continuous help with ac–lacZ transgenic flies, Roger Brent and Lauren Ha for kindly providing us with the material as well as Zeev Paroush for his help in setting up the yeast two-hybrid assay, Mariann Bienz for making the UAS–Pnr flies, Irwin Davidson for the gift of plasmids, the sequencing, oligonucleotide synthesis, and cell culture services of the IGBMC, Stuart Orkin for sharing unpublished information with us, our colleagues at the IGBMC for their help and discussions, and Irwin Davidson for comments on the manuscripts. This work was supported by the the INSERM, the CNRS, the Centre Hospitalier Universitaire Régional, grant 92N60/0694 from the MRE, the Association pour la Recherche Contre le Cancer, the European Community (contract ERBCHRXCT940692), and the Action Concertées-Sciences du Vivant No4 du Ministère de l’Education Nationale de l’Enseignement Supérieur et de la Recherche.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL psimpson@titus.u-strasbg.fr; phr@titus.u-strasbg.fr; FAX (33) 3 88 65 32 01.
References
- Abel T, Michelson AM, Maniatis T. A Drosophila GATA family member that binds to Adh regulatory sequences is expressed in the developing fat body. Development. 1993;119:623–633. doi: 10.1242/dev.119.3.623. [DOI] [PubMed] [Google Scholar]
- Andrews NC, Orkin SH. Transcriptional control of erythropoiesis. Curr Opin Hematol. 1994;1:119–124. [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Calleja M, Moreno E, Pelaz S, Morata G. Visualization of gene expression in living adult Drosophila. Science. 1996;274:252–255. doi: 10.1126/science.274.5285.252. [DOI] [PubMed] [Google Scholar]
- Campuzano S, Modolell J. Patterning of the Drosophila nervous system: The achaete-scute gene complex. Trends Genet. 1992;8:202–208. doi: 10.1016/0168-9525(92)90234-u. [DOI] [PubMed] [Google Scholar]
- Chatton B, Bahr A, Acker J, Kedinger C. Eukaryotic GST fusion vector for the study of protein-protein interactions in vivo: Application to interaction of ATFa with Jun and Fos. BioTechniques. 1995;18:142–145. [PubMed] [Google Scholar]
- Crossley M, Orkin SH. Regulation of the β-globin locus. Curr Opin Genet Dev. 1993;3:232–237. doi: 10.1016/0959-437x(93)90028-n. [DOI] [PubMed] [Google Scholar]
- Crossley M, Merika M, Orkin SH. Self-association of the erythroid transcription factor GATA-1 mediated by its zinc finger domains. Mol Cell Biol. 1995;15:2448–2456. doi: 10.1128/mcb.15.5.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubadda, Y., P. Heitzler, R.P. Ray, M. Bourouis, P. Ramain, W. Gelbart, P. Simpson, and M. Haenlin. 1997. u-shaped encodes a zinc finger protein that regulates the proneural genes achaete and scute during the formation of bristles in Drosophila. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Cubas P, Modolell J. The extramacrochaetae gene provides information for sensory organ patterning. EMBO J. 1992;11:3385–3393. doi: 10.1002/j.1460-2075.1992.tb05417.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, de Celis JF, Campuzano S, Modolell J. Proneural clusters of achaete-scute expression and the generation of sensory organs in the Drosophila imaginal disc. Genes & Dev. 1991;5:996–1008. doi: 10.1101/gad.5.6.996. [DOI] [PubMed] [Google Scholar]
- Cunninngham TS, Cooper TG. Expression of the DAL80 gene, whose product is homologous to the GATA factors and is a negative regulator of multiple nitrogen catabolic genes in Saccharomyces cerevisiae, is sensitive to nitrogen catabolic repression. Mol Cell Biol. 1991;11:6205–6215. doi: 10.1128/mcb.11.12.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- deBoer E, Antoniou M, Mignotte V, Wall L, Grosveld F. The human β-globin promoter: Nuclear protein factors and erythroid specific induction of transcription. EMBO J. 1988;7:4203–4212. doi: 10.1002/j.1460-2075.1988.tb03317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T, Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci. 1988;85:5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— The erythroid-specific transcription factor Eryf1: A new finger protein. Cell. 1989;58:877–885. doi: 10.1016/0092-8674(89)90940-9. [DOI] [PubMed] [Google Scholar]
- ————— Trans-activation of a globin promoter in non erythroid cells. Mol Cell Biol. 1991;11:843–853. doi: 10.1128/mcb.11.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley, R.L., Jr. and R.Brent. 1994. Interaction trap cloning with yeast. In Gene probes: a pratical approach. (ed. D. Hames and D. Glover).
- Ghysen A, Dambly-Chaudière C. From DNA to form: The achaete-scute complex. Genes & Dev. 1988;2:495–501. doi: 10.1101/gad.2.5.495. [DOI] [PubMed] [Google Scholar]
- Gyuris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1-phase and S-phase protein phosphatase that associates with cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- Gomez-Skarmeta JL, Rodriguez I, Martinez C, Culi J, Ferres-Marco D, Beamonte D, Modolell J. Cis-regulation of achaete and scute: Shared enhancer-like elements drive their expression in proneural clusters of the imaginal discs. Genes & Dev. 1995;9:1869–1882. doi: 10.1101/gad.9.15.1869. [DOI] [PubMed] [Google Scholar]
- Heitzler, P. 1993. “La détermination des précurseurs nerveux chez la Drosophile: Interactions cellulaires et contrôle génétique”. Thesis. Université Louis Pasteur, Strasbourg, France.
- Heitzler P, Haenlin M, Ramain P, Calleja M, Simpson P. A genetic analysis of pannier, a gene necessary for viability of dorsal tissues and bristle positioning in Drosophila. Nature Genet. 1996;143:1271–1286. doi: 10.1093/genetics/143.3.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgs DR, Wood WG. Understanding erythroid differentiation. Curr Biol. 1993;3:548–550. doi: 10.1016/0960-9822(93)90054-r. [DOI] [PubMed] [Google Scholar]
- Hwang JJ, Chambon P, Davidson I. Characterization of the transcription activation domain function and the DNA binding domain of transcriptional enhancer factor-1. EMBO J. 1993;12:2337–2348. doi: 10.1002/j.1460-2075.1993.tb05888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla B, Caddick MX, Langdon T, Martinez-Rossi NM, Bennett CF, Sibley S, Davies RW, Arst HN., Jr The regulatory gene areA mediating nitrogen metabolite repression in Aspergillus nidulans. Mutations affecting specificity of gene activation alter a loop residue of a putative zinc finger. EMBO J. 1990;9:1355–1364. doi: 10.1002/j.1460-2075.1990.tb08250.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laverierre AC, MacNeill C, Mueller RE, Poelmann JB, Burch E, Evans T. GATA 4/5/6: A subfamily of three transcription factors expressed in developing heart and gut. J Biol Chem. 1994;269:23177–23184. [PubMed] [Google Scholar]
- Lin WH, Huang LH, Yeh JY, Hoheisel J, Lehrach H, Sun YH, Tsai SF. Expression of a Drosophila GATA transcription factor in multiple tissues in developing embryos. J Biol Chem. 1995;270:25150–25158. doi: 10.1074/jbc.270.42.25150. [DOI] [PubMed] [Google Scholar]
- Merika M, Orkin SH. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Krüppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignotte V, Eleouet JF, Raich N, Roméo PH. Cis- and trans-acting elements involved in the regulation of the erythroid promoter of the human porphobilinogen deaminase gene. Proc Natl Acad Sci. 1989a;86:6548–6552. doi: 10.1073/pnas.86.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignotte V, Wall L, deBoer E, Grosveld F, Roméo PH. Two tissue-specific factors bind the erythroid promoter of the human porphobilinogen deaminase. Nucleic Acids Res. 1989b;17:37–54. doi: 10.1093/nar/17.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller IJ, Bieker JJ. A novel erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol Cell Biol. 1993;13:2776–2786. doi: 10.1128/mcb.13.5.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- Nicolis S, Bertini C, Ronchi A, Crotta S, Lanfranco L, Moroni E, Giglioni B, Ottolenghi S. An erythroid specific enhancer upstream to the gene encoding the cell-specific transcription factor GATA-1. Nucleic Acids Res. 1991;19:5285–5291. doi: 10.1093/nar/19.19.5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedone PV, Omichinski JG, Nony P, Trainor C, Gronenborn AM, Clore GM, Felsenfeld G. The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. EMBO J. 1997;16:2874–2882. doi: 10.1093/emboj/16.10.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D’Agati Y, Orkin SH, Constantini F. Erythroid differentiation in chimeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- Philipsen S, Pruzina S, Grosveld F. The minimal requirement for activity in transgenic mice of hypersensitive site 3′ of the β-globin locus control region. EMBO J. 1993;12:1077–1085. doi: 10.1002/j.1460-2075.1993.tb05749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumb MA, Frampton J, Wainwright H, Walker M, Macleod K, Goodwin G, Harrison P. GATAAG: A cis control region binding an erythroid-specific nuclear factor with a role in globin and non globin gene expression. Nucleic Acids Res. 1989;17:73–91. doi: 10.1093/nar/17.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahuel C, Vinit MA, Lemarchandel V, Cartron JP, Roméo PH. Erythroid specific activity of the glycophorin promoter requires GATA-1 mediated displacement of a repressor. EMBO J. 1992;11:4095–4102. doi: 10.1002/j.1460-2075.1992.tb05502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramain P, Heitzler P, Haenlin M, Simpson P. Pannier, a negative regulator of achaete and scute in Drosophila, encodes a zinc finger protein with homology to the vertebrate transcription factor GATA-1. Development. 1993;119:1277–1291. doi: 10.1242/dev.119.4.1277. [DOI] [PubMed] [Google Scholar]
- Romani S, Campuzano S, Macagno ER, Modolell J. Expression of achaete and scute genes in Drosophila imaginal discs and their function in sensory organ development. Genes & Dev. 1989;3:997–1007. doi: 10.1101/gad.3.7.997. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Simon MC, Pevny L, Wiles MV, Keller G, Constantini F, Orkin SH. Rescue of erythroid development in gene targeted GATA-1 mouse embryonic stem cells. Nature Genet. 1992;1:92–98. doi: 10.1038/ng0592-92. [DOI] [PubMed] [Google Scholar]
- Skeath JB, Carroll S. Regulation of achaete-scute gene expression and sensory organ pattern formation in the Drosophila wing. Genes & Dev. 1991;5:984–995. doi: 10.1101/gad.5.6.984. [DOI] [PubMed] [Google Scholar]
- Spieth JY, Shim H, Lea K, Conrad R, Blumenthal T. Elt-1, an embryonically expressed Caenorhabditis elegans gene homologous to the GATA transcription factor family. Mol Cell Biol. 1991;11:4651–4659. doi: 10.1128/mcb.11.9.4651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SF, Martin DIK, Zon LI, D’Andrea AD, Wong GG, Orkin SH. Cloning of cDNA for the major DNA-binding protein of the erythroid lineage through expression cloning in mammalian cells. Nature. 1989;339:446–451. doi: 10.1038/339446a0. [DOI] [PubMed] [Google Scholar]
- Tsai SF, Strauss E, Orkin SH. Functional analysis and in vivo footprinting implicate the erythroid transcription factor GATA-1 as a positive regulator of its own promoter. Genes & Dev. 1991;5:919–931. doi: 10.1101/gad.5.6.919. [DOI] [PubMed] [Google Scholar]
- Tsang, A.P., J.E. Visvader, C.A. Turner, Y. Fujiwara, C. Yu, M.J. Weiss, M. Crossley, and S.H. Orkin. 1997. FOG, a novel multitype zinc finger protein, acts as a co-factor for transcription factor GATA-1 in erythroid and megakaryotic differentiation. Cell (in press). [DOI] [PubMed]
- Villares R, Cabrera C. The achaete-scute gene complex of D. melanogaster: Conserved domains in a subset of genes required for neurogenesis and their homology to myc. Cell. 1987;50:415–424. doi: 10.1016/0092-8674(87)90495-8. [DOI] [PubMed] [Google Scholar]
- Wall L, deBoer E, Grosveld F. The human β-globin gene 3′ enhancer contains multiple binding sites for an erythroid-specific protein. Genes & Dev. 1988;2:1089–1100. doi: 10.1101/gad.2.9.1089. [DOI] [PubMed] [Google Scholar]
- Whyatt DJ, deBoer E, Grosveld F. The two zinc finger-like domains of GATA-1 have different DNA binding specificities. EMBO J. 1993;12:4993–5005. doi: 10.1002/j.1460-2075.1993.tb06193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winick J, Abel T, Leonard MW, Michelson AM, Chardon-Loriaux I, Holmgren RA, Maniatis T, Engel JD. A GATA family transcription factor is expressed along the embryonic dorso-ventral axis in Drosophila melanogaster. Development. 1993;119:1055–1065. doi: 10.1242/dev.119.4.1055. [DOI] [PubMed] [Google Scholar]
- Xiao JH, Davidson I, Garnier JM, Chambon P. Cloning, expression and transcriptional properties of the human enhancer factor TEF-1. Cell. 1991;65:551–568. doi: 10.1016/0092-8674(91)90088-g. [DOI] [PubMed] [Google Scholar]
- Yamamoto M, Ko LJ, Leonard MW, Beug H, Orkin SH, Engel JD. Activity and tissue-specific expression of the transcription factor Nf-E1 multigene family. Genes & Dev. 1990;4:1650–1662. doi: 10.1101/gad.4.10.1650. [DOI] [PubMed] [Google Scholar]
- Yang HY, Evans T. Distinct roles for the two cGATA-1 finger domains.Mol. Cell Biol. 1992;12:4562–4570. doi: 10.1128/mcb.12.10.4562. [DOI] [PMC free article] [PubMed] [Google Scholar]