Abstract
Protection against infection is the hallmark of immunity and the basis of effective vaccination. For a variety of reasons there is a great demand to develop new, safer and more effective vaccine platforms. In this regard, while ‘first-generation’ DNA vaccines were poorly immunogenic, new genetic ‘optimization’ strategies and the application of in vivo electroporation (EP) have dramatically boosted their potency. We developed a highly optimized plasmid DNA vaccine that expresses the lymphocytic choriomeningitis virus (LCMV) nucleocapsid protein (NP) and evaluated it using the LCMV challenge model, a gold standard for studying infection and immunity. When administered intramuscularly with EP, robust NP-specific cellular and humoral immune responses were elicited, the magnitudes of which approached those following acute LCMV infection. Furthermore, these responses were capable of providing 100% protection against a high-dose, normally lethal virus challenge. This is the first non-infectious vaccine conferring complete protective immunity up to eight weeks after vaccination and demonstrates the potential utility of ‘next-generation’ DNA vaccines.
Keywords: DNA vaccine, electroporation, protection, LCMV, T cell, antibody
INTRODUCTION
We were interested in testing the immunogenicity and protective efficacy of a genetically optimized DNA vaccine delivered with in vivo EP using the mouse model of LCMV infection. LCMV was one of the first human pathogenic viruses to be isolated [1], and the use of this virus in mice has provided a landmark model for characterizing cellular and humoral immune responses during acute and persistent viral infections [2–7]. This enveloped virus is a prototype member of the arenavirus family and has two negative-stranded RNA segments [8], the short genomic segment of which encodes two major protein products: the nucleocapsid protein (NP) and glycoprotein (GP) [9]. The NP is a structural protein involved in viral replication and transcription, is the most abundant viral protein expressed in infected cells, and is known to interfere with IFN-β production by host innate cells. The immune response against LCMV has been extensively studied and the role of virus-specific CTL, in which the response has been precisely mapped to epitopes in the GP, NP and L polymerase, has been well established [10–16]. Infection of mice with the Armstrong strain of LCMV induces a strong CD8+ CTL response which mediates control of the infection within approximately 14 days. When inoculated intracranially (i.c.), this virus induces a massive lymphocytic response in the choriomeninges which typically results in death at approximately 7 – 10 days p.i. [17]. However, following an acute infection with LCMV, mice are completely protected against i.c. inoculation, which is mediated by virus-specific CTL [18].
To date, the efficacies of numerous vaccine strategies have been tested using the lethal LCMV challenge model (Table 1). Those conferring protection against normally lethal LCMV challenge have consisted of both infectious and non-infectious vaccines. The former group consists of recombinant viral (vaccina virus expressing full-length, truncated, or poly-epitopes from NP and/or GP [18–22]; adenoviral vectors encoding NP proteins and epitopes [23, 24]; influenza [25] and Mengo [26] viruses expressing an NP epitope) or bacterial vectors (L. monocytogenes expressing full-length NP [27]; recombinant strains of S. typhimurium expressing an NP epitope [28, 29] or a full-length Lassa NP [22]. Non-infectious vaccines have consisted of genetically detoxified CyaA toxoid proteins from B. pertussis containing an NP epitope [30, 31], recombinant Bluetongue virus (BTV) tubules containing a single NP epitope [32], hybrid recombinant parvovirus-like particles (VLP) vaccines expressing an NP epitope [33–35], listeriolysin O-containing liposomes with truncated NP [36], bacterial minicells derived from a non-pathogenic E. coli K-12 strain capable of the simultaneous delivery of both recombinant NP protein and the corresponding NP-encoding DNA vaccine [37], and various DNA vaccines expressing NP or GP [38–44] including one that was adjuvanted by encapsulation into liposomes [45].
Table 1.
Summary of vaccines providing protection against lethal LCMV challenge*
| Safety | Vaccine Platform |
Type | LCMV Ag | Ag length | Adjuvant | Regimen | Chall- enge |
Challenge dose |
Survival | Refs |
|---|---|---|---|---|---|---|---|---|---|---|
| Infectious | Viral | rVV | NP | Full | - | 1X 2×107 | 6 wks | (101 PFU) 20XLD50 | H-2d (100%) H-2b (84%) | [18] |
| PFU i.p. | 2.6 wks | (102.2 PFU) ?LD50 | H-2b (100%) | [20] | ||||||
| GP1–271 | Truncated | - | 1X 2×107 PFU i.p. | 6wks | 20XLD50 | H-2b (88%) | [19] | |||
| NP121–132 and GP33–50 | Poly-epitopic | - | 1X 2×107 PFU i.p. | 6 wks | 20XLD50 | H-2d (100%) H-2b (88) | [21] | |||
| LAS NP | Full | - | 2X 5×109 CFU i.g. | 0.6 wks | (102.3–3 PFU) ?LD50 | H-2d (33%) | [22] | |||
| rAd | NP118–126 NP396–404 | Epitopic | B2M-linked | 1X 2×107 i.f.u. s.c. | 8 – 12 wks | (20 PFU) ?LD50 | H-2d (70%) H-2b (50%) | [23] | ||
| NP | Full/fusion | li-linked | 1X 2×107 i.f.u. s.c. | 13 wks | (20 PFU) ?LD50 | H-2b (100%) | [24] | |||
| rFlu | NP116–127 | Epitopic | - | 1X i.n. and i.p. | 20 wks | (103 PFU) ?LD50 | H-2d (100%) | [25] | ||
| rMengo | NP117–130 | Epitopic | - | 1X 1×105 PFU i.p. | 6 wks | (102.5 PFU) ?LD50 | H-2d (100%) | [26] | ||
| Bacterial | rLM | NP | Full | Cytosolic secretion | 1X i.v. | 1 wk | (103.5 PFU) ?LD50 | H-2d (100%) | [27] | |
| rSalmonella | LAS NP | Full | - | 2X 5×109 CFU i.g | 0.6 wks | (102.3–3 PFU) ?LD50 | H-2d (37%) | [22] | ||
| NP118–126 | Epitopic | Cytosolic secretion | 3X 1–2×108 CFU i.g. | 6 wks | 10XLD50 | H-2d (100%) | [28, 29] | |||
| Non-infectious | Protein | Detoxified CyaA toxoid | NP135–149 | Epitopic | Alum | 2X i.p. | 9 wk | (101.7 foci) ?LD50 | H-2d (93%) | [30, 31] |
| BTV tubules | NP118–132 | Epitopic | - | 2X i.p. | 1 wk | (101.7 PFU) ?LD50 | H-2d (75%) | [32] | ||
| VLP | rPPV:VLP | NP118–132 | Epitopic | - | 2X i.p. | 7 wks | (101.7 PFU) ?LD50 | H-2d (80%) | [33–35] | |
| Protein/Oil | LLO-liposomes | NP82–173 | Truncated | LLO-liposomes | 2X s.c. | 2 wks | (102 PFU) ?LD50 | H-2d (100%) | [36] | |
| Bacterial/Protein/DNA | E. coli minicells | NP | Full | - | 3X i.m. | 5 wks | (103 PFU) 20XLD50 | H-2b (89%) | [37] | |
| DNA/Oil | pDNA | NP | Full | Lipid complexes | 1X i.v. | 6 wks | 20XLD50 | H-2d (50%) | [45] | |
| DNA | pDNA | GP | Full/fusion | DNA-li/Gene- gun | 2X i.d. | 4 wks | (101.3 PFU) ?LD50 | H-2b (15%) | [38] | |
| NP or GP | Full | - | 3X i.m. | 6 wks | 20XLD50 | H-2d (54%) H-2b (50%) | [42] | |||
| NP | Full/fusion | Ubiquitin tag | 3X i.m. | 6 wks | (? PFU) 20XLD50 | H-2d (100%) H-2b (100%) | [40] | |||
| NP118–126 | Epitopic | PTD-fusion | 1X i.m. | 1.1 wks | 40XLD50 | H-2d (100%) | [44] | |||
| NP | Full | 21 - | 2X i.m. | 1 wk | (101.6 PFU) ?LD50 | H-2b (100%) | [41] | |||
| 3 wks | (100.8 PFU) 30XLD50 | H-2d (50%) | [39] | |||||||
| Gene-gun | 3X i.d. | 2 wks | (104.1PFU) ?LD50 | H-2b (35%) | [43] |
Table displays vaccines and conditions yielding the best level of protection per report; VLP – virus-like particle (rPPV:VLP); li –MHC class II-associated invariant chain; B2M - β2-microglobulin; BTV – Bluetongue virus; pDNA – plasmid DNA; rVV –recombinant vaccina virus; rAd – recombinant adenovirus; rFlu – recombinant influenza; rLM – recombinant L. monocytogenes; LLO – L. monocytogenes listeriolysin O; LAS – Lassa Fever virus; PTD – HIV-1 protein transduction domains; CS – current study
However, no non-infectious vaccine has conferred complete protection against a high dose challenge (≥ 20XLD50) of lethal LCMV when administered during the long-term immunological memory phase at least 8 weeks post-immunization. Memory T cells are considered to be long-lived [46] if they can be maintained following their differentiation during contraction of the acutely proliferating lymphocyte pool in response to an antigenic prime. The peak of the lymphocyte response to acute infection with LCMV occurs 8 days after infection [47] and is followed by a period of approximately three to four weeks in which this activated and proliferating pool of CD8+ T cells contracts and gives rise to memory cells [48]. While a hallmark of memory CD8+ T cells is their ability for speedy activation and proliferation upon restimulation [49], which is critically dependent on CD4+ T cell help during the prime [50], they also must be able to persist long after vaccine administration [51]. Although, previous studies evaluating non-infectious vaccines using the LCMV model have either administered the challenge virus after only a short period of time (< 8 weeks after the final immunization), used a low-dose challenge virus in which the lethal dosage was unclear or not previously determined (< 20XLD50), or were not completely protective (yielding 100% protection in the H-2b background). For example, DNA vaccines previously reporting to have yielded 100% protection in H-2b mice were either challenged only one week post-immunization [41] or the lethality of the challenge dosage was not stated and/or previously determined [40] as PFU per dose does not predict lethality among different virus stocks and preparations.
In this report we show that a genetically optimized pLCMV DNA vaccine is highly immunogenic and induces robust T and B cell responses that approach or surpass those during acute infection with LCMV. Furthermore, this vaccine-generated NP-specific immunity confers complete (100%) protection against a high-dose (20XLD50 lethal LCMV challenge administered at least 8 weeks post-immunization; DNA vaccination yielded 67%, 84%, and 100% protection against lethal challenge after one, two, and three immunizations, respectively. Therefore, the pLCMV-NP vaccine described herein is the first non-infectious vaccine capable of providing complete (100%) protection for up to 8 weeks in the high-dose LCMV challenge model. These data improve upon previous studies characterizing immunity against ‘first-generation’ DNA vaccines and demonstrate the potential utility of ‘next-generation’ DNA vaccines for eliciting protective immunity against infectious disease.
RESULTS
Construction and expression of the pLCMV-NP DNA vaccine
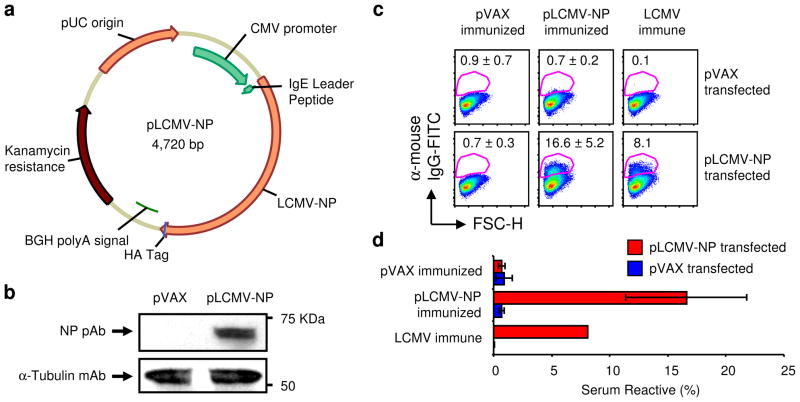
To characterize ‘next-generation’ DNA vaccines that incorporate advanced optimization and delivery strategies, we constructed a genetically optimized DNA vaccine that expresses the LCMV NP protein (Fig 1). The full-length gene was cloned into the pVAX1 mammalian expression vector as displayed in Figure 1a in addition to codon and RNA optimization with the human IgE leader peptide [52], and an HA tag was added to the 3′ terminus for immunodetection. Following construction, protein expression was confirmed by immunoblotting and immunofluorescence; 293T cells were transfected with pLCMV-NP or empty pVAX vector (negative control) and samples were harvested 48 h later and analyzed by Western immunoblotting (Fig 1b) and flow cytometry (Fig 1c and d). The presence of a ~66 KDa protein was detected in the cell lysates of pLCMV-NP-transfected 293T cells using anti-HA tag Abs (data not shown) and NP-specific polyclonal serum (Fig. 1b), while control pVAX empty vector-transfected lysates were negative for Ag expression. Samples were normalized for total protein by Bradford protein assay and contained equivalent amounts of globular α-tubulin protein. Furthermore, pLCMV-NP-transfected 293T cells were reactive with serum from LCMV immune and pLCMV-NP immunized mice (n=5), but not from pVAX immunized (n=5) animals (Fig1c); hyper-immune serum pooled from mice immunized five times with pLCMV-NP reacted with 16.6% of pLCMV-NP-transfected cells on average as compared with 8.1% from LCMV immune animals and 0.7% from pVAX-transfected mice (Fig1d). Non-specific binding was not detected as the positive sera did not react with pVAX-transfected 293T cells. Altogether, transfection of 293T cells using the pLCMV-NP plasmid DNA construct was sufficient for the production of NP protein in vitro that was specifically reactive with Abs recognizing the protein tag and with those generated from repeat immunization of mice.
Figure 1. Plasmid LCMV-NP construction and in vitro expression.
(a) Cartoon of pLCMV-NP displaying the LCMV NP gene cloned into the pVAX1 mammalian expression vector. The CMV promoter, LCMV NP gene with the N-terminal human IgE leader peptide and C-terminal HA tag, BGH polyA signal, kanamycin resistance gene, and pUC origin are shown. (b) Expression of the NP protein in 293T cells transfected with either the experimental (pLCMV-NP) or control (pVAX) plasmids as analyzed by SDS-PAGE and Western immunoblotting using a polyclonal mouse serum specific for LCMV NP reagent for detection. Also shown is a loading control by staining for tubulin and relative sizes are indicated (KDa). (c) Flow cytometric detection of pLCMV-NP encoded NP protein in permeabilized 293T cells using serum from LCMV immune (positive control), pVAX-immunized (negative control; n=5), and pLCMV-NP-immunized (n=5) mice. A representative pseudocolor plot from each group of mice is shown and data are displayed as average ± STDEV. (d) Bar graph summarizing serum reactivity from (c) and error bars represent STDEV. Results were similar from two independent experiments.
pLCMV-DNA vaccine is immunogenic
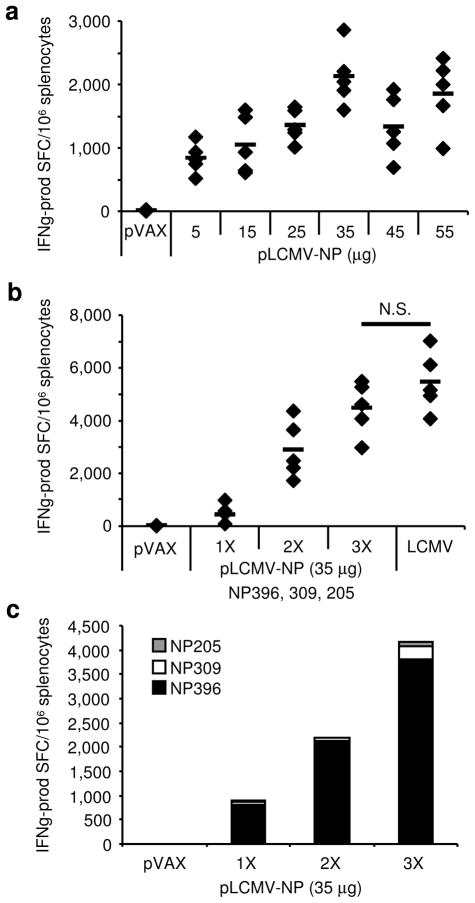
As the optimized pLCMV-NP DNA vaccine was confirmed to express NP protein in vitro we next aimed to evaluate its immunogenicity in vivo. Mice were immunized twice, two weeks between i.m. injections each immediately followed by EP, and IFNγ ELISpot assay was performed to determine whether plasmid immunization was capable of generating NP-specific cellular responses. Firstly, in order to identify the amount of pLCMV-NP required to yield an optimal T cell response following only two immunizations, we titrated the plasmid in a dose optimization study. The pLCMV-NP was administered twice at doses ranging between 5 – 55μg in 10 μg increments (n=5 mice per group) and data are displayed in Figure 2a. NP-specific T cell responses were measured and are in response to peptide stimulation using a pool of three separate peptides known to contain immunodominant epitopes in the H-2b background: DbNP396–404 (NP396), I-AbNP309–328 (NP309), and KbNP205–212 (NP205). While injection of empty pVAX vector at the highest concentration did not elicit an NP-specific response, all doses of the pLCMV-NP plasmid generated responses over 750 IFNγ-producing SFC per million splenocytes. Furthermore, NP-specific T cell responses increased in a dose-dependent manner up to a dose of approximately 35 μg, where from that amount responses no longer increased with dose. Therefore, these data show that DNA vaccination using the pLCMV-NP in combination with in vivo EP yielded measurable NP-specific T cell responses that were specific to the plasmid encoded vaccine Ag.
Figure 2. Immunogenicity of pLCMV-NP.
(a) Immunogenicity of pLCMV-NP and pVAX (55 μg) in mice (n=5 per group). Splenocytes were harvested seven days following the second of two immunizations, two weeks apart, administered i.m. with EP at various doses (μg). (b) Boosting of the NP-specific T cell response following repeat homologous immunizations of 35 μg of pLCMV-NP with EP, two weeks apart. Negative control immunized animals receiving three injections of empty vector control plasmid (pVAX 35 μg) and positive control animals acutely infected with 2×105 PFU LCMV i.p. at 7 d.p.i. are shown. (c) Epitopic composition of the NP-specific response as determined by stimulation with individual peptides (NP396 (black bars), NP205 (grey bars), NP309 (white bars)) after one, two, and three immunizations, two weeks apart. NP-specific T cell responses in (a and b) were measured against a pool of three peptides containing known immunodominant NP epitopes (NP396, NP205, NP309) in the standard IFN γ ELISpot assay. Responses for individual mice (diamonds) and group averages (lines) are shown, as well as the NP-specific response in empty vector control-immunized (pVAX) animals. N.S. is not statistically significant and experiments were performed independently at least three times with similar results.
After determining that the pLCMV-NP vaccine was immunogenic in vivo and that a dose of 35 μg yielded optimal NP-specific T cell responses, we next wanted to determine the contribution of boosting in a homologous prime-boost regimen. The plasmid vaccine was given up to three times with two weeks in between immunizations at a dose of 35 μg and IFNγ ELISpot was performed 7 days post-immunization for each group of vaccinated mice (n=5 per group), as well as for samples from LCMV-infected animals 7 days post-infection (Fig. 2b). The pLCMV-NP prime alone yielded 433 IFNγ-producing SFC/million splenocytes on average while homologous boosting yielded 2,891 and 4,481 after two and three immunizations, respectively. Homologous boosting using the pLCMV-NP with EP resulted in a ~6.7-fold increase in NP-specific T cell responses after the second immunization and a ~1.6-fold increase from the second to the third. Altogether, vaccine-specific T cell responses after 3 immunizations were not statistically different than those generated 7 days post-LCMV acute infection (p=0.244). Furthermore, the epitopic composition of the NP-specific response to the three peptides used for stimulation was individually determined by ELISpot in splenocytes from experimental mice (Fig. 2c). While all three of the epitopes have been described as being immunodominant in the H-2b background, the vast majority of the NP-specific response was specific for the NP396 epitope; > 90% of the NP-specific response was for NP396 while it was approximately 5.0% and 2.2% on average for NP309 and NP205, respectively. Therefore, these data show that multiple immunizations with the pLCMV-NP at an optimized dose of 35 μg were capable of generating NP-specific T cell responses that were predominantly NP396-specific and that were not statistically different from acute LCMV infection.
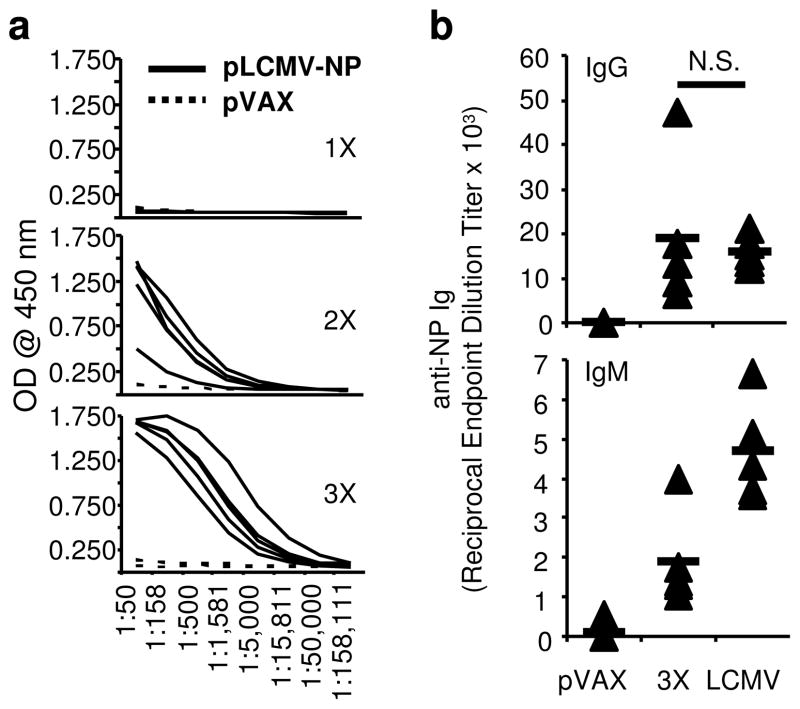
Since ‘first-generation’ DNA vaccines were poorly immunogenic and rarely generated remarkable levels of antibodies, we next evaluated B cell responses after pLCMV-NP vaccination. Serum samples were collected 7 days following each of the three immunizations spaced two weeks apart and used in the ELISA assay to determine the production of NP-specific antibodies (Fig. 3). Assay plates were coated with LCMV NP protein and serum samples were tested for each animal in triplicate for levels of protein-specific immunoglobulin. While little NP-specific IgG was observed directly following the first immunization, significant levels of Ab were detected after the second and third (Fig. 3a). Moreover, repeat immunization by homologous boost resulted in the progressive enhancement of IgG responses as they were the greatest following the third immunization. When compared with the level of IgG induced by acute LCMV infection, reciprocal endpoint dilution titers from vaccinated animals were not significantly different from those from infected ones (p=0.862) (Fig. 3b). Furthermore, levels of NP-specific IgM were almost half of those in LCMV infected animals on average; IgM levels were greater than 40% of those than in infected mice. Thus, these data demonstrate that pLCMV-NP DNA vaccination in combination with EP generated robust B cell responses that approached those generated during acute LCMV infection. Altogether, data herein show that the pLCMV-NP DNA plasmid is immunogenic in mice and generated robust T and B cell responses that approach in magnitude those induced during acute LCMV infection.
Figure 3. DNA immunization with EP induces robust antibody responses.
(a) NP-specific IgG responses in serum from (n=5 per group) pVAX-immunized mice (dotted lines) or mice immunized with pLCMV-NP plus EP (solid lines) seven days after one, two, or three injections two weeks apart as measured by ELISA. Each line represents results from a single animal. (b) Reciprocal endpoint dilution titers in sera from mice immunized three times with pLCMV-NP 7 days post-immunization or infected once with an acute dose of LCMV 60 d.p.i. Serum was collected seven days following the final immunization or infection and NP-specific IgG and IgM responses are shown for individual mice (diamonds) and as group averages (lines) in comparison to negative (pVAX) and positive (LCMV) control serum. N.S. is not statistically significant and experiments were performed independently at least twice with similar results.
DNA vaccination is completely protective against LCMV challenge
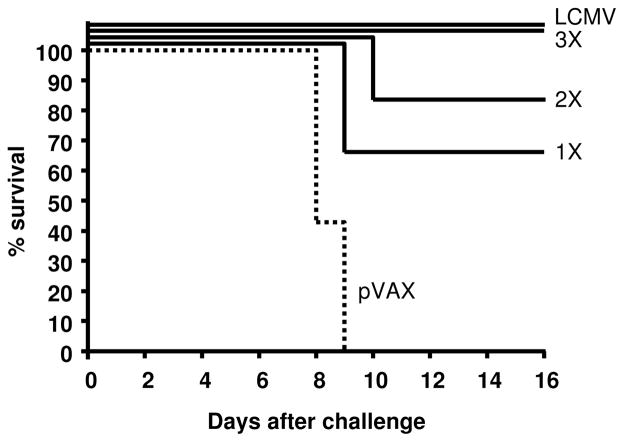
Since three immunizations with the pLCMV-NP DNA vaccine elicited immune responses that were similar in magnitude to those induced by acute LCMV infection, we next aimed to determine whether vaccination was protective against high-dose (20 50% lethal doses (20XLD50)), normally lethal LCMV challenge. Mice were vaccinated three times with 35 μg of pLCMV-NP (n=6), pVAX empty vector (n=7), or infected once with 2×105 PFU LCMV i.p. (n=5). At least eight weeks following the last immunization or infection, mice were challenged with a 20XLD50 dose of LCMV Armstrong by the i.c. route, a dose at which protection in the LCMV model has been commonly assessed [21, 37, 40, 42, 45]. Figure 4 displays the data from the challenge study and survival statistics are shown for each group of mice. While all LCMV-infected animals were completely protected against normally lethal high-dose LCMV challenge, all pVAX empty vector-vaccinated control animals succumbed to infection. Both groups of animals receiving either 1 or 2 immunizations demonstrated a high level of protection against challenge. While only one times vaccination yielded 67% protection, two vaccinations spaced two weeks apart generated greater protection at 83% survival. Moreover, similar to animals that were previously infected acutely with LCMV and are known to be completely protected against lethal challenge, mice immunized three times with pLCMV-NP DNA vaccine exhibited a 100% survival rate. Therefore, three times vaccination with the non-infectious pLCMV-NP induced Ag-specific immunity that approached that following acute LCMV infection and the immunity conferred complete protection at least eight weeks following the final immunization against a high-dose lethal challenge.
Figure 4. DNA vaccination using pLCMV-NP with EP is completely protective against lethal challenge.
Mice (n=6 per group) were immunized three times i.m. using EP with either 35 μg of empty vector control plasmid (pVAX) or 35 μg of pLCMV-NP, with two weeks between injections. Positive control mice were injected i.p. with 2×105 PFU of LCMV. Eight weeks following infection or the final injection of pDNA, mice were challenged i.c. with 20LD50 LCMV and animal survival is displayed in the graph. Mice were observed for 16 days following challenge and results from LCMV immune, pLCMV-NP vaccinated, and pVAX vaccinated (dotted line) are shown. Experiments were performed at least three times in independent experiments and data are representative of the results.
DISCUSSION
Intramuscular administration of plasmid DNA alone has been shown to be immunogenic and even protective in some viral challenge models. However, these ‘first-generation’ DNA vaccines were capable of inducing Ag-specific T cell responses measuring only a fraction of what their infectious, viral-vectored counterparts could achieve. Furthermore, injection of plasmid DNA alone was not previously reported to stimulate significant Ab responses, which are integral for the induction of immunity by an overwhelming majority of currently licensed vaccines in the U.S. Since then, numerous methods for enhancing plasmid DNA immunogenicity and for overcoming some of the other perceived defects of the DNA vaccine approach have been developed. Genetic optimization and DNA delivery strategies aiming to increase transfection efficiency, protein stability, and expression have markedly heightened the immunopotency of DNA vaccination, defining them as ‘next generation’.
In this report, we utilized advanced DNA immunization strategies to improve the immunogenicity of a DNA vaccine expressing a model Ag in mice. The LCMV NP is a structural protein that is the most abundantly expressed in infected cells. Furthermore, the LCMV model is arguably the most studied and best understood mouse model of infection and is commonly used to investigate immunity and to test vaccine strategies. It is one of the first human pathogenic viruses to be isolated and has provided a landmark model for characterizing cellular and humoral immune responses during acute and persistent infection. Thus, we constructed a gene optimized plasmid vaccine expressing the full-length NP protein under control of a CMV promoter in the pVAX1 mammalian expression vector. Expression was first confirmed in vitro, and then in vivo during dosing optimization studies which led us to use a dose of 35 μg of pLCMV-NP in the anterior tibialis muscle followed by EP for all future studies. Since immunity to acute LCMV infection is known to provide full protection against lethal challenge, we aimed to generate similar levels of NP-specific immunity in order to maximize the protective efficacy of the DNA vaccine. Three immunizations of 35 μg plus EP were shown to approach the levels of T cell immunity during acute LCMV protection. Furthermore, we found that this vaccine regimen was capable of inducing robust NP-specific Ab responses which approached (IgM) or surpassed (IgG) those after LCMV infection. These Ab responses were significantly higher than levels previously reported with ‘first-generation’ DNA vaccines without genetic optimization or in vivo EP, which incidentally used three injections of 100 μg of plasmid DNA, almost three times as much DNA per injection [42]. Therefore, current optimization strategies aiming to increase the potency of DNA vaccines were capable of producing a dose-sparing effect while dramatically enhancing humoral immunity. However, it is unlikely that Abs play a major role in protection since B cell-deficient mice can still protect against LCMV challenge [53] and, in the case of our vaccine, that NP-specific Abs target a structural protein that is isolated from the surface of the virus. Nonetheless, these data exemplify the capability of ‘next-generation’ DNA vaccine to generate robust Ab responses in stark contrast to their ‘first-generation’ predecessors.
To date, there have been no reports documenting a non-infectious vaccine strategy that has afforded complete (100%) protection against a 20 50% lethal doses LCMV challenge administered at least 8 weeks post-immunization. Since our fully optimized pLCMV-NP DNA vaccine induced robust B and T cell responses which either approached or surpassed levels of LCMV-specific immunity induced by acute infection, we challenged our vaccinated animal eight weeks following the final immunization, a time frame widely accepted for protection to be mediated by long-lived immunity. Herein, DNA vaccination using the pLCMV-NP DNA vaccine in combination with EP conferred complete (100%) protection against a normally lethal challenge dose of LCMV in H-2b mice after three immunizations. These data demonstrate that optimization strategies including gene optimization and advanced delivery techniques such as in vivo EP have dramatically increased the protective efficacy of a DNA vaccine expressing the NP model Ag. Not only was a dose-sparing effect observed, but three immunizations were completely protective whilst only one vaccination yielded 67% survival. Results presented herein demonstrate the efficacy of highly optimized ‘next-generation’ DNA vaccines and provide proof-of-concept for the development and optimization of the platform as a powerful tool for developing immune therapies and vaccines against infectious diseases.
METHODS
Mice and infections
Adult female C57BL/6 (H-2b) mice 6 – 8 weeks of age were purchased from The Jackson Laboratory (Bar Harbor, ME). Mice were cared for in accordance with Institutional Animal Care and Use Committee-approved protocols at the University Pennsylvania School of Medicine Animal Facility. Acute infection of mice with LCMV Armstrong was described previously [50]. Briefly, 2×105 PFU LCMV was administered i.p. and B and T cell responses were determined 60 and 7 days p.i., respectively. For lethal challenge studies, mice were challenged i.c. with 20XLD50 of LCMV Armstrong as previously described [21, 37, 40, 42, 45] in 30 μl of virus diluent (PBS with 20% FBS and 1X Anti-Anti (Invitrogen, Carlsbad, CA)). Mice were observed daily for 16 days, a time point known to be adequate in the LCMV i.c. challenge model, and all LCMV infected animals were housed in BSL-2 facilities.
Plasmid DNA constructs and in vivo EP
The pLCMV-NP DNA construct encodes the full-length LCMV NP protein (NP_694852; the NP sequence is conserved among strains Armstrong 53b, Armstrong Clone 13, WE, Traub, Docile, Aggressive, and Pasteur). The protein was optimized for expression in mice, including codon and RNA optimization (GeneArt, Regensburg, Germany). It was then further modified by inclusion of the human IgE leader peptide [52] at the N-terminus and an HA tag was added to the C-terminus for immunodetection. The LCMV NP was synthesized and cloned (GeneArt) into the pVAX1 mammalian expression vector (Invitrogen, Carlsbad, CA). For in vivo EP, the anterior tibialis muscle was injected with pLCMV-NP in 30 μl of water and electroporated one time at the same site with a Minimally Invasive Device (MID) as previously described [54] using the CELLECTRA adaptive constant current device (Inovio Pharmaceuticals, Inc., Blue Bell, PA). Briefly, square-wave pulses were delivered through a triangular 3-electrode array consisting of 26-gauge solid stainless steel electrodes and two constant-current pulses of 0.1 Amps were delivered for 52 msec/pulse separated by a 1 sec delay.
Splenocyte isolation and ELISpot assay
Mice were sacrificed 7 days following the final immunization with plasmid DNA or acute LCMV infection and the spleens were harvested and placed in RPMI 1640 medium (Mediatech Inc., Manassas, VA) supplemented with 10% FBS, 1X Anti-anti (Invitrogen), and 1X β-ME (Invitrogen). Splenocytes were isolated by mechanical disruption of the spleen using a Stomacher machine (Seward Laboratory Systems Inc., Bohemia, NY), and the resulting product was filtered using a 40 μm cell strainer (BD Falcon). The cells were treated for 5 min with ACK lysis buffer (Lonza, Switzerland) for lysis of RBCs and then the splenocytes were washed in PBS and then resuspended in RPMI medium.
An IFNγ ELISPOT assay was conducted as previously described [55]. Briefly, ELISPOT 96-well plates (Millipore, Billerica, MA) were coated with anti-mouse IFN-γ capture antibody and incubated for 24h at 4°C (R&D Systems, Minneapolis, MN). The following day, plates were washed with PBS and then blocked for 2 h with blocking buffer (1% BSA and 5% sucrose in PBS). Fifty to two-hundred thousand splenocytes per well and in triplicate from each animal were stimulated overnight at 37°C in 5% CO2 and in the presence of RPMI 1640 (negative control), Concanavalin A (Con A; positive control), or a pool of three peptides (10−7M each) containing immunodominant LCMV epitopes from the H-2b background (DbNP396–404 (NP396), KbNP205–212 (NP205), and I-AbNP309–328 (NP309) (Invitrogen)). After approximately 18 – 24 h of stimulation, the cells were washed in PBS and incubated for 24 h at 4°C with biotinylated anti-mouse IFN-γ mAb (R&D Systems, Minneapolis, MN). The plates were washed in PBS, and streptavidin–alkaline phosphatase (MabTech, Sweden) was added to each well and incubated for 2 h at room temperature. The plates were washed again in PBS, BCIP/NBT Plus substrate (MabTech) was added to each well for 15 – 30 min, and then the plate was rinsed with distilled water and dried at room temperature. Spots were counted with an automated ELISPOT reader (Cellular Technology Ltd., Shaker Heights, OH).
ELISA
To determine sera Ab titers against LCMV NP, Nunc-Immuno MaxiSorp plates (Nunc, Rochester, NY) were coated overnight at 4°C with 5 μg/well of recombinant NP protein that does not include the HA tag sequence (Impact Biologicals, Malvern, PA) or BSA (control) diluted in PBS. The next day, plates were washed with PBS, 0.05% Tween 20 (PBS-T), blocked for 1 h with 10% BSA/PBS-T, and incubated with serial dilutions of serum from immunized/infected animals overnight at 4°C. Plates were then washed six times and bound IgG or IgM was detected using goat anti-mouse IgG or IgM (Santa Cruz, Santa Cruz, CA) at a dilution of 1:5,000. Bound enzyme was detected by SigmaFAST™ O-phenylenediamine dihydrochloride (OPD; Sigma-Aldrich), and the optical density was determined at 450 nm on a Biotek (Winooski, VT) EL312e reader. The reciprocal endpoint titer was reported as the 10% of maximum OD calculated by curve fitting using the sigmoidal dose-response model with a variable slope in GraphPad Prism (GraphPad Software Inc., La Jolla, CA) as described [56].
Statistical Analysis
All values are reported as the mean ± SEM. Analysis between groups was completed by ANOVA with a post-hoc Dunnett’s test to correct for multiple comparisons to one control (LCMV infected). All statistical analysis was carried out using the Statistical Package for the Social Sciences (SPSS, Chicago, IL).
Acknowledgments
We would like to acknowledge J.D. Boyer, M.A. Kutzler, K. A. Kraynyak, and members of the Weiner laboratory for significant contributions and/or critical reading of this manuscript. This work was supported by NIH/NIAID/DAIDS under an HVDDT contract award (HHSN272200800063C) to Inovio Pharmaceuticals, Inc. as well as a HIVRAD grant (P01-AI071739) to DBW, and in part by finding from the NIH to DBW and DJS (T32-AI070099).
Footnotes
COMPETING FINANCIAL INTERESTS
The laboratory of DBW has grant funding and collaborations, advising, or consulting including serving on scientific review committees for commercial entities and therefore notes possible conflicts associated with this work with Pfizer, Inovio, BMS, Virxsys, Ichor, Merck, Althea, VGXI, J&J, Aldevron, and possibly others. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Peters CJ. Lymphocytic choriomeningitis virus--an old enemy up to new tricks. N Engl J Med. 2006 May 25;354(21):2208–11. doi: 10.1056/NEJMp068021. [DOI] [PubMed] [Google Scholar]
- 2.Khanolkar A, Fuller MJ, Zajac AJ. T cell responses to viral infections: lessons from lymphocytic choriomeningitis virus. Immunol Res. 2002;26(1–3):309–21. doi: 10.1385/IR:26:1-3:309. [DOI] [PubMed] [Google Scholar]
- 3.Oldstone MB. Arenaviruses. II. The molecular pathogenesis of arenavirus infections. Introduction. Curr Top Microbiol Immunol. 2002;263:V–XII. [PubMed] [Google Scholar]
- 4.Oldstone MB. Biology and pathogenesis of lymphocytic choriomeningitis virus infection. Curr Top Microbiol Immunol. 2002;263:83–117. doi: 10.1007/978-3-642-56055-2_6. [DOI] [PubMed] [Google Scholar]
- 5.Shin H, Blackburn SD, Blattman JN, Wherry EJ. Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J Exp Med. 2007 Apr 16;204(4):941–9. doi: 10.1084/jem.20061937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slifka MK. Mechanisms of humoral immunity explored through studies of LCMV infection. Curr Top Microbiol Immunol. 2002;263:67–81. doi: 10.1007/978-3-642-56055-2_5. [DOI] [PubMed] [Google Scholar]
- 7.Zinkernagel RM. Lymphocytic choriomeningitis virus and immunology. Curr Top Microbiol Immunol. 2002;263:1–5. doi: 10.1007/978-3-642-56055-2_1. [DOI] [PubMed] [Google Scholar]
- 8.Bishop DH, Auperin DD. Arenavirus gene structure and organization. Curr Top Microbiol Immunol. 1987;133:5–17. doi: 10.1007/978-3-642-71683-6_2. [DOI] [PubMed] [Google Scholar]
- 9.Buchmeier MJ, Oldstone MB. Protein structure of lymphocytic choriomeningitis virus: evidence for a cell-associated precursor of the virion glycopeptides. Virology. 1979 Nov;99(1):111–20. doi: 10.1016/0042-6822(79)90042-4. [DOI] [PubMed] [Google Scholar]
- 10.Buchmeier MJ, Welsh RM, Dutko FJ, Oldstone MB. The virology and immunobiology of lymphocytic choriomeningitis virus infection. Adv Immunol. 1980;30:275–331. doi: 10.1016/s0065-2776(08)60197-2. [DOI] [PubMed] [Google Scholar]
- 11.Kotturi MF, Peters B, Buendia-Laysa F, Jr, Sidney J, Oseroff C, Botten J, et al. The CD8+ T-cell response to lymphocytic choriomeningitis virus involves the L antigen: uncovering new tricks for an old virus. J Virol. 2007 May;81(10):4928–40. doi: 10.1128/JVI.02632-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehmann-Grube F, Assmann U, Loliger C, Moskophidis D, Lohler J. Mechanism of recovery from acute virus infection. I. Role of T lymphocytes in the clearance of lymphocytic choriomeningitis virus from spleens of mice. J Immunol. 1985 Jan;134(1):608–15. [PubMed] [Google Scholar]
- 13.Volkert M, Bro-Jorgensen K, Marker O, Rubin B, Trier L. The activity of T and B lymphocytes in immunity and tolerance to the lymphocytic choriomeningitis virus in mice. Immunology. 1975 Sep;29(3):455–64. [PMC free article] [PubMed] [Google Scholar]
- 14.Volkert M, Lundstedt C. The provocation of latent lymphocytic choriomeningitis virus infections in mice by treatment with antilymphocytic serum. J Exp Med. 1968 Feb 1;127(2):327–39. doi: 10.1084/jem.127.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zinkernagel RM, Doherty PC. Restriction of in vitro T cell-mediated cytotoxicity in lymphocytic choriomeningitis within a syngeneic or semiallogeneic system. Nature. 1974 Apr 19;248(450):701–2. doi: 10.1038/248701a0. [DOI] [PubMed] [Google Scholar]
- 16.Zinkernagel RM, Welsh RM. H-2 compatibility requirement for virus-specific T cell-mediated effector functions in vivo. I. Specificity of T cells conferring antiviral protection against lymphocytic choriomeningitis virus is associated with H-2K and H-2D. J Immunol. 1976 Nov;117(5 Pt 1):1495–502. [PubMed] [Google Scholar]
- 17.Gledhill AW. Protective effect of anti-lymphocytic serum on murine lymphocytic choriomeningitis. Nature. 1967 Apr 8;214(5084):178–9. doi: 10.1038/214178c0. [DOI] [PubMed] [Google Scholar]
- 18.Klavinskis LS, Whitton JL, Oldstone MB. Molecularly engineered vaccine which expresses an immunodominant T-cell epitope induces cytotoxic T lymphocytes that confer protection from lethal virus infection. J Virol. 1989 Oct;63(10):4311–6. doi: 10.1128/jvi.63.10.4311-4316.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klavinskis LS, Whitton JL, Joly E, Oldstone MB. Vaccination and protection from a lethal viral infection: identification, incorporation, and use of a cytotoxic T lymphocyte glycoprotein epitope. Virology. 1990 Oct;178(2):393–400. doi: 10.1016/0042-6822(90)90336-p. [DOI] [PubMed] [Google Scholar]
- 20.Schulz M, Aichele P, Vollenweider M, Bobe FW, Cardinaux F, Hengartner H, et al. Major histocompatibility complex--dependent T cell epitopes of lymphocytic choriomeningitis virus nucleoprotein and their protective capacity against viral disease. Eur J Immunol. 1989 Sep;19(9):1657–67. doi: 10.1002/eji.1830190921. [DOI] [PubMed] [Google Scholar]
- 21.Whitton JL, Sheng N, Oldstone MB, McKee TA. A “string-of-beads” vaccine, comprising linked minigenes, confers protection from lethal-dose virus challenge. J Virol. 1993 Jan;67(1):348–52. doi: 10.1128/jvi.67.1.348-352.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Djavani M, Yin C, Lukashevich IS, Rodas J, Rai SK, Salvato MS. Mucosal immunization with Salmonella typhimurium expressing Lassa virus nucleocapsid protein cross-protects mice from lethal challenge with lymphocytic choriomeningitis virus. J Hum Virol. 2001 Mar-Apr;4(2):103–8. [PMC free article] [PubMed] [Google Scholar]
- 23.Holst PJ, Bartholdy C, Stryhn A, Thomsen AR, Christensen JP. Rapid and sustained CD4(+) T-cell-independent immunity from adenovirus-encoded vaccine antigens. J Gen Virol. 2007 Jun;88(Pt 6):1708–16. doi: 10.1099/vir.0.82727-0. [DOI] [PubMed] [Google Scholar]
- 24.Holst PJ, Sorensen MR, Mandrup Jensen CM, Orskov C, Thomsen AR, Christensen JP. MHC class II-associated invariant chain linkage of antigen dramatically improves cell-mediated immunity induced by adenovirus vaccines. J Immunol. 2008 Mar 1;180(5):3339–46. doi: 10.4049/jimmunol.180.5.3339. [DOI] [PubMed] [Google Scholar]
- 25.Castrucci MR, Hou S, Doherty PC, Kawaoka Y. Protection against lethal lymphocytic choriomeningitis virus (LCMV) infection by immunization of mice with an influenza virus containing an LCMV epitope recognized by cytotoxic T lymphocytes. J Virol. 1994 Jun;68(6):3486–90. doi: 10.1128/jvi.68.6.3486-3490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altmeyer R, Girard M, van der Werf S, Mimic V, Seigneur L, Saron MF. Attenuated Mengo virus: a new vector for live recombinant vaccines. J Virol. 1995 May;69(5):3193–6. doi: 10.1093/benz/9780199773787.article.b00034516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goossens PL, Milon G, Cossart P, Saron MF. Attenuated Listeria monocytogenes as a live vector for induction of CD8+ T cells in vivo: a study with the nucleoprotein of the lymphocytic choriomeningitis virus. Int Immunol. 1995 May;7(5):797–805. doi: 10.1093/intimm/7.5.797. [DOI] [PubMed] [Google Scholar]
- 28.Russmann H, Shams H, Poblete F, Fu Y, Galan JE, Donis RO. Delivery of epitopes by the Salmonella type III secretion system for vaccine development. Science. 1998 Jul 24;281(5376):565–8. doi: 10.1126/science.281.5376.565. [DOI] [PubMed] [Google Scholar]
- 29.Shams H, Poblete F, Russmann H, Galan JE, Donis RO. Induction of specific CD8+ memory T cells and long lasting protection following immunization with Salmonella typhimurium expressing a lymphocytic choriomeningitis MHC class I-restricted epitope. Vaccine. 2001 Nov 12;20(3–4):577–85. doi: 10.1016/s0264-410x(01)00363-2. [DOI] [PubMed] [Google Scholar]
- 30.Fayolle C, Osickova A, Osicka R, Henry T, Rojas MJ, Saron MF, et al. Delivery of multiple epitopes by recombinant detoxified adenylate cyclase of Bordetella pertussis induces protective antiviral immunity. J Virol. 2001 Aug;75(16):7330–8. doi: 10.1128/JVI.75.16.7330-7338.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saron MF, Fayolle C, Sebo P, Ladant D, Ullmann A, Leclerc C. Anti-viral protection conferred by recombinant adenylate cyclase toxins from Bordetella pertussis carrying a CD8+ T cell epitope from lymphocytic choriomeningitis virus. Proc Natl Acad Sci U S A. 1997 Apr 1;94(7):3314–9. doi: 10.1073/pnas.94.7.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh MK, Deriaud E, Saron MF, Lo-Man R, Henry T, Jiao X, et al. Induction of protective antiviral cytotoxic T cells by a tubular structure capable of carrying large foreign sequences. Vaccine. 2002 Jan 31;20(9–10):1369–77. doi: 10.1016/s0264-410x(01)00467-4. [DOI] [PubMed] [Google Scholar]
- 33.Rueda P, Martinez-Torrecuadrada JL, Sarraseca J, Sedlik C, del Barrio M, Hurtado A, et al. Engineering parvovirus-like particles for the induction of B-cell, CD4(+) and CTL responses. Vaccine. 1999 Sep;18(3–4):325–32. doi: 10.1016/s0264-410x(99)00202-9. [DOI] [PubMed] [Google Scholar]
- 34.Sedlik C, Dadaglio G, Saron MF, Deriaud E, Rojas M, Casal SI, et al. In vivo induction of a high-avidity, high-frequency cytotoxic T-lymphocyte response is associated with antiviral protective immunity. J Virol. 2000 Jul;74(13):5769–75. doi: 10.1128/jvi.74.13.5769-5775.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sedlik C, Saron M, Sarraseca J, Casal I, Leclerc C. Recombinant parvovirus-like particles as an antigen carrier: a novel nonreplicative exogenous antigen to elicit protective antiviral cytotoxic T cells. Proc Natl Acad Sci U S A. 1997 Jul 8;94(14):7503–8. doi: 10.1073/pnas.94.14.7503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mandal M, Kawamura KS, Wherry EJ, Ahmed R, Lee KD. Cytosolic delivery of viral nucleoprotein by listeriolysin O-liposome induces enhanced specific cytotoxic T lymphocyte response and protective immunity. Mol Pharm. 2004 Jan 12;1(1):2–8. doi: 10.1021/mp034021m. [DOI] [PubMed] [Google Scholar]
- 37.Giacalone MJ, Zapata JC, Berkley NL, Sabbadini RA, Chu YL, Salvato MS, et al. Immunization with non-replicating E. coli minicells delivering both protein antigen and DNA protects mice from lethal challenge with lymphocytic choriomeningitis virus. Vaccine. 2007 Mar 8;25(12):2279–87. doi: 10.1016/j.vaccine.2006.11.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grujic M, Holst PJ, Christensen JP, Thomsen AR. Fusion of a viral antigen to invariant chain leads to augmented T-cell immunity and improved protection in gene-gun DNA-vaccinated mice. J Gen Virol. 2009 Feb;90(Pt 2):414–22. doi: 10.1099/vir.0.002105-0. [DOI] [PubMed] [Google Scholar]
- 39.Hassett DE, Slifka MK, Zhang J, Whitton JL. Direct ex vivo kinetic and phenotypic analyses of CD8(+) T-cell responses induced by DNA immunization. J Virol. 2000 Sep;74(18):8286–91. doi: 10.1128/jvi.74.18.8286-8291.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodriguez F, Zhang J, Whitton JL. DNA immunization: ubiquitination of a viral protein enhances cytotoxic T-lymphocyte induction and antiviral protection but abrogates antibody induction. J Virol. 1997 Nov;71(11):8497–503. doi: 10.1128/jvi.71.11.8497-8503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rottembourg D, Filippi CM, Bresson D, Ehrhardt K, Estes EA, Oldham JE, et al. Essential role for TLR9 in prime but not prime-boost plasmid DNA vaccination to activate dendritic cells and protect from lethal viral infection. J Immunol. 2010 Jun 15;184(12):7100–7. doi: 10.4049/jimmunol.0803935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yokoyama M, Zhang J, Whitton JL. DNA immunization confers protection against lethal lymphocytic choriomeningitis virus infection. J Virol. 1995 Apr;69(4):2684–8. doi: 10.1128/jvi.69.4.2684-2688.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zarozinski CC, Fynan EF, Selin LK, Robinson HL, Welsh RM. Protective CTL-dependent immunity and enhanced immunopathology in mice immunized by particle bombardment with DNA encoding an internal virion protein. J Immunol. 1995 Apr 15;154(8):4010–7. [PubMed] [Google Scholar]
- 44.Leifert JA, Lindencrona JA, Charo J, Whitton JL. Enhancing T cell activation and antiviral protection by introducing the HIV-1 protein transduction domain into a DNA vaccine. Hum Gene Ther. 2001 Oct 10;12(15):1881–92. doi: 10.1089/104303401753153938. [DOI] [PubMed] [Google Scholar]
- 45.Yokoyama M, Zhang J, Whitton JL. DNA immunization: effects of vehicle and route of administration on the induction of protective antiviral immunity. FEMS Immunol Med Microbiol. 1996 Jul;14(4):221–30. doi: 10.1111/j.1574-695X.1996.tb00290.x. [DOI] [PubMed] [Google Scholar]
- 46.Lau LL, Jamieson BD, Somasundaram T, Ahmed R. Cytotoxic T-cell memory without antigen. Nature. 1994 Jun 23;369(6482):648–52. doi: 10.1038/369648a0. [DOI] [PubMed] [Google Scholar]
- 47.Butz EA, Bevan MJ. Massive expansion of antigen-specific CD8+ T cells during an acute virus infection. Immunity. 1998 Feb;8(2):167–75. doi: 10.1016/s1074-7613(00)80469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murali-Krishna K, Altman JD, Suresh M, Sourdive DJ, Zajac AJ, Miller JD, et al. Counting antigen-specific CD8 T cells: a reevaluation of bystander activation during viral infection. Immunity. 1998 Feb;8(2):177–87. doi: 10.1016/s1074-7613(00)80470-7. [DOI] [PubMed] [Google Scholar]
- 49.Ahmed R, Gray D. Immunological memory and protective immunity: understanding their relation. Science. 1996 Apr 5;272(5258):54–60. doi: 10.1126/science.272.5258.54. [DOI] [PubMed] [Google Scholar]
- 50.Shedlock DJ, Shen H. Requirement for CD4 T cell help in generating functional CD8 T cell memory. Science. 2003 Apr 11;300(5617):337–9. doi: 10.1126/science.1082305. [DOI] [PubMed] [Google Scholar]
- 51.Williams MA, Holmes BJ, Sun JC, Bevan MJ. Developing and maintaining protective CD8+ memory T cells. Immunol Rev. 2006 Jun;211:146–53. doi: 10.1111/j.0105-2896.2006.00389.x. [DOI] [PubMed] [Google Scholar]
- 52.Yang JS, Kim JJ, Hwang D, Choo AY, Dang K, Maguire H, et al. Induction of potent Th1-type immune responses from a novel DNA vaccine for West Nile virus New York isolate (WNV-NY1999) J Infect Dis. 2001 Oct 1;184(7):809–16. doi: 10.1086/323395. [DOI] [PubMed] [Google Scholar]
- 53.Shen H, Whitmire JK, Fan X, Shedlock DJ, Kaech SM, Ahmed R. A specific role for B cells in the generation of CD8 T cell memory by recombinant Listeria monocytogenes. J Immunol. 2003 Feb 1;170(3):1443–51. doi: 10.4049/jimmunol.170.3.1443. [DOI] [PubMed] [Google Scholar]
- 54.Morrow MP, Pankhong P, Laddy DJ, Schoenly KA, Yan J, Cisper N, et al. Comparative ability of IL-12 and IL-28B to regulate Treg populations and enhance adaptive cellular immunity. Blood. 2009 Jun 4;113(23):5868–77. doi: 10.1182/blood-2008-11-190520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kraynyak KA, Kutzler MA, Cisper NJ, Khan AS, Draghia-Akli R, Sardesal NY, et al. Systemic immunization with CCL27/CTACK modulates immune responses at mucosal sites in mice and macaques. Vaccine. 2010 Feb 23;28(8):1942–51. doi: 10.1016/j.vaccine.2009.10.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu X, Zhou T, O’Dell S, Wyatt RT, Kwong PD, Mascola JR. Mechanism of human immunodeficiency virus type 1 resistance to monoclonal antibody B12 that effectively targets the site of CD4 attachment. J Virol. 2009 Nov;83(21):10892–907. doi: 10.1128/JVI.01142-09. [DOI] [PMC free article] [PubMed] [Google Scholar]