Abstract
Motivation: A critical task in high-throughput sequencing is aligning millions of short reads to a reference genome. Alignment is especially complicated for RNA sequencing (RNA-Seq) because of RNA splicing. A number of RNA-Seq algorithms are available, and claim to align reads with high accuracy and efficiency while detecting splice junctions. RNA-Seq data are discrete in nature; therefore, with reasonable gene models and comparative metrics RNA-Seq data can be simulated to sufficient accuracy to enable meaningful benchmarking of alignment algorithms. The exercise to rigorously compare all viable published RNA-Seq algorithms has not been performed previously.
Results: We developed an RNA-Seq simulator that models the main impediments to RNA alignment, including alternative splicing, insertions, deletions, substitutions, sequencing errors and intron signal. We used this simulator to measure the accuracy and robustness of available algorithms at the base and junction levels. Additionally, we used reverse transcription–polymerase chain reaction (RT–PCR) and Sanger sequencing to validate the ability of the algorithms to detect novel transcript features such as novel exons and alternative splicing in RNA-Seq data from mouse retina. A pipeline based on BLAT was developed to explore the performance of established tools for this problem, and to compare it to the recently developed methods. This pipeline, the RNA-Seq Unified Mapper (RUM), performs comparably to the best current aligners and provides an advantageous combination of accuracy, speed and usability.
Availability: The RUM pipeline is distributed via the Amazon Cloud and for computing clusters using the Sun Grid Engine (http://cbil.upenn.edu/RUM).
Contact: ggrant@pcbi.upenn.edu; epierce@mail.med.upenn.edu
Supplementary Information:The RNA-Seq sequence reads described in the article are deposited at GEO, accession GSE26248.
1 INTRODUCTION
The ongoing high-throughput sequencing (HTS) revolution in biology is placing significant demand on the informatics community. Being a sequence based technology, alignment algorithms are critical for most applications. Genome alignment algorithms such as Bowtie and BWA rely on Burrows–Wheeler indexing for very fast genome alignment, but they have difficulties with transcriptome alignment due to splicing, RNA editing and variations from the reference such as substitutions, insertions and deletions (Burrows and Wheeler, 1994; Langmead et al., 2009; Li and Durbin, 2009). Additional complications arise from poorly annotated genomes, or from samples with significant polymorphisms from the sequenced organism or from aberrant splicing found in cells with mutations in components of the spliceosome (Meyerson et al., 2010).
There are a number of programs available for RNA-Seq alignment. Table 1 gives a breakdown of the alignment algorithms used in a random sample of 130 papers listed on PubMed that have ‘RNA-Seq’ in the abstract (see Supplementary Table 1 for detailed information). The most commonly cited algorithm is ELAND, which is part of the analysis pipeline bundled by Illumina with its sequencing instruments. But to be viable for RNA-Seq, an algorithm must satisfy three basic criteria: (i) it must align single reads across splice junctions de novo; (ii) it must handle paired-end reads; and (iii) it must run in a reasonable amount of time. Currently, five algorithms are available that satisfy these three criteria: TopHat (Trapnell et al., 2009) GSNAP (Wu and Nacu, 2010), MapSplice (Wang et al., 2010), SpliceMap (Au et al., 2010) and Soap/Soapals (Li et al., 2009). We further desire algorithms be as robust as possible to polymorphisms and sequencing error. Based on our analyses, described below, only GSNAP and MapSplice from this list satisfy this additional criterion. Further, none of the published algorithms attempt to map against both a genome and a transcriptome and to merge the results into one alignment. As will be shown below, there is an advantage to merging genome and transcriptome alignments to achieve better disambiguation, in particular for reads that extend into introns.
Table 1.
Algorithms used in a random sample of RNA-Seq publications
| Algorithm | No. of times used |
|---|---|
| ELAND | 14 |
| SOAP | 5 |
| BLAST | 4 |
| MAQ | 3 |
| BLAT | 3 |
| BWA | 2 |
| NOVOALIGN | 2 |
| TOPHAT | 2 |
| CORONA LITE | 2 |
| BOWTIE | 2 |
| SOLID PIPELINE | 2 |
| SSAHA2 | 1 |
| ERANGE | 1 |
| SEGEMEHL | 1 |
| GSNAP | 1 |
| SPLICEMAP | 1 |
| SEQMAP | 1 |
| PASS | 1 |
| SUPERSPLAT | 1 |
| SOCS | 1 |
| ARACHNE | 1 |
| NUCMER | 1 |
Complete information regarding this literature is provided in Supplementary Table 1.
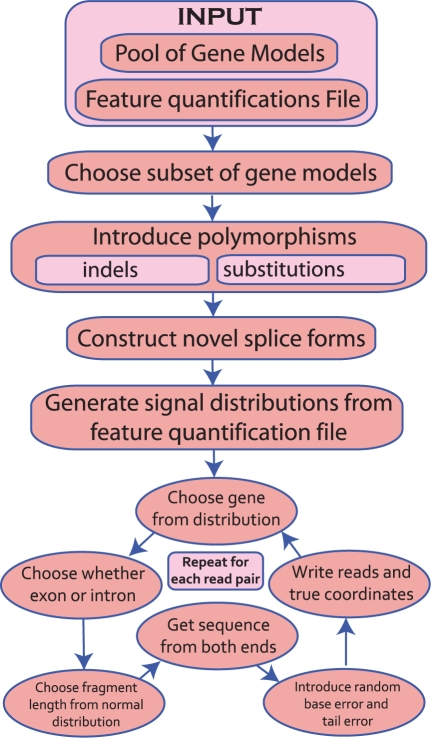
In order to evaluate the accuracy of the various RNA-Seq alignment algorithms, we developed an RNA-Seq simulator that produces paired-end sequence reads with configurable rates for substitutions, indels, novel splice forms, intron signal and random error, including a decrease of the quality in the tails of the reads, as is typically observed in Illumina data. RNA-Seq data are discrete in nature; therefore, as long as good gene models are available, it is possible to simulate RNA-Seq data that is sufficiently realistic to allow for meaningful benchmarking of alignment algorithms. For our purposes, we require paired-end reads with polymorphisms, alternative splice forms, partial retention of introns, and which follows an error model reflective of Illumina data. We also require there be no bias toward any particular set of gene annotations. As far as we are aware, there is no published RNA-Seq simulation software available. However, there are a few simulators available online, e.g. FLUX (flux.sammeth.net, Howard and Heber, 2010), (USeq, useq.sourceforge.net), (simNGS, www.ebi.ac.uk/goldman-srv/simNGS/).However, none of them satisfy the specific requirements for our benchmarking goals. In particular, we require strict control over the sources of polymorphisms: indels, SNPs, errors and alternate splicing. And we require detailed logging. Neither FLUX, USeq nor simNGS provide these capabilities. To meet the necessary criteria, we developed a framework called Benchmarker for Evaluating the Effectiveness of RNA-Seq Software (BEERS) (Fig. 1). The BEERS simulator uses information from a filtered set of the annotated genes from 11 different annotation efforts, to generate simulated sequence read pairs with characteristics similar to those observed in Illumina sequence reads. The details are given in the Section 2 and Supplementary Material.
Fig. 1.
BEERS simulator workflow. Genes are chosen at random from a master pool, polymorphisms and novel splice forms are introduced, and then reads are generated in a six step cycle, as shown.
To evaluate RNA-Seq alignment, we developed a set of metrics to compare an inferred alignment to the true alignment of a BEERS dataset. Accuracy is evaluated on the level of the individual bases and splice junction calls. Neither metric alone indicates which method is superior and it is not clear how to define a single metric that would. For example, BFAST achieves a very high base-wise accuracy, because it handles polymorphisms well and therefore rarely fails to align a read. However, BFAST does not make junction calls and the accuracy at or near splice junctions is quite low, consistent with its original purpose of DNA resequencing (Homer et al., 2009). In contrast, GSNAP, MapSplice and RUM (described below) have a reasonably high base-wise accuracy and very accurate junction detection and so should be preferable overall.
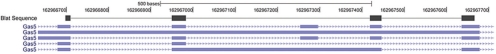
Alignment of transcriptome sequences is not a new problem, as mapping EST's to the genome has been an informatics challenge long before the advent of HTS sequencing. A number of solutions for alignment of ESTs are available, the most popular of which is BLAT (Blast Like Alignment Tool) (Kent, 2002). BLAT has been criticized as inappropriate for short read lengths and is viewed by many as too slow for mapping tens of millions of reads (Dimon et al., 2010). However, as computational resources have become cheaper, and read lengths have increased, these issues can reasonably be resolved. BLAT can efficiently map short reads across exon–exon junctions and identify novel splice junctions (Fig. 2); however, BLAT does not take advantage of related query sequences, such as those from paired-end reads, so it does not, without modification, satisfy the three criteria necessary for an RNA-Seq aligner. BLAT also requires significant post-processing to decrease the false positive rate at both the base and junction levels. But, with code wrappers to handle these issues, our benchmarking shows that this solution is at least as effective as the other existing RNA-Seq aligners in terms of accuracy and speed. In what follows we will refer to this method as the RNA-Seq Unified Mapper (RUM) (Fig. 3).
Fig. 2.
Gapped alignment using BLAT. BLAT alignments (segments in black) of a mouse retina 108 base read that spans three exon/exon junctions. The second junction is unannotated, according to the USCS annotation track shown in blue.
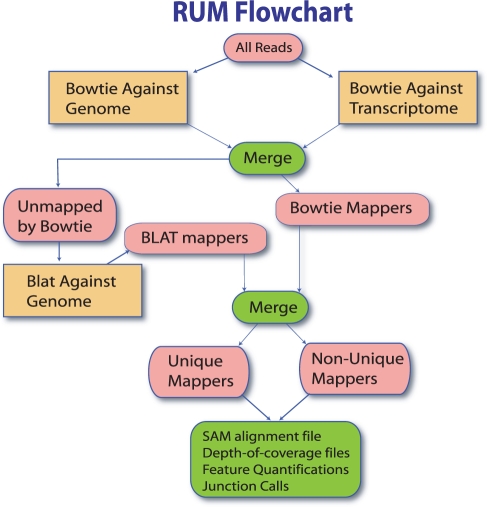
Fig. 3.
The RUM workflow. Reads are first mapped with Bowtie against the genome and transcriptome. This information is merged and non-mappers are sent to BLAT. BLAT and Bowtie mappings are merged for the final alignments. Features are quantified and coverage and junction files are produced.
RUM is implemented as a three-stage pipeline that takes advantage of the speed of Burrows–Wheeler based algorithms, sensitivity of BLAT, information coming from paired-end sequencing and information from both genome and transcriptome alignments. The pipeline first aligns reads with Bowtie to a reference genome and to a reference transcriptome, and then applies BLAT to the reads Bowtie could not align. Significant complexity arises in post-processing the BLAT output to reduce the number of false alignments, to utilize paired-end information and to merge the information from the various mappings. The details are given in the Section 2 and Supplementary Material.
In addition to the simulated data analysis, we validated the RUM pipeline empirically, using data from RNA-Seq analyses of mouse retina, and compared with the other algorithms. Most algorithms were able to accurately identify novel splice variants, including splicing events detected novel junctions with low read depth. However, TopHat performed very poorly on this dataset.
2 METHODS
2.1 RNA-Seq benchmarking
To compare methods and to evaluate the accuracy of options and parameter settings, we have developed a benchmarking framework called Benchmarker for Evaluating the Effectiveness of RNA-Seq Software (BEERS) consisting of a data simulation engine and a set of comparative metrics for measuring the accuracy of an inferred alignment (Fig. 1). In order not to bias for or against any particular set of gene models, 11 different sets of annotation were merged (AceView, Ensembl, Geneid, Genscan, NSCAN, Other RefSeq, RefSeq, SGP, Transcriptome, UCSC, Vega), which produced 672 490 distinct gene models with 1 720 769 exons and 1 052 525 introns. These models were filtered to remove most of the junctions that had uncharacterized splice signals, most of which came from the OtherRefSeq track (Supplementary Table 4), resulting in 538 991 final gene models with <0.0003 of the splice signals being uncharacterized. The characterized splice signals are as follows: GTAG, GCAG, GCTG, GCAA, GCGG, GTTG, GTAA, ATAC, ATAA, ATAG and ATAT. In the first step, in a simulation, a number of the 538 991 gene models are chosen at random, with a default of N = 30 000. This is done in order to not bias toward any particular set of gene models. Alternate splice forms are then created for each gene by preferentially leaving in exons, where the number of alternate forms per gene is a parameter with a default of two. The percentage of signal coming from alternate splice forms is a parameter with a default of 20%. Polymorphisms (indels and substitutions) are introduced into the exons, according to independent rates. A gene quantification file (generated in our case from wild-type mouse retina data) is used to assign an empirical distribution of signal that mimics real data. This file is further used to determine the distribution of intronic signal, so that preferential intron inclusion can be simulated. Reads are then produced by choosing a gene at random, possibly leaving in an intron, choosing a fragment of normally distributed length, introducing random base and tail error, and then reporting the M bases of the fragment from either end, where M is the read length. Random base error is set according to one parameter and tail error is set according to three parameters: percent of low-quality tails; length of the low-quality tail; and quality of the low-quality tail. The reads generated are reported to a fasta file. The true coordinates of each real and the true junctions spanned are reported to text files. The set of gene models used, the alternate splice forms and the polymorphisms are reported to log files. See Supplementary Material for code availability.
Datasets with 10 000 000 paired-end 100-base reads were generated for each of two types of data, one with low polymorphism and error rate (Test 1) and another with moderate polymorphism and error rate (Test 2); details are given in Supplementary Material. The human polymorphism rate is roughly one base in 10 000 (Sachidanandam et al., 2000) and the error rate for a clean run of an Illumina machine is less than one base in 200. So Test 1 was designed with those specifications. Model organisms should be reasonably well represented by this case. Test 2 allowed for quite a bit more polymorphism and error, with fairly low-quality tails, which should present more than the average challenge to alignment. Datasets were generated in triplicate to assess the variability of the accuracy metrics. An example of simulated data with intron signal and a two base deletion is shown in Supplementary Figure 1. Three basic metrics were calculated to compare the inferred alignment to the true alignment. The most straightforward is the percent of bases which map uniquely, and to the right location. A second natural metric is the percent of correct junction calls.
In the first (base-wise) metric, some misalignment of indels must be measured as accurate. Suppose, for example, that the reference sequence is ‘CCCACCC’ and that in the sample being sequenced it is ‘CCCAACCC’ due to an insertion. Logically, there is no way an alignment algorithm could determine which ‘A’ was inserted and in fact it does not make sense biologically to consider it an insertion instead of a duplication. Our benchmarking metrics, therefore, judge an algorithm as correct on this insertion as long as it chooses one or the other of these two possibilities, and a general strategy is employed to handle all cases of indel ambiguity.
A second natural metric is the percent of correct junction calls. This is complicated, however, by the many different ways algorithms report junctions. Some of the algorithms being evaluated do not map reads across junctions at all, so they essentially have an FP rate of 0% and an FN rate of 100%. Among the algorithms that do map across junctions, some of them go further to filter the junctions to produce a final set of reported junctions. Other algorithms report all junctions but attach various scores to them, leaving it to the user to decide which to consider. If reads are aligned across junctions by an aligner, but no extra processing is done by the aligner to report a special junctions file, then we simply use as junctions the gaps indicated by an N in the CIGAR string of the SAM record. On the other hand, if the program produces a final set of junctions that are supposed to be the most reliable, then that set was used for benchmarking. If junctions were attached with scores, we adjusted the scores as best as possible to achieve the best performance. Once a final set of junctions is determined, the false-positive rate is the percent of inferred junctions that are not represented in the database of transcripts used for the simulation. The false-negative rate is the percent of junctions in the database that are crossed by at least one read, but which are not represented in the set of junctions inferred by the algorithm.
2.2 Alignment pipeline
The RUM workflow is given in Figure 3. Bowtie is first run against the genome. A read which is contained entirely in one exon, except for a few bases that align to an adjacent exon, will often be erroneously aligned by Bowtie to the start of the intron. Bowtie is therefore also run against a given transcriptome. The genome and transcriptome alignments are compared for consistency and in most cases the transcriptome alignment is preferred, unless there is a paired-end read that indicates to do otherwise. However, determining which alignment to preference is not always straightforward and in any set of merging rules there will be ambiguous cases.
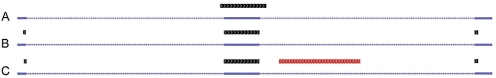
Consider the read alignment in Figure 4. There are three exons, one in the middle and two at each ends. Alignment (A) shows the genome alignment and (B) shows the transcriptome alignment. The correct alignment is uncertain for the few bases on each side that ambiguously align to both the intron and the adjacent exons. Three natural merging strategies arise: (i) preference the transcriptome mapping, (ii) preference the genome mapping or (iii) truncate the alignment and do not report the ambiguous bases. The RUM pipeline preferences the transcriptome alignment in this case. If, however, the paired-end maps as shown in (C), in red, then the genome mapping would be preferred.
Fig. 4.
This illustrates a hypothetical case where it is difficult to resolve the transcriptome and genome mappings. (A) Shows how a read aligns to the genome. It spans an exon and erroneously extends one or two bases on either side into the intron. (B) Shows how the same read maps to the transcriptome. In this case, the few terminal bases map to the adjacent exons. (C) Shown in red is the alignment of the paired-end read, which has aligned to the intron. Even if all bases of all alignments are identities, if all we had was the information in (A) and (B) we would likely preference the transcriptome alignment (B). If we have the information in (C) then we would preference the genome mapping in (A) on the right, but it becomes a difficult judgment on the left, given that the retention of selective introns and partial introns is frequently observed.
The merging rules are guided by a number of cases, which are given in detail in Supplementary Material with a brief description given here. Information between two mapping is joined when possible. So for example, if one mapping aligns the forward read and another the reverse, and they are consistent with being ends of the same fragment, then the two single-end alignments are merged to give the paired-end alignment. If a read (or read pair) has a unique alignment to the transcriptome and a unique alignment to the genome, but the two alignments disagree, then, if they agree on a sufficiently long overlap, just that overlap is reported; otherwise the read is considered a non-unique mapper. In general, RUM tries to resolve ambiguities that are minor by either giving preference to the transcriptome alignment or by just reporting a subalignment consisting of the common spans where both alignments agree.
In the third stage of the RUM pipeline, reads that were not able to be aligned by Bowtie are aligned to the reference genome using BLAT (Kent, 2002). BLAT typically produces many spurious alignments, either because of low complexity sequence or because of partial homology to other locations. Inspection of the false positives gives rise to a number of filters which achieve alignments with an apparently low occurrence of false positives. We then validated and refined these filters using simulated benchmarks. For example, the read in Figure 5 aligned incorrectly due to a majority of the read being low complexity sequence (i.e. containing short repeated elements). However, we do not want to filter out low complexity sequence, because they often represent real signal. Instead we identify the low complexity reads and require more stringent alignment parameters for them. Once filtered and parsed for consistency, BLAT alignments are merged with Bowtie alignments via similar rules to the first merging step; however, in this case both mappings can involve junctions, so the rules are somewhat more complex. Details are given in the Supplementary Material.
Fig. 5.
A false positive BLAT alignment of a 120 base read of mouse retina. BLAT has excessively fragmented the read and aligned it to a low complexity region.
A file of unique aligners and another file of non-unique aligners is output. These are human readable and contain basic alignment information for each read (pair). Also output is a SAM file with all alignments unique and non-unique. Depth-of-coverage files are generated from the final set of unique and non-unique aligners, which give the number of reads mapping to each genome location. A feature quantification file is generated that assigns quantified values to genes, exons, introns and junctions using the RPKM measure (Bullard et al., 2010). However, two quantified values are generated, one assuming no non-unique mappers map to the feature, and another assuming all the non-unique mappers aligning to that feature actually do map to the feature. RPKM values are normalized for feature length and number of reads mapped, and so are appropriate to use for comparisons between samples, as long as expression is reasonably well balanced. If data are unbalanced, the data can be normalized, as described in Supplementary Material. The pipeline does not try to adjust for this effect, however, and assumes such normalization, if necessary, will take place downstream.
Junctions are determined by reads that span gaps long enough to be introns (15 bases or more, by default). A bed file is produced with the junctions that have known splice signals and uniquely mapping reads with at least eight bases on each side of the junction. Increasing this beyond eight bases does not significantly affect the FP rate but does start to affect the FN rate (Supplementary Fig. 2). Another bed file is generated with all junctions. Junctions are colored by whether they have known signal, whether they exist in the supplied transcript database and whether or not the signal is canonical. A spreadsheet is also produced that breaks down the different kinds of evidence in separate columns.
RUM is also enabled for strand-specific mapping, variable length reads and DNA mapping.
2.3 Implementation, availability and cloud distribution
RUM is implemented in Perl and is built on top of Bowtie, BLAT and the low complexity filter mdust. RUM requires 64 bit operating systems with at least 6 GB of RAM to handle genomes as large as mouse or human, and is best run on a cluster or multicore machine, while genomes such as Drosophila can be run on single processor 32 bit systems with 4 GB of RAM. A typical mouse or human alignment of 10 million read pairs requires ~ 200 GB of temporary disk space and ~ 20 GB for the final output files, uncompressed. A 100 million read dataset, as is typical for one lane from a HiSeq machine, requires ~ 500 GB of temp space. RUM is also enabled for compute clusters that use the SUN Grid engine. RUM and the simulator are available as open source under the standard GNU agreement to academic institutions.
The RUM pipeline installs on a stand-alone machine or on a cluster running the SUN Grid Engine. In order to distribute software that requires massive compute, a new paradigm is emerging called cloud computing. In one implementation of cloud computing, infrastructure as a service (IAAS), a user can ‘rent’ a virtual machine in a large data center elsewhere (e.g. Amazon Web services). We developed RUM and optimized its use on AWS using the ‘high memory, quadruple extra large instance’, which provides eight virtual cores and 68.4 GB of RAM. Using this instance, a single paired-end lane (25 million, 120 bp reads) is mapped to a mammalian genome in ~ 5–6 h. However, this is dependent on read quality. With lower quality reads, fewer reads will be aligned by Bowtie and more by BLAT, increasing the run time several fold in the worst case.
Instructions to install RUM on various platforms, including on the Amazon Cloud, are provided at cbil.upenn.edu/RUM/.
2.4 RNA-Seq analysis
Animal research was approved by the Institutional Animal Care and Use Committee at the University of Pennsylvania. Five micrograms of total RNA from neural retinas of 2-month-old C57BL/6J mice was used to prepare a cDNA library. The library was generated using the Illumina mRNA-Seq Sample Prep Kit, with an average insert size of 350 bp (± 25 bp) (Illumina, San Diego, CA, USA). The cDNA library was sequenced using four channels of a flow cell on a Genome Analyzer IIx, with 120 bp paired-end reads. Base calls were generated using the CASAVA v1.6 (Illumina) software, and output unfiltered and unaligned in fasta format. These sequence reads are deposited at GEO, accession GSE26248.
2.5 RT-PCR and sequence validation
Reverse transcription polymerase chain reaction (RT–PCR) was performed from total RNA using primers designed to flank the region of interest, and the products electrophoresed on a 2% agarose gel. Bands were excised and sequenced on an ABI 3730xl DNA Analyzer (ABI, Carlsbad, CA, USA).
3 RESULTS
3.1 Simulated data and comparison to other methods
To evaluate the performance of RNA-Seq aligners, we used BEERS to generate two simulated datasets. For the initial test, designated Test 1, we generated data from 30 000 mouse build mm9 transcript models with low indel (0.0005), substitution (0.001) and error frequency (0.005), with no tail error and with only 20% of the signal coming from novel splice forms. For Test 2, we introduced moderate indel (0.0025), substitution (0.005) and error (0.01) frequency, with 25% of the trailing 10 bases having 50% error and 35% of the signal coming from novel splice forms. The gene models include gene families and highly repetitive intron signal. With 100 base paired-end reads, the non-uniqueness issue affects only 2–3% of reads on average. Datasets, each of 10 million pairs of 100 base reads, were generated in triplicate for each of the two tests. Replication in triplicate allows for assessment of the variability of the metrics. See the Supplementary Material for availability of the simulated data.
The reads in the two simulated datasets described above were aligned using RUM, TopHat, BWA, NovoAlign, Soap/Soapals, MapSplice, SpliceMap, GSNAP and BFAST. Additionally, to evaluate the contributions to accuracy of the transcript database to the RUM pipeline, we ran RUM against the genome without the benefit of the transcript database. We also evaluated Bowtie against the genome alone, against the transcriptome alone and a merging of those two alignments. We also evaluated the BLAT module from the RUM pipeline as a stand-alone aligner. As shown in Figure 6, Bowtie alone has relatively low accuracy with regard to alignment. BLAT provides more accurate alignment, and the combination of the two in RUM is better. Similarly, Bowtie and BLAT alone have high false positive and negative rates, respectively, while their combined use in RUM provides much lower false positive and negative rates. We attempted to optimize the performance of each algorithm. The parameters and processing used are given in the Supplementary Material. Some of these algorithms were included in these comparisons because of their common use in practice, even though they do not satisfy all three of the basic requirements for RNA-Seq alignment stated above.
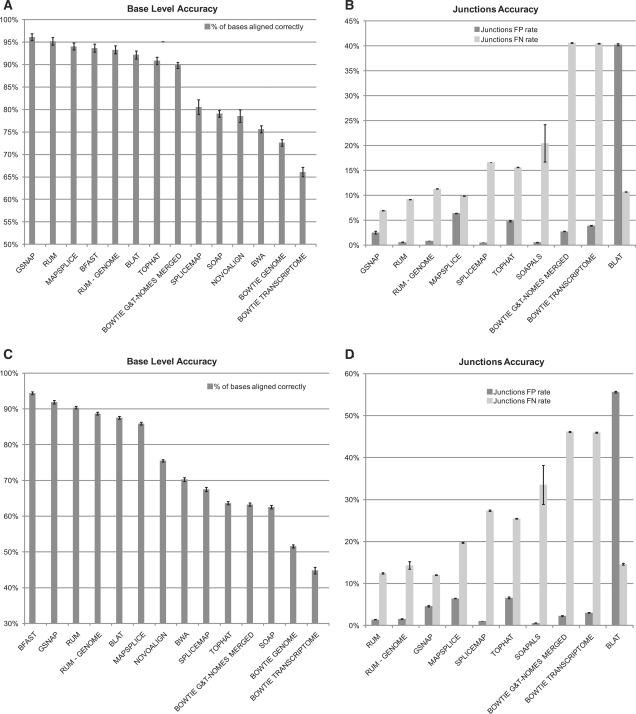
Fig. 6.
Accuracy statistics for analyses of simulated datasets. (A and B) Simulated dataset 1. (C and D) Simulated dataset 2. Test 1 has low polymorphism and error rates, while Test 2 has moderate polymorphism and error rates. In (A) and (C), the bars show the base-wise accuracy (the percent of bases that aligned and to the right location). (B) and (D) Show the accuracy of the junction calls, dark bars show the false positive (FP) rate and light bars show the false negative (FN) rate. The algorithms are sorted in (A) and (C) by accuracy and in (B) and (D) by the sum of the FP and FN rates. Results are mean ± SEM over the three replicate simulated datasets for each test. There is a considerable drop-off in accuracy seen in Test 2 for the algorithms that do not align across indels (SpliceMap, TopHat and Bowtie). The base-wise accuracy and the FP and FN rates on junction calls are taken in conjunction to determine the overall effectiveness of an algorithm. Based on these results, we conclude that GSNAP, MapSplice and RUM are the ones that are most viable for RNA-Seq alignment.
The base-by-base and splice junction accuracies for the analyses of the two tests are given in Figure 6, and Supplementary Table 2. As shown in Figure 6, GSNAP, RUM and MapSplice achieved the most accurate alignment and junction detection on the data in Test 1. RUM and GSNAP also did better on the more complex data in Test 2, with high base-wise and splice junction accuracy. The lower accuracy of SpliceMap, SOAP/SOAPals and TopHat is exacerbated by indels (Fig. 7), with decreased robustness of these algorithms evident by comparing Test 1 to Test 2.
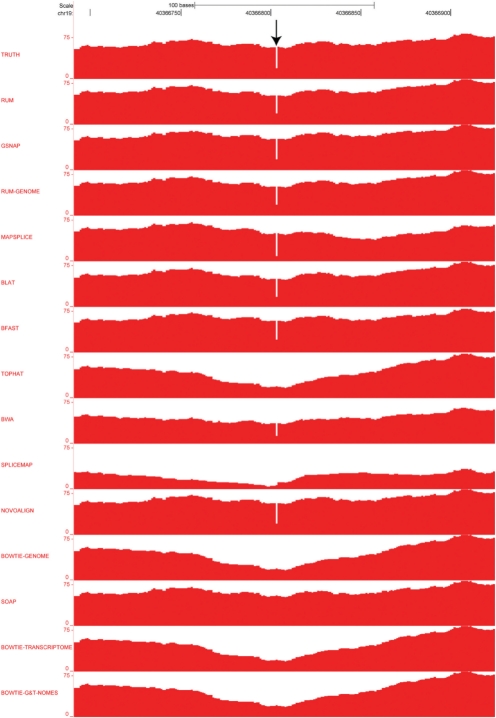
Fig. 7.
Representative coverage plots demonstrating the effect of a two base deletion on alignments with the algorithms indicated. Reads were aligned using RUM, the individual BLAT and Bowtie components of RUM, and 10 currently available alignment algorithms. The TRUTH coverage plot (top) represents the true alignment of the reads containing the two-base deletion (arrow). RUM and several other algorithms were able to correctly align these reads. Note that TopHat, SpliceMap, Bowtie and Soap, which do not identify indels, fail to accurately align reads to these regions.
Figure 8 shows a region with five splice junctions where BLAT- and the BLAT-based algorithms RUM and RUM-Genome properly resolve several of the junctions, as compared with the other algorithms. In contrast, BFAST achieves a high base-wise accuracy, but it does not attempt to make junction calls and in fact has very low accuracy near junctions. Of the three most accurate algorithms, GSNAP, RUM and MapSplice, RUM has the lowest false positive rate on junctions and the junction calls most robust to polymorphisms. Algorithms varied considerably in their false positive (FP) and false negative (FN) rates on junction calls. Ranking junction calls by the sum FP + FN indicates GSNAP and RUM to be the most accurate overall.
Fig. 8.
Comparison of accuracies near junctions on BEERS-generated data. The true junctions are shown in black at the bottom of the figure. Reads mapping to the region of the simulated annotation track (bottom) were aligned using RUM, the individual components of RUM and the 10 currently available alignment algorithms indicated. The TRUTH coverage plot represents the true alignment of the simulated reads. There are five characteristic splice junction sites (1–5) that indicate varying accuracy of the alignment algorithms. BLAT- and the BLAT-based algorithms RUM and RUM-Genome provide the most accurate resolution of the depicted junctions. GSNAP detects the five junctions, and also displays inaccurate alignment of reads in the intron near junction #2.
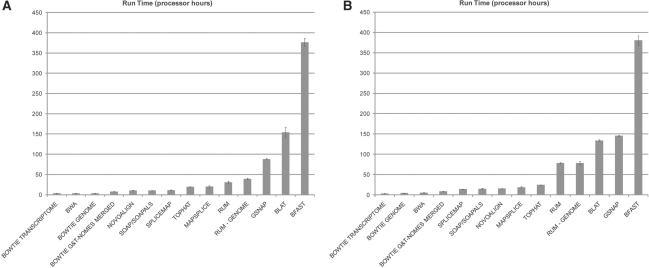
As the number of reads per lane increases upwards to 100 million read pairs, sequence analysis run time becomes an increasing concern. MapSplice cannot, as of yet, be parallelized and on a 94 million read retina dataset, it required 16 days to process, while generating over 2.5 TB of temporary files. In contrast, GSNAP and RUM are both designed for parallel processing and put a much lower demand on the mass storage device. But, GSNAP requires significantly more computational resources: 5 days to process the retina dataset using 300 processors, compared with RUM which required 50 processors and ~ 2 days (Fig. 9). Based on these data, we believe that RUM is currently the most attractive option for RNA-Seq alignment of such large datasets.
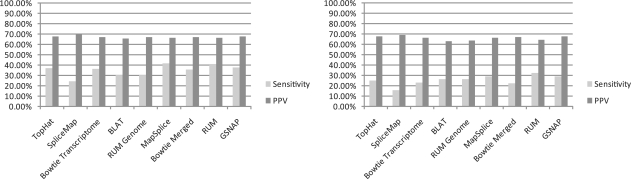
Fig. 9.
The sensitivity and positive predictive value (PPV) at the individual read level. MapSplice and RUM have the highest overall sensitivity, while all algorithms have PPV ~ 65%.
In Test 2, the algorithms that do not attempt to call indels appeared at the bottom of the accuracy list, with a dramatic decrease seen between Test 1 and Test 2. For example, TopHat's overall base-wise accuracy went from 90.86% to 63.67%, while RUM's accuracy only went from 95.19% to 90.39%. In Test 2, RUM achieved the lowest FP + FN rate on junctions with a FP of 1.41% and a FN of 2.48%. In contrast, TopHat's FP rate is 6.62% and FN rate is 25.46%.
For each read that crosses a junction, an algorithm either calls it correctly or not. This allows us to calculate the sensitivity and positive predictive value (PPV) at the individual read level, which is shown in Figure 10. The PPV is ~ 65% in all cases, while MapSplice and RUM have the highest sensitivity, with RUM being the algorithm more robust to polymorphisms, in this case.
Fig. 10.
Processor time required for analysis of simulated datasets. The processor time required for each of the algorithms tested to analyze the first (A) and second (B) simulated datasets is shown. Data are mean ± SEM. The values from which these graphs are derived are shown in Supplementary Table 2. Algorithms were run on 64 bit Linux Debian with 2.6 GHz processors.
3.2 Analysis of a real RNA-Seq library
HTS offers the unprecedented ability to identify novel splice forms, both alternative and aberrant. From empiric RNA-Seq data, we have observed a large number of unannotated splicing events, a majority of which are expressed at a low level compared with annotated variants. We set out to assess the rates at which these represent true and biologically replicable events, and to compare the ability of different RNA-Seq alignment algorithms to detect them. To do this, we prepared an RNA-Seq library from mouse retinal RNA, and sequenced it using four channels of an Illumina Genome Analyzer IIx flow cell to generate 94 million paired-end 120 bp reads. We analyzed these RNA-Seq data using RUM, GSNAP, MapSplice, SpliceMap, Soapals and TopHat, and performed RT–PCR and Sanger sequencing validation studies on biologically independent RNA samples to assess how reliably RUM and the other algorithms detected novel junctions.
Of the 94 million sequence reads in the mouse retina RNA-Seq dataset, ~ 41 million cross exon–exon junctions, including 35 435 507 reads that aligned cleanly (uniquely with at least eight bases on each side, read depth ≥2) to 172 521 known junctions. An additional 290 203 reads aligned cleanly across 47 078 novel junctions with characterized splice signals. ‘Novel’ here means that the junctions are not represented in any of the 11 annotation tracks for the mouse genome currently available from the UCSC genome browser. Many of the novel junctions that were detected fell into three canonical categories: (i) skipping of annotated exons (6001; 12.75%); (ii) inclusion of novel exons in known genes (3207; 6.81%); and (iii) alternate 5′ and 3′ splice sites (≤50 bases from known site, 3802; 8.08%).
We selected 25 examples randomly, from each of the three categories described above, for validation in independent retinal RNA samples. For each category, this includes five cases present in ‘high’ abundance (>33% of reads crossing the novel junction(s), as compared with the annotated junction(s)) and 20 cases present in ‘low’ abundance (<10% of reads crossing the novel junction(s) as compared with the annotated junction(s)). In total, these 75 splicing events involve 100 novel junctions, since the novel exon inclusion events involve two junctions for each example. RT–PCR verified the presence of transcript variants with the novel junctions in 81% of these cases (Table 2 and Supplementary Table 3). Sanger Sequencing verification of the predicted novel junctions was achieved for 55% of these cases. Of the high abundance cases, 100% PCR validated and 95% sequence validated. The detection rates for these novel junctions by GSNAP, MapSplice, SpliceMap, Soapals and TopHat are also listed in Table 2 and detailed in Supplementary Table 3.
Table 2.
The detection rates for RUM novel junctions, by algorithm
| GSNAP | SOAPALS | MAPSPLICE | SPLICEMAP | TOPHAT |
|---|---|---|---|---|
| (%) | (%) | (%) | (%) | (%) |
| 92 | 77 | 98 | 81 | 27 |
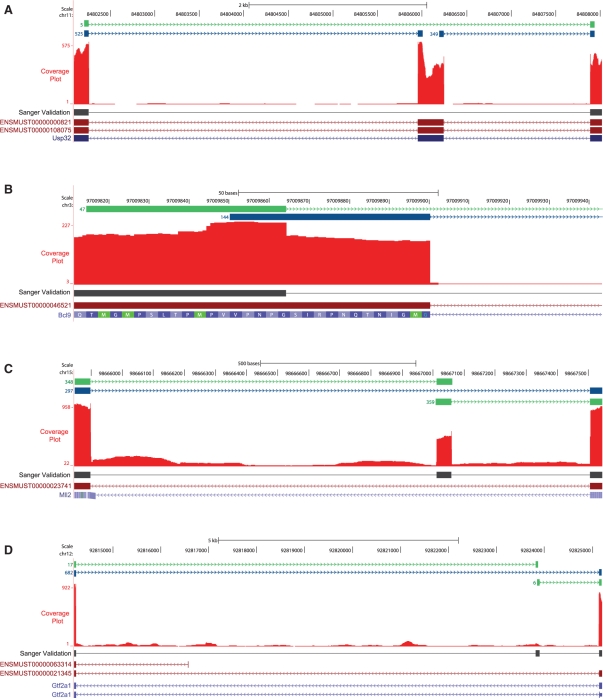
Sequence-verified examples from each of the three novel junction categories are shown in Figure 11 (the full list is available at www.cbil.upenn.edu/RUM/validation). In the first example shown in Figure 11A, five RNA-Seq reads detected a novel junction between exons 29 and 31 of the Usp32 gene, compared with 525 and 349 reads that detected the 5′ and 3′ ends of the annotated exon 30, respectively. RT–PCR and Sanger sequencing confirmed the presence of the mRNA lacking the 289 bp exon 30. In Figure 11B, 47 out of 191 reads detected a novel alternate splice junction at the 5′ end of the 7th and final exon of Bcl9. This novel junction removes 36 bases and 12 amino acids in frame from the coding sequence. RT–PCR and sequencing confirmed the presence of the mRNA with this novel junction. In Figure 11C, an abundant novel exon was detected between exons 50 and 51 of Mll2 gene. In addition to detection by the RUM junction track, this exon is also evident in the coverage plot, shown in red. The 48 bp novel exon is predicted to add 16 amino acids in frame to the Mll2 protein. The presence of this novel transcript in the retina was confirmed by RT–PCR and sequencing. In Figure 11D, 17 and 6 reads, respectively, detected a novel exon between exons 2 and 3 of the Gtf2a1 gene, compared with 682 reads for the known exon–exon junction. RT–PCR and sequencing validated the expression of the novel Gtf2a1 transcript containing this 81 bp exon. This novel exon is located at the 5′ end of the coding sequence, and contains three stop codons in the normal reading frame. In contrast to the novel exon in Mll2, this exon was detected by the identification of novel junctions, and is not evident from the coverage plot (Fig. 11D).
Fig. 11.
Validation of novel splice junctions detected by RUM. Exon junctions detected by RUM are displayed as a track using the UCSC Genome Browser. The reads with annotated junctions are displayed in blue; reads with novel junctions are shown in green. The depth of uniquely mapped sequence reads is shown in the Coverage Plot in red. The BLAT aligned Sanger sequenced reads from RT–PCR products are shown in black under the coverage plot. Annotated Ensemble and UCSC genes are indicated at the bottom of the images. (A) RUM aligned five RNA-seq reads cleanly across a putative novel junction between exons 29 and 31 of the Usp32 gene, compared with 525 and 349 reads that detected the 5′ and 3′ ends of annotated exon 30, respectively. RT–PCR and Sanger sequencing in independent biological samples confirmed the presence of the mRNA lacking exon 30. (B) The 47 reads aligned to a putative novel alternate splice junction at the 5′ end of 7th and final exon of Bcl9, while 144 reads aligned to the known junction. The novel junction removes 36 bases, and 12 amino acids in frame from the coding sequence. RT–PCR and Sanger sequencing in independent biological samples confirmed the presence of the mRNA with this novel junction. (C) An abundant putative novel exon was detected between exons 50 and 51 of Mll2 gene. In addition to detection by the RUM junction track, this exon is also evident in the coverage plot. The 48-bp novel exon is predicted to add 16 amino acids in frame to the Mll2 protein. RT–PCR and Sanger sequencing in independent biological samples confirmed the presence of this novel transcript. (D) A low abundance putative novel exon was detected between exons 2 and 3 of the Gtf2a1 gene. RT–PCR and Sanger sequencing in independent biological samples validated the expression of the novel Gtf2a1 transcript containing this 81 bp exon.
4 DISCUSSION
Robustness of the alignment process to novel splice forms and sequence polymorphisms is a key to wide application of RNA-Seq. Therefore, it is important to test alignment systems with datasets that have varying degrees of such effects, and for which the truth regarding correct alignment is known. BEERS simulates RNA-Seq data with variable levels of polymorphisms, alternative splice forms, partial retention of introns and sequence error. Of these kinds of effects, only sequence error is enabled in the Flux simulator (Howard and Heber, 2010). The ability to simulate alternate splice forms means BEERS can also be used to benchmark the various algorithms that aim to annotate the transcriptome or to reconstruct full splice forms from RNA-Seq data (Guttman et al., 2010; Martin et al., 2010; Trapnell et al., 2010).
We used two configurations of the BEERS parameters, referred to as Test 1 and Test 2, to evaluate 14 alignment algorithms. These simulation analyses indicate that BLAT offers a powerful tool for RNA-Seq alignment that has not been fully explored for RNA-Seq analysis, and as such we have added necessary filters and a paired-end parser to implement this approach in RUM. The analyses performed using the simulated data showed that among the appropriate RNA-Seq alignment algorithms, RUM, GSNAP and MapSplice provide reasonably accurate and robust alignment. Although computing resources are expanding, compute time is still a relevant issue for the analysis of large RNA-Seq datasets. This is underscored by the dramatic increase in the number of reads per lane now generated with the Illumina Hi-Seq and ABI Solid 5500 instruments. All the viable and most accurate alignment algorithms require significant computing resources. For example, none of them can handle a 100 million read dataset on one processor in reasonable time. Therefore, the number of processor hours required for each of the algorithms, as shown in Figure 9, tells only part of the story. What is more important is the number of real hours required for running analyses using the desired alignment software on a reasonably sized compute cluster or multi-processor machine. MapSplice, as yet, cannot be parallelized, and therefore is the least convenient for 100 million read datasets. We use RUM on the High-Performance Computing Facility at the Penn Genome Frontiers Institute consisting of a 400 node cluster of 64 bit Linux machines each with 2.8 GHz quad processors and 16 GB of RAM, managed with the Sun Grid Engine. Using 50 nodes on this cluster, RUM can process clean mouse or human RNA-Seq reads at a rate of ~ 2–3 million read pairs per hour. The run time, however, depends on the error and polymorphism rate, with BLAT taking roughly twice as long on the second simulated data as compared with the first (Fig. 9 and Supplementary Table 2). For small genomes such as microorganisms, run time is considerably faster. Since powerful computational resources may not be available to all investigators, we have taken advantage of the availability of cloud computing to make RUM universally available through the Amazon Elastic Compute Cloud (Amazon EC2: aws.amazon.com/ec2). More generally, RUM should run on any Unix system, and simple installation scripts will place RUM on any of the platforms mentioned above. Further, RUM is designed to work well with default settings in all situations.
When applied to an RNA-Seq dataset from mouse retina, RUM detected 47 078 novel splice junctions with a read depth of ≥2. To explore the reliability of detection for these novel events, we used RT–PCR and Sanger sequencing to validate 75 of them with a focus on the less abundant cases in order to achieve a lower bound on the true occurrence of such novel splicing. We were able to empirically validate 81% of a subset of selected novel junctions in independent RNA samples, indicating the accurate identification of novel junctions by RUM. We believe that the true accuracy of RNA-Seq and RUM are higher than indicated by the RT–PCR and sequencing validation studies we performed due to technical reasons. For example, it is possible that some transcript variants produced by the novel junctions detected by RUM are present at too low a concentration to be detected on agarose gels following RT–PCR. Further, low abundance transcripts detected by RT–PCR are more difficult to isolate for sequencing. We have also found that RUM works well with RNA-Seq data from other species, including human, zebrafish and microorganisms (data not shown).
Perhaps the most important output of RNA-Seq analyses is the identification of novel transcript variants and novel transcripts. Indeed, RNA-Seq data are already being used to improve annotation of the human and mouse transcriptomes (Werner, 2010). The ability to accurately detect the complete complement of transcripts expressed in a given cell or tissue type is especially important for identification of genes, which harbor mutations that cause inherited disorders, and for accurate genetic diagnostic testing of patients with these disorders. A pertinent example of this is the recent identification of a novel, retina-specific isoform of the Bardet-Biedl syndrome 8 (BBS8) gene. Mutations in BBS8 typically cause a multi-system cilia disorder characterized by cystic renal disease, polydactyly, mental retardation, retinal degeneration, obesity, gonadal malformations, diabetes and situs inversus (Ansley et al., 2003; Badano et al., 2006). In contrast, mutations in the retina-specific isoform of BBS8, which was not annotated in the human genome, have recently been identified to cause the retina-specific disorder retinitis pigmentosa (RP) (Riazuddin et al., 2010). The retina-specific isoform of Bbs8, including exon 2a, was readily detected by RUM (Supplementary Fig. 2).
Several of the novel junctions detected in the retina RNA-Seq dataset and validated in these studies also demonstrate the importance of complete characterization of transcriptomes. For example, BCL9 is a component of the Wnt signaling cascade, and is aberrantly expressed in several malignancies. It is hypothesized that deregulation of BCL9 is an important contributing factor to tumor progression (Mani et al., 2009). The variation in splicing of exon 7 of Bcl9 detected in our studies could be relevant to protein function. As a further example, the novel isoform of Mll2 identified in these studies may also have biologic importance, given the known role of the Mll2 protein in histone methylation and regulation of gene expression (Andreu-Vieyra et al., 2010) (Demers et al., 2007). In addition, mutations in MLL2 were recently identified to cause Kabuki syndrome, a form of congenital mental retardation syndrome characterized by post-natal dwarfism, peculiar facies characterized by long palpebral fissures with eversion of the lateral third of the lower eyelids (reminiscent of the makeup of actors of Kabuki, a Japanese traditional theatrical form) and other features (Ng et al., 2010; Niikawa et al., 1981). A complete knowledge of the isoforms of MLL2 expressed in different tissues will be important for investigations of the genetics and pathogenesis of Kabuki syndrome. This idea also applies to other genes and other disorders, and demonstrates the importance of an accurate alignment to the analysis of RNA-Seq and other HTS data.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Lifeng Tian (UPenn), Michael Hughes (Yale), Elisabetta Manduchi (UPenn), Mark Consugar (UPenn), Wei Li (UPenn), John Brestelli (UPenn) and Aaron Goodman (UPenn) for discussions and feedback.
Funding: National Institutes of Health (EY020902 and EY12910 to E.A.P.; F32 EY020747 to M.H.F.); Foundation Fighting Blindness, USA; Rosanne Silbermann Foundation; Penn Genome Frontiers Institute; Institute for Translational Medicine and Therapeutics; EuPath DB Project.
Conflict of Interest: none declared.
REFERENCES
- Andreu-Vieyra C.V., et al. MLL2 is required in oocytes for bulk histone 3 lysine 4 trimethylation and transcriptional silencing. PLoS. Biol. 2010;8 doi: 10.1371/journal.pbio.1000453. pii:e1000453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansley S.J., et al. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- Au K.F., et al. Detection of splice junctions from paired-end RNA-seq data by SpliceMap. Nucleic Acids Res. 2010;38:4570–4578. doi: 10.1093/nar/gkq211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano J.L., et al. The Ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 2006;7:125–144. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Bullard J.H., et al. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC. Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrows M., Wheeler D.J. SRC Research Report 124. Palo Alto, CA: 1994. A block-sorting lossless data compression algorithm; p. 124. Digital Equipment Corporation. [Google Scholar]
- Demers C., et al. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol. Cell. 2007;27:573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimon M.T. et al. HMMSplicer: a tool for efficient and sensitive discovery of known and novel splice junctions in RNA-Seq Data. PLoS One. 2010;5:e13875. doi: 10.1371/journal.pone.0013875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M., et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homer N., et al. BFAST: an alignment tool for large scale genome resequencing. PLoS One. 2009;4:e7767. doi: 10.1371/journal.pone.0007767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard B.E., Heber S. Towards reliable isoform quantification using RNA-SEQ data. BMC Bioinformatics. 2010;11:S6. doi: 10.1186/1471-2105-11-S3-S6. Suppl. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., et al. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Mani M., et al. BCL9 promotes tumor progression by conferring enhanced proliferative, metastatic, and angiogenic properties to cancer cells. Cancer Res. 2009;69:7577–7586. doi: 10.1158/0008-5472.CAN-09-0773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J., et al. Rnnotator: an automated de novo transcriptome assembly pipeline from stranded RNA-Seq reads. BMC. Genomics. 2010;11:663. doi: 10.1186/1471-2164-11-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M. et al. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- Ng S.B., et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat. Genet. 2010;42:790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niikawa N., et al. Kabuki make-up syndrome: a syndrome of mental retardation, unusual facies, large and protruding ears, and postnatal growth deficiency. J. Pediatr. 1981;99:565–569. doi: 10.1016/s0022-3476(81)80255-7. [DOI] [PubMed] [Google Scholar]
- Riazuddin S.A., et al. A splice-site mutation in a retina-specific exon of BBS8 causes nonsyndromic retinitis pigmentosa. Am. J. Hum. Genet. 2010;86:805–812. doi: 10.1016/j.ajhg.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachidanandam R., et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2000;409:928–933. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- Trapnell C., et al. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., et al. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res. 2010;38:e178. doi: 10.1093/nar/gkq622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T. Next generation sequencing in functional genomics. Brief. Bioinform. 2010;11:499–511. doi: 10.1093/bib/bbq018. [DOI] [PubMed] [Google Scholar]
- Wu T.D., Nacu S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics. 2010;26:873–881. doi: 10.1093/bioinformatics/btq057. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.