Abstract
Motivation: Dasty3 is a highly interactive and extensible Web-based framework. It provides a rich Application Programming Interface upon which it is possible to develop specialized clients capable of retrieving information from DAS sources as well as from data providers not using the DAS protocol. Dasty3 provides significant improvements on previous Web-based frameworks and is implemented using the 1.6 DAS specification.
Availability: Dasty3 is an open-source tool freely available at http://www.ebi.ac.uk/dasty/ under the terms of the GNU General public license. Source and documentation can be found at http://code.google.com/p/dasty/.
Contact: hhe@ebi.ac.uk
1 INTRODUCTION
The Distributed Annotation System (DAS) defines a communication protocol used to exchange annotations on genomic or protein sequences (Jenkinson et al., 2008). Implicit in the conception of DAS is the notion of decentralization; annotations should not be provided by single centralized databases, but should instead be spread over multiple sites. DAS relies on client–server architecture; DAS servers manage data distribution by following a standard protocol, while manipulation and visualization are on the client side –thus facilitating the development of tailored tools addressing specific requirements. Several clients are available; for instance, SPICE (Prlic et al., 2005) is a browser that displays protein sequences, structures and their corresponding annotations. Another client, Dasty2, provides a Web-based interface that facilitates visualizing and comparing protein sequence annotations from DAS servers (Jimenez et al., 2008). These clients are addressing specific needs; however, none of them is providing a framework upon which tools can be built and integrated into a cohesive web environment.
2 RESULTS
Dasty3 relies on a modular architecture, which facilitates development of specialized plug-ins. Dasty3 like Dasty2 is a web-based DAS client. However, Dasty3 is not just a client, but a platform delivering a rich API over which tailored plug-ins can be built. Dasty3 inherits from Dasty2 the modularity of its Graphical User Interface (GUI). Additionally, it provides a Web-based framework for visualizing and manipulating information from DAS sources as well as other third-party data providers. It facilitates delivering unified views upon which several manipulation facilities can be implemented. It offers a public Application Programming Interface (API) that delivers methods for integrating, visualizing and manipulating data sources.
2.1 User experience
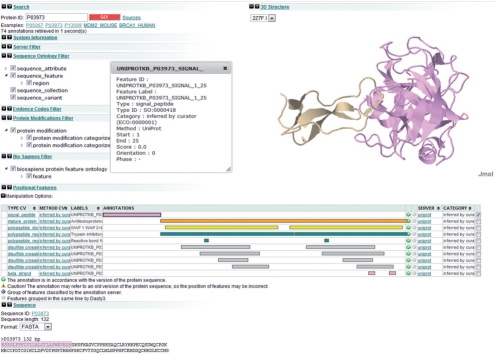
As illustrated in Figure 1, Dasty3 preserves the look and feel of Dasty2; in this way, the learning curve is minimized. Dasty3 enhances some of the previous functionalities available in Dasty2 and adds new ones. For instance, the search plug-in allows users to define the sources against which the query should be executed.
Fig. 1.
A view of Dasty3 and its plug-ins.
Dasty3 uses PICR (Côté et al., 2007) for matching the query string to the corresponding UniProt accession or identifier; the connection to the UniProt DAS reference server (Jones et al., 2005) is then established and the sequence retrieved. Finally, it queries all the selected sources and merges the retrieved sequence annotations. The set of predefined plug-ins provides the user with a unified, organized and interactive view of the available data. A query retrieving information for protein P03973 (antileukoproteinase) is illustrated in Figure 1. The 3D view of the protein structure, supported by Jmol (http://jmol.sourceforge.net), is displayed on the right-hand side while the features, organized in tracks, are presented at the bottom. The pop-up window presents a detailed view of the annotation that is highlighted on the sequence.
New data sources can be manually added including the URL of a valid DAS source. DAS sources can also be selected from a list of sources provided by the DAS registry. The filtering plug-in toggles the display of protein annotations. New filters based on DAS-related ontologies, such as those available from the Ontology Look-up service (Côté et al., 2008), have been included to consistently filter annotations from different sources. A new interaction plug-in retrieves a summary for molecular interaction data. We are using the DAS Writeback (Salazar et al., 2011) to support community annotation. The DAS Writeback is an extension to the DAS protocol. It allows adding, editing and deleting annotations. These functionalities have been implemented in Dasty3 over the DAS Writeback plug-in. Dasty3 also makes it possible to save snapshots of user's sessions in the form of a JSON file. For instance, users may search, select and highlight a feature in the sequence; then, they can generate the corresponding snapshot of the session for future analysis or for sharing it with collaborators.
2.2 Architecture
Dasty3 is based on a plug-in architecture. It emphasizes extensibility, plug-in customization and interoperability e.g. data exchange among plug-ins. For instance, information is exchanged between the sequence and the 3D structure plug-in; as amino acids are selected, these are highlighted in the 3D structure. The architecture makes it possible for plug-ins to listen events from other plug-ins. Furthermore, the architecture also makes it easy for developers to integrate Dasty3 into existing Web applications as well as to configure the look-and-feel by defining customized templates.
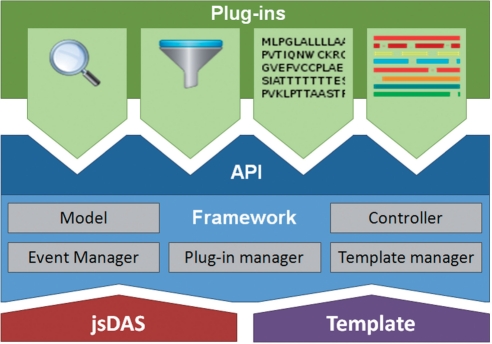
As illustrated in Figure 2, Dasty3 brings together several components that can be accessed by using the API. jsDAS (http://code.google.com/p/jsdas/) is being used as a bridge between the framework and DAS servers; it automatically generates JavaScript objects representing, in concordance with the DAS 1.6 specification, the DAS responses. The framework has five major components, namely: (i) the Model, defining the DAS objects according to the DAS 1.6 specification (http://www.biodas.org); (ii) the Controller, managing the retrieval, organization and storage of the data; (iii) the Plug-in Manager, making it possible to load plug-ins; (iv) the Event Manager, managing and triggering events so that plug-ins can communicate with each other; and, (v) the Template Manager, rendering the plug-ins in the available area according to the selected template. Plug-ins are components that extend the capabilities of the framework by delivering a particular functionality. They are defined as a tightly coupled collection of JavaScript and CSS files. Templates add flexibility to the Graphical User Interface (GUI) by simplifying the display of plug-ins and customization of the layout. For instance, moving and reorganizing plug-ins according to end user preferences can be defined in a template. The modularization upon which Dasty3 relies facilitates sharing; in the same way, as developers share modules and themes in Content Management Systems (CMSs) such as Drupal (http://drupal.org) and Joomla (http://www.joomla.org), they are able to share templates and plug-ins in Dasty3.
Fig. 2.
Dasty3 architecture.
3 FINAL REMARKS
Dasty3 is the first DAS client fully compliant with the 1.6. DAS specification. It has been tested on Internet Explorer, Firefox, Chrome and Safari. Dasty3 is highly modular and extensible, and new components can be easily added to the framework. The interoperability delivered by the API facilitates the flow of data across plug-ins. The architecture also facilitates the organization of the front-end by means of templates; easing in this way the definition of the layout and improving the user experience. Dasty3 separates the functional components, being managed by the Plug-in manager, from those related to the user experience and graphical layout, being managed by the Template manager. This separation makes it easier for developers to deliver richer applications built over the Dasty3 framework; in the same way, end users receive more personalized task-oriented tools. In addition, Dasty3 facilitates mechanisms for integrating resources beyond the DAS ecosystem; by doing so, the retrieved information presents a broader view to the end user.
Funding: NHLBI Proteomics Center Award HHSN268201000035C, as well as the European Commission grants ENFIN (LSHG-CT-2005-518254); APO-SYS (FP7-HEALTH-2007-200767) in part.
Conflict of Interest: none declared.
REFERENCES
- Côté R., et al. The Ontology Lookup Service: more data and better tools for controlled vocabulary queries. Nucleic Acids Res. 2008;36:W372–W376. doi: 10.1093/nar/gkn252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté R., et al. The Protein Identifier Cross-Referencing (PICR) service: reconciling protein identifiers across multiple source databases. BMC Bioinformatics. 2007;8:401. doi: 10.1186/1471-2105-8-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson A., et al. Integrating biological data - the distributed annotation system. BMC Bioinformatics. 2008;9:S3. doi: 10.1186/1471-2105-9-S8-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez R.C., et al. Dasty2, an Ajax protein DAS client. Bioinformatics. 2008;24:2119–2121. doi: 10.1093/bioinformatics/btn387. [DOI] [PubMed] [Google Scholar]
- Jones P., et al. Dasty and UniProt DAS: a perfect pair for protein feature visualization. Bioinformatics. 2005;21:3198–3199. doi: 10.1093/bioinformatics/bti506. [DOI] [PubMed] [Google Scholar]
- Prlic A., et al. Adding some SPICE to DAS. Bioinformatics. 2005;1:ii40–ii41. doi: 10.1093/bioinformatics/bti1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar G., et al. DAS Writeback: a collaborative annotation system. BMC Bioinformatics. 2011;12:143. doi: 10.1186/1471-2105-12-143. [DOI] [PMC free article] [PubMed] [Google Scholar]