Abstract
Summary: Current genome browsers are designed for linear browsing of individual genomic regions, but the high-throughput nature of experiments aiming to elucidate the genetic component of human disease makes it very important to develop user-friendly tools for comparing several genomic regions in parallel and prioritizing them based on their functional content. We introduce VISTA Region Viewer (RViewer), an interactive online tool that allows for efficient screening and prioritization of regions of the human genome for follow-up studies. The tool takes as input genetic variation data from different biomedical studies, determines a number of various functional parameters for both coding and non-coding sequences in each region and allows for sorting and searching the results of the analysis in multiple ways.
Availability and implementation: The tool is implemented as a web application and is freely accessible on the Web at http://rviewer.lbl.gov
Contact: rviewer@lbl.gov; ildubchak@lbl.gov
1 INTRODUCTION
The recent technological advances have allowed researchers to discover a wide range of variations on the scale of a complete human genome (Redon et al., 2006). The role of single nucleotide polymorphisms (SNPs) and genomic copy number variations (CNVs) in human disease is becoming increasingly evident (Abrahams and Geschwind, 2008; Barkovich et al., 2009; Bucan et al., 2009; Mefford et al., 2009), but it is often difficult to move from associations to the identification of causative variations.
The number of genes in CNVs may be large, and non-coding regions may also contribute to the phenotype. On the other hand, the identification of functional SNPs requires predicting what impact a certain polymorphism might have on gene function. This is relatively straightforward with sequence variants leading to amino acid changes in protein coding regions, since these are more likely to disrupt gene function. However, a clear result of genome wide association studies (GWASs) is that functional SNPs with association to disease are as likely to be found in the non-coding portion of the genome (Frazer et al., 2009). Thus data obtained by several types of studies require an informatics approach to organize biological knowledge and prioritize genomic intervals for further analysis.
The current, widely used genome browsers, such as UCSC Browser (Fujita et al., 2011). Ensembl (Flicek et al., 2011) and GBrowse (Donlin, 2009) use individual intervals on a reference genome to display the tracks of functional elements such as genes or RNA. Investigating genomic intervals obtained through CNV or other experiments requires looking at each interval individually, while comparison of functional features in multiple intervals can bring new insights into genome studies.
We describe an interactive online genome analysis tool VISTA RViewer, a new addition to the VISTA suite of tools for comparative genomics (Frazer et al., 2004). RViewer is designed to accept datasets produced by clinical geneticists [either CNVs identified in copy number array comparative genomic hybridization (aCGH) studies or genomic neighborhoods of SNPs of interest from GWASs]. It calculates a set of important parameters for both coding and non-coding regions, and provides investigators with capabilities to compare the intervals and identify those that are likely to be significant in a particular study. This tool also links to mouse phenotype and gene expression data, thus allowing for comparisons and prioritization of significant genes and non-coding regions across a region.
RViewer presents several functionalities not found in currently available tools. First, multiple genomic intervals are displayed simultaneously, allowing for quicker visual inspection. Second, direct links to gene ontology, pathway and mouse phenotype data allow for more rapid prioritization of genes and regions of likely biological significance. And third, the annotation of highly conserved non-coding intervals brings an important component into the analysis, and is likely to be useful for whole genome sequencing studies. To our knowledge, software providing such functions is not currently available.
2 DESCRIPTION
2.1 Software
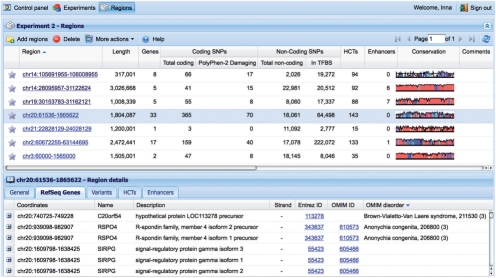
RViewer allows users to create individual accounts and upload their data in the form of genomic regions from either hg18 or hg19 builds of the human genome in the UCSC BED format, analyze the data using various tools, deposit the data for further analysis and export the results. Currently, results of CNV studies and GWASs are supported. Each experiment may include multiple regions visualized in parallel and compared with each other. Given a region location on the chromosome, RViewer integrates a variety of annotations and presents the data in the browser (Fig. 1).
Fig. 1.
VISTA RViewer displaying regions analyzed in this case study (top panel), with annotation of region length, number of RefSeq genes, coding and non-coding SNPs, as well as HCTs and enhancers. A conservation plot is displayed in the last column. The region details view (bottom panel) displays RefSeq genes contained within a selected region, with links to KEGG and mouse phenotype data.
The upper panel of the interface includes general information about each region; the lower panel displays detailed information when one region is selected. Coding and non-coding SNPs within each region are derived from the dbSNP database (Sherry et al., 2001). For coding non-synonymous SNPs, the PolyPhen (Ramensky et al., 2002) annotation is available. For each non-coding region, the homotypic clusters of transcription binding sites (HCT) (Gotea et al., 2010) are provided, as well as the number and location of experimentally verified enhancers from the VISTA Enhancer browser (Visel et al., 2007). The VISTA conservation icon represents the alignments of the human and mouse genomes and gives access to VISTA Point with a wide range of comparative genomics information based on different pair-wise and multiple alignments (Dubchak et al., 2009). For any gene within a region, its Gene Ontology (GO) (Ashburner et al., 2000) terms, KEGG (Kanehisa and Goto, 2000) pathway and mouse phenotype are provided wherever available. Access to relevant databases, such as GenBank, OMIM and the UCSC genome browser is also provided.
The tool is highly interactive. Information in each column can be sorted and filtered, providing flexibility for users to compare and analyze the data. Thus, a user can prioritize the regions based on the number of the coding SNPs predicted deleterious, the number of enhancers, conservation across different species, the GO terms or KEGG pathways of the genes, etc. The total number of SNPs predicted to be deleterious in an interval does not affect the probability that a SNP in that interval is a true causative variant: this requires experimental work to demonstrate. However, the presence of several potentially deleterious SNPs on the same haplotype suggests that that haplotype should be given priority consideration as potentially causative.
The tool is implemented as a Java-based web application. The technologies used include Google Web Toolkit, EXT library, Spring Framework and MySQL database server.
In summary, the RViewer is an analytical platform for genomic regions that allows for visualization, comparison and analysis of large-scale genomic data from current biomedical research.
2.2 Case study
To demonstrate the functionality of RViewer, we analyzed nine de novo copy number variants identified by aCGH in patients with infantile spasms (ISS), a form of epilepsy associated with severe developmental outcome. Preliminary analysis suggested involvement of at least two biological networks—ventral forebrain development and synaptic function. The regions of CNVs (hg18) were uploaded into RViewer and analyzed (Fig. 1). Based on the annotations provided by RViewer, 23 genes with brain expression patterns and GO terms similar to known ISS-associated genes were identified. This allowed for identification of the best candidates for follow-up confirmatory studies much faster than would have been possible using currently available tools. Annotation of non-coding intervals also identified highly conserved regions of possible biological significance for further study.
3 DISCUSSION
The high-throughput nature of modern genomic experiments makes the availability of efficient analytical tools critical. The presented novel software RViewer has been utilized in several biomedical test cases, and showed significant improvement in the speed and quality of analysis allowing for fast retrieval and visualization of relevant biological information. The tool is designed in a modular way allowing for customization and expansion such as taking results of other genomic experiments (for example resequencing, whole exome and genome sequencing), adding annotations relevant to a particular disease, analyzing novel variant data and introducing new complex searches. This work is currently in progress.
ACKNOWLEDGEMENTS
We are grateful to Drs Natalia Maltsev, William Dobyns and Elliott Sherr for fruitful ideas and discussions, to Dr Ivan Ovcharenko for providing the HCT data, to Alexander Poliakov and Igor Ratnere for technical support and to Tatyana Smirnova for designing the RViewer Web site.
Funding: National Institutes of Health (R01 HL091495). The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. (DE-AC02-05CH11231).
Conflict of Interest: none declared.
REFERENCES
- Abrahams B.S., Geschwind D.H. Advances in autism genetics: on the threshold of a new neurobiology. Nat. Rev. Genet. 2008;9:341–355. doi: 10.1038/nrg2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkovich A.J., et al. A developmental and genetic classification for midbrain-hindbrain malformations. Brain. 2009;132(Pt 12):3199–3230. doi: 10.1093/brain/awp247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucan M., et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donlin M.J. Using the Generic Genome Browser (GBrowse) Curr. Protoc. Bioinformatics. 2009 doi: 10.1002/0471250953.bi0909s28. Chapter 9Unit 9.9. [DOI] [PubMed] [Google Scholar]
- Dubchak I., et al. Multiple whole-genome alignments without a reference organism. Genome Res. 2009;19:682–689. doi: 10.1101/gr.081778.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P., et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., et al. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., et al. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- Fujita P.A., et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotea V., et al. Homotypic clusters of transcription factor binding sites are a key component of human promoters and enhancers. Genome Res. 2010;20:565–577. doi: 10.1101/gr.104471.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mefford H.C., et al. A method for rapid, targeted CNV genotyping identifies rare variants associated with neurocognitive disease. Genome Res. 2009;19:1579–1585. doi: 10.1101/gr.094987.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V., et al. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redon R., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry S.T., et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A., et al. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res. 2007;35:D88–D92. doi: 10.1093/nar/gkl822. [DOI] [PMC free article] [PubMed] [Google Scholar]