Abstract
Thymine glycol (Tg) is the most common oxidation product of thymine and is known to be a strong block for replicative DNA polymerases. A previously solved structure of the bacteriophage RB69 DNA polymerase (RB69 gp43) in complex with Tg in the sequence context 5’-G-Tg-G shed light on how Tg blocks primer elongation: The protruding methyl group of the oxidized thymine displaces the adjacent 5’-G which can no longer serve as a template for primer elongation. [Aller, P., Rould, M.A., Hogg, M, Wallace, S.S., & Doublié S. (2007) PNAS 104, 814–818]
Several studies showed that in the 5’-C-Tg-Purine sequence context Tg is more likely to be bypassed by Klenow fragment, a family A DNA polymerase. We set out to investigate the role of sequence context on Tg bypass in a B family polymerase and solved the crystal structures of the bacteriophage RB69 DNA polymerase in complex with Tg containing DNA in the three remaining sequence contexts: 5’-N-Tg-G with N=A, T, or C. A combination of several factors influence Tg bypass, including the associated exonuclease activity, the nature of the 3’and 5’ bases surrounding Tg and the cis/trans interconversion of Tg. We also visualized for the first time the structure of a well-ordered exonuclease complex, allowing us to identify and confirm the role of key residues (Phe123, Met256, and Tyr257) in strand separation and the stabilization of the primer strand in the exonuclease site.
Keywords: Bacteriophage RB69 gp43, DNA replication, translesion synthesis, lesion bypass
Introduction
Thymine glycol (5,6-dihydroxy-5,6-dihydrothymine, Tg) is the most common oxidation product of thymine. Tg is produced endogenously by aerobic metabolism and by exogenous factors such as ionizing radiation and chemical oxidants. It has been estimated that within each cell about 400 Tg lesions are generated per day 1; 2; 3. Tg lesions in DNA are recognized and processed by DNA glycosylases from the base excision repair pathway such as human NTH1 and NEIL1 4; 5; 6. Even though Tg can be repaired, DNA polymerases do encounter unrepaired Tg lesions while replicating DNA. Since replicative DNA polymerases preferentially incorporate an adenine opposite Tg, this lesion is not mutagenic 7. On the other hand, Tg has been shown to constitute a strong block to replication in vitro 8; 9; 10, which can be lethal in vivo in the absence of translesion synthesis or recombination 7; 11; 12; 13. Biochemical and structural studies have shown that the replication block arises not at the insertion step, but rather during extension past the Tg•A base pair 8; 10; 14; 15; 16. Most of the studies of DNA polymerases encountering Tg lesions were performed with the proofreading Klenow fragment from Escherichia coli DNA polymerase I, a polymerase of the A family 8; 9; 10; 15; 16; 17; 18. These studies showed that Klenow fragment can bypass Tg in the specific sequence context 5’-C-Tg-Purine in the DNA template strand. In contrast T4 DNA polymerase, a B family replicative DNA polymerase, cannot bypass Tg regardless of the sequence context 15; 16. In fact, the ability of a polymerase to bypass replication blocking lesions, including Tg, is inversely correlated with its exonuclease activity 8; 10; 15; 16.
We recently solved the crystal structure of a homolog of the T4 replicative DNA polymerase, bacteriophage RB69 gp43, in complex with Tg-containing DNA in the sequence context: 5’-G-Tg-G 14. The structure revealed how the non-planar Tg displaces its 5’-neighbor guanine and thereby blocks elongation after incorporation of an adenine opposite the lesion. In order to further investigate the molecular mechanism underlying Tg bypass by a B family DNA polymerase, we performed elongation assays of RB69 gp43 lacking exonuclease activity in four different template sequence contexts (5’-N-Tg-G with N=A, G, T or C) and solved the crystal structures of polymerase/DNA complexes in the same contexts. A comparison of the different structures suggests that both the 5’ and 3’-neighbor bases of Tg influence bypass. In addition, the 5’-T-Tg-G ternary complex structure, which contains four polymerase/DNA complexes per asymmetric unit, revealed a polymerizing complex with the fingers domain opened and an editing complex in which the primer and the lesion-containing template are visualized for the first time in their entirety interacting with the β-hairpin structure of the exonuclease domain.
Materials and Methods
Enzyme and oligonucleotides
The exonuclease deficient variant (D222A and D327A) of RB69 gp43 (RB69 exo−) was expressed and purified as described previously 19. The oligodeoxyribonucleotides were purchased from the Midland Certified Reagent Co. (Midland, TX) and purified on a 16% (w/v) polyacrylamide gel containing 8 M urea and desalted on a Sep-Pak (Millipore) cartridge. The 18 mer template sequence is 5’-CGN(Tg)GAATGACAGCCGCG with N=A, T, C or G named 5’-A-Tg-G, 5’-T-Tg-G, 5’-C-Tg-G and 5’-G-Tg-G, respectively. The 13 mer primer (5’-GCGGCTGTCATTC) was annealed with each of the 18 mer templates in a buffer containing 10 mM Tris-HCl (pH 7.5) and 50 mM NaCl, heated to 90°C for 5 min and slowly cooled to room temperature. The primer used in primer extension assays was 5’-labeled with tetrachlorofluorescein.
Primer extension assays
In a volume of 20 µL, 200 nM of RB69 exo− was pre-incubated with 100 nM duplex oligodeoxyribonucleotide in a buffer containing 35 mM NaCl, 10 mM Hepes pH 7.5 and 2.0 mM DTT. The reaction was started by adding 20 µL of solution containing 20 mM MgCl2 and 250 µM of either dATP or a dNTP mixture. The final concentrations in the reaction mixture were: 100 nM enzyme, 50 nM oligodeoxyribonucleotides, 125 µM dATP or dNTP, and 10 mM MgCl2. At three different times (15s, 60s and 300s), 10 µL aliquots were quenched with 10 µL formamide. Extended primers were separated from the template on 16% polyacrylamide gels containing 8 M urea. DNA bands were visualized on a Bio-Rad (Hercules, CA) Molecular Imager FX at 532 nm to excite the tetrachlorofluorescein fluorophore.
Crystallization of the protein/DNA complexes
5’-C-Tg-G complex: The exonuclease deficient polymerase RB69 exo− was mixed in an equimolar ratio (0.1 mM) with annealed DNA duplex 5’-C-Tg-G and 2 mM acyclic ATP (New England Biolabs). Hanging drops were made by mixing 0.5 µL of reaction mixture and 0.5 µL of reservoir solution [6.5% (v/v) PEG 2000 MME, 100 mM Na acetate, 150 mM MgSO4, 100 mM Hepes (pH 7.2), 6% (v/v) glycerol and 2 mM β-mercaptoethanol] and equilibrated against 1 mL of reservoir solution at 24°C. The crystals (140 × 60 × 60 µm3) grew within two weeks and belong to the P21 space group with unit cell parameters a = 133.53 Å, b = 123.44 Å, c = 164.29 Å, β = 96.84° (Table 1) and contain four molecules per asymmetric unit. Crystals were cryoprotected by increasing the concentration in the drop of PEG 2000 MME and glycerol to 10 % and 17 % (v/v), respectively and flash cooled in liquid nitrogen.
Table 1.
Data collection and refinement statistics.
| 5’-C-Tg-G | 5’-A-Tg-G | 5’-T-Tg-G-bin | 5’-T-Tg-G-ter | |
|---|---|---|---|---|
| PDB ID | 3RMB | 3RMA | 3RMC | 3RMD |
| Data collection | ||||
| Space group | P21 | P21 | P21 | P21 |
| Cell dimensions | ||||
| a, b, c (Å) | 133.53, 123.44, 164.29 | 132.70, 123.00, 163.94 | 133.99, 123.77, 163.64 | 132.84, 123.02, 168.77 |
| β (°) | 96.84 | 96.07 | 96.57 | 95.87 |
| Resolution (Å) | 50-2.65 (2.74-2.65)a | 50-2.84 (2.94-2.84)a | 50-3.0 (3.11-3.0)a | 50-2.98 (3.09-2.98)a |
| Rmerge | 7.6 (45.8)a | 17.0 (76.4)a, b | 8.5 (69.5)a | 9.2 (40.9)a |
| R Friedel (I) | 7.2 (55.9)a | 8.4 (64.0)a, b | 7.7 (75.3)a | 7.7 (46.8)a |
| I / αI | 15.5 (2.2)a | 10.7 (2.0)a | 12.0 (2.3)a | 15.7 (2.3)a |
| Completeness (%) | 98.4 (87.9)a | 99.8 (98.5)a | 99.9 (99.9)a | 98.1 (82.5)a |
| Redundancy | 3.9 (2.6)a | 7.9 (6.3)a | 4.2 (4.0)a | 4.6 (3.1)a |
| Refinement | ||||
| Resolution (Å) | 50-2.65 | 50-2.84 | 50-3.0 | 50-2.98 |
| No. reflections | 595569 (154468)c | 991917 (125453)c | 454618 (107108)c | 501756 (111177)c |
| Rwork / Rfree (%) | 22.42 / 27.63 | 21.79 / 27.35 | 22.16 / 28.20 | 21.99 / 27.54 |
| No. atoms | 32166 | 31450 | 31561 | 31873 |
| Protein | 29065 | 28890 | 28935 | 29076 |
| DNA | 2596 | 2236 | 2505 | 2549 |
| Water | 505 | 324 | 121 | 188 |
| Average B-factors (Å2) | ||||
| Protein (A, B, C, D) | 63.5, 66.5, 50.7, 96.7 | 52.9, 84.0, 58.4, 114.4 | 73.4, 93.6, 71.0, 128.1 | 60.5, 61.4, 61.6, 76.6 |
| DNA (A, B, C, D) | 105.4, 66.5, 51.2, 127.9 | 95.6, 109.9, 64.6, 101.9 | 117.8, 117.3, 66.3, 155.5 | 114.9, 84.0, 66.8, 110.4 |
| Water | 43.5 | 41.7 | 47.2 | 32.4 |
| R.m.s deviations | ||||
| Bond lengths (Å) | 0.0071 | 0.0071 | 0.0079 | 0.0082 |
| Bond angles (°) | 1.217 | 1.227 | 1.307 | 1.278 |
Highest resolution shell is shown in parentheses.
Two isomorphous data sets were merged.
Number of all reflections. Number of unique reflections is in parentheses.
5’-A-Tg-G complex: RB69 exo− was mixed in an equimolar ratio (0.1 mM) with DNA duplex 5’-A-Tg-G, 1.0 mM dideoxyATP (Sigma Aldrich). Hanging drops were made as described above, albeit with a different reservoir solution [10–11 % (v/v) PEG 2000 MME, 100 mM Na acetate, 150 mM MgSO4, 100 mM Hepes (pH 7.2), 6–10 % (v/v) glycerol and 2 mM β-mercaptoethanol]. The crystals (200 × 40 × 40 µm3) grew within two weeks in space group P21 with unit cell parameters a = 132.70 Å, b = 123.00 Å, c = 163.94 Å, β = 96.07° (Table 1) and contain four molecules per asymmetric unit. Crystals were cryoprotected by increasing or maintaining the concentration in the drop of PEG 2000 MME and glycerol to 12 % and 18 %, respectively then flash cooled in liquid nitrogen.
5’-T-Tg-G binary complex (5’-T-Tg-G-bin): RB69 exo− was mixed in an equimolar ratio (0.1 mM) with DNA duplex 5’-T-Tg-G and 15 mM dideoxyATP. Hanging drops were made by mixing 1 µL of reaction mixture with 1 µL of reservoir solution [4.5 % (v/w) PEG 20,000, 150 mM Na acetate, 125 mM Mg acetate, 100 mM Tris-HCl (pH 6.8), 1 % (v/v) glycerol, 10 mM phenol and 2 mM β-mercaptoethanol] and equilibrated against 1 mL of reservoir solution at 24°C. Crystals (140 × 60 × 60 µm3) grew within 10 days in space group P21 with unit cell parameters a = 133.99 Å, b = 123.77 Å, c = 163.64 Å, β = 96.57° (Table 1) and contained four molecules per asymmetric unit. Crystals were cryoprotected by increasing the concentration of glycerol in the drop to 13 % (v/v), and flash cooled in liquid nitrogen.
5’-T-Tg-G ternary complex (5’-T-Tg-G-ter): RB69 exo− was mixed in equimolar ratio (0.1 mM) with DNA duplex 5’-T-Tg-G and 15 mM dATP. Hanging drops were made by mixing 0.5 µL of reaction mixture with 0.5 µL of reservoir solution [4.0 % (v/w) PEG 20,000, 100 mM Na acetate, 125 mM Mg acetate, 100 mM Tris-HCl (pH 6.8), 0.1 % (v/v) glycerol, 10 mM phenol and 2 mM β-mercaptoethanol)] and equilibrated against 1 mL of reservoir solution at 24°C. Crystals (100 × 40 × 40 µm3) grew within 10 days in space group P21 with unit cell parameters a = 132.84 Å, b = 123.02 Å, c = 168.77 Å, β = 95.87° (Table 1) and also contained four molecules per asymmetric unit. Crystals were cryoprotected by increasing the concentration of glycerol to 13% (v/v) and flash cooled in liquid nitrogen.
Data collection
X-Ray data were collected on a MAR m300 CCD detector at the Advanced Photon Source at beamline 23 ID-D for the complexes 5’-C-Tg-G and 5’-A-Tg-G, and beamline 23 ID-B for 5’-T-Tg-G-bin and 5’-A-Tg-G. For 5’-T-Tg-G-ter, the data were collected at beamline BL-17a of the Photon Factory (Tsukuba, Japan). Data sets were collected at 100 K at λ = 0.99187 Å for 5’-C-Tg-G, λ = 1.000 Å for 5’-T-Tg-G-ter and λ = 1.0332 Å for 5’-T-Tg-G-bin and 5’-A-Tg-G. All data sets were processed and scaled using HKL 2000 20. Data collection statistics are reported in Table 1.
Structure determination and refinement
Our previously determined structure of a binary complex between RB69 gp43 exo− and Tg-containing DNA (PDB ID: 2DY4) 14 devoid of all non-protein atoms was used for rigid body refinement of the different complexes. The structure refinement consisted of successive cycles of rebuilding with Coot 21 followed by positional refinement with NCS restraints, simulated annealing and individual B-factor refinement with CNS 1.21 22; 23. The Rfree value was calculated with 10 % of the reflections that were omitted during the refinement. The water molecules were added during the last refinement cycles with Coot.
The final model of the complex 5’-C-Tg-G includes residues 1 – 903 for molecules A and B, whereas residues 901 – 903 and 898 – 903 are missing for molecule C and D respectively. Primer and template DNA strands are complete in all four molecules per asymmetric unit.
To obtain the final model of 5’-A-Tg-G complex, two isomorphous data sets were merged and scaled together. Molecules A and B include residues 1 – 903, except residues 252 – 260 and 254 – 259 for molecules A and B, respectively; the missing residues belong to the β-hairpin loop 24. Molecules C and D include residues 1 – 899 and 1 – 898, respectively. Residues 252 – 260 and 253 – 260 are missing in molecules C and D, respectively. The first four, five and six bases at the 5’-end of the template are missing in molecules A, B and D respectively. Only the first base at the 5’-end of the template is missing in molecule C. In molecules B and D the incorporated ddAMP at the 3’-end of the primer was not built because of poor density.
The final model 5’-T-Tg-G-bin includes residues 1 – 902 for molecules A and B, 1 – 901 for molecule C and 1 – 897 for molecule D. Residues 254 – 260 are missing in molecule D. The first four bases at the 5’-end of the template and the incorporated ddAMP at the 3’-end of the primer are missing in molecule D.
In the final model of the complex 5’-T-Tg-G-ter, molecules A and B include residues 1 – 902, molecule C residues 1 – 901 and molecule D residues 1 – 897. In molecule A the first five bases on the 5’-end of the template and the incorporated dAMP at the 3’-end of the primer are missing.
The quality of the crystal structures was assessed with PROCHECK 25: All non glycine residues are found in the allowed regions of the Ramachandran plot except Thr622 (in all molecules of the different complexes): this residue adopts a distorted geometry due to its proximity to the active site aspartates Asp621 and Asp623 14; 19; 24; 26; 27; 28. The density for the three N-terminal histidine residues was missing in all molecules. The refinement statistics are shown in Table 1. All molecular structure figures were created using PyMol 29. Atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank (www.pdb.org) and are available under the XXXX accession code for 5’-C-Tg-G, XXXX for 5’-A-Tg-G, XXXX for 5’-T-Tg-G-bin and XXXX for 5’-T-Tg-G-ter.
Results
RB69 gp43 exo− is able to bypass Tg in vitro
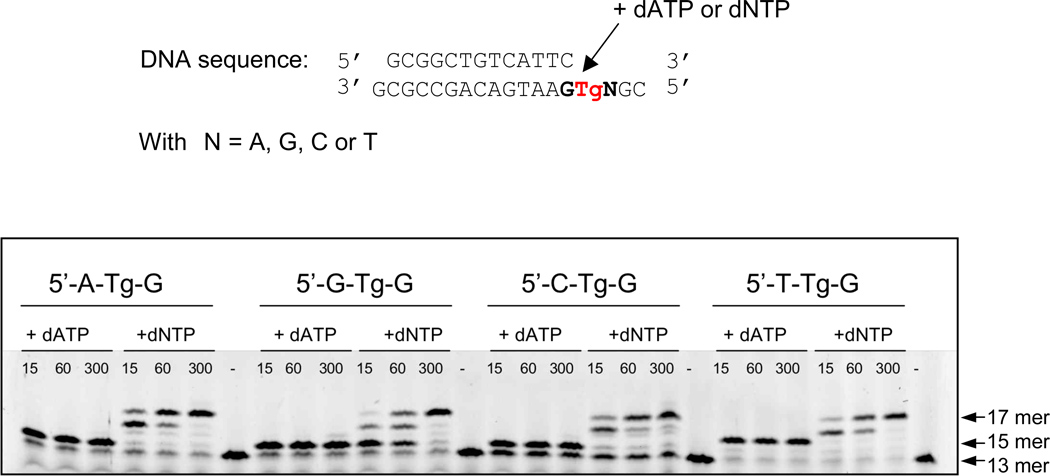
We investigated the ability of a processive DNA polymerase of the B family to bypass Tg in different sequence contexts (5’-N-Tg-G with N=A, T, G or C) using primer extension assays. With an exonuclease-deficient variant of the bacteriophage RB69 DNA polymerase (RB69 exo−), we found that like other DNA polymerases 8; 9; 10; 15; 16 RB69 exo− incorporates an adenine opposite Tg (Figure 1, lanes +dATP), independently of the sequence context. In the sequence context 5’-T-Tg-G and in the presence of dATP, two adenines are added.
Figure 1.
RB69 gp43 exo− can bypass Tg. Primer extension assays were performed for the four sequence contexts by adding either dATP only or a mix of the four deoxyribonucleotides (dNTP). The incubation times were varied from 15 s, 60 s and 300 s. The initial length of the primer before the reaction is a 13 mer. When dNTP is added DNA synthesis goes to completion (17 mer).
When all four deoxynucleoside triphosphates were added to the reaction mix (Figure 1, lanes +dNTP), RB69 exo− extended the primer to completion (17 mer). However for the 5’-G-Tg-G sequence (Figure 1, lane +dNTP, 5’-G-Tg-G), the polymerase stalled for a longer time after the incorporation of presumably an adenine opposite Tg (14 mer on the gel, figure 1) than for the other three sequences. The previously solved crystal structure of RB69 exo- in complex with DNA containing this sequence context showed how the methyl group of Tg displaces the 5’ G and thereby prevents further elongation (Figure 2A) 14. Tg is stabilized by hydrogen bond interactions with both the 5’ and 3’ G and with the newly incorporated adenine.
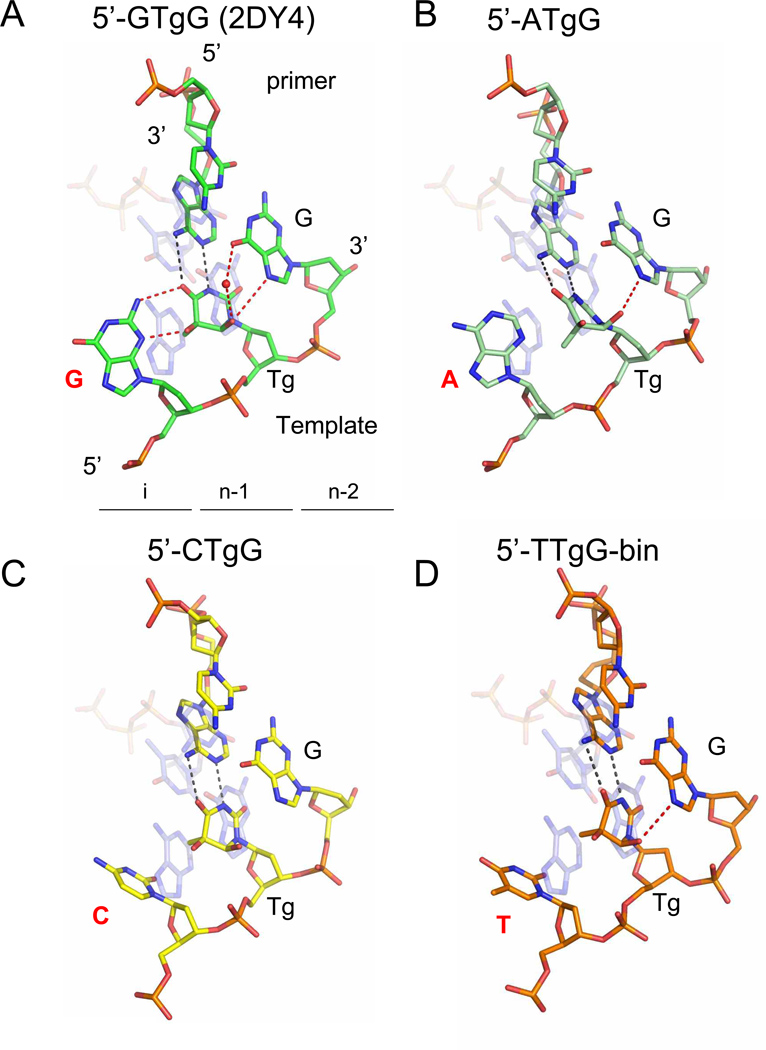
Figure 2.
Snapshots of RB69 exo− with Tg in different sequence contexts. The four sequence contexts are represented: (A) 5’-G-Tg-G (PDB ID code 2DY4) 14, (B) 5’-A-Tg-G, (C) 5’-C-Tg-G and (D) 5’-T-Tg-G-bin. All structures are superimposed 56 with a ternary complex obtained with normal DNA in which A•dTTP is the incipient base pair (transparent light blue) (PDB ID code 1IG9) 27. Tg is located in the post insertion site (n-1) whereas the 5’-neighboring base lies in the insertion site (i). Watson-Crick hydrogen bonds in the Tg•A base pair are represented with black dashed lines, whereas other hydrogen bonds involving Tg are shown as red dashed lines. Water molecules are shown as red spheres.
The results of the primer extension assays prompted us to seek the crystal structures of RB69 exo− in all four sequence contexts in order to shed light on the molecular mechanisms that influence Tg bypass.
Structure determination and overall structure description
Crystals were obtained with RB69 gp43 exo− bound to a 13-mer primer and an 18-mer template containing the Tg lesion, in all four sequence contexts (5’-G-Tg-G, 5’-A-Tg-G, 5’-C-Tg-G and 5’-T-Tg-G), and in the presence of a dATP analog 14. Binary complexes were obtained after incorporation of the chain terminating ddATP in the sequence contexts 5’-A-Tg-G and 5’-T-Tg-G, and with acyclic ATP (AcyATP) in the other sequence contexts. In all binary complexes, including the one solved previously (PDB ID: 2DY4, 5’-G-Tg-G) 14, the enzyme was able to translocate the DNA after incorporating an adenine opposite Tg thus the lesion lies in the n-1 insertion site (Figure 2A–2D). In the 5’-T-Tg-G sequence context we also obtained a ternary complex (5’-T-Tg-G-ter) when dATP was used instead of ddATP. In all cases the fingers domain is in the open conformation.
All complexes crystallized in the monoclinic space group P21 and there are four polymerase/DNA complexes (molecules A, B, C and D) per asymmetric unit. In previously reported monoclinic structures of RB69 gp43, the primer strand of the DNA duplex can be observed either in the polymerase site (generally molecules A and C) or in the exonuclease site (molecules B and D) 19; 28. In some cases the DNA primer strand lies in the polymerase site for the four complexes per asymmetric unit 14; 24. An important factor influencing the formation of a polymerizing vs. an exonuclease complex is the presence of the β-hairpin structure (residues 248 to 265) of the exonuclease domain 24; 30. The β hairpin has been shown to be important for forming stable exonuclease complexes in T4 and RB69 DNA polymerases 24; 31; 32. When the tip of the β-hairpin loop (residues 253 to 261) is deleted the equilibrium is shifted towards the formation of the polymerizing complex 24. The presence of a lesion in the template strand can also influence the formation of an exonuclease complex 19. All Tg complexes described below have an asymmetric unit composed of four polymerizing complexes with the primer strand in the polymerase active site, with the exception of the 5’-T-Tg-G ternary complex, which has three polymerizing complexes (molecules A, B, and C) and one exonuclease complex (molecule D). For each of complexes, the description and discussion will focus solely on molecule C, as it is the most ordered of the four molecules per asymmetric unit, with the lowest B factors (Table 1). We will also describe molecule D of the 5’-T-Tg-G-ter complex in which the DNA was found in the exonuclease active site.
5’ purines are forced into an extrahelical position by Tg
The structure of the 5’-G-Tg-G complex was solved previously 14 (PDB ID: 2DY4). We showed that Tg was able to establish Watson-Crick base pair hydrogen bonds with the incorporated adenine (Figure 2A) resembling a regular T•A base pair. The C5 methyl group of the oxidized thymine protrudes axially from the damaged pyrimidine ring and hinders stacking with the 5’ adjacent guanine in the template. This guanine is stabilized in an extrahelical position by two hydrogen bonds with Tg (figure 2A) and cannot serve as a template for further elongation. Tg is also stabilized by hydrogen bonds (direct or mediated by a water molecule) with the G on the 3’-side on the template (figure 2A).
The 2.84 Å structure of the 5’-A-Tg-G complex was solved by isomorphous replacement using the previously solved binary complex of RB69 exo− with Tg- containing DNA (PDB ID: 2DY4) devoid of all non-protein atoms 14 and refined to Rwork and Rfree values of 21.79% and 27.35%, respectively (Table 1). Although there is also a purine (adenine) on the 5’-side of Tg, we observed a significant difference when compared to the 5’-G-Tg-G complex. Tg can still establish a Watson-Crick base pair with the opposite adenine in the primer strand (Figure 2B), and the 5’ purine is extrahelical, but in contrast to what was observed in the 5’-G-Tg-G structure, the 5’-A does not make any hydrogen bond with Tg. The oxidized thymine is stabilized by a hydrogen bond with the G on the 3’-side.
5’ pyrimidines are less affected by Tg than 5’ purines
A 5’-C-Tg-G complex was obtained with acyclic ATP (acyATP). AcyATP was added instead of dATP because the crystal quality was slightly better with acyATP as shown previously 14 and family B polymerases can use the acyclic form of nucleotides quite efficiently 33. The 2.65 Å structure was solved as described above and refined to Rwork and Rfree values of 22.42% and 27.63%, respectively (Table 1). The 3.0 Å structure of the 5’-T-Tg-G binary complex (5’-T-Tg-G-bin) obtained with dideoxyATP was solved as described above and refined to Rwork and Rfree values of 22.16% and 28.20%, respectively (Table 1).
In both the 5’-C-Tg-G and 5’-T-Tg-G-bin complexes, we observe similar Watson-Crick base pairing between Tg and the newly incorporated adenine (Figures 2C and 2D), as seen in the 5’-Purine-Tg-G complexes. The pyrimidine group in the insertion site (i) is less bulky than a purine, and as a result, the pyrimidine adopts a configuration that differs from that of G or A (Figure 2). Although the C5 methyl of Tg still hinders proper stacking with the pyrimidine in (i), the pyrimidine is not completely rotated out of the helix. The pyrimidine therefore adopts an “intermediate” conformation, neither stacked nor completely extra-helical. In the 5’-T-Tg-G binary complex, we note that a hydrogen bond is formed between Tg and G in the (n-2) post-insertion site (Figure 2D) like in the 5’-purine-Tg-G complexes (Figures 2A and 2B).
An open ternary complex of 5’-T-Tg-G is revealed
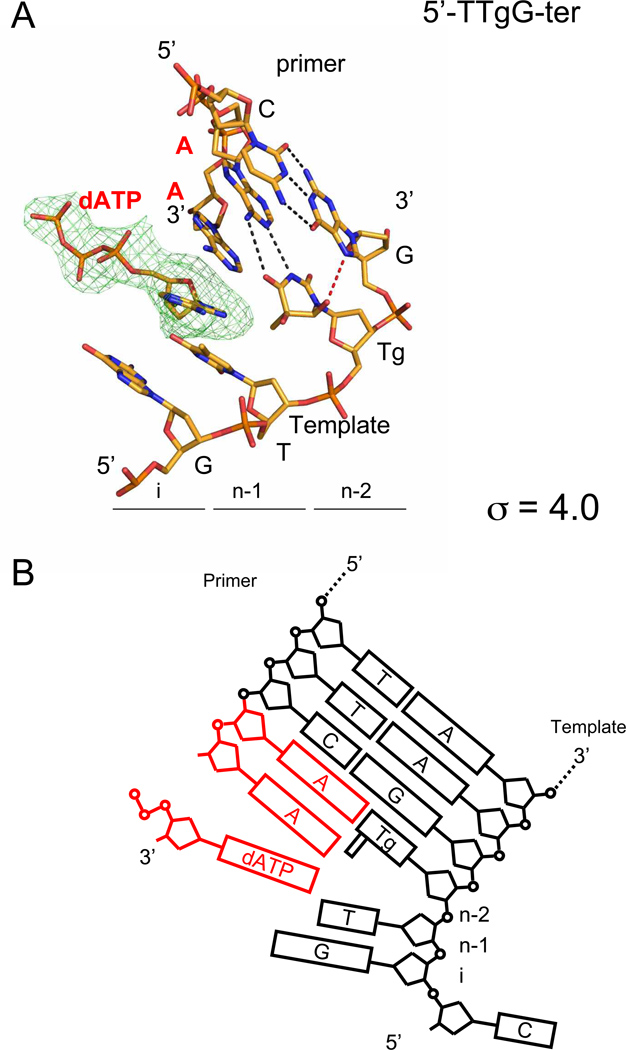
The 2.98 Å structure of the 5’-T-Tg-G ternary complex (5’-T-Tg-G-ter) with dATP was solved as described above, and refined to Rwork and Rfree values of 21.99% and 27.54%, respectively (Table 1). The DNA for molecules A, B and C was found in the polymerase active site, and in the exonuclease active site for molecule D. In molecules B and C we clearly observe the electronic density for an incoming dATP in the polymerase active site, yet the fingers are open. This complex is similar to an open ternary complex reported previously with complexes of RB69 gp43 exo− and a non-natural purine triphosphate analog, 5-nitro-1-indolyl-2'-deoxyriboside-5'-triphosphate (5-NITP)28. We note that we were unable to obtain ternary complexes in the other three sequence contexts, even in the presence of a large excess of incoming nucleotide. Interestingly in molecule D of the 5’-T-Tg-G-ter complex, we are now able to visualize for the first time both the DNA primer and template strands in their entirety in the exonuclease active site, along with the β hairpin motif. This structure, which will be described in further detail below, provides useful insight into strand separation and the proofreading reaction.
In the 5’-T-Tg-G-ter complex, we observe that Tg is also able to form a Watson-Crick base pair with the opposite adenine (Figure 3A). Like in the 5’-G-Tg-G, 5’-A-Tg-G and 5’-T-Tg-G-bin complexes, Tg interacts with the 3’G via a direct hydrogen bond interaction. Tg sits in the (n-2) post-insertion site (Figure 3A), meaning that two translocation events happened. In fact, two adenines were incorporated in the primer strand (Figures 3A and 3B). This observation agrees with the results of the primer extension assay (Figure 1, 5’-T-Tg-G lane +dATP). Surprisingly, the second adenine added is not paired with any base from the template, including the thymine at (n-1) (Figures 3A and 3B). Another unusual feature of this structure is the presence of an incoming dATP in the (n-1) post-insertion site (Figures 3A and 3B) in spite of the fact that the fingers domain is in the open conformation. The dATP is in syn conformation (torsion angle O4’-C1’-N9-C4 = 54.3°) 34 and does not pair with any opposite base. In the regular ternary complex with fingers closed 27; 28; 35, the triphosphate moiety interacts with the conserved basic residues Arg482, Lys486, Lys560, and with Asp411 and Asp623 via divalent ions. The interactions with Arg482, Lys486, and Lys560 are only possible if the fingers are in the closed conformation. In our open ternary complex, we did not find any divalent cation coordinated with the triphosphate tail of dATP and no interaction with the residues mentioned above was observed. Only one non-bridging oxygen of the β-phosphate makes a hydrogen bond with Asn564 of the fingers domain. A superposition with a closed ternary complex 27 reveals that the α-P is far from the 3’-end of the primer (5.6 Å compared to 4.5 Å in a closed ternary complex) and the 3’-end of the primer in the Tg complex is oriented differently compared to a regular ternary complex. The misalignment of the primer 3’end and α-P does not allow completion of nucleophilic attack 27.
Figure 3.
Open ternary complex with Tg in the sequence context 5’-T-Tg-G. (A) Tg is located in the post-insertion site (n-2) meaning that two translocation events occurred. The simulated annealing omit map is centered on the incoming dATP in the insertion site (i) and contoured at 4.0 σ. Watson-Crick hydrogen bonds are represented by black dashed lines, whereas the hydrogen bond involving Tg is shown as a red dashed line. (B) Diagram representation of the DNA in the open ternary complex. The two adenines added to the primer by RB69 exo− and the incoming dATP are shown in red.
An editing complex confirms the roles of key amino acid residues in the proofreading process
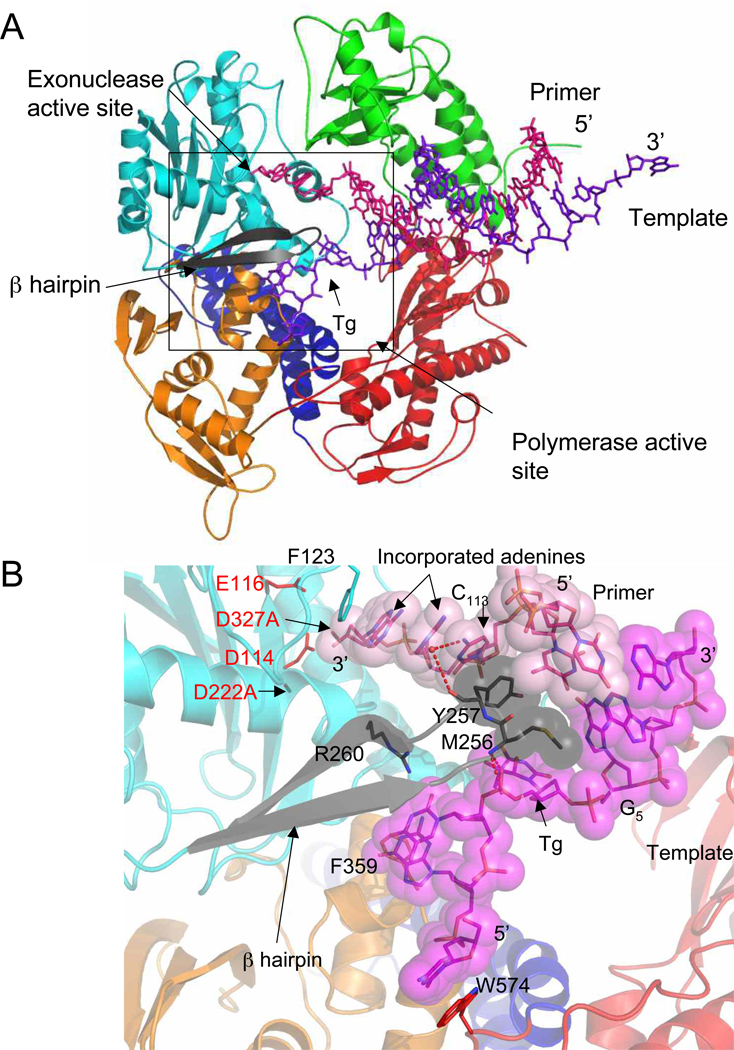
Two crystal structures of an RB69 gp43 exonuclease complex were previously described: One structure was obtained with DNA without a damaged base or mismatch 36 and the other was visualized after incorporation of an adenine opposite an abasic site analog19. In these structures parts of the DNA or protein are disordered and therefore absent from the model. A region of the exonuclease domain, the β-hairpin structure, was identified as important in the stability of the exonuclease complex formation 24; 31. In the crystal structure of a variant in which the tip of the β-hairpin loop is deleted, the lesion-containing DNA remained in the polymerase active site 24, whereas in complexes in which the β-hairpin is intact we observed an equilibrium where half of the polymerase/DNA complexes had primer DNA in the exonuclease site and the other half in the polymerase site 19. Until now we could not characterize precisely the interactions between the β-hairpin and the DNA strands (primer and lesion-containing template) because of disorder in the electron density maps. In contrast, the whole DNA duplex is seen in the 5’-T-Tg-G open ternary complex, including the damaged base, as well as the entire β–hairpin. As mentioned above, two adenines were incorporated in the primer strand opposite thymine glycol, after which the primer was transferred into the exonuclease active site. The polymerase melts three base pairs (G5•C113; Tg4•A114; T3•A115) of double stranded DNA, as shown before 36 (Figure 4A). The single stranded 3’-end of the primer is stabilized in the exonuclease site via stacking interactions with Phe123 and Tyr257 (Figure 4b) and hydrogen bonds with two residues from the thumb domain (Asn786 and Gly827 contacts A114 and A115, respectively) (SI figure 1A). G5 of the template no longer participates in a Watson Crick base pair with C113, which instead stacks against Tyr257 of the β hairpin loop. The backbone carbonyl of Tyr257 also interacts with C113 via a water-mediated hydrogen bond. The torsion angle (O4’-C1’-N9-C4) 34 of G5 is ~ −120° instead of −100°. The modification of the torsion angle is caused by Met256, which nudges the guanine toward the minor groove (Figure 4B and SI Figure 1B). Tg itself makes van der Waals contact with Lys278. In addition the phosphate moiety of Tg participates in two hydrogen bonds with the backbone amide of Met256 (Figure 4B and SI Figure 1B). Ile253 stacks with T3, the thymine 5’ to Tg. The two bases at the 5’-end of the template (C1 and G2) are stabilized via stacking interactions with Phe359 and Trp574, as observed previously 14; 19; 28 and a hydrogen bond network involving water molecules and residues Glu219, Val270, and Asp275 (SI figure 1B).
Figure 4.
Exonuclease complex. (A) Overall view of the exonuclease complex of RB69 gp43 exo− with Tg containing DNA. The primer strand (red) lies in the exonuclease site and is separated from the template strand (purple) by the β-hairpin loop (black). The polymerase domains are colored in orange for the N-terminal domain, red for the palm, green for the thumb, blue for the fingers, and cyan for the exo domain. (B) Close up view of the exonuclease active site with the β-hairpin (black) and DNA represented as stick models. Two residues from the β-hairpin loop (Met256 and Tyr257) and the DNA strands are also shown as van der Waals surface (primer in light pink and template in magenta). The hydrogen bonds are shown as red dashed lines. The four carboxylate residues participating in the exonuclease activity are highlighted in red.
Discussion
Tg bypass and sequence context
Almost all earlier studies of Tg bypass were performed with a Klenow fragment containing exonuclease activity 8; 9; 10; 15; 16; 17; 18 and showed that this polymerase can preferentially bypass Tg in the sequence context 5’-C-Tg-G or 5’-C-Tg-Purine 9; 16; 17 with bypass being more efficient with longer template lengths 16. On the other hand, it was reported that T4 DNA polymerase, which has a very potent exonuclease activity, cannot bypass Tg regardless of the template length or sequence context 15; 16. Since DNA polymerases lacking exonuclease activity readily bypass Tg 8; 15; 16, the inability of T4 DNA polymerase to bypass Tg was presumed to be due to its vigorous exonuclease activity. Here we show that, as expected, Tg bypass can occur with RB69 gp43 if its exonuclease activity is abrogated (Figure 1). When wild type RB69 gp43 is used, an adenine can be incorporated opposite Tg (with a small amount of bypass) but it is concomitant with primer degradation 14. For this reason we used an exonuclease deficient enzyme for our crystallization experiments.
The primer extension assays and the crystal structures of RB69 exo− in complex with Tg in the four different sequence contexts (5’-N-Tg-G with N=A, T, G or C) helped us evaluate different factors influencing Tg bypass, including the nature of the 5’ and 3’ bases, and the fact that Tg exists as diastereomeric pairs. We have previously shown 14 that RB69 exo− stalls when it encounters Tg in the sequence context 5’-G-Tg-G (Figure 1). The structure showed that the C5 methyl group of Tg prevents stacking of the 5’-adjacent template guanine, which instead participates in two hydrogen bonds with Tg (Figure 2A). In such a conformation the 5’-G cannot be used as a template for primer extension 14. Based on the 5’-G-Tg-G crystal structure we predicted that an adenine at the 5’ position would similarly hinder Tg bypass whereas a pyrimidine in the same position might facilitate Tg bypass. A primer extension assay (Figure 1) hinted that the situation was not necessarily that clearcut. While it appears that the 5’-G-Tg-G stalls RB69 gp43 exo− to the greatest extent, the other three sequence contexts appear to be readily bypassed9; 10; 15; 17.
After solving the structures of the other three sequence contexts we observe different positions and orientations of the 5’-adjacent template base depending upon whether the base is a purine or pyrimidine (Table 2). Purines are more extrahelical than pyrimidines and, accordingly, the twist angle diverges more for purines than for pyrimidines compared to regular B-DNA (Table 2). The protruding C5 methyl of Tg pushes the larger purine in a more extrahelical position compared to a pyrimidine. The 5’- pyrimidine therefore adopts an intermediate orientation between a well-stacked base and extrahelical purine, which may explain why RB69 exo− bypasses 5’-T-Tg-G more easily than 5’-G-Tg-G. The structures do not, however, explain why 5’-A-Tg-G and 5’-C-Tg-G are bypassed equally as well as 5’-T-Tg-G. On the other hand, only 5’-G-Tg-G shows hydrogen bonds between Tg and the 5’-base, which help stabilize and presumably reduce the flexibility of the 5’-G. The decreased flexibility of the 5’-base in the 5’-G-Tg-G sequence context may explain why this sequence is less easily bypassed by RB69 exo− in the primer extension assay.
Table 2.
Tilt and roll angles between Tg and adjacent bases
| Sequence context |
Inter-base pair |
Tilt (°) |
Roll (°) |
Inter-base pair |
Tilt (°) |
Roll (°) |
|---|---|---|---|---|---|---|
| 5′-G-Tg-G | G-Tg | 14.9 | −16.2 | Tg-G | −9.1 | −3.5 |
| 5′-A-Tg-G | A-Tg | 60.4 | −44.7 | Tg-G | −11.0 | 12.7 |
| 5′-C-Tg-G | C-Tg | −46.1 | 11.8 | Tg-G | 9.6 | −6.7 |
| 5′-T-Tg-G | T-Tg | −45.6 | −6.5 | Tg-G | 15.0 | 1.3 |
| B-DNA | 4.5 | −2 | 4.5 | −2 |
Tilt and roll angles were calculated with Curves.31 In the 5′-A-Tg-G sequence context, Tg adopts an orientation that ismore extrahelical than in other complexes, which explains the marked difference in the angle values. The tilt and roll angles for B-DNAwere calculated and averaged from the structure of a B-form DNA dodecamer (PDB ID code: 1BNA).32
The 3’-neighbor base also plays an important role in the stabilization of Tg. It was previously shown for Klenow fragment that bypass can occur preferentially if the 3’-base is a purine 9; 15; 17. The in silico model proposed in 1987 by Clark and collaborators 37 pointed out that Tg could interact via a hydrogen bond with a 3’-G. This interaction was confirmed recently with NMR 38 and the authors proposed that the hydrogen bond between O6 (Tg) and N7 (3’-G) could alleviate the steric clash with the 5’ neighbor base pair. In all but one of the structures presented here the distance between O6 (Tg) and N7 (3’-G) is less than 3.5 Å14 (It is 3.7 Å in the 5’-C-Tg-G structure). The formation of this hydrogen bond is a consequence of the non-planar structure of Tg and the presence of N7 in the adjacent purine. When a cytosine is modeled as the 3’ base no stabilizing hydrogen bond is possible between O6 (Tg) and C5 (3’-C) and the situation worsens with a 3’-thymine, since the C5 methyl group creates a steric clash with O6 (Tg). The 3’-base is therefore important to stabilize Tg and keeping it intrahelical for interaction with the incoming dATP during replication.
Another factor that might influence the rate of bypass is the fact that Tg exists as two diastereoisomeric pairs of epimers: the 5S cis trans pair (5S,6R; 5S,6S) and the 5R cis trans pair (5R,6S; 5R,6R). The 5R pair is more stable and abundant and composed of cis (5R,6S) Tg (87%) and trans (5R,6R) Tg (13%) 39. The epimerization rate between the two is 5.8 × 10−3 min−1 39. When Tg is opposite A in a DNA duplex, the equilibrium ratio cis (5R,6S) : trans (5R,6R) is 7:3 40. In our structure, Tg is in the cis (5R,6S) configuration and the protruding methyl group is axial which prevents stacking interactions with the 5’-adjacent base. The NMR structure of trans (5R,6R) Tg-containing DNA 38 showed that the methyl group becomes equatorial. The epimerization between cis and trans Tg could therefore influence Tg bypass. Indeed, trans (5R,6R) Tg is not predicted to prevent stacking interactions with the 5’-adjacent base and could facilitate bypass of Tg by the DNA polymerase.
The open ternary complex
The work on the effect of sequence context on thymine glycol bypass serendipitously allowed us to capture two new complexes within the same 5’-T-Tg-G ternary crystal: an open ternary complex captured after translocation and an editing complex with noteworthy interactions between the DNA and the β-hairpin motif.
We define a complex as an open ternary complex when the fingers are opened and an incoming nucleotide is observed in the polymerase active site. This complex differs from other ternary complexes in which the fingers are in a closed conformation 27; 28; 35. An open ternary complex of RB69 exo− with an abasic site analog and 5-NITP has already been described 28. In this case the DNA was not translocated. Our open ternary complex differs from the abasic site complex because two translocation events occurred (two adenines were added to the primer strand) and the position of the incoming nucleotide is different. The structure of the open ternary complex (Figure 3A) is consistent with the primer extension assay in which the addition of two adenines was observed (Figure 1). In addition to the two incorporated dAMP nucleotides the structure also revealed the presence of an incoming dATP, which is primarily stabilized by stacking interactions with the terminal dAMP on the primer strand (Figure 3A). The presence of the dATP is probably due to the large excess of dATP used during crystallization. However, in the other sequence contexts, in which the same excess of dATP was used, we only observe one dAMP addition and no extra nucleoside triphosphate. This finding suggests an additional effect due to sequence context. In fact, in the binary complex 5’-T-Tg-G-bin obtained with the chain terminator ddATP, we observed some residual electronic density within van der Waals distance of the ddAMP incorporated opposite Tg.
The exonuclease complex
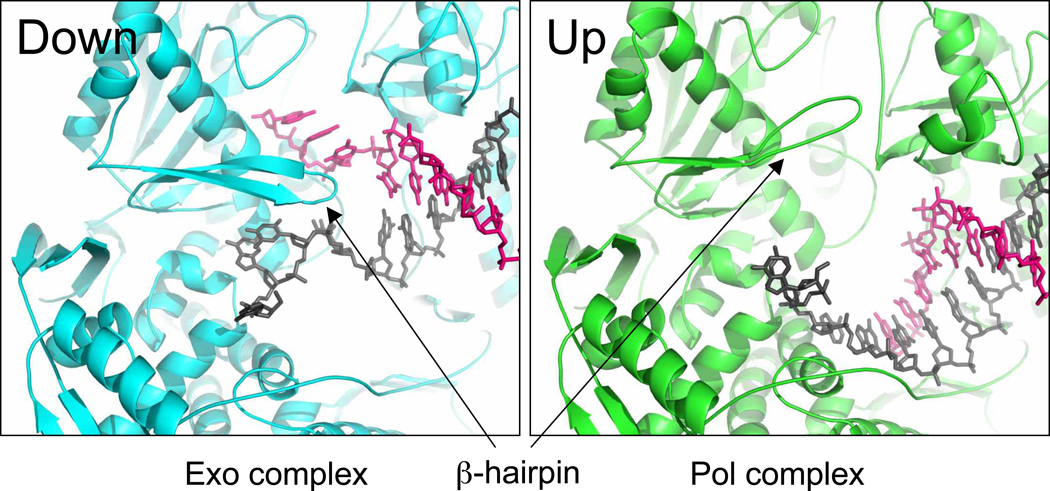
The editing function has been studied extensively in both T4 and RB69 DNA polymerases 24; 30; 41; 42. This reaction is essential for replicative DNA polymerases since it increases the fidelity of the enzyme by up to two orders of magnitude. The exonuclease mechanism involves two magnesium ions 43; 44; 45; 46 and four carboxylates, D114, D222, D327, and E116 30. Two of these aspartates are mutated in the constructs used in crystallography (D222A/D327A) and the metals are therefore absent from the exonuclease active site. The two active sites (polymerase and exonuclease) in RB69 gp43 are located 40 Å apart. How the primer migrates from the polymerase site (pol) to the exonuclease site (exo) is still a matter of debate. The primer strand could either slide from pol to exo or, after dissociation of the enzyme from the DNA, the primer strand could bind to the exo site. In the second hypothesis, an intermolecular reaction, is favored in the case of T4 DNA polymerase 30 and Klenow fragment 47. A β-hairpin structure from the exonuclease domain plays an important role in the stability of the DNA in the exonuclease complex 24; 32; 36. Our new structure of the complete exonuclease complex reveals the β-hairpin and the DNA in their entirety and shows key residues playing a part in the stabilization of the DNA. The β-hairpin loop can adopt two conformations “up” and “down” 19; 24; 27 (Figure 5). The up conformation is routinely observed when the primer DNA is in the polymerase site 27. In the 5’-T-Tg-G editing complex and two other complexes obtained with tetrahydrofuran, an abasic site analog 19; 28, the loop adopts a down conformation. Unlike the two tetrahydrofuran complexes, in which the tip of the β hairpin was not visible in the electron density map, the β hairpin structure is ordered in the 5’-T-Tg-G exonuclease complex and it makes important contacts with both DNA strands.
Figure 5.
Two conformations of the β-hairpin conformation. The β-hairpin loop can adopt two conformations, “down” in the exonuclease complex (sequence context 5’-T-Tg-G) and “up” in the ternary complex with normal DNA and incoming nucleotide (PDB ID code 1IG9) 27. The primer is colored in pink and the template in dark gray.
Before the primer strand migrated to the exonuclease site, two adenines were incorporated (Figure 4). In the exonuclease site, the 3’-end of the primer (consisting of the two newly incorporated adenines and the cytosine originally at the 3’-end) is sandwiched between two aromatic residues, Phe123 and Tyr257 from the exonuclease domain (Figure 4 and SI figure 1A). The same phenylalanine residue was shown to intercalate between the 3′-primer terminus and the penultimate base of a polydT single strand primer bound in the exonuclease site of T4 DNA polymerase 48; 49. The mutation of two residues from the β-hairpin loop in the homologous T4 DNA polymerase, Tyr254 and Gly255 (Tyr257 and Gly258, respectively in RB69), leads to a substantial decrease of DNA replication fidelity 31; 48; 50; 51; 52. Tyr257 and Met256 are wedged at the junction between the primer and template strands, the tyrosine stacking with the primer, and the methionine with Tg and nudging the 3’ guanine toward the minor groove (Figure 4B and SI Figure 1B). This finding is in good agreement with recent fluorescence data showing that the conserved Tyr257 52 plays a key role in strand separation. The first unpaired base in the template is the 3’ neighbor (G5) of thymine glycol and the opposite base in the primer (C113) is switched in the exonuclease site and stacks with Tyr257 (Figure 4B). The β-hairpin also interacts with the template strand via hydrogen bonds and stacking interactions (Figure 4B). The exonuclease complex illustrates the role of the β-hairpin in strand separation and in stabilizing the DNA in the exonuclease site, as proposed earlier 42; 52, and highlights Met256 and Tyr257 as key residues. In addition to the β hairpin, aspartate residues (Asp222 and Asp327) in the exonuclease active site were shown to participate in the formation of a stable exonuclease complex 52. The crystals described in this study were obtained with an exonuclease deficient version of the enzyme in which Asp222 and Asp327 are replaced with alanines. Modeling an aspartate at position 222 suggests that the carboxylate could hydrogen bond with the 3’OH of the terminal primer base and therefore stabilize the primer 3’ end. A third catalytic carboxylate, Asp114, is within van der Waals distance of the terminal primer base. The other two carboxylates, Asp327 and Glu116 are in the vicinity but not close enough to interact with the 3’ primer.
In a previously reported structure of an RB69 gp43 editing complex, Arg260 of the β hairpin stacked against the template strand and interacted with a primer base 36. In other structures of the editing complex 19; 28, the side chain of Arg260 could not be built because of disorder in the electronic density in this region. The corresponding arginine (Arg247) in the archaeal Thermococcus gorgonarius (Tgo) DNA polymerase (also a member of the B family of DNA polymerases) was proposed to act as a wedge separating the primer and template strands 53. These results are in contradiction with our new structural data in which Arg260 is clearly seen and does not contact the DNA (5’-T-Tg-G-ter exonuclease complex). Our conclusion that Arg260 does not appear to participate in strand separation is consistent with the negligible effect on replication fidelity resulting from amino acid substitutions at this position 52. These contradicting results warrant a further evaluation of the role of this arginine in archaeal DNA polymerases of the B family.
In summary, we have investigated the role of sequence context on the ability of RB69 gp43 to bypass Tg and showed that a combination of several factors play a part in this event. The nature of the 5’ and 3’ base plays a role, as do the associated exonuclease activity of the polymerase and the structure of the lesion itself (cis-trans interconversions in the case of Tg). Moreover, this work allowed us to visualize for the first time a well-ordered structure of an exonuclease complex and identify residues critical for DNA strand separation and stability. While these two results may appear at first glance to be unrelated topics the formation of an exonuclease complex is a natural response from a DNA polymerase with proofreading capability when it encounters a DNA lesion.
Supplementary Material
β-hairpin interactions with primer and template strands in the 5’-T-Tg-G exonuclease complex. The β-hairpin is shown in black, primer in pink, template in purple, N-terminal domain in orange, exo in cyan, palm in red and thumb in green. The hydrogen bonds are shown as red dashed lines. A simulated annealing map contoured at 3 σ is shown as a green mesh. A) Polymerase interactions with the primer strand B) Polymerase interactions with the template strand
Acknowledgments
We thank K. E. Zahn for help with data collection at the APS synchrotron and Dr. H. Unno for collecting diffraction data at the Photon Factory (Tsukuba, Japan). This work was supported by NIH RO1 CA52040 awarded by the National Cancer Institute. The beamline GM/CA CAT has been funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357.
Bibliography
- 1.Adelman R, Saul RL, Ames BN. Oxidative damage to DNA: relation to species metabolic rate and life span. Proc Natl Acad Sci U S A. 1988;85:2706–2708. doi: 10.1073/pnas.85.8.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cathcart R, Schwiers E, Saul RL, Ames BN. Thymine glycol and thymidine glycol in human and rat urine: a possible assay for oxidative DNA damage. Proc Natl Acad Sci U S A. 1984;81:5633–5637. doi: 10.1073/pnas.81.18.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saul RL, Ames BN. In: Mechanisms of DNA Damage and Repair. Simic MG, Grossman L, Upton AC, editors. Plenum, NY: 1986. [Google Scholar]
- 4.Bandaru V, Sunkara S, Wallace SS, Bond JP. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 2002;1:517–529. doi: 10.1016/s1568-7864(02)00036-8. [DOI] [PubMed] [Google Scholar]
- 5.Aspinwall R, Rothwell DG, Roldan-Arjona T, Anselmino C, Ward CJ, Cheadle JP, Sampson JR, Lindahl T, Harris PC, Hickson ID. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc Natl Acad Sci U S A. 1997;94:109–114. doi: 10.1073/pnas.94.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dizdaroglu M, Karahalil B, Senturker S, Buckley TJ, Roldan-Arjona T. Excision of products of oxidative DNA base damage by human NTH1 protein. Biochemistry. 1999;38:243–246. doi: 10.1021/bi9819071. [DOI] [PubMed] [Google Scholar]
- 7.Hayes RC, Petrullo LA, Huang HM, Wallace SS, LeClerc JE. Oxidative damage in DNA. Lack of mutagenicity by thymine glycol lesions. J Mol Biol. 1988;201:239–246. doi: 10.1016/0022-2836(88)90135-0. [DOI] [PubMed] [Google Scholar]
- 8.Clark JM, Beardsley GP. Thymine glycol lesions terminate chain elongation by DNA polymerase I in vitro. Nucleic Acids Res. 1986;14:737–749. doi: 10.1093/nar/14.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayes RC, LeClerc JE. Sequence dependence for bypass of thymine glycols in DNA by DNA polymerase I. Nucleic Acids Res. 1986;14:1045–1061. doi: 10.1093/nar/14.2.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ide H, Kow YW, Wallace SS. Thymine glycols and urea residues in M13 DNA constitute replicative blocks in vitro. Nucleic Acids Res. 1985;13:8035–8052. doi: 10.1093/nar/13.22.8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kow YW, Faundez G, Melamede RJ, Wallace SS. Processing of model single-strand breaks in phi X-174 RF transfecting DNA by Escherichia coli. Radiat Res. 1991;126:357–366. [PubMed] [Google Scholar]
- 12.Laspia MF, Wallace SS. Excision repair of thymine glycols, urea residues, and apurinic sites in Escherichia coli. J Bacteriol. 1988;170:3359–3366. doi: 10.1128/jb.170.8.3359-3366.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moran E, Wallace SS. The role of specific DNA base damages in the X-ray-induced inactivation of bacteriophage PM2. Mutat Res. 1985;146:229–241. doi: 10.1016/0167-8817(85)90063-x. [DOI] [PubMed] [Google Scholar]
- 14.Aller P, Rould MA, Hogg M, Wallace SS, Doublie S. A structural rationale for stalling of a replicative DNA polymerase at the most common oxidative thymine lesion, thymine glycol. Proc Natl Acad Sci U S A. 2007;104:814–818. doi: 10.1073/pnas.0606648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark JM, Beardsley GP. Functional effects of cis-thymine glycol lesions on DNA synthesis in vitro. Biochemistry. 1987;26:5398–5403. doi: 10.1021/bi00391a027. [DOI] [PubMed] [Google Scholar]
- 16.Clark JM, Beardsley GP. Template length, sequence context, and 3'–5' exonuclease activity modulate replicative bypass of thymine glycol lesions in vitro. Biochemistry. 1989;28:775–779. doi: 10.1021/bi00428a054. [DOI] [PubMed] [Google Scholar]
- 17.Evans J, Maccabee M, Hatahet Z, Courcelle J, Bockrath R, Ide H, Wallace S. Thymine ring saturation and fragmentation products: lesion bypass, misinsertion and implications for mutagenesis. Mutat Res. 1993;299:147–156. doi: 10.1016/0165-1218(93)90092-r. [DOI] [PubMed] [Google Scholar]
- 18.Rouet P, Essigmann JM. Possible role for thymine glycol in the selective inhibition of DNA synthesis on oxidized DNA templates. Cancer Res. 1985;45:6113–6118. [PubMed] [Google Scholar]
- 19.Hogg M, Wallace SS, Doublie S. Crystallographic snapshots of a replicative DNA polymerase encountering an abasic site. Embo J. 2004;23:1483–1493. doi: 10.1038/sj.emboj.7600150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Emsley P, Lohkamp B, Scott W, Cowtan K. Features and Development of Coot. Acta Crystallogr D Biol Crystallogr. 2010 doi: 10.1107/S0907444910007493. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- 23.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 24.Hogg M, Aller P, Konigsberg W, Wallace SS, Doublie S. Structural and biochemical investigation of the role in proofreading of a beta hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J Biol Chem. 2007;282:1432–1444. doi: 10.1074/jbc.M605675200. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 26.Aller P, Ye Y, Wallace SS, C JB, Doublie S. Crystal Structure of a Replicative DNA Polymerase Bound to the Oxidized Guanine Lesion Guanidinohydantoin. Biochemistry. 2010 doi: 10.1021/bi902195p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol alpha family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- 28.Zahn KE, Belrhali H, Wallace SS, Doublie S. Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]
- 29.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010. [Google Scholar]
- 30.Reha-Krantz LJ. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim Biophys Acta. 2010;1804:1049–1063. doi: 10.1016/j.bbapap.2009.06.012. [DOI] [PubMed] [Google Scholar]
- 31.Reha-Krantz LJ, Marquez LA, Elisseeva E, Baker RP, Bloom LB, Dunford HB, Goodman MF. The proofreading pathway of bacteriophage T4 DNA polymerase. J Biol Chem. 1998;273:22969–22976. doi: 10.1074/jbc.273.36.22969. [DOI] [PubMed] [Google Scholar]
- 32.Stocki SA, Nonay RL, Reha-Krantz LJ. Dynamics of bacteriophage T4 DNA polymerase function: identification of amino acid residues that affect switching between polymerase and 3' --> 5' exonuclease activities. J Mol Biol. 1995;254:15–28. doi: 10.1006/jmbi.1995.0595. [DOI] [PubMed] [Google Scholar]
- 33.Gardner AF, Jack WE. Acyclic and dideoxy terminator preferences denote divergent sugar recognition by archaeon and Taq DNA polymerases. Nucleic Acids Res. 2002;30:605–613. doi: 10.1093/nar/30.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saenger W. In: Principles of nucleic acid structure. Cantor CR, editor. New York, NY: Springer-Verlag; 1995. [Google Scholar]
- 35.Hogg M, Rudnicki J, Midkiff J, Reha-Krantz L, Doublie S, Wallace SS. Kinetics of mismatch formation opposite lesions by the replicative DNA polymerase from bacteriophage RB69. Biochemistry. 2010;49:2317–2325. doi: 10.1021/bi901488d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shamoo Y, Steitz TA. Building a replisome from interacting pieces: sliding clamp complexed to a peptide from DNA polymerase and a polymerase editing complex. Cell. 1999;99:155–166. doi: 10.1016/s0092-8674(00)81647-5. [DOI] [PubMed] [Google Scholar]
- 37.Clark JM, Pattabiraman N, Jarvis W, Beardsley GP. Modeling and molecular mechanical studies of the cis-thymine glycol radiation damage lesion in DNA. Biochemistry. 1987;26:5404–5409. doi: 10.1021/bi00391a028. [DOI] [PubMed] [Google Scholar]
- 38.Brown KL, Roginskaya M, Zou Y, Altamirano A, Basu AK, Stone MP. Binding of the human nucleotide excision repair proteins XPA and XPC/HR23B to the 5R-thymine glycol lesion and structure of the cis-(5R,6S) thymine glycol epimer in the 5'-G-TgG-3' sequence: destabilization of two base pairs at the lesion site. Nucleic Acids Res. 2010;38:428–440. doi: 10.1093/nar/gkp844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lustig MJ, Cadet J, Boorstein RJ, Teebor GW. Synthesis of the diastereomers of thymidine glycol, determination of concentrations and rates of interconversion of their cis-trans epimers at equilibrium and demonstration of differential alkali lability within DNA. Nucleic Acids Res. 1992;20:4839–4845. doi: 10.1093/nar/20.18.4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brown KL, Adams T, Jasti VP, Basu AK, Stone MP. Interconversion of the cis-5R,6S- and trans-5R,6R-thymine glycol lesions in duplex DNA. J Am Chem Soc. 2008;130:11701–11710. doi: 10.1021/ja8016544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fidalgo da Silva E, Reha-Krantz LJ. DNA polymerase proofreading: active site switching catalyzed by the bacteriophage T4 DNA polymerase. Nucleic Acids Res. 2007;35:5452–5463. doi: 10.1093/nar/gkm591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trzemecka A, Plochocka D, Bebenek A. Different behaviors in vivo of mutations in the beta hairpin loop of the DNA polymerases of the closely related phages T4 and RB69. J Mol Biol. 2009;389:797–807. doi: 10.1016/j.jmb.2009.04.055. [DOI] [PubMed] [Google Scholar]
- 43.Beese LS, Steitz TA. Structural basis for the 3'–5' exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism. EMBO J. 1991;10:25–33. doi: 10.1002/j.1460-2075.1991.tb07917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Curley JF, Joyce CM, Piccirilli JA. Functional Evidence That the 3‘–5‘ Exonuclease Domain of Escherichia coli DNA Polymerase I Employs a Divalent Metal Ion in Leaving Group Stabilization. J Am Chem Soc. 1997;119:12691–12692. [Google Scholar]
- 45.Elisseeva E, Mandal SS, Reha-Krantz LJ. Mutational and pH studies of the 3' --> 5' exonuclease activity of bacteriophage T4 DNA polymerase. J Biol Chem. 1999;274:25151–25158. doi: 10.1074/jbc.274.35.25151. [DOI] [PubMed] [Google Scholar]
- 46.Freemont PS, Friedman JM, Beese LS, Sanderson MR, Steitz TA. Cocrystal structure of an editing complex of Klenow fragment with DNA. Proc Natl Acad Sci U S A. 1988;85:8924–8928. doi: 10.1073/pnas.85.23.8924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Joyce CM. How DNA travels between the separate polymerase and 3'–5'-exonuclease sites of DNA polymerase I (Klenow fragment) J Biol Chem. 1989;264:10858–10866. [PubMed] [Google Scholar]
- 48.Beechem JM, Otto MR, Bloom LB, Eritja R, Reha-Krantz LJ, Goodman MF. Exonuclease-polymerase active site partitioning of primer-template DNA strands and equilibrium Mg2+ binding properties of bacteriophage T4 DNA polymerase. Biochemistry. 1998;37:10144–10155. doi: 10.1021/bi980074b. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol alpha family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 50.Marquez LA, Reha-Krantz LJ. Using 2-aminopurine fluorescence and mutational analysis to demonstrate an active role of bacteriophage T4 DNA polymerase in strand separation required for 3' --> 5'-exonuclease activity. J Biol Chem. 1996;271:28903–28911. doi: 10.1074/jbc.271.46.28903. [DOI] [PubMed] [Google Scholar]
- 51.Reha-Krantz LJ. Regulation of DNA polymerase exonucleolytic proofreading activity: studies of bacteriophage T4 "antimutator" DNA polymerases. Genetics. 1998;148:1551–1557. doi: 10.1093/genetics/148.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Subuddhi U, Hogg M, Reha-Krantz LJ. Use of 2-aminopurine fluorescence to study the role of the beta hairpin in the proofreading pathway catalyzed by the phage T4 and RB69 DNA polymerases. Biochemistry. 2008;47:6130–6137. doi: 10.1021/bi800211f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Killelea T, Ghosh S, Tan SS, Heslop P, Firbank SJ, Kool ET, Connolly BA. Probing the interaction of archaeal DNA polymerases with deaminated bases using X-ray crystallography and non-hydrogen bonding isosteric base analogues. Biochemistry. 2010;49:5772–5781. doi: 10.1021/bi100421r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J Biomol Struct Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 55.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
β-hairpin interactions with primer and template strands in the 5’-T-Tg-G exonuclease complex. The β-hairpin is shown in black, primer in pink, template in purple, N-terminal domain in orange, exo in cyan, palm in red and thumb in green. The hydrogen bonds are shown as red dashed lines. A simulated annealing map contoured at 3 σ is shown as a green mesh. A) Polymerase interactions with the primer strand B) Polymerase interactions with the template strand