Abstract
Murine noroviruses (MNV) are currently the most prevalent viruses infecting mouse research colonies. Concurrent infection of research mice with these viruses can dramatically alter the experimental outcome in some research models, but not others. In this report, we investigated the effect of MNV1 and MNV4 on a murine model of intestinal inflammation and fibrosis induced by Salmonella typhimurium infection in C57BL/6 mice. Subsequent co-infection of these mice with MNV1 or MNV4 did not lead to major changes in histopathology, the inflammatory response, or the fibrotic response. Thus, MNV does not substantially alter all gastrointestinal research models, highlighting the importance of investigating potential alterations in the research outcome by MNV on an individual basis. We hypothesize that this is particularly important in cases of research models that use immunocompromised mice, which could be more sensitive to MNV infection-induced changes.
Keywords: murine norovirus, Salmonella typhimurium, animal model, fibrosis, large intestine, inflammatory bowel disease
Introduction
Murine noroviruses (MNV) are non-enveloped, positive-strand RNA viruses that belong to the Norovirus genus in the Caliciviridae family. Most noroviruses infect humans and cause the vast majority of sporadic cases and outbreaks of infectious nonbacterial gastroenteritis worldwide in people of all ages [1–3]. However, noroviruses infecting mice [4–6], cattle [7–9], pigs [10, 11], sheep [12], a lion cub [13], and a dog [14] have also been described. The first murine norovirus, MNV1, was isolated in 2003 from immunocompromised mice [4] and shown to replicate in macrophages and dendritic cells in vitro and in vivo [15–18]. We and others have since shown that MNV1 also causes a transient infection in immunocompetent inbred and outbred mice [5, 6, 18, 19]. Peak viral titers are seen between 1 and 3 days post-infection (dpi) in the intestine, mesenteric lymph nodes, and spleen, with viral clearance by day 7 [5, 18]. Like the human noroviruses, multiple MNV strains have been isolated. Of the ones that are characterized (e.g. MNV4, CR6), many persistently infect wild-type animals and viral genomes can be detected in the small intestine, mesenteric lymph nodes and feces of infected animals 1–2 months post-infection [5, 6]. Despite the different biological phenotypes, of the 15 strains analyzed in one report, all cluster in one genogroup and form one serogroup [5]. MNV is the most prevalent virus in research mice [20]. Serologic analysis of mice from research colonies in North America and Europe demonstrated that 22–64% of all mice had antibodies against MNV1 [19–22]. Similar prevalence rates (22–23%) are seen in Japan and South Korea after RT-PCR analysis of murine fecal samples [23, 24] with a lower rate of genome positive samples (13%) found in the cecum [25]. Eradication of MNV from research facilities requires complete depopulation, a potentially large financial burden for the research community [26]. Therefore, the impact of MNV for biomedical research is actively being investigated. Early indications suggest that MNV has the ability to alter the outcome of some research investigations but will likely require evaluation on a case-by-case basis. To date, striking abnormalities in Paneth cell function were observed in mice with gene-trap-mediated disruptions of Atg16L1, a Crohn’s disease susceptibility marker, when infected with the CR6 MNV strain but not MNV1 [27]. More subtle histopathologic changes have been described in immunocompromised [16, 17] and immunocompetent mice [27]. In one report, MNV4 altered antigen presentation by dendritic cells and was associated with exacerbation of a bacterial-induced inflammatory bowel disease model in Mdr1a / mice [28]. In another model, previous infection of BALB/cByJ mice with MNV-G led to an increased mouse parvovirus genome load in tissues and increased duration of mouse parvovirus shedding [29]. In contrast, previous infection of C57BL/6 mice with MNV1 and/or CR6 had no effect on vaccinia virus and influenza A virus-induced CD8+ T-cell and antibody responses or lethality [30]. Similarly, MNV infection did not impact murine cytomegalovirus (MCMV) titers or reactivation, but decreased CD8+ T-cell responses to immunodominant MCMV epitopes [31]. Furthermore, CR6 infection does not alter Friend retrovirus-induced immune responses (T cell, NK cell and antibody responses) or the course of infection (viremia, titers, pathology) [32]. These varying responses of MNV in research outcomes led Ammann et al. [32] to suggest that MNV co-infections might be more likely to alter responses to infections in the gastrointestinal tract. To better understand the impact of MNV on a research model of inflammation and intestinal fibrosis, we investigated the effect of the transient MNV1 and persistent MNV4 strains on a Salmonella typhimurium-induced intestinal fibrosis model in C57BL/6 mice. Treatment of mice with streptomycin followed by infection with S. typhimurium ΔaroA produces a persistent bacterial infection of the cecum [33]. Subsequent intestinal colitis, characterized by transmural inflammation, hypertrophy of the muscle layers, and excessive extracellular matrix deposition, produces an inflammatory disease phenotype similar to inflammatory bowel disease in humans [33]. However, no major effect of MNV1 or MNV4 on the histopathology and inflammatory response were observed in this model. Thus, we propose that MNV is more likely to affect research outcomes in intestinal models using immunocompromised mice.
Materials and Methods
Mice, Bacterial Strains, Virus stocks
Specific pathogen-free female 8–10 week old C57BL/6 mice were purchased from Jackson Laboratories (Bar Harbor, ME).
Salmonella typhimurium strain ΔaroA (a kind gift from G. Grassl, University of British Columbia, Vancouver, Canada) which is naturally resistant to streptomycin was grown in LB broth containing 100 μg/ml streptomycin at 37°C. Infectious titers were determined by plating serial dilutions of the inoculum onto LB/streptomycin plates. The plaque purified MNV1 isolate MNV1.CW3 [5] (referred herein as MNV1) was used at passage 6 for all experiments. MNV4 was a kind gift from R. Livingston (University of Missouri - Columbia) and was used at passage 2. A concentrated virus stock and corresponding mock lysate was generated as described [34] for both MNV strains and used in all experiments.
Animal Studies
In three independent experiments, female C57BL/6 mice were divided into six groups. Animals in the three S. typhimurium infection groups received 20 mg streptomycin by oral gavage followed by infection with 2×106 colony forming units (cfu) of S. typhimurium ΔaroA in 100 μl 0.1M HEPES buffer (pH = 8.0) 24 hrs later. Two days after the S. typhimurium infection, one group (St+M1) was co-infected orally with 2×107 plaque forming units (PFU) of MNV1 per mouse; a second group (St +M4) was co-infected 2×107 PFU of MNV4 per mouse; and a third group (St, or positive control) was orally given an equal volume of concentrated mock lysate. The additional three groups were not infected with S. typhimurium ΔaroA but received 20 mg streptomycin. The negative control group (no Tx) and MNV only groups were gavaged with 0.1 M HEPES buffer at the same time point as the S. typhimurium infection. Two days later, the MNV only groups were infected orally with 2×107 PFU MNV1 per mouse (MNV1 group) or 2×107 PFU MNV4 per mouse (MNV4 group), while the negative control group was orally given the same volume of mock lysate. Mice were euthanized 21 days post S. typhimurium infection. All animal experiments were conducted with the approval and oversight of the University of Michigan Committee on Use and Care of Animals in accordance with all federal guidelines.
Gross Pathology and Tissue Collection
Serum, fecal pellets, small intestine, mesenteric lymph nodes, cecum and distal colon samples were collected 21 days post S. typhimurium infection, the time point of harvest. Cecum and distal colon were photographed, measured, and weighed. Cecal area was determined from digital photographs using NIH ImageJ in a region of interest (ROI) that included only the cecum. To control for differences in images, the cecal area was normalized against a 1 cm marker in the photographic image. Tissues were snap-frozen in liquid nitrogen and stored at −80°C prior to analysis. Cecal contents were collected and serially diluted before plating onto LB-streptomycin plates to determine bacterial titers in the cecum.
Histology
Formalin-fixed and paraffin-embedded tissues were stained with Masson’s trichrome by the University of Michigan Comprehensive Cancer Center Research Histology and Immunoperoxidase Laboratory (Ann Arbor, MI). Digital photomicrographs of tissue sections were taken with an Olympus BX microscope at the University of Michigan Microscopy and Image Analysis Laboratory. Tissue sections were scored separately for inflammation and fibrosis in a blinded fashion by a pathologist (DM). Inflammation was scored based on the Wirtz scale [35]: (0) no inflammation, (1) low level of inflammation with scattered infiltrating mononuclear cells, (2) moderate inflammation with multiple foci, (3) high level of inflammation with increased vascular density and marked wall thickening, (4) maximal severity of inflammation with transmural leukocyte infiltration and loss of goblet cells. Fibrosis was scored as (0) no fibrosis, (1) mild fibrosis (focal mucosal/submucosal collagen deposition without architectural distortion), (2) moderate fibrosis (significant mucosal/submucosal collagen deposition with modest distortion of mucosal/submucosal architecture but without obscuring of the mucosal/submucosal border), and (3) severe fibrosis (extensive mucosal submucosal collagen deposition with marked architectural distortion obscuring the mucosal/submucosal border).
Tissue measurements were quantitated with NIH ImageJ analysis software (NIH, Bethesda, MD, version 1.38). Image analysis and quantification of collagen density was performed using color segmentation analysis performed with Matlab software (R2007a; copyright 2007, The MathWorks, Inc., Natick, Mass.) by the method of Koga et al. [36]. Briefly, full thickness trichrome stained sections of mouse cecum were scanned using a Pathscan Enabler IV (Meyer Instruments, Houston, TX) slide scanner and digitized. The image background was removed with Adobe Photoshop prior to analysis with Matlab. Each pixel was categorized to its likely tissue type (muscle, blood, and collagen) based on color as determined by nearest neighbor classification. Collagen density (blue pixel count) was quantitated and normalized to the length of the tissue section. Where possible, analysis was performed with a full cross-section of cecum.
Quantitative Real-time PCR for host genes
RNA was extracted from the cecum using the RNeasy kit (Qiagen, Valencia, CA). Reverse transcription of 2 μg of total RNA was performed with the Superscript First Strand RT kit (Invitrogen). Quantitative real-time PCR (qRT-PCR) of interleukin (IL) 1β, tumor necrosis factor (TNF) α, IL-12p40, interferon (IFN) γ, IL-17, IL-13, IL-4, IL-10, and GAPDH was performed with commercial TaqMan gene expression assays (ABI, Foster City, CA). Q-PCR was performed using a Stratagene Mx3000P real-time PCR system (Stratagene, La Jolla, CA). Cycling conditions were 95°C 10 minutes, followed by 40 cycles of 95°C 15 seconds and 62°C 60 seconds. Gene expression was normalized to GAPDH as the endogenous control and fold-changes (RQ) relative to uninfected controls (no Tx) were calculated using the ΔΔCt-method [37].
Determination of MNV infection status
The infection status was first determined by measuring seroconversion by ELISA. Sera of all animals were evaluated for anti-MNV antibodies as previously described [4] with the following modifications. Cesium chloride purified MNV1 was diluted in phosphate buffered saline (2 μl/well) and wells were coated overnight at 40C. Sera were diluted 1:100 in ELISA buffer (0.15 M NaCl, 0.001 M EDTA, 0.05 M Tris-HCl, 0.05% Tween 20, 0.1% BSA, pH to 7.4).
To distinguish between animals infected at the time of harvest (i.e. day 21 post-S. tymphimurium infection) or those that had cleared the infection, a combination of quantitative (TaqMan®) and qualitative RT-PCR and/or plaque assay of cecum or mesenteric lymph nodes was used. In general, viral loads were very low, but animals with detectable virions and/or genome were considered persistently infected. Quantitative PCR (TaqMan®) was performed and confirmed by qualitative RT-PCR and/or plaque assay. Total RNA was extracted from homogenized tissue using TRIZOL (Invitrogen, CA) and resuspended in 15 μl DNase/RNase free water. Viral cDNA was reverse transcribed from equal amounts of total RNA using Moloney Murine Leukemia Virus reverse transcriptase (Invitrogen, CA) in a reaction volume of 35 μl as previously described [38]. Quantitative PCR (TaqMan®) was carried out on an Applied Biosystems 7500 Fast Real-Time PCR System in a reaction volume of 10 μl using previously described primers (targeting a conserved region in open reading frame 1), probe, and PCR conditions [38]. Qualitative PCR was performed using Taq DNA polymerase (NEB, CA) according to the manufacturer’s recommendations. Briefly, 5 μl cDNA, 500 nM sense and antisense oligo and probe, 3 mM MgCl2, and 1 U Taq polymerase (NEB) were used in the reaction with cycling conditions of 1× (95°C for 2 min) and 35× (94°C for 15 sec, 55°C for 30 sec, 72°C for 15 sec), and products were analyzed by gel electrophoresis. Plaque assay was performed as described previously [15].
Statistical Analysis
Statistical differences were determined by a two-sided, unpaired Student’s t-test. Results with a p value of < 0.05 were considered statistically significant.
Results
Both MNV1 and MNV4 infections can be transient or persistent
The MNV1.CW3 strain used in this study, and referred herein as MNV1, is reported to cause a transient infection in C57BL/6 mice with high levels of viral genome and infectious virus in the small intestine and mesenteric lymph nodes three days postinfection but undetectable levels of viral genome in the feces or mesenteric lymph nodes seven days postinfection [5]. In contrast, MNV4 genomes are still detectable in 100% (10/10) of the feces and mesenteric lymph nodes of outbred CD1 mice eight weeks postinfection [6]. Thus, MNV1 is considered an acute strain, while MNV4 is a persistent strain. To determine the MNV infection status in the absence or presence of S. typhimurium, anti-MNV1 serum antibody titers were measured to verify the serological status of all animals. MNV1 antigen can be used for the detection of both viruses since MNV1 and MNV4 belong to the same serogroup [5, 6]. In addition, the presence of viral genome in the cecum or mesenteric lymph nodes was determined by reverse transcriptase-PCR at the time of harvest (i.e. 21 days post-bacterial infection). In some cases viral titers were also determined by plaque assay in the mesenteric lymph nodes to confirm these results. These data are summarized in Table 1. All uninfected and exclusively S. typhimurium infected animals had no evidence of MNV infection; i.e. they did not seroconvert in our rodent facility, nor did they have any measurable levels of viral genome or infectious virus in the mesenteric lymph nodes. All MNV infected animals (alone and co-infected) seroconverted. Interestingly, while the majority of the MNV1 single and co-infected animals showed evidence of a transient infection, up to one third of the animals were persistently infected and had detectable levels of infectious virus or genomes in the mesenteric lymph nodes. Similarly, while many of the MNV4 singly or co-infected animals were persistently infected, up to half of the MNV4 infected animals had no detectable infectious virus or viral genome at harvest in the mesenteric lymph nodes. This suggested that either MNV strain can cause transient or persistent infections in C57BL/6 mice. However, the infection status (Table 1) was unaffected by S. typhimurium co-infection.
Table 1. Summary of the infection status of all animals.
The numbers of animals that are seropositive by ELISA (seropositive) and have detectable genome by PCR and/or infectious virus by plaque assay (MNV-positive) in the mesenteric lymph nodes or cecum at the time of harvest (i.e. day 21 post-S. tymphimurium infection) from each group are listed compared to the total number of animals in each group. Animals were defined as transiently or persistently infected with MNV based on the following criteria. Transient infections were defined as positive by ELISA but negative by plaque assay and PCR. Persistently infected animals were positive by ELISA and positive by either plaque assay or PCR or both. Data shown are from three independent animal experiments. Numbers are expressed as number of animals per total animals in each of the six groups: uninfected (no Tx), MNV1 infected (MNV1), MNV4 infected (MNV4), S. typhimurium infected (St), S. typhimurium and MNV1 co-infected (St+MNV1), and S. typhimurium and MNV4 co-infected (St+MNV4).
| seropositive | MNV-positive Day 21 | |
|---|---|---|
| No Tx | 0/9 | 0/9 |
| MNV1 | 9/9 | 3/9 |
| MNV4 | 10/10 | 4/10 |
| St. typh | 0/6 | 0/6 |
| St+MNV1 | 9/9 | 1/9 |
| St+MNV4 | 15/15 | 7/15 |
Transient or persistent norovirus co-infection does not affect S. typhimurium bacterial load in the cecum
To evaluate whether MNV infection affected S. typhimurium infection, we measured bacterial loads in the cecal contents at harvest. Infection with an oral dose of 2 x 106 cfu of S. typhimurium efficiently colonized the mouse cecum and by day 21 post S. typhimurium infection, bacterial loads in the cecal content in the absence of viral infection were on average 8.8 x 105 cfu/ml (Fig. 1). Co-infection with MNV1 or MNV4 did not significantly alter S. typhimurium colonization (Fig. 1). This was regardless of whether the MNV strains were still present at day 21 (see Table 1) or not. S. typhimurium colonization of transiently MNV4 infected animals did not differ from either the MNV4 persistently infected animals (1.6×106 cfu/ml vs 3.7×105 cfu/ml, p = 0.07) or S. typhimurium singly infected animals (1.6×106 cfu/ml vs. 8.8×105 cfu/ml, p = 0.30). Similarly, persistent MNV4 infection did not alter S. typhimurium colonization (3.7×105 cfu/ml vs 8.8×105 cfu/ml, p = 0.23). Although MNV1 persisted in only 2 of 10 co-infected animals by day 21, again viral persistence did not impact S. typhimurium colonization (6.0×103 cfu/ml vs 8.8×105 cfu/ml, p = 0.21). Taken together, these data demonstrate that neither MNV nor S. typhimurium impacted the infection status of the other.
Figure 1. S. typhimurium colonization of the mouse cecum in the presence or absence of MNV1 or MNV4.
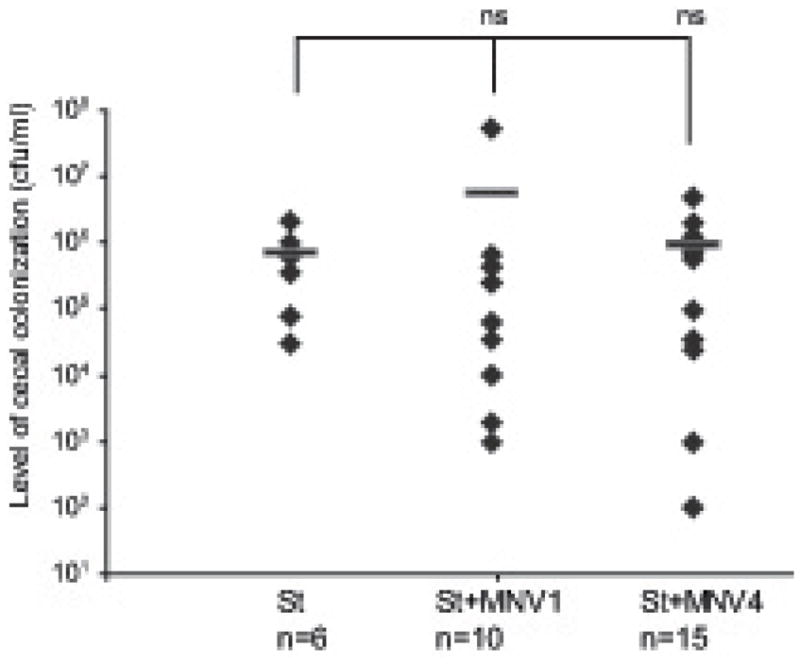
The S. typhimurium loads were determined by plating 100 μl of cecal contents onto LB/streptomycin plates. S. typhimurium colonization is expressed as cfu/ml volume of cecal contents and the mean cfu of each experimental group is indicated by a horizontal line. Each diamond represents the data from an individual animal and the total number (n) of animals per experimental group is indicated. St = S. typhimurium infected animals; St + MNV1 = S. typhimurium and MNV1 co- infected animals; St + MNV4 = S. typhimurium and MNV4 co-infected animals; ns = not statistically significant (p≥0.05) compared to no St.
Co-infection with either norovirus strain does not affect the gross pathology, area and weight of the cecum and distal colon
Chronic infection with S. typhimurium is characterized by a reduction in cecal size with a concurrent increase in combined cecal/colon weight reflecting both tissue edema and increased collagen deposition in the mucosa and submucosa (i.e. fibrosis) [33]. These findings were confirmed in our experiments. Mice infected with S. typhimurium alone but not mice infected with either MNV strain alone exhibited changes in the gross pathology of the cecum, had significantly reduced cecal area (65.1 vs. 94.5 pixels, p < 0.0001 vs. no Tx) and significantly increased cecum and colon weight (0.71 vs. 0.40 g, p < 0.001 vs. no Tx) compared to uninfected controls (Fig. 2).
Figure 2. Gross pathology of the cecum and distal colon.
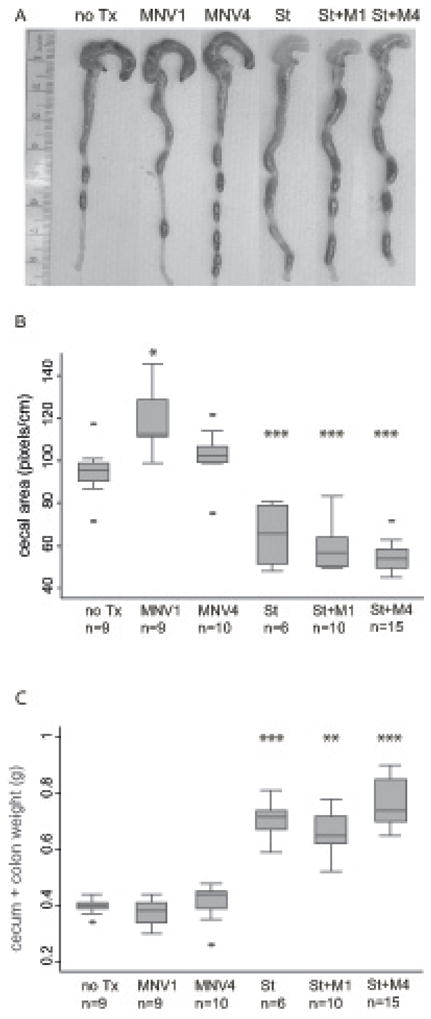
(A) Representative photograph of the cecum and colon of uninfected mice (no Tx) compared to MNV1 infected (MNV1), MNV4 infected (MNV4), S. typhimurium infected (St), S. typhimurium and MNV1 co-infected (St + M1), and S. typhimurium and MNV4 co- infected (St + M4) animal. A 1 cm ruler is shown as reference. (B). Cecal area (in pixels) from animals in each experimental group described in A were calculated using NIH ImageJ and normalized against a 1 cm reference ruler. (C) Combined cecum and colon tissue weight was determined after the cecum and colon was longitudinally cut, rinsed of fecal contents, and blotted dry. For B) and C), the horizontal line in the box plot represents the median, the upper and lower box boundaries represent the interquartile range, and the ends of the whiskers represent the 5% and 95% confidence limits. Extreme values (> 2 standard deviations) are represented as individual dots.
n = number of animals per experimental group. * p < 0.05, ** p < 0.01, *** p < 0.001 compared to no Tx.
Co-infection with either MNV strain alone did not alter the gross cecal appearance compared to S. typhimurium infection alone (Fig. 2A). However, MNV1 infection alone caused an increase in cecal area compared to uninfected mice (102.3 vs. 94.5 pixels, p = 0.002). The biological significance of this finding is unknown, particularly as co-infection of MNV1 and S. typhimurium did not alter the S. typhimurium-induced cecal contraction (59.0 vs. 65.1, p = 0.34, MNV1/S. typhimurium vs. S. typhimurium) (Fig. 2B). MNV4 alone did not alter cecal area (100.2 vs. 94.5, p = 0.70 vs. no Tx) and MNV4/S. typhimurium co-infection resulted in a small additional contraction in cecal size (55.0 vs. 65.1, p = 0.036, MNV4/S. typhimurium vs. S. typhimurium alone) (Fig. 2B). Similar to previous work [33], a reduction in cecal size after S. typhimurium infection alone was accompanied by a marked increase in combined cecum and colon weight compared to uninfected controls (Fig. 2C). A similar increase occurred with co-infection of S. typhimurium and MNV1 (0.60 vs. 0.41g, p < 0.001 vs. MNV1 alone) or MNV4 (0.76 vs. 0.40 g, p < 0.0001 vs. MNV4 alone) (Fig. 2C). However, cecal and colon weight of mice co-infected with either MNV strain and S. typhimurium was indistinguishable from S. typhimurium infection alone (0.60 g [MNV1+St] vs. 0.71 g [S. typhimurium], p = 0.26, 0.76 g [MNV4+St] vs. 0.71 g [S. typhimurium], p = 0.15). Overall, the gross pathology, cecal area and weight of the large intestine of mice infected with MNV1 or MNV4 and S. typhimurium was similar to S. typhimurium infected mice. Single MNV infection resulted in a larger cecal area in case of MNV1 but no change in case of MNV4 compared to untreated mice. Whether this is a general feature of MNV1 infection and occurs in the absence of antibiotic treatment remains unclear.
Co-infection with MNV1 or MNV4 does not affect the overall histopathology of the cecum
Chronic S. typhimurium infection of the mouse cecum produces extensive transmural inflammation, massive cellular infiltration, epithelial destruction, disruption of tissue architecture, and increased extracellular matrix deposition [33]. Histological analysis of cecal tissue sections showed extensive crypt loss, submucosal edema, transmural inflammation and collagen deposition, and hypertrophy in the muscularis propria in all animals infected with S. typhimurium and co-infected with MNV1 or MNV4 and S. typhimurium but not in the animals infected with MNV alone or untreated controls (Fig. 3A-F).
Figure 3. Histopathology and submucosal thickness of the cecum.
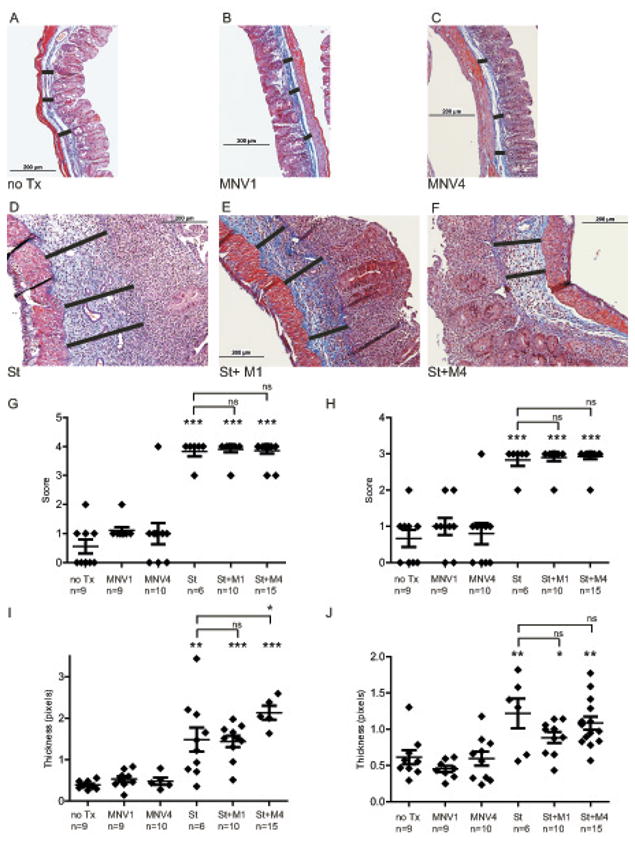
Representative trichrome stained sections of mouse cecum from uninfected (A) compared to MNV1 infected (B), MNV4 infected (C), S. typhimurium infected (D), S. typhimurium and MNV1 co-infected (E), and S. typhimurium and MNV4 co-infected (F) mice. Photomicrographs were taken at 100× magnification with a 200 μM scale. (G) Inflammatory scoring (blinded) of cecal sections using the 0 to 4 point Wirtz scale as described in Materials and Methods. (H) Fibrosis scoring (blinded) of cecal sections was determined from trichrome stained cecal sections: (0) no fibrosis, (1) mild focal fibrosis, (2) moderate fibrosis, and (3) severe fibrosis. (I) Submucosal expansion was measured from 3 reference points (black bars in Fig.A-F) for each tissue section and averaged to determine submucosal thickness. (J) Muscularis propria thickness was measured from 3 reference points from each tissue section and averaged to determine the extent of muscularis propria hypertrophy. In graphs G-J diamonds represent the average thickness of cecal submucosa from individual animals while the horizontal bar represents the mean submucosal thickness for each experimental group. Results are from three independent animal experiments.
n= number of animals per experimental group, not significant (ns) = p≥0.05, * p < 0.05, ** p < 0.01, *** p < 0.001 compared to no Tx.
To determine the extent of intestinal inflammation, cecal tissue sections were blindly scored for inflammation by a pathologist on the Wirtz 0 to 4 scale for mononuclear infiltration, vascular density, wall thickening, transmural leukocyte infiltration, and loss of goblet cells as described[35]. S. typhimurium induced profound cecal inflammation compared to uninfected animals (3.83 vs 0.56, p <0.0001) while co- infection with either MNV1 or MNV4 and S. typhimurium induced a nearly identical inflammatory response of S. typhimurium alone (3.9 vs 0.56, p <0.0001 and 3.87 vs 0.56, p <0.0001) (Fig. 3G). However, single infection with MNV1 or MNV4 did not cause significant inflammation in the cecum (1.0 vs 0.56, p = 0.053, 1.0 vs 0.56, p =0.33).
Similarly, trichrome stained cecal tissue sections were blindly scored by a pathologist for the extent of fibrosis using a four tier scale. Infection with S. typhimurium, induced marked tissue fibrosis compared to uninfected mice (2.83 vs 0.67, p <0.0001). Co-infection with S. typhimurium and either MNV1 or MNV4 induced similar tissue fibrosis compared to S. typhimurium alone (2.9 vs 0.67, p <0.0001 and 2.93 vs 0.67, p <0.0001) (Fig. 3H). In contrast, single infections of either MNV1 or MNV4 did not cause significant fibrosis in the cecum (0.80 vs 0.67, p = 0.33, 0.89vs 0.67, p = 0.73).
Another indicator for the extent of fibrosis and tissue edema is the thickness of the cecal submucosa and muscularis propria. Cecal submucosa thickness was increased in S. typhimurium singly or co-infected animals compared to untreated and MNV-infected mice (Fig. 3I). Specifically, mice infected with S. typhimurium exhibited a 3.4 fold expansion of the submucosa compared to uninfected animals (1.27 vs. 0.37 pixels, p = 0.009). Co-infection with either MNV1 or MNV4 and S. typhimurium induced a 3.9-fold (1.44 vs. 0.37 pixels, p < 0.0001) and 5.7-fold (2.13 vs 0.37 pixels, p < 0.0001) increase, respectively, in submucosal thickness compared to uninfected animals. However, cecal submucosal thickness did not differ between the S. typhimurium single infection and MNV1 co-infection (1.27 vs. 1.44 pixels, p = 0.53) (Fig. 3I). Co-infection with MNV4 augmented submucosal expansion compared to S. typhimurium alone (2.13 vs. 1.27 pixels, p = 0.03). Yet single infections with either MNV1 or MNV4 did not result in changes in cecal histopathology or increases in the thickness of the cecal submucosa compared to untreated mice (MNV1 = 0.48 pixels, MNV4 = 0.49 pixels, uninfected = 0.37 pixels, p = 0.11, p = 0.27 vs. uninfected, respectively).
Measurements of the muscularis propria demonstrated an approximately 2-fold increase in muscularis propria thickness in S. typhimurium-infected mice compared to uninfected mice (1.22 vs 0.61 pixels, p<0.01). Co-infection with S. typhimurium and either MNV1 or MNV4 induced a similar muscularis propria hypertrophy (0.88 vs 0.61, p = 0.04) and (1.08 vs 0.61, p <0.01) (Fig. 3J). However, single MNV infection with either MNV1 or MNV4 did not stimulate an increase the muscularis propria thickness as compared to uninfected controls (0.59 vs 0.61 pixels, p = 0.17, 0.62 vs 0.61 pixels, p = 0.89).
The histopathology of the distal colon was also evaluated in a blinded fashion. However, the histopathology of the distal colon was unaffected by infection with S. typhimurium, MNV1 alone, or MNV4 alone, or co-infection with S. typhimurium and either MNV strain compared to untreated mice (data not shown).
Taken together these data demonstrated that MNV infection alone does not significantly alter the histological inflammation or fibrosis of the cecum or colon in C57BL/6 mice. Furthermore, MNV co-infection did not significantly alter the S. typhimurium-induced histological inflammation or fibrosis of the cecum in this model, with the exception of a modest submucosal expansion in the MNV4 co-infection.
Inflammatory Th1 cytokine levels in the cecum are largely unaffected by norovirus co- infection
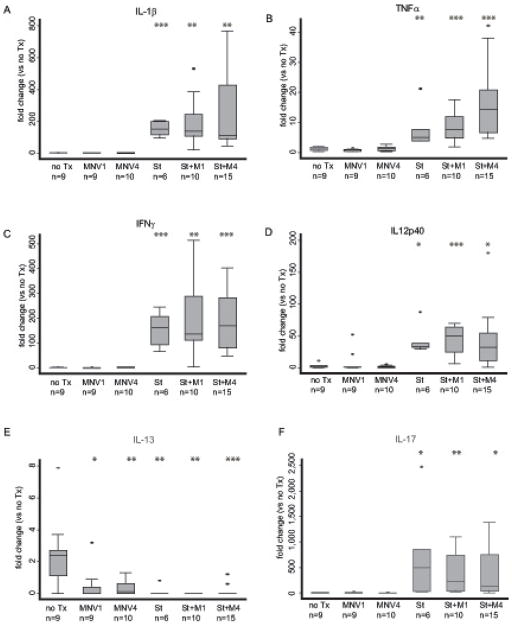
Infection with S. typhimurium in the fibrosis model reportedly leads to a marked increase in the Th1 inflammatory cytokines TNFα and IFNγ [33]. Therefore, quantitative RT-PCR analysis of RNA extracted from cecal tissue was performed for a panel of Th1 cytokines; IL-1, TNFα, IL-12p40, and IFNγ (Fig. 4A-D). As previously reported [33], infection with S. typhimurium alone led to an increase in the expression level of TNFα and IFNγ compared to uninfected controls. In addition, increases in the expression of IL-1β and IL-12p40 were also seen. Specifically, infection with S. typhimurium induced IL-1β 146-fold (p < 0.0001 vs. no Tx [Fig. 4A]), TNFα 5-fold (p < 0.0001 vs. no Tx [Fig. 4B]), IFNγ 107-fold (p < 0.0001 vs. no Tx [Fig. 4C]), and IL-12p40 57-fold (p < 0.0001 vs. no Tx [Fig. 4D]). Co-infection with S. typhimurium and either MNV1 or MNV4 resulted in IL-1β, and IFNγ expression levels similar to S. typhimurium infection alone. Co-infection with either MNV1 or MNV4 dramatically induced IL-1β (138-fold and 160-fold, respectively) but was indistinguishable from S. typhimurium alone (p = 0.90, p = 0.81, respectively). A similar robust IFNγ response was induced by either MNV1/S. typhimurium (132-fold) or MNV4/S. typhimurium (118-fold) co-infection compared to the 107-fold induction by S. typhimurium infection alone (p = 0.57, p = 0.76, respectively). MNV1/S. typhimurium co-infection induced TNFα 7-fold, comparable to the 5-fold induction by S. typhimurium alone (p = 0.25). However, MNV4/S. typhimurium co-infection induced an 11-fold response, which is significantly increased from S. typhimurium alone (p = 0.033). In case of IL-12p40, co-infection of S. typhimurium and either MNV1 or the MNV4 induced IL-12p40 to a lesser extent (34-fold and 35-fold, respectively) compared to S. typhimurium alone (57-fold) but this was not significantly different (MNV1, p = 0.3; MNV4, p = 0.27 vs. S. typhimurium).
Figure 4. Th1, Th2, and Th17 cytokine responses in the mouse cecum.
Cytokine expression of (A) IL-1β, (B) TNFα, (C) IFNγ, (D) IL-12p40, IL-13 (E) and IL-17 (F) in the cecum of uninfected mice (no Tx), MNV1 infected (MNV1), MNV4 infected (MNV4), S. typhimurium infected (St), S. typhimurium and MNV1 co-infected (St+M1), and S. typhimurium and MNV4 co-infected (St+M4) mice were determined by qRT-PCR. Cytokine expression was normalized to GAPDH expression and compared to expression in the no Tx group using the the ΔΔCT method [37]. Fold-change (RQ) was calculated (RQ=2-ΔΔCt) and graphed. The RQ is represented by the horizontal line in the box plot and the ends of the whiskers represent RQ-min and RQ-max limits, based on the standard error (SEM) of the individual ΔCt (2(– ΔCt ± SEM) / 2(– ΔCt reference)). Outliers are defined as exceeding two SEM from the RQ and are represented as individual dots. Results are from three independent animal experiments.
n = number of animals per experimental group, ** p < 0.01, *** p < 0.001 compared to no Tx.
In contrast, MNV1 or MNV4 infection alone did not lead to increases in IL-1β, TNFα and IL-12p40 expression levels compared to uninfected controls (Fig. 4B and C). Compared to uninfected mice, MNV1 and MNV4 IL-1β expression was 0.83-fold and 0.76-fold (p = 0.63, p = 0.43, respectively), TNFα was 0.62-fold and 1.07-fold (p = 0.039, p = 0.79, respectively), and IL-12p40 was 0.8-fold and 0.53-fold (p = 0.81, p = 0.46, respectively). Though norovirus infection triggers an innate immune response characterized by induction of serum IFNγ [39, 40], cecal IFNγ gene expression 19 days post norovirus challenge was only induced by MNV4 (1.8-fold, p = 0.029), likely reflecting differences in the persistent versus acute phenotype of MNV4 and MNV1, respectively. Together, these data demonstrate that MNV infection alone and MNV/S.typhimurium co-infection lead to subtle changes in selected cecal Th1 cytokine responses in this model.
IL-13 mRNA expression in the cecum is repressed by both S. typhimurium and MNV
The Th2 immune response, characterized by induction of IL-13, IL-4, and IL-10, supports humoral immunity, is essential for eliminating intestinal helminth infections and involved in allergic reactions [41]. We determined the mRNA expression levels of these three cytokines in the cecum of animals from our experimental groups. S. typhimurium infection did not induce a Th2 response with respect to IL-4 and IL-10 expression (data not shown). Neither MNV1 nor MNV4 alone or co-infected with S. typhimurium induced IL-4 or IL-10 expression (data not shown).
However, S. typhimurium infection significantly repressed IL-13 expression compared to uninfected controls (>350-fold, p <0.0001) (Fig. 4E). Similarly, infection with either norovirus strain alone also markedly repressed IL-13 expression (~70 and 53-fold, p = 0.002 [MNV1], p = 0.001 [MNV4] vs. uninfected) (Fig. 4E). Co-infection of S. typhimurium and either norovirus strain did not further reduce IL-13 expression (St+MNV1, p = 0.21 vs. S. typhimurium; St+MNV4, p = 0.85 vs. S. typhimurium) beyond S. typhimurium alone. Specifically, IL-13 was repressed >1,000-fold by MNV1/S. typhimurium co-infection (p < 0.0001 vs. uninfected) and 450-fold by MNV4/S. typhimurium co-infection (p < 0.0001 vs. uninfected). Thus, cecal IL-13 expression was reduced by either S. typhimurium, or MNV infection but co-infection did not significantly change the response in this model.
The Th17 response to S. typhimurium infection is not significantly reduced by norovirus co-infection
IL-17 is a cytokine critical for activating the innate immune response [42] and is induced by S. typhimurium infection in the fibrosis model [33]. Analysis of mRNA levels of IL-17 in the cecal tissue of our experimental animals demonstrated that IL-17 was profoundly induced by S. typhimurium (232-fold, p < 0.0001 vs. uninfected) but was not induced by either MNV1 (1.12-fold, p = 0.89 vs. uninfected) or MNV4 (0.35-fold, p = 0.08 vs. uninfected) norovirus infection (Fig. 4F). Co-infection of MNV1/S. typhimurium induced IL-17 to a lesser extent (174 vs. 232 fold, p = 0.76) but this was still significantly increased over uninfected controls (p < 0.0001). Similarly, co-infection of MNV4/S. typhimurium induced a numerically lower (107 vs. 232-fold, p = 0.33) but non-significant change in IL-17 response compared to S. typhimurium infected animals. This IL-17 response in MNV4/S.typhimurium co-infected mice remained significantly greater than the IL-17 expression in uninfected controls (p < 0.0001). Thus, MNV co-infection slightly (but not significantly) dampens the strong cecal IL-17 response to S. typhimurium.
Collagen deposition in the cecum was unaffected by norovirus co-infection
Intestinal fibrosis is characterized by deposition of extracellular matrix in the bowel wall [43]. In the S. typhimurium model, infection with S. typhimurium triggers extensive collagen deposition in the mucosa and submucosa (see blue staining in Fig. 3A-F). Therefore, we determined the collagen density in the cecum of all animals by measuring the intensity of blue (i.e. trichrome) staining as described in Methods. Collagen density in trichrome stained cecal sections from S. typhimurium infected animals was 2.5-fold higher than both uninfected controls (p = 0.003) and animals infected with either MNV1 or MNV4 (Fig. 5). Co-infection of either norovirus strain and S. typhimurium did not significantly alter collagen deposition compared to S. typhimurium infected animals. Co-infection of MNV1 and S. typhimurium induced a 1.8-fold increase in collagen density, while MNV4/S. typhimurium co-infected induced a 1.5-fold increase compared to uninfected controls. However, both co-infections were similar to S. typhimurium infection alone (MNV1, p = 0.13; MNV4, p = 0.14 vs. S. typhimurium). These data demonstrate that MNV co-infection does not alter collagen deposition in this model.
Figure 5. Collagen deposition in the cecum.
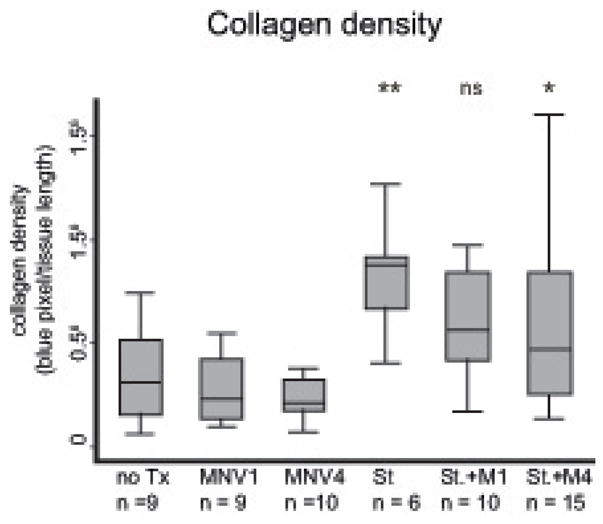
Collagen density was determined from digital images of an entire cross-section of cecal tissue from each animal and normalized to the length of tissue. Results are from three independent animal experiments.
n = number of animals per experimental group, not significant (ns) = p≥0.05, * p < 0.05, ** p < 0.01, *** p < 0.001 compared to no Tx.
Discussion
MNV is the most prevalent virus in mouse research colonies worldwide [20] and can alter the experimental outcome in some mouse research models, particularly in studies that involve the gastrointestinal tract [28, 29]. Therefore, we tested whether concurrent MNV infection altered the experimental outcome in a model of intestinal fibrosis. We chose the prototypical MNV1 [4] and MNV4 strains [6], the latter of which was shown to exacerbate inflammatory bowel disease in a mouse model [28]. Analysis of gross pathology, histology, and collagen deposition in norovirus co-infection with S. typhimurium did not result in significant changes in the major phenotypes of this model. The fibrosis, cecal contraction, and collagen deposition that are the signature of this model were unchanged by co-infection with MNV. MNV4 did slightly reduce the submucosal expansion in the co-infected animals but whether this is strain specific or true for other persistent MNV strains remains unknown. In addition, Th1, Th2, and Th17 cytokine responses were largely unaltered, with slight but not statistically significant decreases in the IL-12p40 and IL-17 expression levels in MNV1 or MNV4 co-infected animals compared to S. typhimurium alone. Therefore, simultaneous MNV infection does not significantly alter the overall phenotype of this model of intestinal inflammation and fibrosis.
MNV4 but not MNV1 co-infection resulted in a significant increase in TNFα levels compared to S. typhimurium alone. Significant exacerbation of the inflammatory bowel disease phenotype by MNV4 co-infection was also observed in the Mdr1a−/− mouse model [28]. This difference between virus strains in outcomes is likely attributable to biological differences (transient versus persistent infection) between strains. A similar difference in experimental outcomes between MNV1 and another persistent MNV strain was recently described by Cadwell [27]. Infection with the persistent CR6 strain but not MNV1 in mice with mutations in the Crohn’s disease susceptibility gene Atg16L1HM exacerbated DSS colitis. However, in a genetically uncompromised host, the S. typhimurium intestinal fibrosis model is not dramatically affected by MNV co-infection, regardless of whether transient or persistent infections occur.
Our analysis of the MNV infection status based on the presence of infectious virions and viral genome in C57BL/6 mice demonstrated that MNV1 and MNV4 can cause transient or persistent infections in either a single infection or with S. typhimurium co-infection. It is unclear whether this is a result of the antibiotic treatment which removes most of the gut-associated indigenous microbiota (and which these mice received as part of this model) or whether this is a typical feature of MNV infections. The first possibility appears less likely, since previously published data suggests that the transient or persistent infection phenotype of MNV1 and MNV4, respectively, is not absolute [5, 6]. Infection of C57BL/6 mice with MNV1 resulted in low levels of detectable virus detectable in the mesenteric lymph nodes and distal ileum in ~10% of mice at 7 days postinfection [5]. MNV4 genomes are detectable in 100% (10/10) of mesenteric lymph nodes but in only 90% (9/10) of jejunal samples and in 40% (4/10) of spleens of outbred CD1 mice 8 weeks postinfection [6]. Taking current data and published data together, we conclude that viral shedding in feces is not a reliable predictor of the presence of infectious virus or viral genomes in mouse tissues.
Little is known regarding the cytokine response to norovirus infections. Our studies provide the first data on cecal cytokine responses to norovirus infection by demonstrating that MNV1 or MNV4 infection alone did not cause major changes in cecal IL-1β, IL-12p40, and IL-17 cytokine expression while reducing IL-13 (MNV1 and MNV4) and TNFα (MNV1 only) expression compared to uninfected controls. This suggests that MNV infection alters only selected Th1 responses in the large intestine and that this is dependent on the virus strain. The cytokine response to human norovirus infection in the gut is thought to be dominated by a Th1 immune response [44]. Fecal IL-2 and IFNγ were significantly increased in international travelers with norovirus diarrhea, with TNFα and IL-5 slightly increased (but not statistically significant) [44]. Furthermore, IL-12 and IFNγ concentrations in small intestinal contents of gnotobiotic pigs infected with a human norovirus were also increased although they did not reach statistical significance [45]. In case of Th2 cytokine responses, the picture appears to be somewhat similar between MNV and human norovirus infections as IL-4 and IL-10 responses in the cecum (our study) and in the feces [44], respectively, were not significantly different from controls. IL-4 but not IL-10 responses in the small intestinal contents of gnotobiotic pigs infected with a human norovirus were slightly but not significantly increased [45]. No studies have addressed the role of IL-13 and IL-17 in norovirus infection.
This study has two significant limitations. First, the effect of chronic, pre-existing MNV on the S. typhimurium intestinal fibrosis model was not tested. We chose to focus on the new acquisition of MNV after induction of the fibrosis model, as we are most concerned about mice acquiring new infections with MNV in our facility shortly after arrival, and during the induction of fibrosis. Second, only two strains of MNV were tested in this model. An exhaustive study of all isolated MNV strains would not be practical, and thus we chose to focus on two strains, the MNV-1 strain that is usually considered to cause transient infections, and the MNV4 strain that is considered to cause persistent infections and is known to alter at least one intestinal phenotype [28].
In summary, the data presented herein demonstrate that co-infection of C57BL/6 mice with either MNV1 or MNV4 do not cause major alterations in the histopathology and only subtle change in the inflammatory response in a research model of intestinal inflammation and fibrosis. Therefore, contrary to previous findings [27, 28] not all intestinal phenotypes are dramatically altered by MNV. Although not all MNV strains have been tested, in the case of this intestinal fibrosis model, elimination of an entire mouse research colony to eradicate norovirus infection is most likely not necessary, but requires careful interpretation of the data to account for possibly subtle changes in cytokine levels. Thus, in biomedical research facilities with known norovirus contamination, the effects of norovirus infections need to be examined on a model-to- model basis particularly in case of mouse models of gastrointestinal infection and inflammation.
Acknowledgments
This work was funded by NIH K08DK080172-01 to P. H. and start-up funds from the University of Michigan to C. E. W. We thank Drs. G. Grassl (University of British Columbia, Vancouver, Canada; S. typhimurium ΔaroA) and R. Livingston (University of Missouri – Columbia, USA; MNV4) for their generous gifts of reagents.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mead PS, et al. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5(5):607–25. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med. 2009;361(18):1776–85. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siebenga JJ, et al. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. J Infect Dis. 2009;200(5):802–12. doi: 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- 4.Karst SM, et al. STAT1-dependent innate immunity to a Norwalk-like virus. Science. 2003;299(5612):1575–8. doi: 10.1126/science.1077905. [DOI] [PubMed] [Google Scholar]
- 5.Thackray LB, et al. Murine noroviruses comprising a single genogroup exhibit biological diversity despite limited sequence divergence. J Virol. 2007;81(19):10460–73. doi: 10.1128/JVI.00783-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu CC, et al. Persistent infection with and serologic cross-reactivity of three novel murine noroviruses. Comp Med. 2006;56(4):247–51. [PubMed] [Google Scholar]
- 7.Oliver SL, et al. Complete genomic characterization and antigenic relatedness of genogroup III, genotype 2 bovine noroviruses. Arch Virol. 2007;152(2):257–72. doi: 10.1007/s00705-006-0856-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oliver SL, et al. Molecular characterization of bovine enteric caliciviruses: a distinct third genogroup of noroviruses (Norwalk-like viruses) unlikely to be of risk to humans. J Virol. 2003;77(4):2789–98. doi: 10.1128/JVI.77.4.2789-2798.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wise AG, et al. Molecular characterization of noroviruses detected in diarrheic stools of Michigan and Wisconsin dairy calves: circulation of two distinct subgroups. Virus Res. 2004;100(2):165–77. doi: 10.1016/j.virusres.2003.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang QH, et al. Porcine noroviruses related to human noroviruses. Emerg Infect Dis. 2005;11(12):1874–81. doi: 10.3201/eid1112.050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mauroy A, et al. Noroviruses and sapoviruses in pigs in Belgium. Arch Virol. 2008 doi: 10.1007/s00705-008-0189-4. [DOI] [PubMed] [Google Scholar]
- 12.Wolf S, et al. Molecular detection of norovirus in sheep and pigs in New Zealand farms. Vet Microbiol. 2008 doi: 10.1016/j.vetmic.2008.06.019. [DOI] [PubMed] [Google Scholar]
- 13.Martella V, Campolo M, Lorusso E, Cavicchio P, Camero M, Bellacicco AL, Decaro N, Elia G, Greco G, Corrente M, Desario C, Arista S, Banyai K, Koopmans M, Buonavoglia C. Norovirus in captive lion (Panthera leo) Emerg Infect Dis. 2007;13:1071–1073. doi: 10.3201/eid1307.070268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martella V, et al. Detection and molecular characterization of a canine norovirus. Emerg Infect Dis. 2008;14(8):1306–8. doi: 10.3201/eid1408.080062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wobus CE, et al. Replication of Norovirus in cell culture reveals a tropism for dendritic cells and macrophages. PLoS Biol. 2004;2(12):e432. doi: 10.1371/journal.pbio.0020432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perdue KA, et al. Naturally occurring murine norovirus infection in a large research institution. J Am Assoc Lab Anim Sci. 2007;46(4):39–45. [PubMed] [Google Scholar]
- 17.Ward JM, et al. Pathology of immunodeficient mice with naturally occurring murine norovirus infection. Toxicol Pathol. 2006;34(6):708–15. doi: 10.1080/01926230600918876. [DOI] [PubMed] [Google Scholar]
- 18.Mumphrey SM, et al. Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J Virol. 2007;81(7):3251–63. doi: 10.1128/JVI.02096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu CC, et al. Development of a microsphere-based serologic multiplexed fluorescent immunoassay and a reverse transcriptase PCR assay to detect murine norovirus 1 infection in mice. Clin Diagn Lab Immunol. 2005;12(10):1145–51. doi: 10.1128/CDLI.12.10.1145-1151.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson KS. Murine norovirus, a recently discovered and highly prevalent viral agent of mice. Lab Anim (NY) 2008;37(7):314–20. doi: 10.1038/laban0708-314. [DOI] [PubMed] [Google Scholar]
- 21.Muller B, et al. Genetic diversity and recombination of murine noroviruses in immunocompromised mice. Arch Virol. 2007;152(9):1709–19. doi: 10.1007/s00705-007-0989-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahler M, Kohl W. A serological survey to evaluate contemporary prevalence of viral agents and Mycoplasma pulmonis in laboratory mice and rats in western Europe. Lab Anim (NY) 2009;38(5):161–5. doi: 10.1038/laban0509-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kitajima M, et al. Development of a broadly reactive nested reverse transcription-PCR assay to detect murine noroviruses, and investigation of the prevalence of murine noroviruses in laboratory mice in Japan. Microbiol Immunol. 2009;53(9):531–4. doi: 10.1111/j.1348-0421.2009.00152.x. [DOI] [PubMed] [Google Scholar]
- 24.Kim M, et al. Molecular characterization of murine norovirus isolates from South Korea. Virus Res. 2010;147(1):1–6. doi: 10.1016/j.virusres.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 25.Goto K, et al. Molecular detection of murine norovirus from experimentally and spontaneously infected mice. Exp Anim. 2009;58(2):135–40. doi: 10.1538/expanim.58.135. [DOI] [PubMed] [Google Scholar]
- 26.Kastenmayer RJ, Perdue KA, Elkins WR. Eradication of murine norovirus from a mouse barrier facility. J Am Assoc Lab Anim Sci. 2008;47(1):26–30. [PMC free article] [PubMed] [Google Scholar]
- 27.Cadwell K, et al. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell. 2010;141(7):1135–45. doi: 10.1016/j.cell.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lencioni KC, et al. Murine norovirus: an intercurrent variable in a mouse model of bacteria-induced inflammatory bowel disease. Comp Med. 2008;58(6):522–33. [PMC free article] [PubMed] [Google Scholar]
- 29.Compton SR, Paturzo FX, Macy JD. Effect of murine norovirus infection on mouse parvovirus infection. J Am Assoc Lab Anim Sci. 2010;49(1):11–21. [PMC free article] [PubMed] [Google Scholar]
- 30.Hensley SE, et al. Murine norovirus infection has no significant effect on adaptive immunity to vaccinia virus or influenza A virus. J Virol. 2009 doi: 10.1128/JVI.00623-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doom CM, Turula HM, Hill AB. Investigation of the impact of the common animal facility contaminant murine norovirus on experimental murine cytomegalovirus infection. Virology. 2009;392(2):153–61. doi: 10.1016/j.virol.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ammann CG, et al. Effects of acute and chronic murine norovirus infections on immune responses and recovery from Friend retrovirus infection. J Virol. 2009;83(24):13037–41. doi: 10.1128/JVI.01445-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grassl GA, et al. Chronic enteric salmonella infection in mice leads to severe and persistent intestinal fibrosis. Gastroenterology. 2008;134(3):768–80. doi: 10.1053/j.gastro.2007.12.043. [DOI] [PubMed] [Google Scholar]
- 34.Chachu KA, et al. Antibody is critical for the clearance of murine norovirus infection. J Virol. 2008;82(13):6610–7. doi: 10.1128/JVI.00141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wirtz S, et al. Chemically induced mouse models of intestinal inflammation. Nat Protoc. 2007;2(3):541–6. doi: 10.1038/nprot.2007.41. [DOI] [PubMed] [Google Scholar]
- 36.Koga H, et al. Transanal delivery of angiotensin converting enzyme inhibitor prevents colonic fibrosis in a mouse colitis model: development of a unique mode of treatment. Surgery. 2008;144(2):259–68. doi: 10.1016/j.surg.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 38.Taube S, et al. Ganglioside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J Virol. 2009;83(9):4092–101. doi: 10.1128/JVI.02245-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Souza M, et al. Pathogenesis and immune responses in gnotobiotic calves after infection with the genogroup II.4-HS66 strain of human norovirus. J Virol. 2008;82(4):1777–86. doi: 10.1128/JVI.01347-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lindesmith L, et al. Cellular and humoral immunity following Snow Mountain virus challenge. J Virol. 2005;79(5):2900–9. doi: 10.1128/JVI.79.5.2900-2909.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paul WE. What determines Th2 differentiation, in vitro and in vivo? Immunol Cell Biol. 2010;88(3):236–9. doi: 10.1038/icb.2010.2. [DOI] [PubMed] [Google Scholar]
- 42.O'Quinn DB, et al. Emergence of the Th17 pathway and its role in host defense. Adv Immunol. 2008;99:115–63. doi: 10.1016/S0065-2776(08)00605-6. [DOI] [PubMed] [Google Scholar]
- 43.Danese S. Nonimmune cells in inflammatory bowel disease: from victim to villain. Trends Immunol. 2008;29(11):555–64. doi: 10.1016/j.it.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 44.Ko G, et al. Fecal cytokines and markers of intestinal inflammation in international travelers with diarrhea due to Noroviruses. J Med Virol. 2006;78(6):825–8. doi: 10.1002/jmv.20630. [DOI] [PubMed] [Google Scholar]
- 45.Souza M, et al. Cytokine and antibody responses in gnotobiotic pigs after infection with human norovirus genogroup II.4 (HS66 strain) J Virol. 2007;81(17):9183–92. doi: 10.1128/JVI.00558-07. [DOI] [PMC free article] [PubMed] [Google Scholar]