Abstract
The Nijmegen breakage syndrome gene product (Nbs1) was shown recently to associate in vivo with the Mre11 and Rad50 proteins, which play pivotal roles in eukaryotic DNA double-strand break repair, meiotic recombination, and telomere maintenance. We show in this work that the triple complex of recombinant Nbs1, Mre11, and Rad50 proteins binds cooperatively to DNA and forms a distinct protein–DNA species. The Mre11/Rad50/Nbs1 complex displays several enzymatic activities that are not seen without Nbs1, including partial unwinding of a DNA duplex and efficient cleavage of fully paired hairpins. Unwinding and hairpin cleavage are both increased by the presence of ATP. On nonhairpin DNA ends, ATP controls a switch in endonuclease specificity that allows Mre11/Rad50/Nbs1 to cleave a 3′-protruding strand at a double-/single-strand transition. Mutational analysis demonstrates that Rad50 is responsible for ATP binding by the complex, but the ATP-dependent activities are expressed only with Nbs1 present.
Keywords: Nijmegen breakage syndrome, Mre11/Rad50 protein, ATP binding, DNA unwinding
Among the many threats to the genomic integrity of a cell, one of the most serious is DNA double-strand breakage, produced either by external agents such as ionizing radiation or by internal errors during replication or recombination. In eukaryotic cells, double-strand breaks can cause activation of checkpoint functions at the G1/S and the G2/M boundaries of the cell cycle. The most error-free method to repair a break involves recombination between the broken strands and a homologous chromosome or sister chromatid, which replaces any missing sequence information in addition to reconnecting the chromosome. This system is functionally conserved between the RecA/RecBCD system in bacteria and the Rad51/52/54/55/57 system in eukaryotic cells (Game 1993; Bianco et al. 1998). Alternatively, a double-strand break can be repaired via a pathway that facilitates ligation of the two ends, but is much more error-prone because missing sequence is not recovered, and even more DNA can be lost from the ends during the joining process. This pathway, known as nonhomologous end-joining (NHEJ), is conserved from yeast to humans (Critchlow and Jackson 1998; Tsukamoto and Ikeda 1998), although its importance varies greatly among species. Yeast, for instance, primarily utilizes the homologous recombination system (Petes et al. 1991), whereas mammalian cells favor the NHEJ pathway (Roth and Wilson 1985).
In Saccharomyces cerevisiae, the NHEJ pathway is essential only when members of the homologous pathway, for instance, Rad52, are inactivated (Kramer et al. 1994). In this genetic background, several factors have been shown to be necessary for survival after the introduction of double-strand breaks. Yeast cells lacking the DNA end-binding heterodimer, Ku, exhibit a 100-fold decrease in NHEJ (Boulton and Jackson 1996a,b; Milne et al. 1996; Tsukamoto et al. 1996b), as do strains that are missing DNA ligase IV (Teo and Jackson 1997) or its partner, Lif1, a yeast homolog of Xrcc4 (Herrmann et al. 1998). The few junctions recovered from these cells often exhibit large deletions and joining of the resected ends at regions of short microhomologies. In mammalian cells, Xrcc4, Ku, and the DNA-dependent protein kinase that associates with Ku were first identified from mutations that lower the resistance of cell lines to ionizing radiation and reduce the frequency of a specialized type of double-strand break repair drastically, the joining of V(D)J recombination intermediates (Li et al. 1995; Weaver 1996; Hendrickson 1997; Jeggo 1997). Ligase IV is also required for both processes in mammalian cells (Frank et al. 1998; Grawunder et al. 1998).
Three other factors, Mre11, Rad50, and Xrs2, have been identified in yeast as components of the NHEJ pathway that form a functional group (Ivanov et al. 1992; Johzuka and Ogawa 1995; Moore and Haber 1996; Tsukamoto et al. 1996a). Physical complexes among these factors have also been identified (Johzuka and Ogawa 1995; Ogawa et al. 1995; Usui et al. 1998). Yeast strains lacking Mre11, Rad50, or Xrs2 show ∼100-fold decreased levels of NHEJ in a rad52 background but, unlike cells defective in the factors described above, do not generate deletions in the few ends that are joined (Boulton and Jackson 1998). In addition, the complex is involved in the early stages of homologous recombination in yeast: The three proteins are absolutely necessary for the introduction of double-strand breaks that apparently initiate strand exchange during meiosis (Alani et al. 1990; Ivanov et al. 1992; Johzuka and Ogawa 1995), and increase the rate (although not the overall efficiency) of the 5′ strand resection that precedes homologous recombination in vegetative cells (Ivanov et al. 1994; for review, see Haber 1998).
Homologs of Mre11 and Rad50 have been identified in mammalian cells, and the proteins have been shown to associate with one another, as they do in yeast (Petrini et al. 1995; Dolganov et al. 1996; Trujillo et al. 1998). Exposure of cells to ionizing radiation causes rapid assembly of the complex into foci within the nucleus (Maser et al. 1997), and the foci occur in exactly the irradiated regions, suggesting that this complex localizes directly to the sites of DNA damage (Nelms et al. 1998). Further in vivo characterization of these proteins has been limited, however, by the fact that Mre11 is an essential gene in mammalian cells, so knockout mice or even cell lines are not viable (Xiao and Weaver 1997).
Immunoprecipitated complexes of Mre11 and Rad50 in human cells were found to contain a component the same size as the yeast Xrs2 protein (Dolganov et al. 1996), but the mammalian equivalent of Xrs2 was not identified until recently. This missing partner for Mre11 and Rad50 has been shown now to be the product of the NBS1 gene. Mutations in this gene are responsible for the rare human genetic disorder, Nijmegen breakage syndrome (NBS) (Carney et al. 1998; Varon et al. 1998). All of the mutations of the NBS1 gene found in patients with this disorder cause a premature truncation of the protein, leaving the amino-terminal portion which contains a breast cancer carboxy-terminal (BRCT) domain and a forkhead-associated (FHA) domain, the only identifiable motifs in the open reading frame. Cell lines from NBS patients do not exhibit foci of Mre11 and Rad50 proteins in response to ionizing radiation (Carney et al. 1998), and do not postpone DNA synthesis in response to DNA damage (Shiloh 1997). In addition, these cells show an elevated frequency of chromosomal translocations, especially at V(D)J-related loci, and patients have a higher incidence of malignancies.
To understand the biochemical functions of the Mre11/Rad50/Nbs1 complex that underlie the response of mammalian cells to DNA damage, these proteins must be characterized in vitro. We have shown previously that human Mre11 by itself is a 3′–5′ double-strand DNA exonuclease as well as an endonuclease, and that these activities are increased when Mre11 is present in a complex with Rad50 (Paull and Gellert 1998). The exonuclease activity of Mre11 can also facilitate the joining of two DNA molecules by a ligase through the use of microhomologies. Here we characterize recombinant human Nbs1 protein expressed in combination with Mre11 and Rad50, and show that the triple complex possesses several activities not seen in the absence of the Nbs1 protein. These include the efficient opening of fully paired DNA hairpins, a change in the specificity of the endonuclease activity, and partial unwinding of DNA at a double-/single-strand transition which is induced by ATP.
Results
To investigate the functions of the Nbs1 protein, the coding region of the human gene was amplified from a HeLa cell cDNA library. Compared to the published genomic sequence, our first isolate was missing nucleotides exactly corresponding to exon 13, suggesting that it was an alternatively spliced product. For the purposes of this work we generated a full-length copy of the gene, which was expressed in Sf9 cells using a baculovirus expression system.
Like Rad50 (Paull and Gellert 1998), Nbs1 protein was produced efficiently only when coexpressed with Mre11 and was undetectable in lysates when expressed by itself (data not shown). We coexpressed Nbs1 with Mre11 (M/N) as well as with Mre11 and Rad50 in combination (M/R/N). On a S200 gel-filtration column, the M/N complex eluted at a position equivalent to ∼400 kD, much larger than the sum of the molecular masses of monomeric Mre11 and Nbs1, and significantly larger than Mre11 alone (Paull and Gellert 1998). Like the Mre11/Rad50 preparation, the complex including all three proteins was excluded from the column and therefore is apparently larger than ∼1300 kD (data not shown). At the end of the purification the protein complexes were ≥95% homogeneous, as shown in the Coomassie-stained SDS–polyacrylamide gel in Figure 1A. The ratio of the three proteins in the recombinant triple complex appeared to be very similar to the ratio in the native complex (Trujillo et al. 1998).
Figure 1.
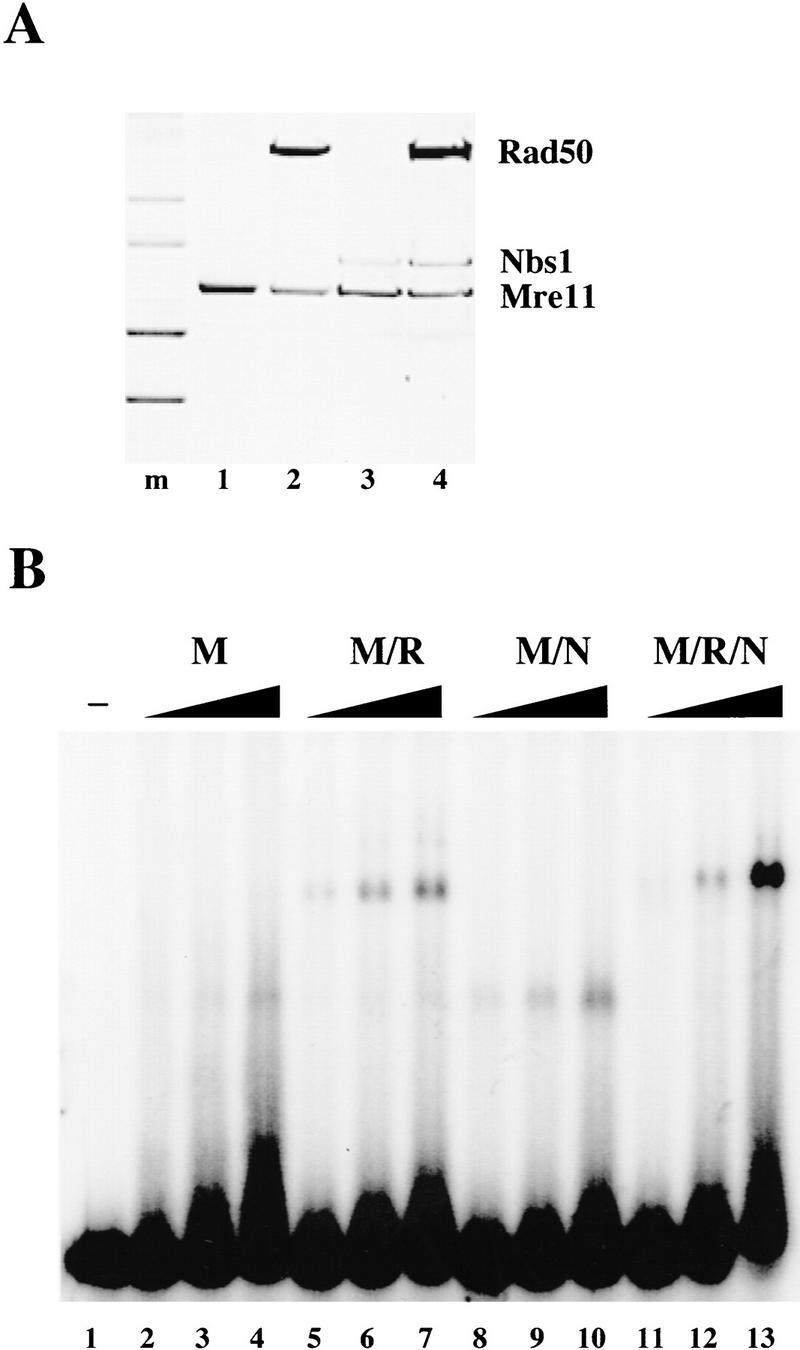
Nbs1 alters the activities of Mre11 and Rad50. (A) Preparations of Mre11 (lane 1), M/R (lane 2), M/N (lane 3), and M/R/N (lane 4) were separated on an SDS–polyacrylamide gel and stained with Coomassie blue. Protein amounts varied between 0.5 and 2 μg, keeping the amount of Mre11 approximately constant at 0.5 μg. (m) The molecular mass markers are 116, 97, 66, and 55 kD. (B) Gel mobility-shift assays were performed with Mre11 (lanes 2–4), M/R (lanes 5–7), M/N (lanes 8–10), and M/R/N (lanes 11–13) as indicated. Approximately 37.5 ng of each protein was added in lanes 2, 5, 8, and 11; 75 ng each in lanes 3, 6, 9, and 12; and 150 ng each in lanes 4, 7, 10, and 13. Proteins were mixed with a 32P-labeled double-stranded DNA substrate containing 3′ overhangs at each end, and incubated for 10 min at room temperature before electrophoresis in a 0.7% 1/2× TBE agarose gel.
Nbs1 stabilizes Mre11/Rad50 protein–DNA complexes
We tested the recombinant protein complexes in gel mobility-shift assays and found that protein preparations containing Nbs1 protein formed more stable protein–DNA complexes than complexes without Nbs1, as shown in Figure 1B. Under these conditions, Mre11 by itself formed a faint protein–DNA complex (lane 4), and Mre11/Rad50 generated two much larger complexes (lanes 5–7). Mre11 plus Nbs1 formed a complex similar in size to the Mre11 band (lanes 8–10), and the combination of all the proteins generated a single complex that appeared to form more cooperatively and at a significantly higher level than any of the other complexes shown on the gel (lanes 11–13).
Mre11/Rad50/Nbs1 complexes cut fully paired DNA hairpins efficiently
The similarity between the Escherichia coli hairpin nuclease SbcC/D and the Rad50/Mre11 proteins, respectively (Sharples and Leach 1995), suggested that the Mre11 complex might also possess endonuclease activity on DNA hairpins. We showed previously that human Mre11 and M/R complexes can cleave DNA hairpins, but are much more active on substrates containing mismatched nucleotides at the tip (hairpin loops) than on hairpins with no mismatches (Paull and Gellert 1998). When Nbs1 was present in complexes with Mre11 and Rad50, however, hairpins with no mismatches could be cleaved very efficiently, as shown in Figure 2A. The M/N complex (lane 4) was much more active on the fully paired hairpin substrate than either Mre11 (lane 2) or M/R (lane 3), and M/R/N (lane 5) was much more active than M/N. Quantitatively, the triple complex was actually at least 60-fold more active in cleaving fully paired hairpins compared to Mre11, and at least 25-fold more active than M/R.
Figure 2.
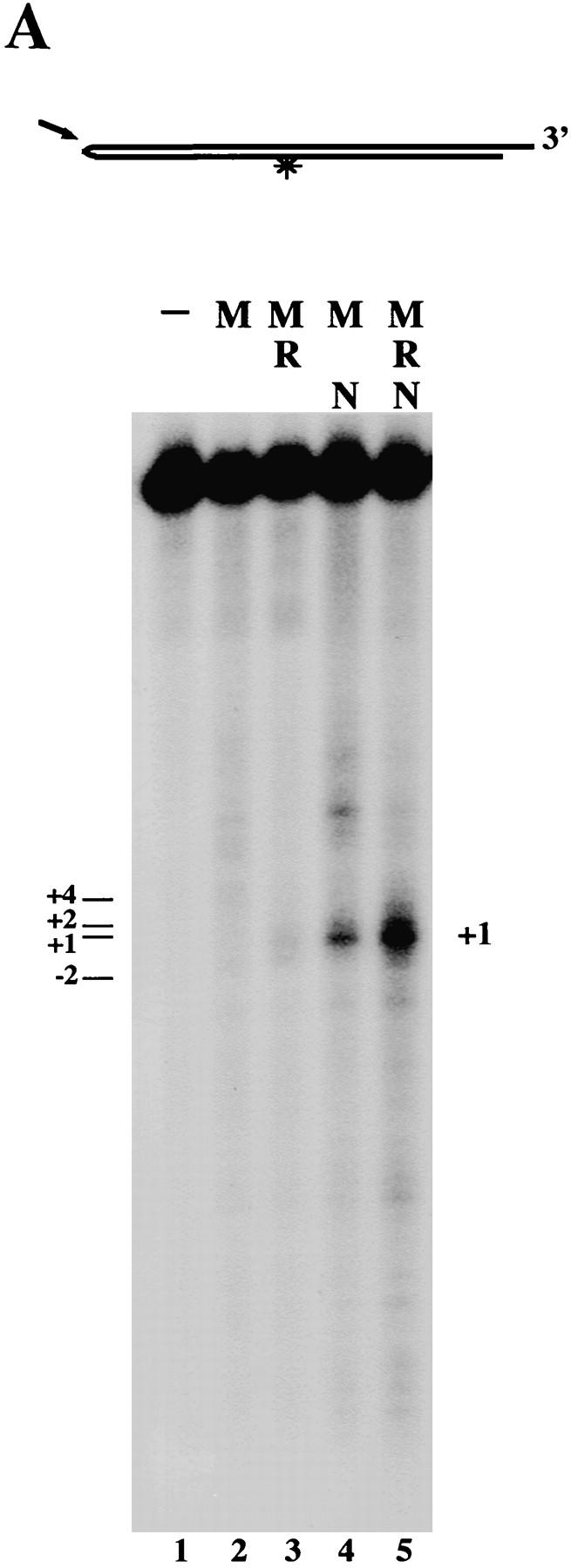
M/R/N opens fully paired hairpins. (A) Nuclease assays were performed with 150 ng each of Mre11 (lane 2), M/R (lane 3), M/N (lane 4), and M/N/R (lane 5) in 1 mm MnCl2 on a 32P-labeled substrate containing a fully paired hairpin on one end, a 4-bp 3′ overhang on the other end, separated by a 50-bp duplex. Reactions were incubated for 30 min at 37°C before separation on a denaturing polyacrylamide gel. (*) The location of the 32P label in the diagram of the substrate. The cleavage sites are numbered such that a cut exactly at the tip is “0”, sites 3′ of this are positive, and sites 5′ of this are negative. The predominant cut made by M/R/N is at +1, as indicated by the arrow in the diagram. (B) Nuclease assays were performed on four different fully paired hairpin substrates that varied in sequence within 10 bp of the hairpin tip. The sequence of each substrate near the tip is shown above the gel. The structure of the substrates is the same as in A, and the hairpin used in A is identical to hairpin I in B. The sequence at the tip of hairpin I is from the coding end of the mouse Jκ1 locus; hairpin II corresponds to the coding end of the JH1 locus; and hairpin III corresponds to the coding end of the JH2 locus. Reactions contained 150 ng M/R/N (as indicated) and were analyzed as in A. The asterisk indicates the location of the 32P label in the diagram of the substrate, and the arrows indicate the predominant cleavage sites on each substrate.
On this hairpin substrate, the cuts were made almost exclusively at positions one or two nucleotides 3′ of the center of the inverted repeat, generating products with 3′ overhangs of one or two nucleotides. Because opened products with 5′ overhangs would be rapidly resected at 37°C by all of the complexes and thus not visible as cleaved hairpins, these assays were also performed at room temperature, which minimizes the exonuclease but not the endonuclease activity of Mre11 (Paull and Gellert 1998). Under these conditions, the same cleaved products were observed (data not shown), demonstrating that all of the cleavage products are visible in Figure 2A. The triple complex was also able to cut three other hairpin substrates containing different sequences at the tip, always on the 3′ side of the hairpin tip (Fig. 2B), indicating that this pattern of cutting is a property of the protein complex and not of a particular hairpin substrate.
Although cutting of the fully paired substrates was increased greatly if the complexes contained Nbs1, cutting of larger hairpin loops by these complexes was not significantly greater than that of Mre11 or M/R (data not shown). Therefore Nbs1 appears to have changed the specificity of the hairpin endonuclease activity and not its overall level of activity.
In contrast to the hairpin substrates, Nbs1 did not alter the exonuclease activity of the Mre11 and Mre11/Rad50 complexes on oligonucleotide substrates with blunt or 3′ recessed ends (data not shown).
M/R/N generates different products in the presence of ATP
The RAD50 gene contains consensus Walker A and Walker B NTP-binding motifs (Alani et al. 1989; Dolganov et al. 1996), and the yeast homolog of Rad50 was shown to bind DNA in an ATP-dependent manner (Raymond and Kleckner 1993), but thus far the human M/R complex has failed to show any kind of response to ATP in DNA binding or exonuclease assays (data not shown). When ATP was added to reactions containing M/R/N and a dumbbell hairpin substrate, however, we observed that much more of the substrate was consumed (Fig. 3A). Forty percent of the dumbbell was degraded in the absence of ATP (Fig. 3A, lane 2), whereas 76% was degraded by M/R/N with ATP in the reaction (Fig. 3A, lane 3). With ATP present, the cleaved dumbbell was further degraded into smaller pieces, as evidenced by the smear of labeled DNA below the singly cut form (lane 3).
Figure 3.
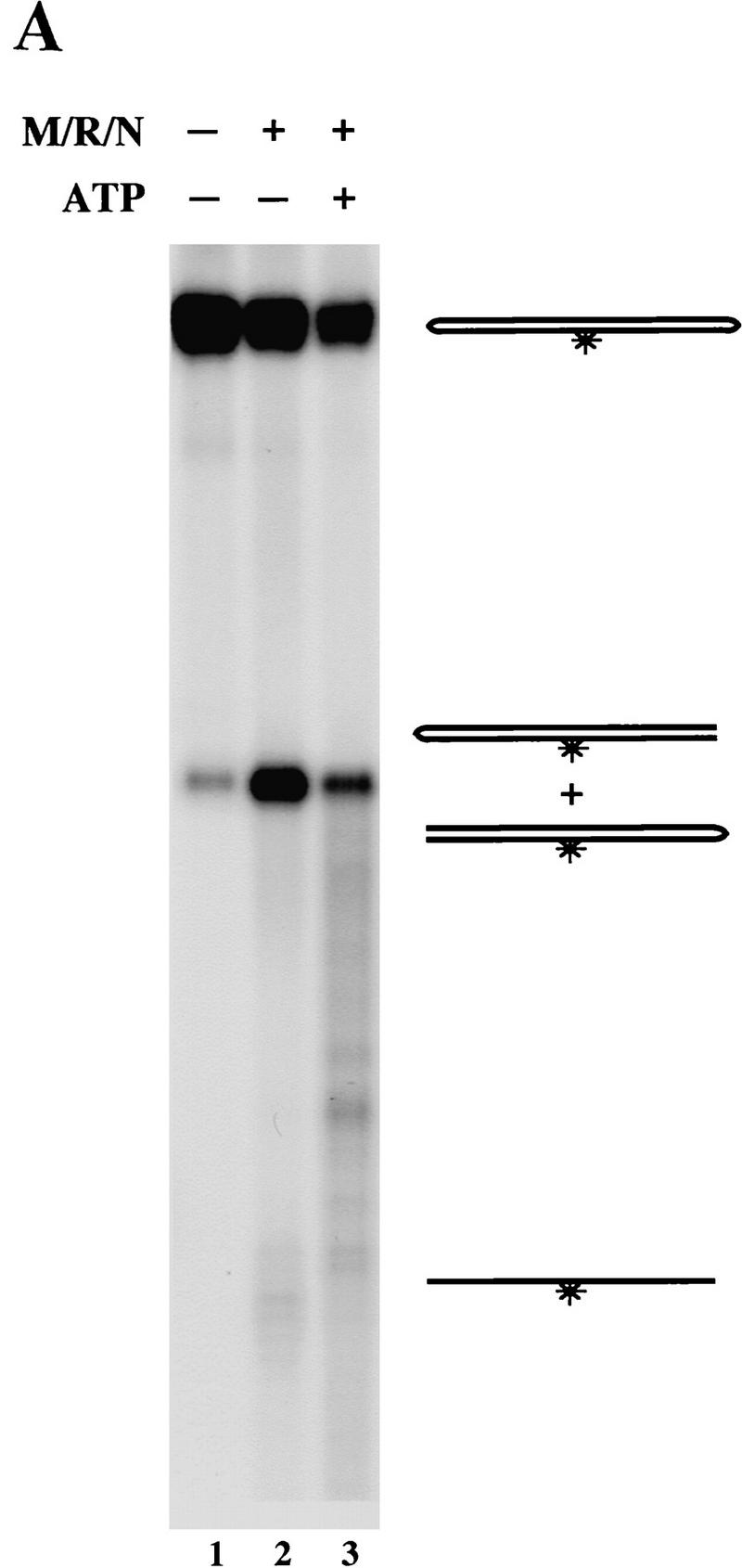
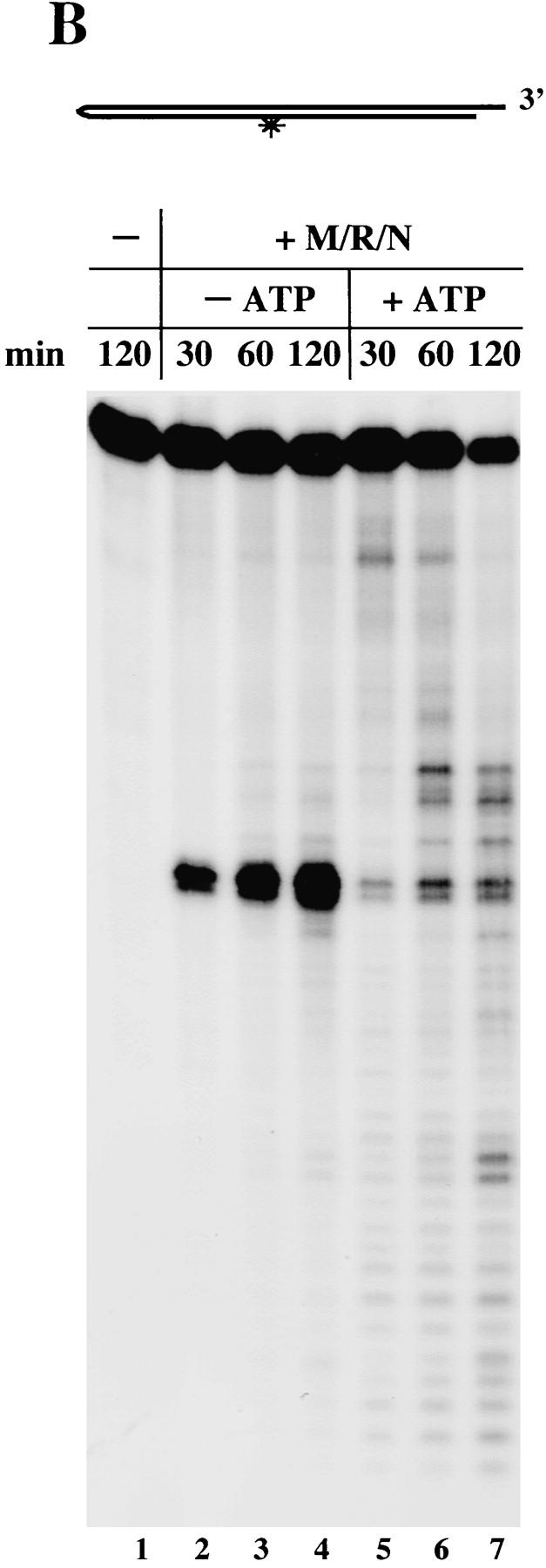
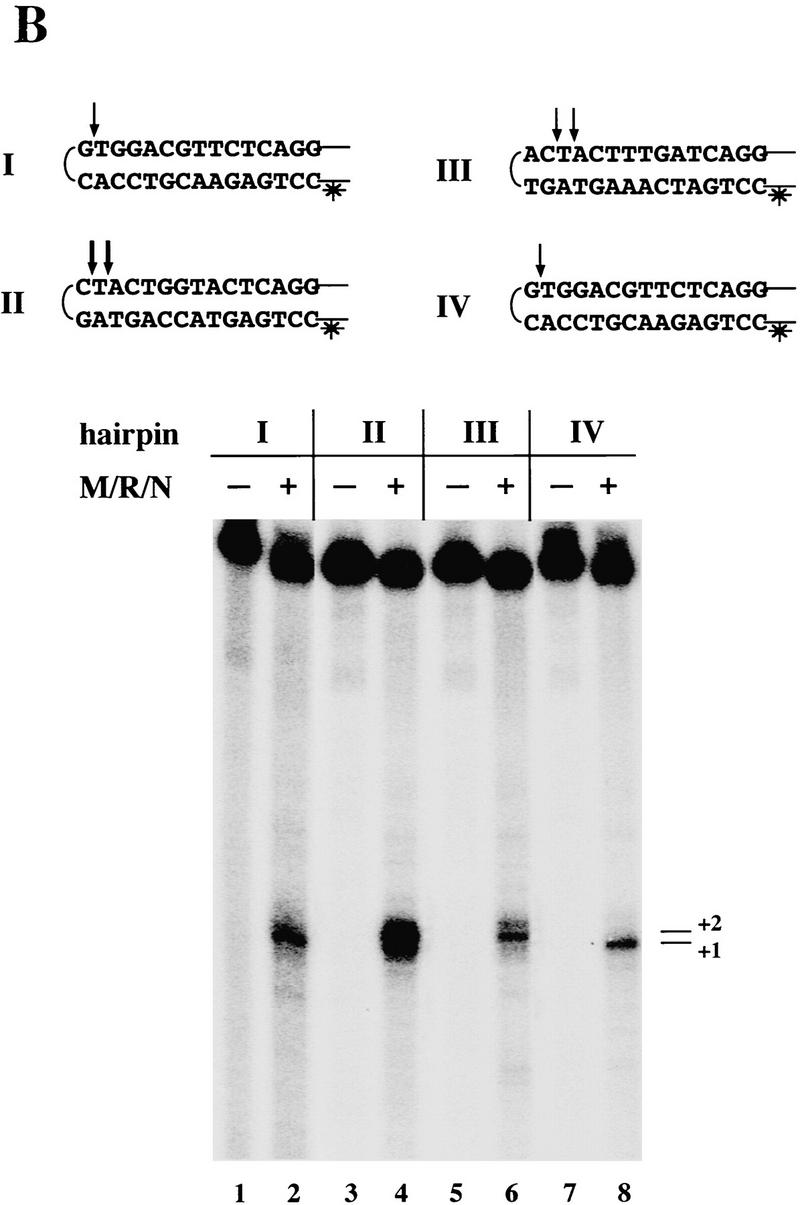
M/R/N generates different hairpin cleavage products in the presence of ATP. (A) Nuclease assays were performed with 150 ng of M/R/N in 1 mm MnCl2 with 0.5 mm ATP as indicated, on a 50-bp 32P-labeled dumbbell substrate containing fully paired hairpins on each end. Reactions were incubated for 30 min at 37°C before separation on a denaturing polyacrylamide gel. The small amount of opened dumbbell in the control lane is because of spontaneous breakage during gel purification of the substrate. (B) M/R/N (150 ng) was incubated as in Fig. 2A with a single-hairpin substrate (hairpin II from Fig. 2B) in the presence of 0.5 mm ATP as indicated. Aliquots were taken at different time points as shown.
When M/R/N was tested on substrates containing only one hairpin and a 3′ overhang on the other end (as in Fig. 2A), we found that the substrate was more rapidly consumed by M/R/N in the presence of ATP, and there was also a change in the size of the major cleavage products (Fig. 3B, lanes 5–7). During the course of the reaction, the larger cleavage products appeared before the smaller ones, and the product of hairpin opening at the +1 position was present, but at a low level (Fig. 3B, lanes 5–7). In contrast, the reaction without ATP showed only increasing production of one species, the hairpin opened at the tip, and did not give rise to any intermediate-size bands (Fig. 3B, lanes 2–4). The change in the size of the products made by M/R/N in the presence of ATP suggested that the substrate was being degraded starting from the open end, as analyzed in more detail below.
M/R/N partially unwinds a DNA duplex
From the results on the hairpin substrates it seemed possible that the ATP-stimulated nuclease activity of M/R/N could be explained if the complex was acting as a helicase and cutting the resulting single-stranded DNA endonucleolytically. We therefore tested whether M/R/N could separate a short labeled strand from a duplex DNA substrate, and we tried both a 3′ overhang and 5′ overhang because many helicases unwind exclusively from only one type of single-/double-strand junction. These assays had to be performed with magnesium as the divalent cation, because the complex degraded the substrate in manganese and prevented analysis of helicase activity. As shown in Figure 4, M/R/N was able to unwind a duplex of 17 bp, but only with a 3′ overhang, not a 5′ overhang. After 30 min at 37°C in the presence of M/R/N and ATP, ∼10% of the input DNA was released as a single-stranded product (Fig. 4A, lane 6). Surprisingly, however, about half this amount was released even in the absence of ATP (lane 5). We found that only M/R/N exhibited this activity, not Mre11, M/R, or M/N (Fig. 4B, lanes 8,9).
Figure 4.
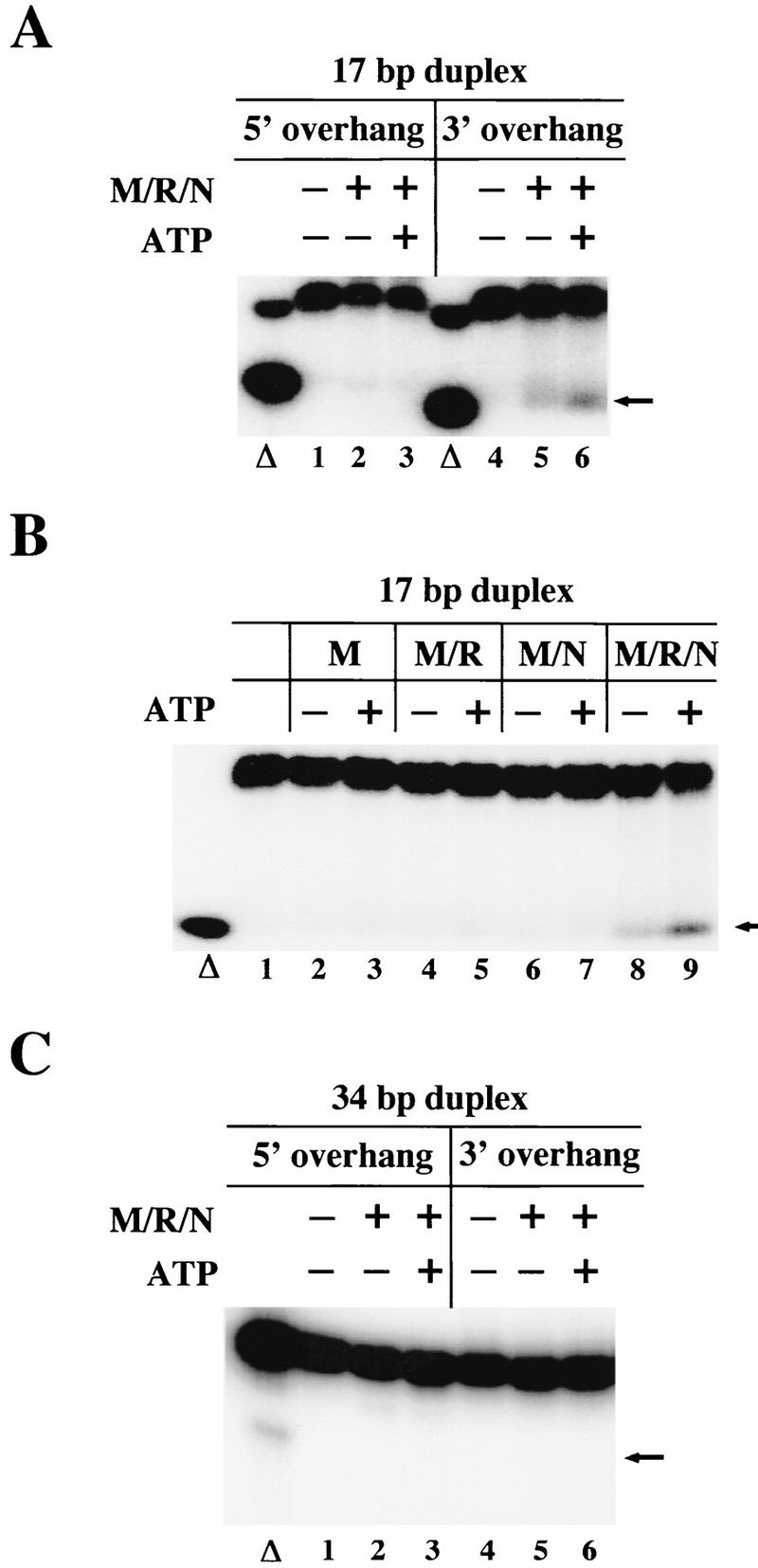
M/R/N partially unwinds short DNA duplexes from a 3′ single-stranded overhang. (A) M/R/N (150 ng) was incubated in 5 mm MgCl2 and 0.5 mm ATP as indicated with a DNA substrate containing a 17-bp duplex and either a 3′ or 5′ single-stranded overhang, with the 17-mer labeled at the 5′ end with 32P. The reactions were incubated at 37°C for 30 min and were stopped with SDS and EDTA before separation in a native polyacrylamide gel. (Δ) Substrate that was heated to denature the duplex. The arrow indicates the position of the free labeled strand. (B) Mre11 (25 ng, lanes 2,3), M/R (60 ng, lanes 4,5), M/N (50 ng, lanes 6,7), and M/R/N (150 ng, lanes 8,9) were incubated with the same 3′ overhang substrate as in A in the presence of 0.5 mm ATP as indicated. The varying protein levels yield approximately the same amount of Mre11 (25 ng) in each reaction. (C) M/R/N (150 ng) was incubated as in A and B except with DNA substrates containing a 34-bp duplex region and either a 3′ or 5′ single-stranded overhang as shown.
The 17-bp duplex (theoretical Tm = 42°C) is not very stable, and if the binding of a protein complex caused partial unwinding of the DNA strands, this could then stimulate release of the single strand even without an active (catalytic) helicase. To address this issue, we used a substrate with a longer duplex (34 bp) and found that M/R/N was not able to release the labeled single strand, even in the presence of ATP (Fig. 4C). The 34-bp duplex was also not released when the single-stranded DNA-binding protein replication protein A (RPA) was added (data not shown). The fact that some unwinding was observed on the 17-bp duplex in the absence of ATP and that no ATP-dependent unwinding occurred with longer duplexes led us to conclude that M/R/N is not functioning as a processive DNA helicase, but that binding of the complex does partially unwind a DNA duplex from a 3′ overhang and that this activity is increased in the presence of ATP.
ATP analogs cannot substitute for ATP in strand opening by M/R/N
We then asked whether alterations at the γ phosphate of ATP were compatible with its effects on the activities of M/R/N. Nonhydrolyzable ATP analogs were found to substitute for ATP in DNA binding by the yeast Rad50 protein, suggesting that hydrolysis was not required in that assay (Raymond and Kleckner 1993). In M/R/N cleavage of the substrate with a single fully paired hairpin, we found that only dATP could substitute for ATP (Fig. 5A, lane 4), whereas CTP, GTP, or UTP were inactive (lanes 6–8). Neither AMP–PNP nor ADP had any effect on hairpin opening by the triple complex (lanes 9,10). The nonhydrolyzable analog ATPγS did not substitute for ATP in the reaction, and even had an inhibitory effect on the hairpin nuclease activity (lane 5).
Figure 5.
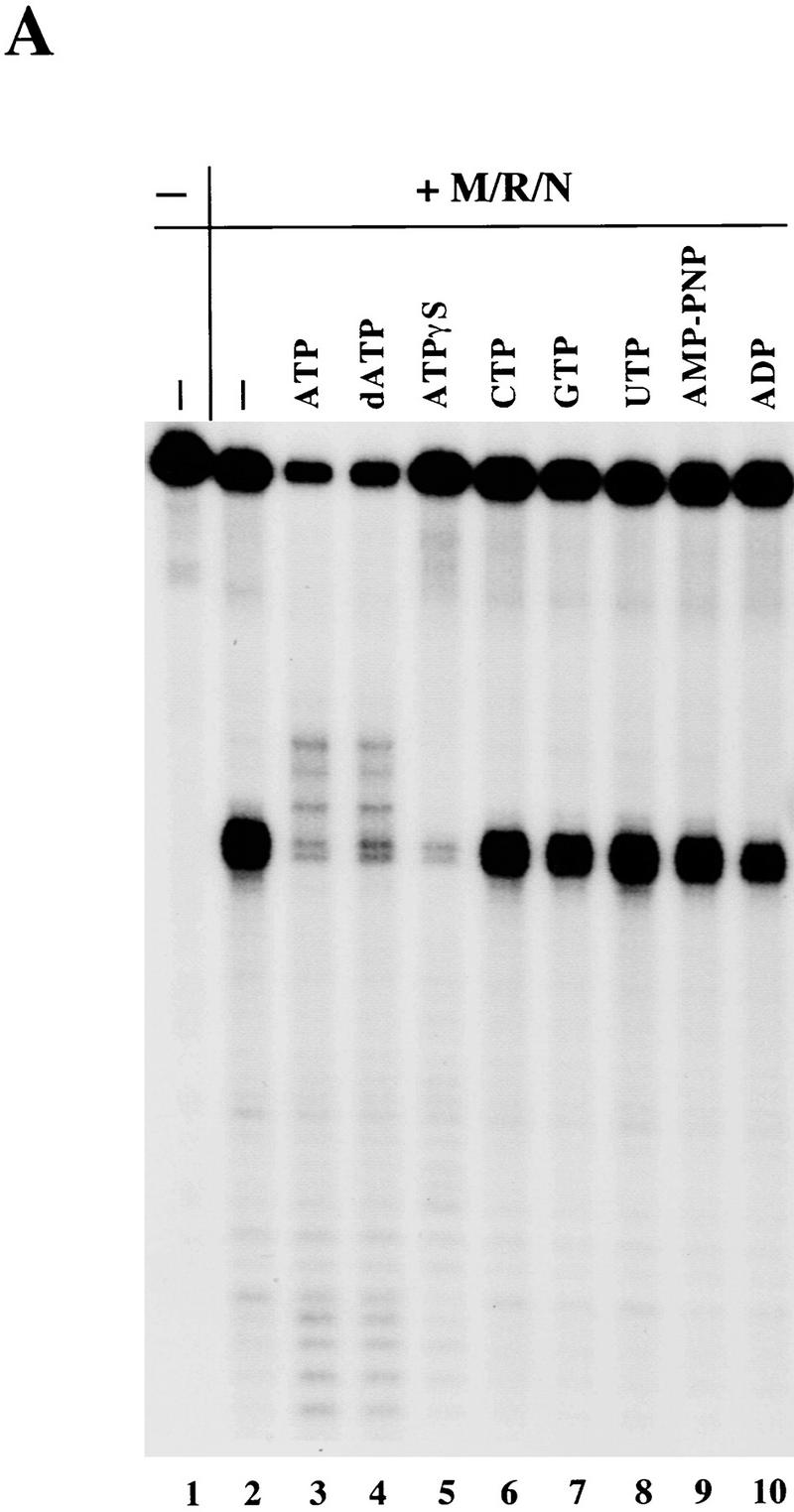
Nonhydrolyzable ATP analogs do not substitute for ATP in strand unwinding. (A) Nuclease assays were performed with 150 ng of M/R/N on a 32P-labeled single-hairpin substrate identical to hairpin II in Fig. 2B. Reactions contained 1 mm MnCl2 and 0.5 mm nucleotides as indicated, and were incubated for 30 min at 37°C before separation on a denaturing polyacrylamide gel. (B) Unwinding assays were performed with a DNA substrate containing a 17-bp duplex and a 3′ overhang as described in Fig. 4A. The ratio of free labeled oligonucleotide to the total amount of labeled substrate in each reaction was determined by quantitation on a PhosphorImager, and is shown here with the standard deviation calculated from two different experiments.
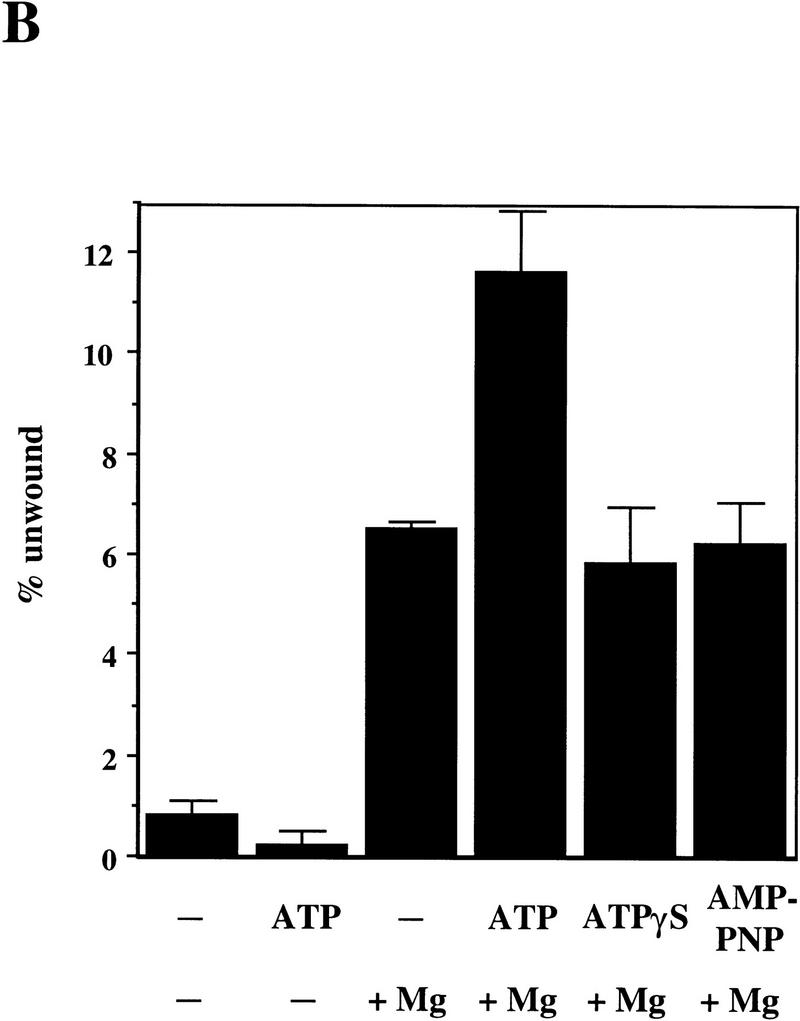
To determine which step of the ATP-stimulated M/R/N nuclease reaction was sensitive to chemical changes at the γ phosphate of ATP, nucleotide analogs were tested in the helicase assay using the short 17-bp duplex. As shown in Figure 4 and represented quantitatively in Figure 5B, the M/R/N complex caused a modest amount (∼6%) of duplex unwinding even without ATP, and this ATP-independent unwinding was dependent on a divalent cation. The extent of unwinding observed in the presence of both magnesium and ATP was approximately double that with magnesium alone, and neither ATPγS nor AMP–PNP could substitute for ATP in generating this stimulation (Fig. 5B).
These data would be consistent with a requirement for ATP hydrolysis in the stimulation of strand unwinding by M/R/N; however, we have not yet been able to detect an ATPase activity associated with the triple complex. Because M/R/N appears not to be a processive DNA helicase, it is possible that the hydrolysis of ATP leads to an altered conformation of the complex on a DNA end, a conformation in which the turnover of ATP is very slow. We do not know at this time whether the hydrolysis of ATP is necessary for its effects on the M/R/N complex.
ATP induces a switch in M/R/N endonuclease specificity
Mre11 is a 3′–5′ exonuclease, but is ∼50-fold less active on a 3′ overhang than on a blunt or recessed 3′ end (Paull and Gellert 1998). Incorporation of a 3′ overhang in a nuclease substrate is thus a useful way to block one end of the molecule from exonuclease attack and was employed, for instance, to map sites of hairpin opening (Figs. 2A,B and 3B). The ATP-dependent nucleolytic degradation by M/R/N presented in Figure 3B, however, appeared to begin at the 3′ overhang and finish at the 5′ end, suggesting that the 3′ overhang was no longer inhibitory. To investigate the ATP-dependent nuclease activities of M/R/N apart from hairpin cutting, we turned to the substrate shown in Figure 6, which contains 3′ overhangs on each end. In the experiment shown in Figure 6A, the 3′ end of the top strand was labeled. On this substrate, the only nucleolytic product visible with Mre11 or M/R was the labeled cordycepin mononucleotide at the bottom of the gel (Fig. 6A, lanes 2–9), showing that all the complexes degrade the substrate from the end of the 3′ overhang. In contrast, the M/R/N complex generated a product of 17 nucleotides in addition to the mononucleotide and this product was only seen when ATP was included in the reaction (Fig. 6A, lanes 12,13). The size of the product indicates that M/R/N made an endonucleolytic cut on the top strand of the substrate, exactly at the border of the duplex region.
Figure 6.
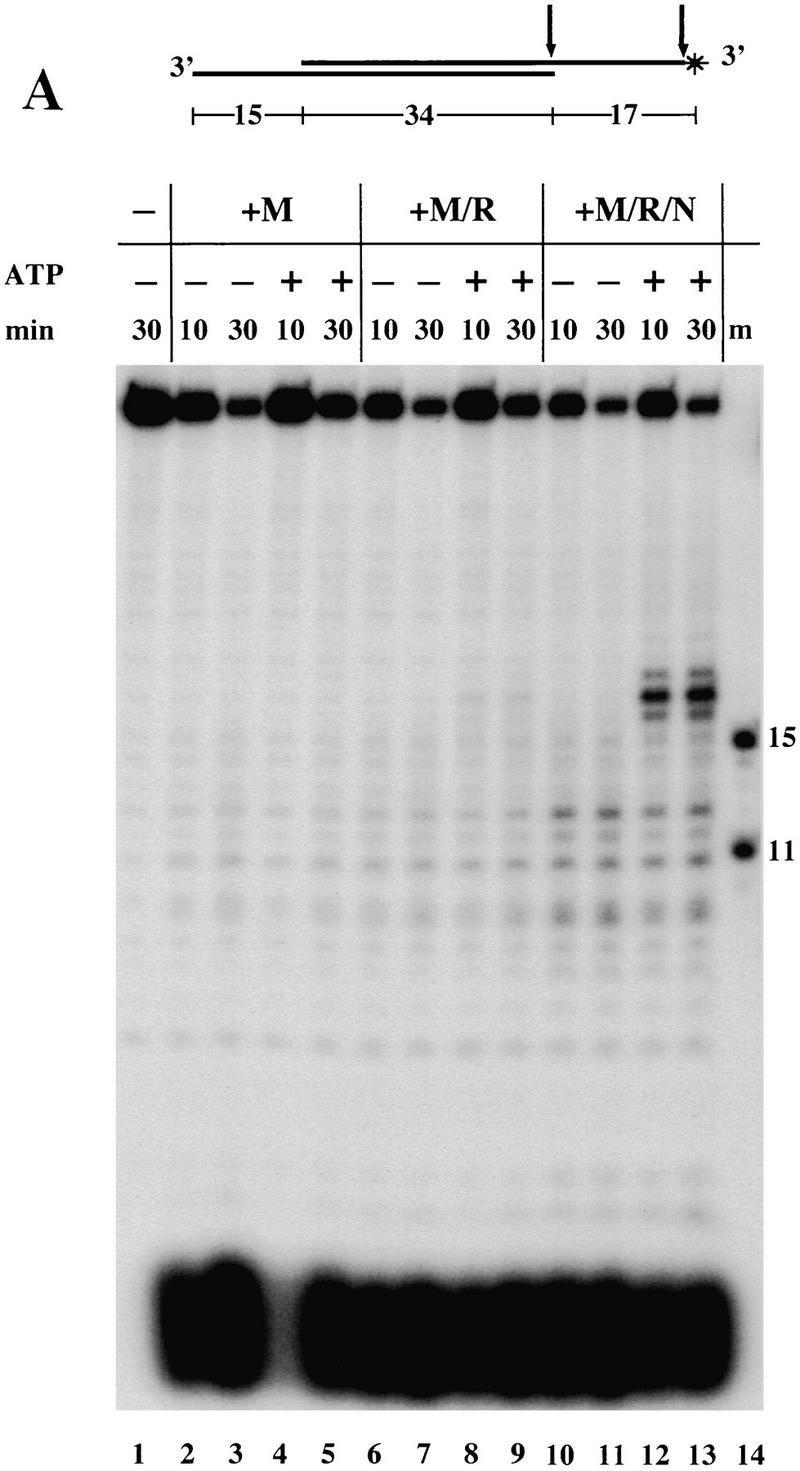
ATP stimulates M/R/N cleavage of a 3′ overhang at the border of the duplex region. (A) Nuclease assays were performed with Mre11 (100 ng, lanes 2–5), M/R (150 ng, lanes 6–9) and M/R/N (150 ng, lanes 10–13) on a double-stranded DNA substrate with 3′ overhangs on each end. The 3′ end of the top strand was labeled with 32P-labeled cordycepin, as diagrammed, which lengthens the 3′ overhang by one nucleotide. Reactions contained 1 mm MnCl2 and 0.5 mm ATP as indicated, and aliquots were taken at 10- and 30-min time points. The locations of the cuts made by the proteins are shown as arrows in the diagram. (m) Size markers of 11 and 15 nucleotides are shown. (B) Nuclease assays were performed as in A with the same substrate, except that the top strand was labeled with 32P at the 5′ end as shown in the diagram.
When the top strand of the double-ended 3′ overhang substrate was labeled on the 5′ end, the consequences of the ATP-stimulated endonuclease activity of M/R/N became apparent, as shown in Figure 6B. Each of the protein complexes removed 1–5 nucleotides from the 3′ end (seen on this gel as a slight shortening of the full-length band in lanes 2–13), similar to that seen previously on longer 3′ overhangs (Paull and Gellert 1998). The activities of Mre11 and M/R did not change with the addition of ATP (Fig. 6B, lanes 4,5,8,9), but ATP induced M/R/N to generate a series of small products ranging from 8 to 17 nucleotides (lane 13). The size of these products indicates that the top strand was resected within the duplex region. Therefore, the ATP-stimulated endonucleolytic cut made at the border of the duplex (as shown in Fig. 6A) allows the triple complex to enter the duplex region and resect the top strand, an activity that Mre11 and M/R do not exhibit. The resection is an expected consequence of the endonucleolytic cut because 3′–5′ exonuclease degradation by Mre11 is rapid on a blunt end.
One concern with this experiment is the fact that an identical reaction is presumably occurring from the opposite end of the molecule and we know of no efficient method to block endonucleolytic attack from the other end. In this case, however, the time points taken were sufficiently early to ensure that only a small fraction of the substrate had undergone endonucleolytic cleavage by M/R/N, so a large majority of labeled duplexes were still unresected on the bottom, unlabeled strand.
Rad50 is responsible for ATP binding in the M/R/N complex
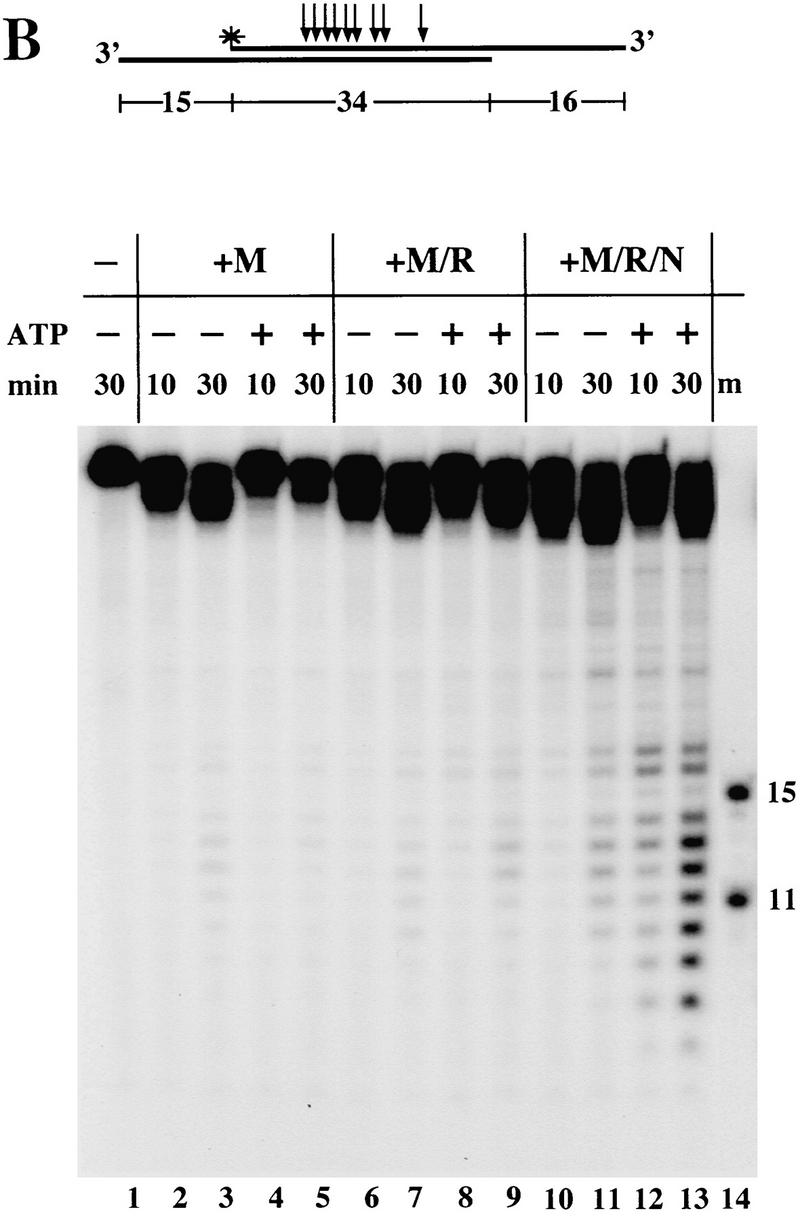
Rad50 is the only component of the triple complex which contains obvious ATP-binding motifs in its sequence. To determine whether the requirement for ATP that we observed with the triple complex originated in Rad50, we made a mutation in a conserved lysine residue within the Walker A nucleotide-binding domain in the Rad50 gene (K42E). The equivalent mutation in the yeast Rad50 gene, K40E, caused the mutant strain to be as sensitive to MMS treatment as a Rad50 null strain (Alani et al. 1990). We also mutated the conserved aspartate residue in the Walker B box of the Rad50 gene, generating Rad50 D1231A. Expression of these mutant Rad50 proteins with wild-type Mre11 and Nbs1 yielded M/R(K42E)/N and M/R(D1231A)/N triple complexes which were expressed at approximately the same levels as the wild-type complex and behaved similarly during purification (data not shown).
On the single hairpin substrate, the triple complexes containing Rad50 D1231A or K42E made the endonucleolytic cuts at the hairpin tip (Fig. 7, lanes 4,6), similarly to the wild-type complex (Fig. 7, lane 2). However, the mutant complexes did not degrade the substrate from the other end when ATP was added (lanes 5,7). Similarly, in the unwinding assay, M/R(K42E)/N and M/R(D1231A)/N did exhibit the Mg-dependent partial unwinding of the short duplex, but the level of unwound duplex did not increase in the presence of ATP (data not shown). Thus, the triple complexes containing mutant Rad50 exhibit all the activities of the wild-type complex in the absence of ATP, but are completely defective in the ATP-dependent activities.
Figure 7.
M/R/N complexes containing Rad50 protein mutated in the Walker A or Walker B motifs do not respond to ATP. Nuclease assays were performed on a substrate containing a fully paired hairpin on one end and a 3′ overhang on the other end, identical to hairpin II in Fig. 2B. Reactions contained 150 ng each of wild-type M/R/N, M/R(D1231A)/N, or M/R(K42E)/N protein, 1 mm MnCl2, and 0.5 mm ATP as indicated.
Ku and RPA inhibit degradation of DNA overhangs by M/R/N
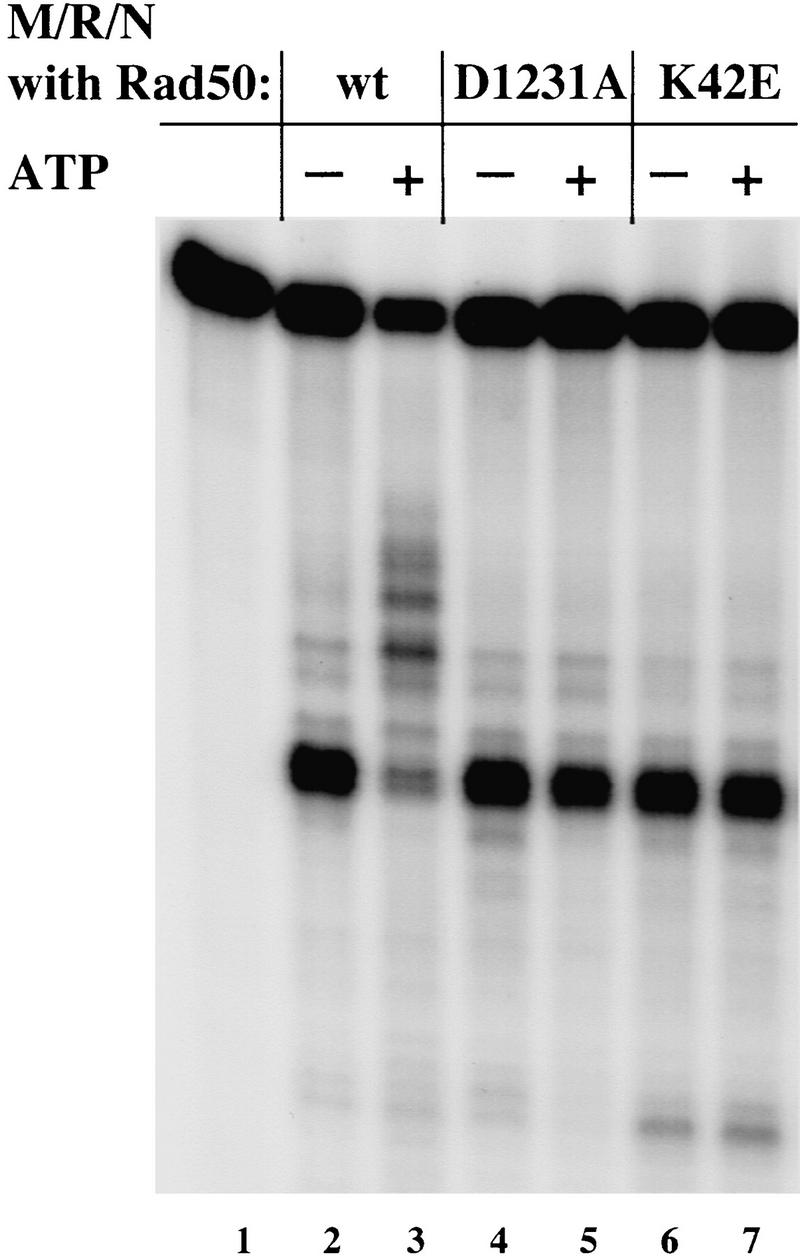
Yeast Mre11 and its associated factors, Rad50 and Xrs2, have been shown to play a critical role in maintaining genomic stability in vivo. It is not clear yet how they accomplish this or how they interact with other DNA repair factors, but in at least some situations the Mre11 complex appears to act in opposition to other DNA end-binding proteins, the Ku heterodimer and the single-stranded DNA-binding protein RPA (Lee et al. 1998). To investigate this possibility, human RPA and Ku proteins were added to nuclease reactions containing the M/R/N complex and the substrate with 3′ overhangs at each end. Both protein complexes effectively inhibited the endonucleolytic degradation of the 3′ overhang by M/R/N in the presence of ATP (Fig. 8, lanes 3,4,6,7), suggesting that these factors may in fact be competing with each other and that Ku and RPA have the potential to regulate the nuclease activities of the M/R/N complex.
Figure 8.
Ku and RPA inhibit nuclease digestion of 3′ overhangs by M/R/N. Nuclease assays were performed as in Fig. 6B with the addition of varying amounts of RPA (30 or 300 ng in lanes 3 and 4, respectively) or Ku protein (5 and 50 ng in lanes 6 and 7, respectively). (m) Size markers of 11, 15, 21, and 28 nucleotides are shown.
Discussion
With the identification of NBS1 as the gene defective in NBS, and the simultaneous discovery that the NBS1 product was the missing member of the Mre11/Rad50 complex, two important branches of double-strand break repair intersected (Carney et al. 1998; Varon et al. 1998). The radiation sensitivity and chromosomal instability seen in cells from NBS patients mirror the defects seen in yeast cells lacking components of the Mre11 complex, confirming that this complex plays a central role in defending eukaryotic cells against the harmful effects of double-strand breaks.
NBS is caused by mutations that prematurely truncate the Nbs1 protein (Varon et al. 1998). In all of the known cases, the truncations are downstream of the FHA domain, near the carboxy-terminal end of the BRCT motif. The truncated protein has been reported to be completely absent from NBS1-defective cells (Carney et al. 1998), suggesting that these cells may be functionally equivalent to a knockout cell line. We expressed an Nbs1 deletion mutant with Mre11 and Rad50 and found that the truncated protein was not made at any significant level and definitely did not bind to Mre11 or Rad50 (data not shown). Our findings confirm the hypothesis that the carboxy-terminal two-thirds of Nbs1 is required for association with the Mre11/Rad50 complex.
The radioresistant DNA synthesis exhibited by NBS1-defective cells (for review, see Shiloh 1997) had suggested that the protein might be involved in signaling DNA damage, especially as the Nbs1 protein does not contain any obvious enzymatic motifs and the BRCT and FHA domains that are present are thought to be involved in protein–protein interactions (Hofmann and Bucher 1995; Bork et al. 1997). We were surprised then to find that Nbs1 had a dramatic effect on the endonuclease activity and specificity of Mre11. Further evidence for an interaction between Nbs1 and the active site of the Mre11 nuclease comes from our observation that four different nuclease deficient mutants of Mre11 do not associate with Nbs1 (data not shown). Thus, although Nbs1 may function as a signaling molecule, it is also associated closely with the active site of the nuclease.
As we were not able to produce triple complexes containing mutated forms of Mre11 and were not able to produce Nbs1 by itself, we cannot conclude definitively whether any of the novel activities can be attributed directly to Nbs1. Because Nbs1 lacks any clear catalytic motifs, however, and because the nuclease activities retain the strand polarity of Mre11 and still require manganese, it is likely that Mre11 is the only nuclease in the complex and that Nbs1 and Rad50 modify and regulate its activities.
Cleavage of fully paired hairpins requires Nbs1
More evidence for a close interaction between Nbs1 and the nuclease domain of Mre11 comes from our observation that the presence of Nbs1 increases the ability of Mre11 to cut fully paired DNA hairpins dramatically. Both M/N and M/R/N cleave these hairpins, although the triple complex is much more efficient than the complex lacking Rad50. DNA hairpins are of interest to us because the RAG1 and RAG2 proteins make the coding flanks of immunoglobulin genes into fully paired hairpin intermediates during the course of V(D)J recombination, and it is not known yet how these hairpins are opened prior to joining (for review, see Lewis 1994). The observation that NBS patients suffer from elevated levels of chromosomal translocations that often involve V(D)J recombination sites suggests that the M/R/N complex may be involved in processing V(D)J intermediates. It may be significant in this regard that the M/R/N complex cleaves fully paired hairpins on the 3′ side of the tip, because broken coding ends in cells undergoing V(D)J recombination have been found to contain short 3′ overhangs (Schlissel 1998). We are currently investigating V(D)J hairpin cleavage by the M/R/N complex, alone and in combination with other factors such as Ku, XRCC4, and DNA ligase IV, which are known to be necessary for V(D)J recombination in vivo. There has been a recent report of V(D)J hairpin opening by the RAG1 and RAG2 proteins themselves (Besmer et al. 1998), but it is not clear yet whether this process occurs in vivo.
The efficient cleavage of both fully paired hairpins and larger hairpin loops by M/R/N may also be critical for the removal of cruciforms or other secondary structures in a nonlymphoid cell. Mre11 and Rad50 show sequence homology with the E. coli proteins SbcD and SbcC, respectively (Sharples and Leach 1995). Loss of SbcC or SbcD activity allows E. coli strains to propagate long inverted repeats; thus these factors were thought to remove cruciform structures in vivo (Leach 1994). The purified SbcC/D enzyme exhibits manganese-dependent endonuclease activity on hairpin loops as well as ATP-dependent exonuclease activity on double-stranded DNA (Connelly et al. 1997, 1999). Interestingly, a recent study of trinucleotide repeat expansion in E. coli found that expansion of CTG repeats required loss of SbcC (Sarkar et al. 1998). The amplification appeared to depend on the size of the repeat relative to the size of Okazaki fragments in E. coli and was postulated to arise from intrastrand secondary structures formed within the Okazaki fragments. If there is functional homology between M/R/N and SbcC/D, the Mre11 complex may act to remove hairpin structures during replication in mammalian cells, a role that might be essential for normal growth.
An ATP-induced switch in substrate specificity: preference for single strands
The presence of Walker A and Walker B NTP-binding motifs in Rad50 (Alani et al. 1989; Dolganov et al. 1996) and the observation of ATP-dependent DNA binding by yeast Rad50 (Raymond and Kleckner 1993) suggested that ATP might play a regulatory role in M/R/N functions. We observed a pronounced effect of ATP on the M/R/N digestion of a DNA substrate containing a hairpin on one end and a 3′ overhang on the other end. The time course of this degradation suggested that the nuclease was initiating its attack at the 3′ end, not the hairpin end (Fig. 3B). This was unexpected because Mre11 and Mre11/Rad50 are much less active on a 3′ overhang compared to a blunt or recessed 3′ end (Paull and Gellert 1998).
When DNA substrates with 3′ overhangs at both ends were tested, it became clear that ATP induced the triple complex to cleave the 3′ overhang at the single-/double-strand transition (Fig. 6). After this initial endonucleolytic cut was made, exonucleolytic degradation of the cut strand (now blunt) proceeded. Thus, with the hairpin substrate, it is very likely that M/R/N cut off the 3′ overhang and degraded the rest of the molecule 3′–5′.
In this process, ATP appears to primarily influence DNA unwinding (although an additional role in the nucleolytic attack has not been ruled out). As shown in Figures 4 and 5, ATP binding (or hydrolysis) stimulates unwinding of duplex DNA from a 3′ overhang. Changes in the γ phosphate linkages of ATP reduced unwinding to the basal level, and these effects correlated with the effects on endonuclease activity.
The ATP-driven unwinding and cleavage by M/R/N on open ends appears to be specific to 3′ overhangs; however, we also observed a strong effect of ATP on degradation of a covalently closed dumbbell DNA (Fig. 3A). On this substrate, ATP increased the rate of the first endonuclease cut of a hairpin as well as the subsequent degradation of the opened end. We are not yet able to measure unwinding of a dumbbell, but we predict that the same unwinding/cleavage event that occurs on the 3′ overhang substrates also takes place with M/R/N on these dumbbell molecules. To link these two observations we postulate that ATP may act to switch the binding preference of M/R/N from double-stranded DNA to unwound duplex DNA. Therefore, on the single hairpin substrate, ATP favored the endonuclease substrate that could be unwound more easily (the 3′ overhang) over one that was more difficult (the hairpin). It may be significant in this respect that a Bacillus subtilis SMC (structural maintenance of chromosomes) protein related to Rad50 in primary sequence exhibits ATP-dependent aggregation of single-stranded DNA (Hirano and Hirano 1998).
The significance of 3′ overhangs
The preference for 3′ protruding strands could be a critical aspect of M/R/N function, considering the importance of 3′ single-stranded ends as recombination intermediates in vivo. Long 3′ single-stranded tails are produced at the onset of meiotic and mitotic homologous recombination in S. cerevisiae, for example, and the presence of the Mre11 complex increases the rate of resection of the 5′ end (Ivanov et al. 1994). In overall efficiency of homologous mitotic recombination, however, mre11 strains are equivalent to wild-type. Since Mre11 is not itself a 5′–3′ exonuclease (Furuse et al. 1998; Paull and Gellert 1998; Trujillo et al. 1998; Usui et al. 1998), we conclude that it is very unlikely that Mre11 performs the resection of 5′ ends prior to homologous recombination. It is possible, however, given the affinity of M/R/N for 3′ overhangs, that the complex may recruit a 5′–3′ nuclease or facilitate its activities there.
The Mre11 complex was also shown recently to be involved in telomere maintenance in S. cerevisiae (Kironmai and Muniyappa 1997; Boulton and Jackson 1998; Nugent et al. 1998), and epistasis analysis of yeast mutants has shown that the Mre11 complex functions in the same pathway as telomerase (Nugent et al. 1998). As the ends of telomeres undergo a transient lengthening of the 3′ G-rich strand during S phase (Wellinger et al. 1993), it is possible that the activities of the Mre11 complex on 3′ overhang structures also form the basis of its role at telomeres. The Mre11/Rad50/Xrs2 complex may be important in preparing a DNA end for the addition of telomeric repeats by presenting the end to telomerase so it can create the G-rich strand. Alternatively, it could initiate the degradation of the opposite strand in the same way that the 3′ overhang is thought to be generated for homologous recombination. In either case, the activities of the Mre11 complex on 3′ overhangs are likely to be important in the mechanism used to replicate telomeric repeats.
A model for the role of M/R/N in double-strand break repair
We propose that the M/R/N complex may function in double-strand break repair as a key organizer that determines the type of processing to be used on double-strand breaks, depending on the structure of the ends at the break and perhaps on the availability of other factors. In this model, M/R/N would bind to the ends and form a specific complex which may entail ATP-dependent DNA unwinding or a conformation change in the complex. In some cases, M/R/N would recruit other factors that would initiate homologous recombination. Alternatively, the factors involved in NHEJ could be enlisted to bring the broken ends together, to prevent Mre11 from degrading more than the few nucleotides necessary to form blunt ends or compatible overhangs, and to facilitate rejoining. Finally, as a last resort, M/R/N could commence a homology search through more extensive 3′–5′ degradation of the ends and could enter a 3′ overhang by the ATP-driven unwinding of the end of the duplex.
A critical component of this model is the nuclease activity of Mre11, which is likely to be important in removing damaged or mismatched base pairs prior to ligation. Several groups have made mutations in the conserved nuclease motifs of yeast Mre11 and all of these mutant strains show reduced survival after radiation or MMS treatment (Bressan et al. 1998; Furuse et al. 1998; Tsubouchi and Ogawa 1998; Usui et al. 1998; Moreau et al. 1999). The sensitivity observed with these catalytic mutants is, in some cases, because of the failure of nuclease-deficient Mre11 proteins to associate with the other factors (Usui et al. 1998, and this work). However, there is at least one instance of an Mre11 mutant which forms the triple complex normally but still is deficient in nuclease activity, and this mutant (Mre11-6) exhibits MMS resistance which is partway between a wild-type strain and a null strain, indicating that nuclease activity does contribute to the efficiency of Mre11-mediated repair (Usui et al. 1998).
The intermediate double-strand break repair phenotypes of some of the Mre11 catalytic mutants in yeast (Bressan et al. 1998; Furuse et al. 1998; Moreau et al. 1999) suggest that the M/R/N complex is likely to have a function separate from the nuclease activity that may require only binding of the complex to double-strand breaks. In agreement with this view, the Mre11 complex has been shown to be necessary for the joining of plasmid ends in vivo, even when the ends are complementary (Boulton and Jackson 1998). In addition, the M/R/N complex may have a role in assembly of other components, for instance Ku, Xrcc4, and ligase IV, onto DNA ends prior to any processing events.
The Ku protein is capable of preventing extensive degradation by M/R/N and thus could play dual roles of bringing ends together (Ramsden and Gellert 1998) and regulating Mre11 nuclease activity. The residual junctions found in yeast strains deficient in Ku contain large deletions and are often joined at microhomologies (Boulton and Jackson 1996a,b, 1998), whereas the few junctions recovered from mre11, rad50, and xrs2 mutants are predominantly intact (Boulton and Jackson 1998). This observation argues for the existence of an Mre11-dependent, deletion-prone pathway which is only seen in the absence of Ku or other NHEJ components. Mre11 and M/R can facilitate joining of mismatched 3′ recessed ends and blunt ends in vitro, creating junctions at short microhomologies (Paull and Gellert 1998) which are similar to junctions seen in Ku− cells in vivo (Boulton and Jackson 1996a,b, 1998). As we show in this work, the triple complex can resect any type of 3′ end that may need to be removed to uncover matching sequences. It is likely that the ATP requirement for removing a 3′ overhang reflects the importance of this type of DNA end structure in eukaryotic cells, thus access of the Mre11 nuclease to a 3′ overhang is tightly regulated by Rad50 and possibly other factors involved in end joining. Further investigation of the interactions between Ku, M/R/N, and RPA in vitro should lead to a fuller understanding of the balance of power between these important protein complexes in double-strand break repair.
Materials and methods
Proteins
An NBS1 cDNA was isolated from a human HeLa cell library (Clontech) and was cloned into the baculovirus transfer vector pFastBac1 (GIBCO-BRL), yielding plasmid pTP36. The cDNA was missing exon 13, according to the genomic sequence (Matsuura et al. 1998), and had several single amino acid substitutions. One of these, Q185E, was present in one of the published cDNA sequences (Varon et al. 1998) so it was left unchanged, but the other mutations were changed back to the correct sequence and the missing exon was replaced. No affinity tags were included with the NBS1 gene. pTP36 was used to make the bacmid pTP40, according to the procedures specified by GIBCO-BRL. In this work, the human MRE11 gene with a carboxy-terminal histidine tag (Paull and Gellert 1998) was also cloned into pFastBac1 yielding pTP17 and this was used to make the corresponding bacmid, pTP19. The MRE11 and RAD50 genes, each with carboxy-terminal histidine tags (Paull and Gellert 1998), were also cloned into pFastBacDual (GIBCO-BRL) for use in coexpression of both proteins, yielding pTP30, and this was used to make the bacmid pTP43. pTP30 was modified to include the K42E mutation in RAD50, yielding pTP50, and was also used to construct the D1231A mutant, yielding pTP48. Bacmids were generated from these constructs, yielding pTP51 and pTP49, respectively. pTP19, pTP40, pTP43, pTP49, and pTP51 were used to make recombinant baculovirus according to the supplier’s instructions (GIBCO-BRL) and gave rise to high-titer virus after one amplification. pTP19, pTP43, pTP19 plus pTP40, pTP40 plus pTP43, pTP40 plus pTP49, and pTP40 plus pTP51 combinations of viral stocks were used to express M, M/R, M/N, M/R/N,M/R(D1231A)/N, and M/R(K42E)/N, respectively. Complexes of Mre11, Rad50, and Nbs1 were purified as described previously for Mre11 and Rad50 (Paull and Gellert 1998). Ku protein and RPA were purified as described (Weinberg et al. 1990; Ramsden and Gellert 1998).
Oligonucleotide substrates
The substrate used for DNA binding in Figure 1B was composed of TP53 (GACCTGGCACGTAGGACAGCATGGGCTGCAGGTCGACTTACACAGTGCTACAGACTGGAACAAAAACCCTGCAG) annealed to TP54 (CTGCAGGGTTTTTGTTCCAGTCTGTAGCACTGTGTAAGTCGACCTGCAGCCCATGCTGTCCTACGTGCCAGGTCACTGA) with both oligos labeled with 32P at the 5′ ends. The substrate used for mapping of endonuclease cleavage sites in Figures 6 and 8 contained 3′ overhangs on each end and was composed of DAR40 (Ramsden et al. 1996) annealed to DG113 (CACAGTGCTACAGACTGGAACAAAAACCCTGCAGTACTCTACTCATCTC) with DAR40 labeled with 32P at the 5′ end in Figures 6B and 8 and labeled with 32P cordycepin at the 3′ end in Figure 6A. The substrate used in Figures 4, A and B, and 5B had a 44-bp 3′ overhang and was composed of DG62 (McBlane et al. 1995) annealed to DG130 (GTACAAAAACCCTCGGG) with DG130 labeled with 32P at the 5′ end. The 5′ overhang substrate used in Figure 4A consisted of DG61 (McBlane et al. 1995) annealed to DG128 (CCCGAGGGTTTTTGTAC) with DG128 labeled with 32P at the 5′ end. The helicase substrate with a 5′ overhang used in Figure 4C was composed of DG9 (McBlane et al. 1995) annealed to DAR39 (Ramsden et al. 1996) with DG9 labeled with 32P at the 5′ end. The substrate with the 3′ overhang used in Figure 3C was composed of DG10 (McBlane et al. 1995) annealed to DAR40 (above) with DG10 labeled with 32P at the 5′ end.
Hairpin oligonucleotide substrates
In Figures 2, A and B, 3B, 5A, and 7, the substrates were composed of a hairpin (TP40 in Fig. 2A; TP40, TP41, TP42, and DAR132 in Fig. 2B; TP41 in Figs. 3B, 5A, and 7) ligated to a linker duplex. TP40 (Paull and Gellert 1998), TP41 (CATCCATGCCTACCTGAGTACCAGTAGCTACTGGTACTCAGGTAGGCATG), TP42 (CATCCATGCCTACCTGATCAAAGTAGTACTACTTTGATCAGGTAGGCATG), and DAR132 (CATCCATGCCTACCTGACAGTCCGATGCATCGGACTGTCAGGTAGGCATG) were each labeled with 32P at the 5′ end. After removal of excess ATP, the oligonucleotides were made into hairpins by boiling and quickly cooling them to 4°C. The linker was composed of TP45 annealed to TP5 (Paull and Gellert 1998) and TP45 was phosphorylated with nonradioactive ATP prior to annealing. The hairpins were each ligated with a twofold excess of linker and T4 DNA ligase (NEB) overnight, precipitated, and separated on a sequencing gel containing 10% acrylamide, 20% formamide, and 7 m urea. The labeled band with the slowest mobility was cut out of the gel, eluted overnight in extraction buffer (500 mm ammonium acetate, 10 mm MgCl2, 1 mm EDTA, and 0.1% SDS), ethanol-precipitated, and resuspended in TE. Before use in the experiment, the hairpins were boiled and cooled again to 4°C.
In Figure 3A, the dumbbell substrate was composed of two hairpins ligated together: DAR133 (Paull and Gellert 1998), which was phosphorylated with cold ATP, and TP41 (above), which was labeled with 32P at the 5′ end. The hairpins were ligated and purified as described above for the single hairpin substrates.
Gel shifts and helicase reactions
Gel mobility shift assays contained 25 mm MOPS (pH 7.0), 100 mm NaCl, 0.1% Triton X-100, 100 μg/ml BSA, 2 mm DTT, 0.1 pmole DNA substrate, and 37.5–150 ng of protein (as indicated in Fig. 1B). The reactions were incubated for 10 min at room temperature, 1 μl of 50% glycerol was added, then each reaction was loaded onto a 1/2× TBE gel containing 0.7% agarose and run at 7 V/cm for ∼2 hr. DNA unwinding reactions contained 25 mm MOPS (pH 7.0), 50 mm NaCl, 2 mm DTT, 5 mm MgCl2, 0.1 pmole DNA substrate, 0.5 mm ATP and varying combinations of proteins as indicated. Reactions were incubated at 37°C for 30 min, then stopped by the addition of EDTA and SDS to final concentrations of 10 mm and 0.2%, respectively. TBE dye (5×) was added before loading half of each sample onto a 10% polyacrylamide gel (Novex) and running for ∼90 min at 100 V.
Nuclease reactions
Reactions contained 25 mm MOPS (pH 7.0), 50 mm NaCl, 2 mm DTT, 1 mm MnCl2, 0.1 pmole of DNA substrate, and protein complexes as stated in the figure legends. Except where noted in the figures, the reactions were performed in a volume of 10 μl, and were incubated at 37°C for 30 min. The amount of protein added in each reaction varied as described in the figure legends. The amount of Mre11 present in 150 ng of the wild-type M/R/N preparation was ∼25 ng. Reactions were stopped by the addition of EDTA and SDS to final concentrations of 10 mm and 0.2%, respectively. Three microliters of each reaction was dried down, resuspended in 5 μl of formamide loading buffer, and then loaded onto a sequencing gel containing 15%–22.5% acrylamide and 7 m urea (gels with hairpin substrates also contained 20% formamide). After the run, each gel was analyzed using a PhosphorImaging system (Molecular Dynamics).
Acknowledgments
We thank Dale Ramsden for the purification of RPA and are grateful to our colleagues at the Laboratory of Molecular Biology for valuable discussions. T.T.P. was supported by a Helen Hay Whitney Postdoctoral Fellowship.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL gellert@helix.nih.gov; FAX (301) 496-0201.
References
- Alani E, Subbiah S, Kleckner N. The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics. 1989;122:47–57. doi: 10.1093/genetics/122.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Besmer E, Mansilla-Soto J, Cassard S, Sawchuk DJ, Brown G, Sadofsky M, Lewis SM, Nussenzweig MC, Cortes P. Hairpin coding end opening is mediated by RAG1 and RAG2 proteins. Mol Cell. 1998;2:817–828. doi: 10.1016/s1097-2765(00)80296-8. [DOI] [PubMed] [Google Scholar]
- Bianco PR, Tracy RB, Kowalczykowski SC. DNA strand exchange proteins: A biochemical and physical comparison. Front Biosci. 1998;3:d570–d603. doi: 10.2741/a304. [DOI] [PubMed] [Google Scholar]
- Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF, Koonin EV. A superfamily of conserved domains in DNA damage-responsive cell cycle checkpoint proteins. FASEB J. 1997;11:68–76. [PubMed] [Google Scholar]
- Boulton SJ, Jackson SP. Identification of a Saccharomyces cerevisiae Ku80 homologue: Roles in DNA double strand break rejoining and in telomeric maintenance. Nucleic Acids Res. 1996a;24:4639–4648. doi: 10.1093/nar/24.23.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Saccharomyces cerevisiae Ku70 potentiates illegitimate DNA double-strand break repair and serves as a barrier to error-prone DNA repair pathways. EMBO J. 1996b;15:5093–5103. [PMC free article] [PubMed] [Google Scholar]
- ————— Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan DA, Olivares HA, Nelms BE, Petrini JH. Alteration of N-terminal phosphoesterase signature motifs inactivates Saccharomyces cerevisiae mre11. Genetics. 1998;150:591–600. doi: 10.1093/genetics/150.2.591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR, 3rd, Hays L, Morgan WF, Petrini JH. The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: Linkage of double-strand break repair to the cellular DNA damage response. Cell. 1998;93:477–486. doi: 10.1016/s0092-8674(00)81175-7. [DOI] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Leach DRF. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 1999;27:1039–1046. doi: 10.1093/nar/27.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JC, de Leau ES, Okely EA, Leach DR. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J Biol Chem. 1997;272:19819–19826. doi: 10.1074/jbc.272.32.19819. [DOI] [PubMed] [Google Scholar]
- Critchlow SE, Jackson SP. DNA end-joining: From yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- Dolganov GM, Maser RS, Novikov A, Tosto L, Chong S, Bressan DA, Petrini JH. Human Rad50 is physically associated with human Mre11: Identification of a conserved multiprotein complex implicated in recombinational DNA repair. Mol Cell Biol. 1996;16:4832–4841. doi: 10.1128/mcb.16.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank KM, Sekiguchi JM, Seidl KJ, Swat W, Rathbun GA, Cheng HL, Davidson L, Kangaloo L, Alt FW. Late embryonic lethality and impaired V(D)J recombination in mice lacking DNA ligase IV. Nature. 1998;396:173–177. doi: 10.1038/24172. [DOI] [PubMed] [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K. Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J. 1998;17:6412–6425. doi: 10.1093/emboj/17.21.6412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game JC. DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces. Semin Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber MR. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- Haber JE. The many interfaces of Mre11. Cell. 1998;95:583–586. doi: 10.1016/s0092-8674(00)81626-8. [DOI] [PubMed] [Google Scholar]
- Hendrickson EA. Cell-cycle regulation of mammalian DNA double-strand-break repair. Am J Hum Genet. 1997;61:795–800. doi: 10.1086/514895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann G, Lindahl T, Schar P. Saccharomyces cerevisiae LIF1: A function involved in DNA double-strand break repair related to mammalian XRCC4. EMBO J. 1998;17:4188–4198. doi: 10.1093/emboj/17.14.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Hirano T. ATP-dependent aggregation of single-stranded DNA by a bacterial SMC homodimer. EMBO J. 1998;17:7139–7148. doi: 10.1093/emboj/17.23.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Bucher P. The FHA domain: A putative nuclear signaling domain found in protein kinases and transcription factors. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- Ivanov EL, Korolev VG, Fabre F. XRS2, a DNA repair gene of Saccharomyces cerevisiae, is needed for meiotic recombination. Genetics. 1992;132:651–664. doi: 10.1093/genetics/132.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov EL, Sugawara N, White CI, Fabre F, Haber JE. Mutations in XRS2 and RAD50 delay but do not prevent mating-type switching in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:3414–3425. doi: 10.1128/mcb.14.5.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo PA. DNA-PK: At the cross-roads of biochemistry and genetics. Mutat Res. 1997;384:1–14. doi: 10.1016/s0921-8777(97)00009-8. [DOI] [PubMed] [Google Scholar]
- Johzuka K, Ogawa H. Interaction of Mre11 and Rad50: Two proteins required for DNA repair and meiosis-specific double-strand break formation in Saccharomyces cerevisiae. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kironmai KM, Muniyappa, K K. Alteration of telomeric sequences and senescence caused by mutations in RAD50 of Saccharomyces cerevisiae. Genes Cells. 1997;2:443–455. doi: 10.1046/j.1365-2443.1997.1330331.x. [DOI] [PubMed] [Google Scholar]
- Kramer KM, Brock JA, Bloom K, Moore JK, Haber JE. Two different types of double-strand breaks in Saccharomyces cerevisiae are repaired by similar RAD52-independent, nonhomologous recombination events. Mol Cell Biol. 1994;14:1293–1301. doi: 10.1128/mcb.14.2.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach DR. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. BioEssays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Saccharomyces Ku70, mre11/rad50 and RPA proteins regulate adaptation to G2/M arrest after DNA damage. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- Lewis SM. The mechanism of V(D)J joining: Lessons from molecular, immunological, and comparative analyses. Adv Immunol. 1994;56:27–150. doi: 10.1016/s0065-2776(08)60450-2. [DOI] [PubMed] [Google Scholar]
- Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, Stamato TD, Taccioli GE, Alt FW. The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination. Cell. 1995;83:1079–1089. doi: 10.1016/0092-8674(95)90135-3. [DOI] [PubMed] [Google Scholar]
- Maser RS, Monsen KJ, Nelms BE, Petrini JH. hMre11 and hRad50 nuclear foci are induced during the normal cellular response to DNA double-strand breaks. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura S, Tauchi H, Nakamura A, Kondo N, Sakamoto S, Endo S, Smeets D, Solder B, Belohradsky BH, Der Kaloustian VM, Oshimura M, Isomura M, Nakamura Y, Komatsu K. Positional cloning of the gene for Nijmegen breakage syndrome. Nat Genet. 1998;19:179–181. doi: 10.1038/549. [DOI] [PubMed] [Google Scholar]
- McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83:387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- Milne GT, Jin S, Shannon KB, Weaver DT. Mutations in two Ku homologs define a DNA end-joining repair pathway in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JK, Haber JE. Cell cycle and genetic requirements of two pathways of nonhomologous end-joining repair of double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelms BE, Maser RS, MacKay JF, Lagally MG, Petrini JH. In situ visualization of DNA double-strand break repair in human fibroblasts. Science. 1998;280:590–592. doi: 10.1126/science.280.5363.590. [DOI] [PubMed] [Google Scholar]
- Nugent CI, Bosco G, Ross LO, Evans SK, Salinger AP, Moore JK, Haber JE, Lundblad V. Telomere maintenance is dependent on activities required for end repair of double-strand breaks. Curr Biol. 1998;8:657–660. doi: 10.1016/s0960-9822(98)70253-2. [DOI] [PubMed] [Google Scholar]
- Ogawa H, Johzuka K, Nakagawa T, Leem SH, Hagihara AH. Functions of the yeast meiotic recombination genes, MRE11 and MRE2. Adv Biophys. 1995;31:67–76. doi: 10.1016/0065-227x(95)99383-z. [DOI] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Petes TD, Malone RE, Symington LS. Recombination in yeast. In: Broach JR, Jones EW, Pringle JR, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1991. pp. 407–521. [Google Scholar]
- Petrini JH, Walsh ME, DiMare C, Chen XN, Korenberg JR, Weaver DT. Isolation and characterization of the human MRE11 homologue. Genomics. 1995;29:80–86. doi: 10.1006/geno.1995.1217. [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Gellert M. Ku protein stimulates DNA end joining by mammalian DNA ligases: A direct role for Ku in repair of DNA double-strand breaks. EMBO J. 1998;17:609–614. doi: 10.1093/emboj/17.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsden DA, McBlane JF, van Gent DC, Gellert M. Distinct DNA sequence and structure requirements for the two steps of V(D)J recombination signal cleavage. EMBO J. 1996;15:3197–3206. [PMC free article] [PubMed] [Google Scholar]
- Raymond WE, Kleckner N. RAD50 protein of S. cerevisiae exhibits ATP-dependent DNA binding. Nucleic Acids Res. 1993;21:3851–3856. doi: 10.1093/nar/21.16.3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth DB, Wilson JH. Relative rates of homologous and nonhomologous recombination in transfected DNA. Proc Natl Acad Sci. 1985;82:3355–3359. doi: 10.1073/pnas.82.10.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar PS, Chang HC, Boudi FB, Reddy S. CTG repeats show bimodal amplification in E. coli. Cell. 1998;95:531–540. doi: 10.1016/s0092-8674(00)81620-7. [DOI] [PubMed] [Google Scholar]
- Schlissel MS. Structure of nonhairpin coding-end DNA breaks in cells undergoing V(D)J recombination. Mol Cell Biol. 1998;18:2029–2037. doi: 10.1128/mcb.18.4.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharples GJ, Leach DR. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- Shiloh Y. Ataxia-telangiectasia and the Nijmegen breakage syndrome: Related disorders but genes apart. Annu Rev Genet. 1997;31:635–662. doi: 10.1146/annurev.genet.31.1.635. [DOI] [PubMed] [Google Scholar]
- Teo SH, Jackson SP. Identification of Saccharomyces cerevisiae DNA ligase IV: Involvement in DNA double-strand break repair. EMBO J. 1997;16:4788–4795. doi: 10.1093/emboj/16.15.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KM, Yuan SS, Lee EY, Sung P. Nuclease activities in a complex of human recombination and DNA repair factors Rad50, Mre11, and p95. J Biol Chem. 1998;273:21447–21450. doi: 10.1074/jbc.273.34.21447. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H, Ogawa H. A novel mre11 mutation impairs processing of double-strand breaks of DNA during both mitosis and meiosis. Mol Cell Biol. 1998;18:260–268. doi: 10.1128/mcb.18.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Ikeda H. Double-strand break repair mediated by DNA end-joining. Genes Cells. 1998;3:135–144. doi: 10.1046/j.1365-2443.1998.00180.x. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Kato J, Ikeda H. Effects of mutations of RAD50, RAD51, RAD52, and related genes on illegitimate recombination in Saccharomyces cerevisiae. Genetics. 1996a;142:383–391. doi: 10.1093/genetics/142.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Hdf1, a yeast Ku-protein homologue, is involved in illegitimate recombination, but not in homologous recombination. Nucleic Acids Res. 1996b;24:2067–2072. doi: 10.1093/nar/24.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usui T, Ohta T, Oshiumi H, Tomizawa J, Ogawa H, Ogawa T. Complex formation and functional versatility of Mre11 of budding yeast in recombination. Cell. 1998;95:705–716. doi: 10.1016/s0092-8674(00)81640-2. [DOI] [PubMed] [Google Scholar]
- Varon R, Vissinga C, Platzer M, Cerosaletti KM, Chrzanowska KH, Saar K, Beckmann G, Seemanova E, Cooper PR, Nowak NJ, Stumm M, Weemaes CM, Gatti RA, Wilson RK, Digweed M, Rosenthal A, Sperling K, Concannon P, Reis A. Nibrin, a novel DNA double-strand break repair protein, is mutated in Nijmegen breakage syndrome. Cell. 1998;93:467–476. doi: 10.1016/s0092-8674(00)81174-5. [DOI] [PubMed] [Google Scholar]
- Weaver DT. Regulation and repair of double-strand DNA breaks. Crit Rev Eukaryot Gene Expr. 1996;6:345–375. doi: 10.1615/critreveukargeneexpr.v6.i4.20. [DOI] [PubMed] [Google Scholar]
- Weinberg DH, Collins KL, Simancek P, Russo A, Wold MS, Virshup DM, Kelly TJ. Reconstitution of simian virus 40 DNA replication with purified proteins. Proc Natl Acad Sci. 1990;87:8692–8696. doi: 10.1073/pnas.87.22.8692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellinger RJ, Wolf AJ, Zakian VA. Saccharomyces telomeres acquire single-strand TG1-3 tails late in S phase. Cell. 1993;72:51–60. doi: 10.1016/0092-8674(93)90049-v. [DOI] [PubMed] [Google Scholar]
- Xiao Y, Weaver DT. Conditional gene targeted deletion by Cre recombinase demonstrates the requirement for the double-strand break repair Mre11 protein in murine embryonic stem cells. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]


