Abstract
In the eight years since phylogenomics was introduced as the intersection of genomics and phylogenetics, the field has provided fundamental insights into gene function, genome history and organismal relationships. The utility of phylogenomics is growing with the increase in the number and diversity of taxa for which whole genome and large transcriptome sequence sets are being generated. We assert that the synergy between genomic and phylogenetic perspectives in comparative biology would be enhanced by the development and refinement of minimal reporting standards for phylogenetic analyses. Encouraged by the development of the Minimum Information About a Microarray Experiment (MIAME) standard, we propose a similar roadmap for the development of a Minimal Information About a Phylogenetic Analysis (MIAPA) standard. Key in the successful development and implementation of such a standard will be broad participation by developers of phylogenetic analysis software, phylogenetic database developers, practitioners of phylogenomics, and journal editors.
INTRODUCTION
Phylogenies have provided a historical framework for interpreting the evolution of form and function since Darwin (1859) and Haeckel (1866) published their iconic tree figures some 150 years ago. In recent years, phylogenetics has come to play a multifaceted role in genomic analyses and interpretation of genomics data. Phylogenetic analyses are now being performed on a genomic scale in order to address issues ranging from the prediction of gene and protein function (Eisen, 1998; Sjölander, 2004; Engelhardt et al., 2005) to organismal relationships (Philippe et al., 2005; Delsuc et al., 2005), to the influences of polyploidy (Bowers et al., 2003; Byrne and Wolfe, 2005) and horizontal gene transfer (Ge et al., 2005; Simonson et al., 2005) on genome content and structure (Wolf et al., 2002), to the reconstruction of ancestral genome characteristics (Blanchette et al., 2004).
The fundamental nature of inferences drawn from all of these applications underscores the growing importance of genomic and sub-genomic investigations of species covering the spectrum of organismal diversity. Phylogenomic analyses, defined here broadly as the integration of phylogenetic and genomic analysis (Eisen and Fraser, 2003), place genome sequence, gene expression (Gu and Gu, 2003; Gu 2004; Gu et al., 2005; Duarte et al., 2006) and functional data in a historical context and thereby help to elucidate those processes shaping the structure and function of genes, genetic systems and whole organisms. The development and refinement of searchable phylogeny databases such as TreeBase (Piel et al., 2003) or gene tree databases (Duret et al., 1994; Sjölander, 2004; Roth et al., 2005; Hartmann et al., 2006; Li et al., 2006) is an important step in the advancement of phylogenomics, but only a miniscule fraction of published phylogenies are currently deposited in a database. What is worse, the alignments for many published phylogenies are not easily accessible, and methods of analysis are not adequately described. These are serious impediments to those wanting to test the robustness of published phylogenies, conduct cross-study comparisons of phylogenetic inferences, or draw new inferences from meta-analyses.
Accurate phylogenetic trees provide a valuable historical context for a variety of comparative analyses, and can be applied to a host of biological questions unforeseen by the original authors. This is particularly true in phylogenomics, where many applications require the investigation of phylogenetic trees for a large number of independent gene/protein families. If inadequately documented, however, even the most carefully constructed phylogenetic analysis will languish in the pages of a journal. Thus, a key step in the continued ability of phylogenomics to take full advantage of the rapidly expanding volume of sequence data will be the development of reporting standards for phylogenetic analyses, along with databases from which these metadata can easily be retrieved. In this paper, we propose a roadmap to develop a set of reporting standards for phylogenetic analyses. Using the MIAME standard (Brazma et al., 2001) as a model, we call for a community-wide effort to develop a Minimal Information About a Phylogenetic Analysis (MIAPA) standard.
CONSIDERATIONS FOR DEVELOPING STANDARDS FOR REPORTING PHYLOGENETIC ANALYSES
The papers in this special issue constitute a series of case studies on the importance of standard practices for reporting the results of various types of experiments in a way that facilitates the ability of scientists to use these data in subsequent studies (Field and Sansone, 2006). The motivating question behind the MIAME standard for microarray experiments was as follows: “What is the minimum information necessary for an independent scientist to carry out an independent analysis of the data?” (Quackenbush, 2005).
The motivation for the MIAPA standard is the same, as is the challenge: minimizing the reporting requirements while maximizing the information available to those interpreting the results of a study (Brazma, 2001; Brazma et al., 2001; Ball and Brazma, this issue). The phylogenetics community is coming together to develop this standard with careful consideration of the types of future analyses that are likely to be performed and the data required. For example, systematists may combine pre-existing phylogenies into supertree analyses (Davies et al., 2004; Page, 2005), while genomicists may combine them to investigate the timing of genome duplication events (Chapman et al., 2004). At the same time, investigators may require access to the alignments and component sequences used to build the selected phylogenies in order to perform independent phylogenetic analyses on single or combined datasets. Thus, just as the MIAME standard was designed to accommodate the nested organization of gene expression levels derived from signal quantification matrices derived in turn from raw image data (Brazma et al., 2001), the MIAPA standard would need to accommodate phylogenies derived from analysis of alignments derived in turn from raw sequence data.
A decision that was integral to development and success of the MIAME standards was that they should be applicable to a wide variety of microarray technologies and no one platform or hybridization protocol was prescribed. Similarly, we suggest that the MIAPA standard should be agnostic concerning methods of alignment and phylogenetic reconstruction. The diversity of methods of phylogenetic inference is perhaps even greater than the diversity of applications to which phylogenies may be applied (Swofford et al., 1996; Felsenstein, 2004; Delsuc et al., 2005) and novel methods are likely to be developed in the future. Parsimony, likelihood, Bayesian and distance-based approaches have all been adapted for analyses of the various data types relevant to phylogenomics, including aligned nucleotide and protein sequences, gene structure (insertions and deletions), gene content, motif frequencies (Qi et al., 2004) and gene order (Moret et al., 2001). Multiple sequence alignment has its own diverse set of methodologies, and, in some approaches, a multiple sequence alignment and phylogenetic tree are constructed simultaneously (Gladstein and Wheeler, 1997; Edgar and Sjölander, 2003; Lunter et al., 2005; Fleissner et al., 2005). The relative performance of these different methods is an area of active research, but it is clear that no single method is optimal for all data sets (Swofford et al., 2001; Spencer et al., 2005). Benchmark datasets have been compiled for comparing the performance of alignment algorithms (van Walle et al., 2004; Thompson et al., 2005) but there are few comparable benchmarks for phylogenetic algorithms, and so comparisons have relied largely on analyses of simulated or contrived data sets (Huelsenbeck 1995; Swofford et al., 2001; Spencer et al., 2005, but see Hillis et al., 1992; Cunningham et al., 1997). Thus, for a variety of reasons, methodological diversity in phylogenetics is likely to be the state of affairs for the foreseeable future. No matter how phylogenies are constructed, however, a comprehensive description of how a set of sequences was aligned, and how phylogenetic trees were derived from an alignment would allow researchers to evaluate their confidence in a phylogeny and run their own analyses if they see fit.
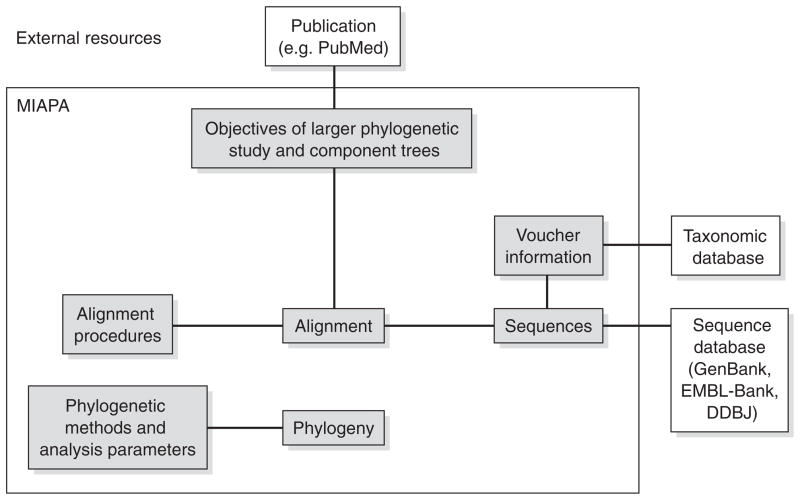
The six required components of the MIAME standards proposed in 2001 (Brazma et al., 2001) included descriptions of (1) the experimental design for a complete study; (2) the design of each array and the identity of each spot on the arrays used in the study; (3) the biological sample extraction preparations and labelling procedures used for each hybridization; (4) the hybridization protocols; (5) the measurements, including imaging and signal quantification parameters; and (6) the normalization and control information. At this stage, it would be premature to specify the details of the MIAPA standard, but Figure 1 offers a starting point for considering MIAPA’s essential components that it might include. By analogy with the MIAME standards, minimum reporting standards for phylogenetic analyses are likely to include (1) a description of the objectives of the phylogenetic analysis and the component trees included in a study (many phylogenetic studies produce multiple trees based on different data sets or analytical methods); (2) the raw sequences or character descriptions; (3) sample voucher information; (4) a description of procedures for establishing orthology of characters (e.g., sequence alignment); (5) the sequence alignment or some other character matrix; (6) detailed description of the phylogenetic analysis, including search strategies and parameter values (specific commands for the analysis program would be optimal); and (7) the phylogenies including branch lengths and support values (e.g., bootstrap). The schematic shown in Figure 1 is likely to be incomplete. For example, it is not clear whether or how to report measures of node support, such as bootstrap values, and phylogenetic analyses are often performed on data matrices other than nucleotide and protein sequence alignments. If the reporting standard were focused on sequence data, referencing an external database for the unaligned and unmasked sequences would require that all sequence identifiers in a database such as GenBank would be stable over the long term. If the standard were to extend to phylogenetic analyses of morphological characters, character descriptions and data matrices could be deposited in MorphBank (〈www.morphbank.com〉) or MorphoBank (〈www.morphobank.org〉). Following the MIAME model (Brazma et al., 2001), the scheme in Figure 1 is reliant on an external database (e.g., the taxonomy database at NCBI) for information about the taxonomic placement of the studied organisms. However, it might be better to require the full taxonomy of the studied organisms to be reported in order to allow a full search of the taxonomic hierarchy (Page, 2005). We suggest that sample voucher information be included in the reporting standard in order to properly synthesize future combined data matrices or build supertrees. The phylogenetics community will have to grapple with these issues and more as we formalize the reporting standard, and we reiterate that Figure 1 is presented simply as a starting point for deeper consideration.
FIG. 1.
A schematic diagram showing the components of a phylogenetic analysis that could be included in a minimal reporting standard.
DEFINING A ROADMAP FOR THE DEVELOPMENT OF MIAPA STANDARDS
The nearly universal adoption of the MIAME standards and their impact on all aspects of microarray-based expression profiling was driven by necessity. The deliberate process by which they were constructed started with an international meeting of what became the Microarray Gene Expression Data Society (〈www.mged.org〉) in 1999 and culminated in an open letter first published in Nature Genetics in 2001. There has been continued refinement of the standards at annual meetings (Ball and Brazma, this issue). Much of the success of MIAME must also be attributed to the fact that MGED engaged commercial interests and database managers. The compliance of microarray databases (Parkinson et al., 2005; Barrett et al., 2005) was facilitated by the development of formal protocols for data exchange, namely the microarray gene expression object model (MAGE-OM) implemented in XML (MAGE-ML) (Spellman et al., 2002). User-friendly systems for submission of expression data and metadata that built upon these protocols (Mukherjee et al., 2005) have further promoted widespread compliance with MIAME guidelines among investigators.
Development of the MIAPA standard must also involve developers of phylogenetic analysis software (Felsenstein, 2005; Goloboff et al., 2004; Kumar et al., 2004; Ronquist and Huelsenbeck, 2003; Swofford, 2001; Roshan et al., 2004; 〈www.phylo.org〉), existing public databases for organismal (Piel et al., 2003) and gene family phylogenies (Duret et al., 1994; Sjölander, 2004; Roth et al., 2005; Hartmann et al., 2006; Li et al., 2006), as well as editors of the journals in which phylogenetic analyses are published. A well-defined protocol for saving and transferring phylogenetic metadata should be considered, one that would complement existing formats such as New Hampshire (or Newick) and PhyloXML (〈www.phyloxml.org〉).
Following the example of MGED, development of the MIAPA standard could be advanced through an international conference of representative stakeholders in conjunction with open discussions across the phylogenetics community. We will be soliciting involvement in an organizational conference at scientific meetings this coming summer and publishing proposals for the MIAPA standard in the journals most read by the phylogenetics community. We anticipate these efforts will culminate in an open letter to the editors of all journals publishing phylogenies in which MIAPA will be described in detail. In addition, the standard would be most viable if accompanied by software and database tools that would facilitate utility and widespread compliance.
CONCLUSION
These are ambitious objectives, but the time is ripe for the development and implementation of minimal reporting standards for phylogenetic analyses. Widespread recognition of the importance of phylogenetics to genome biology comes at a time when recent advances have increased the rate of sequence generation by orders of magnitude (Margulies et al., 2005). Increases in sequencing capacity and concomitant cost decreases are spurring a rapid expansion in the availability of whole genome sequences (Liolios et al., 2006) or subgenomic sequence data (Lee et al., 2005). Beyond doubt, this flood of sequence data will spur a corresponding flood of comparative analyses in which phylogenetic trees play a central role. Indeed, many computational and statistical methods for functional genomic analysis are being developed, which are, more or less, phylogeny-based. When reporting of phylogenetic analyses is brought more fully into the informatics age, it will have manifold beneficial effects on the utility and impact of phylogenomics.
Acknowledgments
We thank Dawn Field and Peter Sterk for organizing the “Cataloguing our Current Genome Collection” workshop at EBI, where some of these ideas were first proposed. We also thank Dawn Field, Susanna Sansone, and Eugene Kolker for their roles in putting together this special issue.
References
- BALL CA, BRAZMA A. OMICS. 2006. MGED standards: work in progress. (this issue) [DOI] [PubMed] [Google Scholar]
- BARRETT T, SUZEK TO, TROUP DB, et al. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BLANCHETTE M, GREEN ED, MILLER W, et al. Reconstructing large regions of an ancestral mammalian genome in silico. Genome Res. 2004;14:2412–2423. doi: 10.1101/gr.2800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOWERS JE, CHAPMAN BA, RONG J, et al. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- BRAZMA A. On the importance of standardisation in life sciences. Bioinformatics. 2001;17:113–114. doi: 10.1093/bioinformatics/17.2.113. [DOI] [PubMed] [Google Scholar]
- BRAZMA A, HINGAMP P, QUACKENBUSH J, et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- BYRNE KP, WOLFE KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHAPMAN BA, BOWERS JE, SCHULZE SR, et al. A comparative phylogenetic approach for dating whole genome duplication events. Bioinformatics. 2004;20:180–185. doi: 10.1093/bioinformatics/bth022. [DOI] [PubMed] [Google Scholar]
- CUNNINGHAM CW, JENG K, HUSTI J, et al. Parallel molecular evolution of deletions and nonsense mutations in bacteriophage T7. Mol Biol Evol. 1997;14:113–116. doi: 10.1093/oxfordjournals.molbev.a025697. [DOI] [PubMed] [Google Scholar]
- DARWIN C. The Origin of Species by Means of Natural Selection. John Murray; London: 1859. [Google Scholar]
- DAVIES TJ, BARRACLOUGH TG, CHASE MW, et al. Darwin’s abdominable mystery: insights from a supertree of the angiosperms. Proc Natl Acad Sci USA. 2004;101:1904–1909. doi: 10.1073/pnas.0308127100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DELSUC F, BRINKMANN H, PHILIPPE H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- DUARTE JM, CUI L, WALL PK, et al. Expression pattern shifts following duplication indicative of sub-functionalization and neofunctionalization in regulatory genes of Arabidopsis. Mol Biol Evol. 2006;23:469–478. doi: 10.1093/molbev/msj051. [DOI] [PubMed] [Google Scholar]
- EDGAR RC, SJOLANDER K. SATCHMO: sequence alignment and tree construction using hidden Markov models. Bioinformatics. 2003;19:1404–1411. doi: 10.1093/bioinformatics/btg158. [DOI] [PubMed] [Google Scholar]
- EISEN JA. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 1998;8:163–167. doi: 10.1101/gr.8.3.163. [DOI] [PubMed] [Google Scholar]
- EISEN JA, FRASER CM. Phylogenomics: intersection of evolution and genomics. Science. 2003;300:1706–1707. doi: 10.1126/science.1086292. [DOI] [PubMed] [Google Scholar]
- ENGELHARDT BE, JORDAN MI, MURATORE KE, et al. Protein molecular function prediction by Bayesian phylogenomics. PLoS Comput Biol. 2005;1:e45. doi: 10.1371/journal.pcbi.0010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FIELD D, SANSONE S-A. A special issue on data standards. OMICS. 2006 doi: 10.1089/omi.2008.0013. (this issue) [DOI] [PubMed] [Google Scholar]
- FELSENSTEIN J. Inferring Phylogenies. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- FELSENSTEIN J. PHYLIP (Phylogeny Inference Package), version 3.6. distributed by author, Department of Genome Sciences, University of Washington; Seattle, Washington: 2005. [Google Scholar]
- FLEISSNER R, METZLER D, VON HAESELER A. Simultaneous statistical multiple alignment and phylogeny reconstruction. Syst Biol. 2005;54:548–561. doi: 10.1080/10635150590950371. [DOI] [PubMed] [Google Scholar]
- GE F, WANG LS, KIM J. The cobweb of life revealed by genome-scale estimates of horizontal gene transfer. PLoS Biol. 2005;3:e316. doi: 10.1371/journal.pbio.0030316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GLADSTEIN DS, WHEELER WC. POY: the optimization of alignment characters. 1997 Available at: 〈 ftp.amnh.org/pub/molecular〉.
- GOLOBOFF PA, FARRIS JS, NIXON KC. TNT Tree Analysis Using New Technology, version 1.0. 2004 Available at: 〈 www.cladistics.com〉.
- GU J, GU X. Induced gene expression in human brain after the split from chimpanzee. Trend Genet. 2003;19:63–65. doi: 10.1016/s0168-9525(02)00040-9. [DOI] [PubMed] [Google Scholar]
- GU X. Statistical framework for phylogenetic analysis of expression profiles. Genetics. 2004;167:531–542. doi: 10.1534/genetics.167.1.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GU X, ZHANG Z, HUANG W. Rapid evolution of expression and regulatory network after yeast gene/genome duplications. Proc Natl Acad Sci USA. 2005;102:707–712. doi: 10.1073/pnas.0409186102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAEKEL F. Generelle Morphologie der Organismen. G. Reimer; Berlin: 1866. [Google Scholar]
- HARTMANN S, LU D, PHILLIPS J, et al. Phytome: a platform for plant comparative genomics. Nucleic Acids Res. 2006;34:D724–D730. doi: 10.1093/nar/gkj045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HILLIS DM, BULL JJ, WHITE ME, et al. Experimental phylogenetics: generation of a known phylogeny. Science. 1992;255:589–592. doi: 10.1126/science.1736360. [DOI] [PubMed] [Google Scholar]
- HUELSENBECK JP. The robustness of two phylogenetic methods: four-taxon simulations reveal a slight superiority of maximum likelihood over neighbor joining. Mol Biol Evol. 1995;12:843–849. doi: 10.1093/oxfordjournals.molbev.a040261. [DOI] [PubMed] [Google Scholar]
- KUMAR S, TAMURA K, NEI M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- LEE Y, TSAI J, SUNKARA S, et al. The TIGR Gene Indices: clustering and assembling EST and known genes and integration with eukaryotic genomes. Nucleic Acids Res. 2005;33:D71–D74. doi: 10.1093/nar/gki064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI H, COGHLAN A, RUAN J, et al. TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 2006;34:D572–D580. doi: 10.1093/nar/gkj118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIOLIOS K, TAVERNARAKIS N, HUGENHOLTZ P, et al. The Genomes On Line Database (GOLD) v.2: a monitor of genome projects worldwide. Nucleic Acids Res. 2006;34:D332–D334. doi: 10.1093/nar/gkj145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUNTER G, MIKLOS I, DRUMMOND A, et al. Bayesian coestimation of phylogeny and sequence alignment. BMC Bioinform. 2005;6:83. doi: 10.1186/1471-2105-6-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MARGULIES M, EGHOLM M, ALTMAN WE, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORET BM, WANG LS, WARNOW T, et al. New approaches for reconstructing phylogenies from gene order data. Bioinformatics. 2001;17:S165–S173. doi: 10.1093/bioinformatics/17.suppl_1.s165. [DOI] [PubMed] [Google Scholar]
- MUKHERJEE G, ABEYGUNAWARDENA N, PARKINSON H, et al. Plant-based microarray data at the European Bioinformatics Institute. Introducing AtMIAMExpress, a submission tool for Arabidopsis gene expression data to ArrayExpress. Plant Physiol. 2005;139:632–636. doi: 10.1104/pp.105.063156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAGE RDM. Towards a taxonomically intelligent phylogenetic database. Technical Reports in Taxonomy 04-01, presented at DBiBD; Edinburgh. 2005. [Google Scholar]
- PARKINSON H, SARKANS U, SHOJATALAB M, et al. ArrayExpress—a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2005;33:D553–D555. doi: 10.1093/nar/gki056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PHILIPPE H, SNELL EA, BAPTESTE E, et al. Phylogenomics of eukaryotes: impact of missing data on large alignments. Mol Biol Evol. 2004;21:1740–1752. doi: 10.1093/molbev/msh182. [DOI] [PubMed] [Google Scholar]
- PIEL WH, SANDERSON MJ, DONOGHUE MJ. The small-world dynamics of tree networks and data mining in phyloinformatics. Bioinformatics. 2003;19:1162–1168. doi: 10.1093/bioinformatics/btg131. [DOI] [PubMed] [Google Scholar]
- QI J, WANG B, HAO BI. Whole proteome prokaryote phylogeny without sequence alignment: a K-string composition approach. J Mol Evol. 2004;58:1–11. doi: 10.1007/s00239-003-2493-7. [DOI] [PubMed] [Google Scholar]
- QUACKENBUSH J. Extracting meaning from functional genomics experiments. Toxicol Appl Pharmacol. 2005;207:195–199. doi: 10.1016/j.taap.2005.04.029. [DOI] [PubMed] [Google Scholar]
- ROSHAN UW, MORET BM, WARNOW T, et al. Rec-I-DCM3: a fast algorithmic technique for reconstructing large phylogenetic trees. Proc IEEE Comput Syst Bioinform Conf. 2004:98–109. doi: 10.1109/csb.2004.1332422. [DOI] [PubMed] [Google Scholar]
- ROTH C, BETTS MJ, STEFFANSSON P, et al. The Adaptive Evolution Database (TAED): a phylogeny based tool for comparative genomics. Nucleic Acids Res. 2005;33:D495–D497. doi: 10.1093/nar/gki090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMONSON AB, SERVIN JA, SKOPHAMMER RG, et al. Decoding the genomic tree of life. Proc Natl Acad Sci USA. 2005;102:6608–6613. doi: 10.1073/pnas.0501996102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SJOLANDER K. Phylogenomic inference of protein molecular function: advances and challenges. Bioinformatics. 2004;20:170–179. doi: 10.1093/bioinformatics/bth021. [DOI] [PubMed] [Google Scholar]
- SPELLMAN PT, MILLER M, STEWART J, et al. Design and implementation of microarray gene expression markup language (MAGE-ML) Genome Biol. 2002;3:RESEARCH0046. doi: 10.1186/gb-2002-3-9-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SPENCER M, SUSKO E, ROGER AJ. Likelihood, parsimony, and heterogeneous evolution. Mol Biol Evol. 2005;22:1161–1164. doi: 10.1093/molbev/msi123. [DOI] [PubMed] [Google Scholar]
- SWOFFORD DL. PAUP*: Phylogeneic Analyses Using Parsimony (* and Other Methods) Sinauer Associates; Sunderland, MA: 2003. [Google Scholar]
- SWOFFORD DL, OLSEN GJ, WADDELL PJ, et al. Phylogenetic inference. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. 2. Sinauer Associates; Sunderland, MA: 1996. [Google Scholar]
- SWOFFORD DL, WADDELL PJ, HUELSENBECK JP, et al. Bias in phylogenetic estimation and its relevance to the choice between parsimony and likelihood methods. Syst Biol. 2001;50:525–539. [PubMed] [Google Scholar]
- THOMPSON JD, KOEHL P, RIPP R, et al. BAliBASE 3.0: latest developments of the multiple sequence alignment benchmark. Proteins. 2005;61:127–136. doi: 10.1002/prot.20527. [DOI] [PubMed] [Google Scholar]
- VAN WALLE I, LASTERS I, WYNS L. SABmark—a benchmark for sequence alignment that covers the entire known fold space. Bioinformatics. 2005;21:1267–1268. doi: 10.1093/bioinformatics/bth493. [DOI] [PubMed] [Google Scholar]
- WOLF YI, ROGOZIN IB, GRISHIN NV, et al. Genome trees and the tree of life. Trends Genet. 2002;18:472–479. doi: 10.1016/s0168-9525(02)02744-0. [DOI] [PubMed] [Google Scholar]