Abstract
Cognitive neuroscience aims to map mental processes onto brain function, which begs the question of what “mental processes” exist and how they relate to the tasks that are used to manipulate and measure them. This topic has been addressed informally in prior work, but we propose that cumulative progress in cognitive neuroscience requires a more systematic approach to representing the mental entities that are being mapped to brain function and the tasks used to manipulate and measure mental processes. We describe a new open collaborative project that aims to provide a knowledge base for cognitive neuroscience, called the Cognitive Atlas (accessible online at http://www.cognitiveatlas.org), and outline how this project has the potential to drive novel discoveries about both mind and brain.
Keywords: ontology, informatics, neuroimaging, cognitive science
“We’re drowning in information and starving for knowledge”
–Rutherford B. Rogers
The field of cognitive neuroscience faces an increasingly critical challenge: How can we integrate knowledge from an exploding number of studies across multiple methodologies in order to characterize how mental processes are implemented in the brain? The creation of neuroimaging databases containing data from large numbers of studies has provided the basis for powerful meta-analyses (Laird et al., 2005). However, the semantic infrastructure for characterizing the psychological aspects of these studies has lagged far behind the technical infrastructure for databasing and analyzing the imaging results. We propose that cumulative progress in cognitive neuroscience requires such a semantic infrastructure, and that this problem must be addressed through the development of knowledge bases of mental processes (Price and Friston, 2005; Bilder et al., 2009). Here we outline a new project called the Cognitive Atlas (CA)1 that aims to develop such a framework through collaborative social knowledge building.
What is the Problem?
The cognitive neuroscientist wishes to answer questions such as: “What are the neural substrates of working memory?” According to PubMed, as of February 2011 there were 2613 published research papers that mentioned “working memory” along with either functional magnetic resonance imaging (fMRI), positron emission tomography (PET), EEG/ERP, or lesion analysis. Despite this substantial body of published research, it remains difficult to integrate across this work in order to understand the concept of “working memory” and how it relates to brain function, for two major reasons: ambiguous terminology and confounding of cognitive processes with the tasks used to measure these.
Ambiguous terminology
There is substantial ambiguity in the way that terms are used in cognitive neuroscience. On the one hand, many terms are used to denote multiple, potentially distinct processes. For example, the term “working memory” has several distinct definitions in the neuroscience literature:
holding information online in memory, as used by Goldman-Rakic (1995) and measured in non-human primates using tasks such as the oculomotor delayed response task
manipulating information held in memory, as used by Baddeley (1992) and measured in humans using tasks such as the letter–number sequencing task
memory for temporally varying aspects of a task (roughly equivalent to the concept of episodic memory), as used by Olton et al. (1979) and measured in rodents using a radial arm maze task with varied food locations
Thus, searching for “working memory” may retrieve papers that are relevant to a range of different specific psychological processes. On the other hand, many processes are described in the literature using several different terms. For example, the first sense of “working memory” listed above is often described using other terms including “short-term memory” or “active maintenance.” For this reason, searches that only include “working memory” will fail to retrieve papers that use those other terms unless the search is expanded to include those other terms, whereas query expansions that do include those other terms may yield unacceptably high numbers of irrelevant documents.
Tasks versus constructs
There is a longstanding tendency within the cognitive neuroscience literature to equate tasks with mental constructs. For example, the “Sternberg item recognition task” is often referred to as the “Sternberg working memory task,” which implies that it measures a specific mental construct (“working memory”). This conflation of tasks and constructs causes a number of difficulties. First, the measurement of a psychological construct requires a comparison between specific task conditions (Sternberg, 1969); thus, whereas the contrast of particular conditions within the Sternberg task (e.g., high load versus low load) may indeed be associated with the construct of working memory, other contrasts may not (e.g., probe match versus probe mismatch). Second, any link between tasks and constructs reflects a particular theory about how the task is performed; thus, equating tasks with constructs makes theoretical assumptions that may not be shared throughout the community (and further, those community assumptions may be incorrect). For example, the color–word Stroop task is sometimes referred to as an “inhibition task” (Donohoe et al., 2006). However, the role of an active inhibitory process in producing the Stroop effect has been questioned by a number of investigators (Cohen et al., 1990). Similarly, while the N-back task is often referred to as the “N-back working memory task,” serious questions have been raised regarding whether it truly measures the construct of “working memory” (Kane et al., 2007). The equation of tasks and processes thus produces substantial confusion about what is actually being measured by cognitive neuroscience studies.
One problem that arises from this is that a single task is often associated with multiple constructs in the literature. As an example, Sabb et al. (2008) used literature mining tools to examine the published literature related to the construct of “cognitive control.” They found that this construct was associated with a number of other constructs (including “working memory,” “response inhibition,” “response selection,” and “task/set switching”), and that there were no tasks that were uniquely associated with the construct of “cognitive control” in the literature; each task was also associated with at least one of those other constructs. Further, the association in the literature between these tasks and constructs changed over time. This lack of consistency in the way that tasks and concepts are treated in the literature makes it difficult to draw meaningful inferences from existing literature and limits the cumulative value of the knowledge represented in this literature.
Toward an Ontology for Cognition
What is urgently needed is an informatics resource that can solve the problems listed above. Such a resource would provide the ability to identify the particular usage of terms, to allow browsing for related concepts, and to allow the identification of relevant evidence from the literature that is related to these concepts. This would allow intelligent aggregation of research findings, which could help overcome the information overload that currently afflicts researchers. We propose that this challenge can be best addressed through the development and widespread implementation of an ontology for cognitive neuroscience. In philosophy, “ontology” refers to the study of existence or being. However, in bioinformatics the term is increasingly used in the sense defined by Gruber (1993) as an “explicit specification of a conceptualization,” or a structured knowledge base meant to support the sharing of knowledge as well as automated reasoning about that knowledge. Ontologies have also provided the basis for effective knowledge accumulation in molecular biology and genomics (Bard and Rhee, 2004). One of the best known examples is the Gene Ontology2 (GO; Ashburner et al., 2000). This ontology provides consistent descriptors for gene products, including cellular components (e.g., “ribosome”), biological processes (e.g., “signal transduction”), and molecular functions (e.g., “catalytic activity”). GO provides the basis on which to annotate datasets regarding their function, which prevents the common problem of different researchers using different names to describe the same biological structure or process across different organisms. It also provides the ability to traverse the ontology in order to discover larger-scale regularities by expanding the search to include the subordinate terms in the ontology. There are increasingly powerful tools that are built around ontologies such as GO; given a dataset (such as a gene expression pattern), these tools provide a broad range of functions such as the comparison of genetic datasets based on the similarity of their GO annotation patterns (Ruths et al., 2009) and the extraction of novel biological facts from the text of articles (Müller et al., 2004). Ontologies have also been developed in a number of other domains in neuroscience (Martone et al., 2004); most relevant to cognitive neuroscience, there are well-developed ontologies of brain structure (Bowden and Dubach, 2003).
A large body of research in cognitive science has developed detailed domain-specific theories of mental processes, but there has been very little work to systematically characterize how these processes are defined and how they fit together into a larger structure. In part this likely reflects the functionalist character of modern psychology, which arose in reaction to the structuralist approach of the nineteenth century (e.g., as seen in the so-called “faculty psychology” that was employed by phrenologists; Boring, 1950). There have been some attempts at larger-scale “unified theories of cognition” such as Anderson’s ACT-R (Anderson et al., 2004) and Newell’s SOAR (Laird et al., 1987), but these approaches have primarily focused on the development of general unifying computational principles rather than on a systematic characterization of the broad range of cognitive processes.
Other extant vocabularies, such as the medical subject headings (MeSH), contain some content relevant to cognitive neuroscience, but suffer from serious limitations. For example, the MeSH hierarchy for “Cognition” includes just the following concepts: Awareness, Cognitive Dissonance, Comprehension, Consciousness, Imagination, and Intuition. These terms possess no meaningful relation to the current conceptual framework of cognitive science. In addition, the MeSH terms are a mixture of mental processes (e.g., “comprehension”), experimental phenomena (e.g., “illusions”), and experimental procedures (e.g., “maze learning”), along with outdated terms such as “neurolinguistic programming” (which is best characterized as a pseudoscience). Given that MeSH is the lexicon used for indexing articles and expanding queries in PubMed, this suggests that searches of this literature could be greatly improved through the use of vocabularies that better reflect current thinking.
The development of formal ontologies of cognition faces a distinct challenge in comparison to other domains in biology, such as neuroanatomy or cellular function: There is precious little consensus across the field regarding the basic units of mental function. Given that a formal ontology is generally meant to express the shared ontological commitments of a group, this poses a difficult challenge to the development of an ontology of mental processes. There are two alternatives in this case. The first would be to forge ahead and develop a single ontology based on the consensus obtained within a small group of individuals. This would have the benefit of providing an ontology approved by consensus of its architects, but it would be useless to anyone who did not share the group’s ontological commitments. An alternative approach, which we adhere to in the present work, is to allow and capture disagreement, in order to represent the range of views that are present in the field. Our approach to this issue is inspired by the success of social collaborative knowledge building projects such as Wikipedia, which allow discussion and the expression of divergent views in service of developing a broader consensus, and one that can be modified flexibly over time as new knowledge emerges.
The Cognitive Atlas
To address the need for a formal knowledge base that captures the broad range of conceptual structure within cognitive science, we have developed the CA (accessible online at http://www.cognitiveatlas.org). The system is under continuous development, and new features will be added in the future, but the current system provides the basic functionality for specification of knowledge about cognitive processes and tasks. The system has been designed with the intention of making interaction with the knowledge base as easy as possible, without requiring users to possess expertise in ontologies or knowledge base development (Miller et al., 2010). In addition, the system uses standard mechanisms to enable programmatic access to the database (such as SPARQL Protocol and RDF Query Language, SPARQL), which allows other sites or databases to use the content in an automated manner.
An important guiding principle in the design of the CA has been the distinction between mental tasks and mental processes. Mental processes are not directly accessible, but psychological tasks can be used to manipulate and measure them, and behavior or brain activity observed during those tasks is interpreted as reflecting those latent mental constructs. The ontological status of psychological tasks is not in question (i.e., nearly everyone will agree on what the “Stroop task” is), but the relation of those tasks to the latent mental constructs is at the center of many debates in cognitive science. For this reason, we propose that it is essential to make a clear distinction between mental processes and psychological tasks, and to develop separate ontologies for those two domains (resulting in two separate but interlinked ontologies that form a bipartite graph).
Structure of the Knowledge Base
Development of the schemas for the CA knowledge base required an analysis of the kinds of knowledge structures that are used in cognitive science. An initial vocabulary of more than 800 terms was identified manually through analysis of a broad set of publications on cognitive psychology and cognitive neuroscience and curated by three of the authors (Russell A. Poldrack, Robert M. Bilder, Fred W. Sabb). These entitles were classified into two broad classes: mental concepts and mental tasks.
Mental concepts
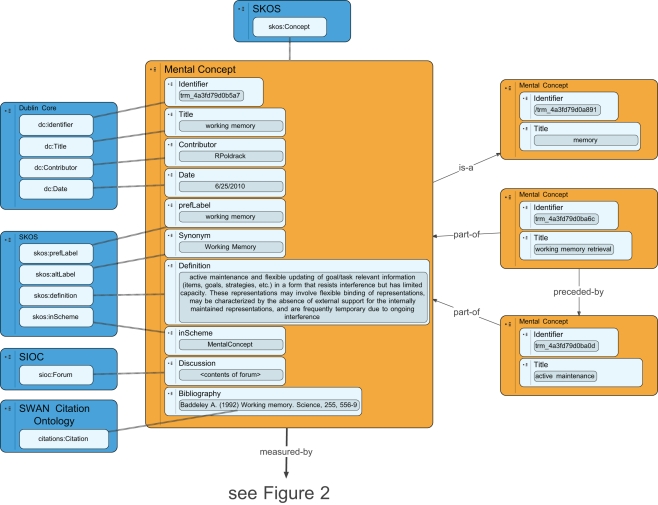
A mental concept is a latent unobservable construct postulated by a psychological theory. Although these mental concepts are ultimately instantiated in brain tissue, the mental concept entity in the knowledge base refers to the latent construct (e.g., at Marr’s computational or algorithmic levels) rather than its physical instantiation. Some potential kinds of mental concepts include (but are not limited to) mental representations and mental processes. Mental representations are mental entities that stand in relation to some physical entity (e.g., a mental image of a visual scene stands in relation to, or is isomorphic with, some arrangement of objects in the physical world) or abstract concept (which could be another mental entity). Mental processes are entitles that transform or operate on mental representations (e.g., a process that searches a mental representation of the visual scene for a particular object). In order to accommodate the widest possible range of theories of cognition (including non-representational theories such as Edelman, 1989), the knowledge base does not require that mental concepts be specified into these subclasses, and indeed it is agnostic about this distinction, permitting but not demanding that cognitive “representations” exist. Mental concepts in the CA knowledge base are modeled using the Concept class from the simple knowledge organization system (SKOS; Bechhofer and Miles, 2009), which describes the basic structure of conceptual entities. An overview of the database schema for mental concepts is presented in Figure 1.
Figure 1.
An overview of the database schema for representation of mental concepts in the Cognitive Atlas. Blue boxes reflect external ontologies, and dashed lines reflect class inheritance, while solid lines reflect ontological relations.
Mental tasks
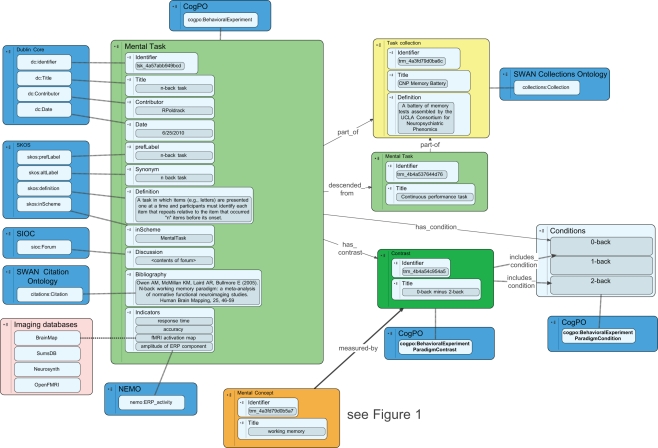
A mental task is a prescribed activity meant to engage or manipulate mental function in an effort to gain insight into the underlying mental processes. The structure of the representation of mental tasks in the CA builds upon the cognitive paradigm ontology (CogPO3; Turner and Laird, 2011), which has a basic class of Behavioral Experimental Paradigm that describes mental tasks. An overview of the database schema for mental tasks is presented in Figure 2.
Figure 2.
An overview of the database schema for representation of mental tasks in the Cognitive Atlas. Blue boxes reflect external ontologies, and dashed lines reflect class inheritance, while solid lines reflect ontological relations. The dashed line connecting activation maps to imaging databases is meant to reflect an empirical relation, as these databases do not currently expose formal ontologies.
Tasks often evoke overt responses (such as motor actions), but this is not necessary; e.g., a brain imaging study could measure the neural responses evoked by a particular form of stimulation (such as watching a movie) without any overt behavior. Any particular task has a number of different features that need to be distinguished and/or measured.
Experimental conditions
Experimental conditions are the subsets of an experiment that define the relevant experimental manipulation. For example, in the color–word Stroop task there are generally three conditions (congruent, incongruent, and neutral trials). This could also be extended to include parametric manipulations as well. These are defined according to the Behavioral Experimental Paradigm Condition class in CogPO.
Indicators
An indicator is a specific quantitative or qualitative variable that is recorded for analysis. These may include behavioral variables (such as response time, accuracy, or other measures of performance) or physiological variables (including genetics, psychophysiology, lesion effects, or neuroimaging data). In the current implementation of the CA, we focus primarily on behavioral indicators, but we intend the system to be generally applicable to any indicators measured in the context of mental function, including physiological measurements, genetics, and imaging data.
Contrasts
Although absolute measures of behavior may occasionally be meaningful (e.g., scores on a standardized test), it is usually the comparison of indicators across different experimental conditions that we associate with particular mental processes, through subtraction logic or other experimental designs. In the CA, we define a contrast as any function over experimental conditions (borrowing this usage from the notion of linear contrasts in the general linear model, as commonly used in the neuroimaging literature). The simplest contrast is the indicator value for a specific condition; In some cases this will be meaningful in absolute terms, whereas in other cases (e.g., with neuroimaging data) this would reflect a comparison with a more basic baseline condition, such as rest or visual fixation. More complex contrasts include linear or non-linear functions of the indicator across different experimental conditions. For example, in the Stroop task, there would be simple contrasts for each of the three conditions, in addition to contrasts such as (incongruent–congruent) and (incongruent–neutral) that index the well-known “Stroop effect.” These are defined according to the Behavioral Experimental Paradigm Contrast class in CogPO.
Relations
While a well-defined vocabulary is critical to our knowledge base, it is the relations between the terms within the vocabulary that are of greatest interest, because they express the theoretical claims of cognitive theories. The CA includes a number of different types of relations (though not all have been implemented in the current release). Where ever possible, we have reused existing ontologies.
Basic ontological relations
A standard set of ontological relations has been codified into the open biomedical ontologies (OBO) Relational Ontology (Smith et al., 2005), which provides guidelines regarding the consistent use of specific ontological relations across different ontologies. We have adopted several of the basic ontological relations from this ontology:
is-a (e.g., “declarative memory is a kind of memory”)
part-of (e.g., “memory retrieval is a part of declarative memory”)
transformation-of (e.g., “consolidated memory is a transformation of encoded memory”)
preceded-by (e.g., “memory consolidation is preceded by memory encoding”)
We have excluded a set of spatial relations defined in the OBO Relational Ontology, because they only apply to entities that have a spatial location. We have also excluded a set of participation relations (has_participant, has_agent); these operators express relations between processes and continuants (things), and in the context of latent mental entities it is not yet clear how to conceptualize those constructs.
Relations between processes and tasks
In addition to the basic relations from the OBO Relational Ontology, we also define a measured by relation, which denotes the relation between a cognitive process and a particular contrast on a task [e.g., “conflict processing is measured by the contrast of (incongruent–congruent) in the Stroop task”]. This is meant to reflect the primary form of theoretical claim made by cognitive psychologists, namely that some particular task manipulation affects a particular mental process.
Relations among tasks
A second new relation introduced in CA is the descended-from relation, which represents historical and/or conceptual relations between tasks. It is clear that within a broad class of tasks (e.g., “Color–Word Stroop task”), there will be a large number of potential variations that could have functional implications, and these develop over time. In order to capture these relationships, we use the concept of “task phylogeny” (Bilder et al., 2009), which treats tasks according to a family tree in which tasks inherit particular features from earlier tasks. Thus, the descended-from relation reflects something like a biological inheritance relationship. In this case, “speciation” is determined by whether the resulting data are commensurate for meta-analysis. If they are not, then one task would be considered as derived from another, rather than being considered slightly different variants of the same task.
Literature relations
All of the entities in the CA knowledge base (including concepts, tasks, and relations) can be associated with literature citations, using a built-in interface to the PubMed literature database. The knowledge base also allows annotation of relations to specific citations, using the relations defined in the citation typing ontology (CiTO; Shotton, 2010). This ontology supports annotation that specifies whether particular citations support or refute a particular claim, as well as many other aspects of citation. Currently, we have only implemented a single literature relation, which is equivalent to the CITO “citesForInformation” relation.
Relations to observed data
The CA does not directly store data; instead, in order to support the annotation of relations between tasks and observed data, the system provides the ability to relate specific task contrasts to entities or data that are stored in external databases. These will include brain regions (as represented in databases of brain structure such as the Foundational Model of Anatomy or Brainmap), genes and genetic variants (as represented in dbSNP and EntrezGene), and cellular functions (as represented in GO).
Technical Infrastructure
The CA knowledge base is stored natively in a custom MySQL relational database. We chose to do this, rather than storing the knowledge natively in an ontology language or resource description format (RDF) triplestore, in order to maximize the flexibility with which the knowledge can be stored. Instead, we have developed a pipeline to generate an Web Ontology Language (OWL) ontology from the database; this allows us to expose the ontological knowledge in a standard format, while still retaining flexibility to store information that may not be easily represented in a formal ontology language. The OWL representation of the CA is available via the NCBO BioPortal4 and the Python code used to generate the OWL representation from the database dumps is available at https://github.com/poldrack/cogat.
In order to maximize the ability to interact directly and automatically with other projects, the CA project is built around a set of Semantic Web (Berners-Lee et al., 2001) technologies (Miller et al., 2010). First, the representation of every concept in the knowledge base is available in RDF, which is a format for representing semantic resources in a machine-understandable way. Second, the CA site exposes a web service known as a SPARQL endpoint, which allows direct queries of the knowledge base by humans or other computer systems and returns results from the knowledge base in a standards-based format that preserves conceptual relationships and valuable contextual information. Together, these services provide other projects with the ability to directly and effectively access the current state of the knowledge base. This allows substantially greater interoperability between systems representing different kinds of information, and supports the automation of such interactions based on common standards for interoperability and knowledge sharing.
In addition to the specific benefits for cognitive neuroscience, the infrastructure developed as part of the CA should also serve as a building block for other projects that aim to build collaborative knowledge bases. Given that the informatics community is still converging on standards for interoperability between projects, we hope that our demonstration of the effectiveness of Semantic Web technologies will provide further impetus for their use in such projects.
The web based interface utilizes HTML 5, CSS 3, and custom JavaScript to generate interactive features. It relies on the jQuery libraries and a large number of jQuery utility plugins. The server side software uses a standard LAMP stack (Linux, Apache, MySQL, and PHP). Graphviz is used for generating RDF visualizations, and Arc2/Semsol for RDF parsing and SPARQL libraries. The front end and back end communicate with AJAX techniques, passing JSON objects from the end-user page to the server and back, as well as with standard POST and GET requests. PDF support is provided by the wkhtmltopdf libraries. Infamous powers the RSS bubble visualizations with Feedburner generating the RSS feed, while PubBrain powers the fMRI imaging references. The PubMed Entrez API is used for bibliographic citations and abstract lookups.
Benefits and Challenges of Collaborative Knowledge Building
The Internet has made it possible to tap into the knowledge of people across the globe on an unprecedented scale. Hundreds of thousands of people have worked together to write the software that runs the Internet, write the largest encyclopedia in human history5, discover new stars and galaxies, and achieve many other goals that would be impossible through either humans or computers working alone. Such systems serve as both existence proofs and design models for collaborative knowledge building in science. For example, thousands of individuals may contribute to a single Wikipedia article, each with different knowledge and points of view, with results rivaling those of expert-written encyclopedias in quality (Giles, 2005) and vastly exceeding them in scope. However, the process through which such high-quality collaborative knowledge building happens is by no means a given, requiring the evolution of community norms, explicit rules, technological features, and significant effort spent on coordination and conflict resolution (Kittur and Kraut, 2008). Our goal in the CA project is to leverage the emerging understanding of crowd-driven collaborative knowledge building to develop a system for scientists that captures a wide variety of viewpoints and builds consensus across fields.
Capturing and resolving disagreement
Though seemingly simple at first glance, Wikipedia is a sophisticated engine for collaborative knowledge building with highly developed mechanisms for lowering participation costs, promoting collaboration, updating information, resolving conflict, and consolidating content. Over a third of all work in Wikipedia goes not to editing articles but instead to coordination activities such as debate about policies and procedures, maintenance activities such as deleting non-conforming pages, and negotiation about content and issues (Kittur et al., 2007). The very large and increasing amount of effort being spent on coordination emphasizes the need for a collaborative knowledge creation system to focus on supporting collaboration at least as much as supporting knowledge creation. Wikipedia supports collaboration through a wide variety of mechanisms. One of the simplest but most important is the presence of a “discussion” section on every page, which contains a record of all current and past conversations regarding a page separate from the content of the page itself. New users can view past discussions in order to take advantage of the information accrued in past conversations and avoid repeating past mistakes. The CA adds to this functionality to deal with relations, capturing discussion not only for concepts but also for relations between them. Furthermore, discussions are integrated directly into concept and relation pages, surfacing them and making them salient to readers as well as contributors. When discussion alone cannot resolve disagreement, concepts can be “forked” or merged as discussed further below.
Curation
Although completely open systems such as Wikipedia work well for general encyclopedic knowledge, some of the greatest successes in scientific knowledge building have come from curated models such as GO. The CA aims to strike a balance between the two extremes, with curation done on an as-needed basis by the core team and volunteer curators. However, curation will not be done in a vacuum; viewpoints and discussion from the community will be elicited for curation decisions, and decisions are expected to reflect the consensus of the community. At the same time, a guiding principle is that curatorial decisions should be “evidence-based” to ensure that questions about terminology, concepts, and relationships are not determined only by popular fiat. As the Atlas grows, it will likely need to develop more sophisticated procedures for dealing with curation (as have been developed in Wikipedia: Forte et al., 2009), which will be developed in collaboration with and by the scientific user community.
Utility of the Cognitive Atlas
The CA will provide a formalization of what are often implicit conceptual schemes in cognitive neuroscience, and in particular will make clear the mapping of particular task contrasts onto particular mental processes. We envision a number of ways in which the CA could impact cognitive neuroscience research.
Clearer vocabulary
The controlled vocabulary of the CA provides a way for researchers to use terms in a more precise way, and to help reduce polysemy, wherein the same term is used by different researchers to mean different things. This occurs surprisingly often within the neuroscience literature. An example is the term “working memory,” which has several distinct meanings in the literature, as discussed above. This can lead to complications in automated processing of the literature, since it is not possible to know which of these senses is implied in any particular usage.
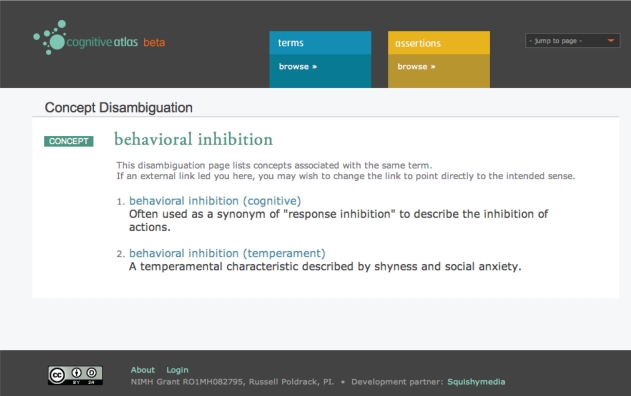
In the CA, each term begins with a single concept definition. Researchers who do not agree with this definition can discuss their disagreement using the built-in discussion feature, similar to the way that conflicts are resolved in Wikipedia. However, if it becomes clear from the discussion that there is an irreconcilable conceptual difference, the concept can be “forked,” in which case the original concept is broken into a number of senses, each of which would have its own separate concept page and participate separately in relations with tasks. For example, in the working memory example, the original concept “working memory” would be broken into separate senses, such as “working memory (maintenance),” “working memory (manipulation),” and “working memory (temporal),” with the original page being converted to a disambiguation page for the different senses. An example of such a page for the concept of “behavioral inhibition” is shown in Figure 3
Figure 3.
A screenshot of the disambiguation page for the concept of “behavioral inhibition,” which points to two separate senses of the term.
Conversely, it is also common that different terms are used to describe the same underlying processes. For example, the terms “declarative memory” and “explicit memory” are often used to refer to the same mental function. Within the CA, it is possible to specify terms as synonyms, such that subsequent analyses using the knowledge base will recognize the terms as synonyms. This is preferred to the merging of the concepts into a single concept, because it retains the original terms in the knowledge base while still noting them as referring to the same process.
Improved query expansion
The precision and recall of literature searches can be greatly improved by the use of ontological knowledge to guide the expansion of search queries. For example, PubMed currently expands queries using the MeSH lexicon, which (as discussed above) is not reflective of the state of the art. For example, the query (sublexical route) is expanded by PubMed as [sublexical[All Fields] AND (“drug administration routes”[MeSH Terms] OR (“drug”[All Fields] AND “administration”[All Fields] AND “routes”[All Fields]) OR “drug administration routes”[All Fields] OR “route”[All Fields])]. The confusion of MeSH regarding the meaning of “route” in this context leads to incorrect query expansion, whereas the use of a database that included “sublexical route” as a concept would more specifically target that particular phrase. In addition, it could potentially expand the query to include other related terms such as “phonological assembly,” leading to search results that are more likely to find relevant literature.
Metadata annotation and meta-analysis
The availability of large databases of neuroimaging data, particularly as the Brainmap.org database (Laird et al., 2005), has enabled powerful meta-analyses. However, the ability to perform meta-analysis is limited by the metadata that are associated with each data set; in order to assess which brain systems are associated with particular mental processes, the data need to be annotated using an ontology of mental processes. The Brainmap database currently uses a relatively coarse ontology of mental processes, which limits the ability to make finer assessments about structure–function associations (Poldrack, 2006). However, the availability of a more detailed cognitive ontology would provide the ability to perform such mappings. As an example, Poldrack et al. (2009) used a set of annotated task-process relations, along with latent variables identified from fMRI data obtained across a set of eight tasks, in order to identify which mental processes were mapped onto brain networks. With larger data sets annotated using a more detailed ontology, this kind of analysis could provide new insights into which ontological distinctions in the mental process ontology are biologically realized and which are not (Lenartowicz et al., 2010).
Theory testing
It is also possible to envision the use of meta-analysis with the CA to test larger theories of cognitive organization. For example, within the psychology of categorization there is a longstanding debate between theories that posit single processes underlying both categorization and recognition memory versus separate processes (Poldrack and Foerde, 2008). Each of these theories would make different ontological claims regarding cognitive processes and their relations with mental tasks. These ontological claims could potentially be translated into claims about the covariance structure in the data obtained on those tasks, and different theories could be compared for their relative fit to the data using covariance structure modeling methods. While the system is not currently able to support such theory testing, it remains an important goal for the future of the system.
Translational mental health research
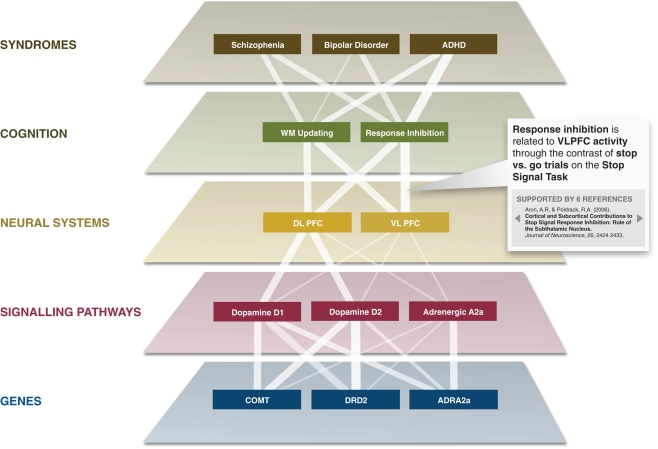
Within psychiatry, there is an increasing movement toward the use of dimensional rather than categorical approaches to characterizing psychiatric disorders (Kraemer, 2007). Ongoing efforts such as the NIH Research Domain Criteria project (Insel et al., 2010) aim to characterize these underlying dimensions in terms of their cognitive and neural bases. The social collaborative knowledge building tools that are provided by the CA offer the ability for such projects to interactively develop their knowledge base and annotate a rich set of links between cognitive processes and data at other levels, such as neural circuits, cellular signaling pathways, or genes. The CA will play an essential link in allowing relationships to be made between the neural level and the level of psychiatric symptoms and syndromes (see Figure 4).
Figure 4.
The Cognitive Atlas provides a framework for relating biological functions and processes to psychiatric symptoms and syndromes. The links between each level in this graph reflect proposed empirical relations; the strength of each link (noted by its width) is proportional to the literature association between each set of terms (defined as the Jaccard coefficient between the two search terms derived from PubMed). Each link can also be associated with specific empirical results, as noted in the box demonstrating a particular annotation for one of the edges.
Present Status and Future Plans
The development of the CA began in 2008. In the design phase, we analyzed the structure of the knowledge to be represented and developed an initial schema for the database. In the initial implementation phase, we worked with the development team to implement the basic functionality for presenting and editing the knowledge base (Miller et al., 2010), and also worked with a small number of investigators to begin to populate the database and refine the interaction design for the site. In the current phase, we are continuing to implement new features as well as refining the existing interface, with a particular focus on scaling to larger amounts of content. We have also begun soliciting open contributions from researchers across the field. The CA currently has entries for 904 terms, including 708 mental constructs, and 196 tasks, with definitions present for 795 of these terms; most of these definitions would not be viewed as sufficient by an expert in the area, but are provided as a starting point for those experts to edit and refine. To date the database has fewer than 900 relations specified; whereas the first phase of the project focused on concept definitions, a major goal in the next phase of the project will be to enlist a wide range of researchers to contribute their knowledge of these relations.
Beyond the addition of content, future development of the site will focus on three areas. First, we plan to add personalization features to allow users to keep better track of relevant information. This will include tracking of their own contributions, tracking of recent changes to topics of interest, and recommendation of content and/or publications that are relevant to the user’s interests (based on their past contributions). Second, we plan to include content based on mining of the published literature. For example, we might include relations in the database that are based on association between terms in the published literature, which could then serve as input to the manual annotation process. Third, we plan to implement a greater degree of integration with other databases. The CA lexicon already is integrated with free web services enabling: mining of literature associations (PubAtlas6); mapping associations of literature with a three-dimensional probabilistic atlas of brain structure (PubBrain7); and collaborative entry of quantitative annotations for meta-analysis of findings from cognitive studies (PhenoWiki8). While the CA currently represents the structure of tasks in a relatively coarse way, the CogPO (Turner and Laird, 2011; see text footnote 2) project is currently developing much more detailed task ontologies. As those become available we will link directly to them, providing a much more thorough way of modeling the fine-grained details of tasks. The CA lexicon will be included in a future version of the NeuroLex database9, and we will also implement greater integration biological ontologies such as the GO (Ashburner et al., 2000).
Finally, it is important to be clear that the CA itself will not contain any data, but we plan to link to empirical databases from within the CA. In the short term, this will include direct links to coordinate-based neuroimaging databases with exposed APIs such as the NeuroSynth project (Yarkoni et al., 2011) or SumsDB10, which can provide automated access to meta-analytic empirical data linking the concepts in the database to brain systems. Currently, the NeuroSynth database includes forward and reverse inference maps for all concept terms in the CA. In addition, if other databases become available (e.g., genome-wide association data, lesion mapping data, patient behavioral data, etc.) these can also be linked directly to the database. It should also be noted that while the CA is currently focused on concepts from the human psychology literature, it will also be important to encompass concepts from non-human animal literatures as well, in order to provide truly systematic coverage of the literature.
Conclusion
The mapping of mental processes to brain systems has to date relied largely upon informal representations of mental processes and the tasks that are used to manipulate them. We propose that this approach is fundamentally limited, and that continued scientific progress in cognitive neuroscience will require the development and adoption of formal knowledge bases that provide a more systematic explication of cognitive theories and their relation to empirical data. The CA aims to provide such a resource that reflects the views of the entire community, and we welcome the contribution of interested researchers through the cognitiveatlas.org web site. By including the contributions of researchers across the field, we hope that the CA can become the standard ontology for mental function.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by NIH grants RO1MH082795 (to Russell A. Poldrack), and the Consortium for Neuropsychiatric Phenomics [NIH Roadmap for Medical Research grants UL1-DE019580, PL1MH083271 (to Robert M. Bilder) and RL1LM009833 (to D. Stott Parker)]. Thanks to Rajeev Raizada for helpful comments on a draft of this paper.
Footnotes
1By “cognitive” we mean to refer to mental processes very broadly, which we take to include domains such as emotion or motivation that have historically been distinguished from cognition.
References
- Anderson J. R., Bothell D., Byrne M. D., Douglass S., Lebiere C., Qin Y. (2004). An integrated theory of the mind. Psychol. Rev. 111, 1036–1060 10.1037/0033-295X.111.4.1036 [DOI] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000). Gene ontology: tool for the unification of biology. the gene ontology consortium. Nat. Genet. 25, 25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baddeley A. (1992). Working memory. Science 255, 556–559 10.1126/science.1736359 [DOI] [PubMed] [Google Scholar]
- Bard J. B. L., Rhee S. Y. (2004). Ontologies in biology: design, applications and future challenges. Nat. Rev. Genet. 5, 213–222 10.1038/nrg1295 [DOI] [PubMed] [Google Scholar]
- Bechhofer S., Miles A. (2009). SKOS Simple Knowledge Organization System Reference. W3C recommendation, W3C. Available at: http://www.w3.org/TR/2009/REC-skos-reference-20090818/
- Berners-Lee T., Hendler J., Lassila O. (2001). The semantic web. Sci. Am. 284, 34–4311396337 [Google Scholar]
- Bilder R. M., Sabb F. W., Parker D. S., Kalar D., Chu W. W., Fox J., Freimer N. B., Poldrack R. A. (2009). Cognitive ontologies for neuropsychiatric phenomics research. Cogn. Neuropsychiatry 14, 419–450 10.1080/13546800902787180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boring E. G. (1950). A History of Experimental Psychology, 2nd Edn Englewood Cliffs, NJ: Prentice Hall [Google Scholar]
- Bowden D. M., Dubach M. F. (2003). Neuronames 2002. Neuroinformatics 1, 43–59 10.1385/NI:1:1:043 [DOI] [PubMed] [Google Scholar]
- Cohen J. D., Dunbar K., McClelland J. L. (1990). On the control of automatic processes: a parallel distributed processing account of the stroop effect. Psychol. Rev. 97, 332–361 10.1037/0033-295X.97.3.332 [DOI] [PubMed] [Google Scholar]
- Donohoe G., Corvin A., Robertson I. H. (2006). Evidence that specific executive functions predict symptom variance among schizophrenia patients with a predominantly negative symptom profile. Cogn. Neuropsychiatry 11, 13–32 10.1080/13546800444000155 [DOI] [PubMed] [Google Scholar]
- Edelman G. M. (1989). The Remembered Present: a Biological Theory of Consciousness. New York: Basic Books [Google Scholar]
- Forte A., Larco V., Bruckman A. (2009). Decentralization in Wikipedia governance. J. Manag. Inf. Syst. 26, 49–72 [Google Scholar]
- Giles J. (2005). Internet encyclopaedias go head to head. Nature 438, 900–901 10.1038/438136b [DOI] [PubMed] [Google Scholar]
- Goldman-Rakic P. S. (1995). Cellular basis of working memory. Neuron 14, 477–485 10.1016/0896-6273(95)90304-6 [DOI] [PubMed] [Google Scholar]
- Gruber T. (1993). A translation approach to portable ontology specifications. Knowl. Acquisition 5, 199–220 [Google Scholar]
- Insel T., Cuthbert B., Garvey M., Heinssen R., Pine D. S., Quinn K., Sanislow C., Wang P. (2010). Research domain criteria (rdoc): toward a new classification framework for research on mental disorders. Am. J. Psychiatry 167, 748–751 10.1176/appi.ajp.2010.09091379 [DOI] [PubMed] [Google Scholar]
- Kane M. J., Conway A. R. A., Miura T. K., Colflesh G. J. H. (2007). Working memory, attention control, and the n-back task: a question of construct validity. J. Exp. Psychol. Learn. Mem. Cogn. 33, 615–622 10.1037/0278-7393.33.3.615 [DOI] [PubMed] [Google Scholar]
- Kittur A., Kraut R. E. (2008). “Harnessing the wisdom of crowds in wikipedia: quality through coordination,” in CSCW 2008: Proceedings of the ACM Conference on Computer-Supported Cooperative Work (New York: ACM Press). [Google Scholar]
- Kittur A., Suh B., Pendleton B. A., Chi E. (2007). “He says, she says: conflict and coordination in wikipedia,” in CHI 2007: Proceedings of the ACM Conference on Human-factors in Computing Systems (New York, NY: ACM Press). [Google Scholar]
- Kraemer H. C. (2007). Dsm categories and dimensions in clinical and research contexts. Int. J. Methods Psychiatr. Res. 16(Suppl. 1), S8–S15 10.1002/mpr.211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird A. R., Lancaster J. L., Fox P. T. (2005). Brainmap: the social evolution of a human brain mapping database. Neuroinformatics 3, 65–78 10.1385/NI:3:1:065 [DOI] [PubMed] [Google Scholar]
- Laird J., Newell A., Rosenbloom P. (1987). Soar: an architecture for general intelligence. Artif. Intell. 33, 1–64 10.1016/0004-3702(87)90050-6 [DOI] [Google Scholar]
- Lenartowicz A., Kalar D., Congdon E., Poldrack R. A. (2010). Towards an ontology of cognitive control. Top. Cogn. Sci. 2, 678–692 [DOI] [PubMed] [Google Scholar]
- Martone M. E., Gupta A., Ellisman M. H. (2004). E-neuroscience: challenges and triumphs in integrating distributed data from molecules to brains. Nat. Neurosci. 7, 467–472 10.1038/nn1229 [DOI] [PubMed] [Google Scholar]
- Miller E., Seppa C., Kittur A., Sabb F., Poldrack R. A. (2010). The cognitive atlas: employing interaction design processes to facilitate collaborative ontology creation. Nat. Proc. [Epub ahead of print]. [Google Scholar]
- Müller H.-M., Kenny E. E., Sternberg P. W. (2004). Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2, e309. 10.1371/journal.pbio.0020309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olton D. S., Becker J. T., Handelmann G. E. (1979). Hippocampus, space, and memory. Behav. Brain Sci. 2, 313–365 10.1017/S0140525X00062713 [DOI] [Google Scholar]
- Poldrack R. A. (2006). Can cognitive processes be inferred from neuroimaging data? Trends Cogn. Sci. (Regul. Ed.) 10, 59–63 10.1016/j.tics.2005.12.004 [DOI] [PubMed] [Google Scholar]
- Poldrack R. A., Foerde K. (2008). Category learning and the memory systems debate. Neurosci. Biobehav. Rev. 32, 197–205 10.1016/j.neubiorev.2007.07.007 [DOI] [PubMed] [Google Scholar]
- Poldrack R. A., Halchenko Y. O., Hanson S. J. (2009). Decoding the large-scale structure of brain function by classifying mental states across individuals. Psychol. Sci. 20, 1364–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price C., Friston K. (2005). Functional ontologies for cognition: the systematic definition of structure and function. Cogn. Neuropsychol. 22, 262–275 10.1080/02643290442000095 [DOI] [PubMed] [Google Scholar]
- Ruths T., Ruths D., Nakhleh L. (2009). Gs2: an efficiently computable measure of go-based similarity of gene sets. Bioinformatics 25, 1178–1184 10.1093/bioinformatics/btp128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabb F. W., Bearden C. E., Glahn D. C., Parker D. S., Freimer N., Bilder R. M. (2008). A collaborative knowledge base for cognitive phenomics. Mol. Psychiatry 13, 350–360 10.1038/sj.mp.4002124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shotton D. (2010). Cito, the citation typing ontology. J Biomed. Semantics 1(Suppl. 1), S6. 10.1186/2041-1480-1-S1-S6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B., Ceusters W., Klagges B., Köhler J., Kumar A., Lomax J., Mungall C., Neuhaus F., Rector A. L., Rosse C. (2005). Relations in biomedical ontologies. Genome Biol. 6, R46. 10.1186/gb-2005-6-9-119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg S. (1969). The discovery of processing stages: Extensions of donders’ method. Acta Psychol. (Amst.) 30, 276–315 10.1016/0001-6918(69)90055-9 [DOI] [Google Scholar]
- Turner J. A., Laird A. R. (2011). The cognitive paradigm ontology: design and application. Neuroinformatics. Available at: http://www.springerlink.com/content/w23070351h220513/ [DOI] [PMC free article] [PubMed]
- Yarkoni T., Poldrack R. A., Nichols T. E., Van Essen D., Wager T. (2011). Large-scale automated synthesis of human functional neuroimaging data. Nat. Methods 8, 665–670 [DOI] [PMC free article] [PubMed] [Google Scholar]