Abstract
High-throughput screening data repositories, such as PubChem, represent valuable resources for the development of small molecule chemical probes and can serve as entry points for drug discovery programs. While the loose data format offered by PubChem allows for great flexibility, important annotations, such as the assay format and technologies employed, are not explicitly indexed. We have previously developed a BioAssay Ontology (BAO) and curated over 350 assays with standardized BAO terms. Here we describe the use of BAO annotations to analyze a large set of assays that employ luciferase- and β-lactamase-based technologies. We identified promiscuous chemotypes pertaining to different sub-categories of assays and specific mechanisms by which these chemotypes interfere in reporter gene assays. Our results show that the data in PubChem can be used to identify promiscuous compounds that interfere non-specifically with particular technologies. Furthermore, we show that BAO is a valuable toolset for the identification of related assays and for the systematic generation of insights that are beyond the scope of individual assays or screening campaigns.
Keywords: compound promiscuity, assay ontology, reporter gene assays, high-throughput screening data analysis, cheminformatics
INTRODUCTION
The field of high throughput screening (HTS) is rapidly advancing through the development of sophisticated robotics and liquid handling systems, sensitive and versatile detection technologies, and powerful informatics systems that enable miniaturization and increased throughput.1 Furthermore, HTS is being used to interrogate increasingly complex biological systems and processes, driven by advancements in molecular and cellular biology in combination with innovative assay designs.
In an effort to find novel entry points for drug discovery programs, countless HTS campaigns comprising large commercial and proprietary compound libraries have produce massive data sets – primarily in pharmaceutical companies. The NIH Molecular Libraries Roadmap Initiative2 and the availability of more affordable “out of the box” screening systems and reagents have facilitated a dissemination of HTS capabilities into academic institutes and universities, where they are now relatively common and available to researchers.
HTS datasets, which consist of experimental results and assay metadata, are typically stored in data warehouses using relational database schemas.3;4 The fast pace of innovation in assay designs and detection technologies, as well as the increasing complexity of the biological targets under investigation, pose challenges to “static” database schemas to capture and manage the diversity of screening experiments and their outcomes. To optimize the value of HTS efforts beyond any individual HTS campaign and to facilitate more informed decision-making as compounds progress in the value chain, systematic knowledge management is receiving increased attention from informatics organizations.5 In this context, a formal, well-structured, knowledge-based, and extensible description of biological assays is required. Expert biocuration to organize and annotate existing data is also a critical component of any HTS knowledge management solution.
PubChem is a public repository of HTS assay descriptions, small molecule compounds, and HTS results (which we refer to as endpoints).6;7 Originally put in place as part of the Molecular Libraries Program (MLP), it serves to host data generated at the MLP centers as well as that from other NIH funded projects. As of September 2010, there were over 2,100 bioassays from the MLP deposited in PubChem. In addition to PubChem, there are several other publically available sources of screening data, including, ChEMBL8, which contains structure activity relationship (SAR) data curated from the medicinal chemistry literature; the Psychoactive Drug Screening Program (PDSP);9;10 and ChemBank.11;12 In addition, private resources, such as Collaborative Drug Discovery (CDD),13;14 also make large screening data sets publically accessible.
Despite recommendations from industry and government work groups, there is currently no agreed upon standard for the representation of HTS assay data. Such a representation is vital for researchers to meaningfully interpret and compare diverse assay results.15 Because HTS data repositories lack detailed annotations using standardized terms, seemingly trivial queries such as “list the biochemical vs. cell-based assays”, or “list assays that use a luciferase reporter construct” are not possible. In addition, the lack of a formal description of biological assays hinders the integration of HTS data from different sources as well as with other life science databases (e.g. biological pathways).
PubChem’s already large and diverse set of deposited assay results along with several other accessible screening data repositories form a large corpus of data that can serve as a starting point to develop a systematic categorization of HTS assays. The exponential growth of public data repositories indicates that we are only beginning to explore the space of possible assay designs. The development of a clearly structured and standardized formal description of concepts that are relevant to interpreting HTS results is therefore very timely.
In this report we demonstrate how such a formalized terminology can facilitate analyses across multiple diverse assays to identify promiscuous compounds. These compounds are traditionally problematic for HTS and it is desirable to identify them as early as possible in a campaign. Compound promiscuity can be related to an assay technology, detection method or interaction with biological targets and often the specific mechanisms of action are not fully understood. There have been attempts at the identification of compound classes that can interfere with specific assay technologies, but these studies are usually focused on a small number of biological assays and did not make use of the large numbers of data sets currently available.16; 17 Here we attempt for the first time to identify promiscuous behavior on a large- scale using a curated data set that allowed us to interrogate compound behavior across certain assay categories and sub-categories.
METHODS
PubChem local mirror database and chemical structures
A local relational mirror of the PubChem bioassay database was created using in-house scripts and a public version of the MySQL database. The details of this database, schema, population and update processes, implementation, and code are reported elsewhere.18 Briefly, the database consisted of several tables, including assay details (such as AID, assay name, description, project category, protocol), panel assay specifications, result definitions (such as IC50, % inhibition or any other observed measurement or statistics), result data (with PubChem Activity Outcome and Score and the most important results, such as IC50, and qualifiers such as <,>,=), cross references (links of assays to other NCBI databases such as protein or nucleotide target, PubMed, taxonomy), and relationships (links between different assays, links to other NCBI Entrez databases, and links between targets and their sequences). The system utilized the PubChem FTP site to access XML assay descriptions and CSV assay data and the NCBI Entrez Utilities (eUtils) to access additional information (including if an assay had changed) to keep the mirror database current. The database included a structure table only as a placeholder. Chemical structures corresponding to the assay data were downloaded by Substance IDs (SIDs) directly from PubChem as SDFiles using the batch download facility.
PubChem Assay Annotation and Assay Clustering
PubChem assays were annotated manually using the mirror database described above, which was fetched from PubChem in April 2010 with 2,299 assays by AID. 172 assays had no data at all (on hold assays). There were 194 summary assays, of which 136 had no substances or activity data. These assays were not considered for annotation. There were 105 assays with no activity outcome method (which is usually assigned as screening, confirmatory, other, or summary) - these are from Ambit Biosciences, DTP/NCI, and SGCO. 1,498 assays were from the screening centers of the NIH Molecular Libraries Probe Center Network (MLPCN) and former Molecular Libraries Screening Center Network (MLSCN) – not including assays without data (on-hold) and summary assays.
To aid the manual annotation process, all assays were clustered based on the assay title, description, protocol, and source. Several assays (other than on hold or summary assays) did not have a protocol or only a minimal description, but all had information about the source. To cluster the assays, first for each assay a text fingerprint was generated from all words used in title, description, procedure, and source after stemming (to consolidate different grammatical forms) using the Pipeline Pilot 8.0 (Accelrys)19 text analytics component collection. The text fingerprints (TXFP_Custom) encode for each individual assay the presence and absence of word tokens from the global corpus of assays. The assay “documents” were then clustered based on the fingerprints using the Tanimoto similarity metric and setting the average cluster size to 5 members. The clustering method is a relocation technique based on maximum dissimilarity partitioning implemented in the Pipeline Pilot text analytics collection. A total of 460 clusters were generated. This method grouped together similar assays very effectively; for example all assays of the same screening campaign by center or assays with the same procedure or assay design (for example many NCGC toxicity assays); as expected, clusters usually included assays from the same source. The method also grouped together related assays with minimal annotations (such as many of the NCI or Chembank assays), summary assays, or assays that were on hold. 299 clusters were generated from the 1,498 MLPCN and MLSCN assays that had data deposited and were not summary assays. To illustrate the similarity relationships of these assays we generated a minimum spanning tree (MST) based on the pair-wise (Tanimoto) similarities of the assays computed from their text fingerprints (the same similarities that were used for clustering above). The MST was computed using an in-house protocol implementing Kruskal’s algorithm. The MST was visualized in Cytoscape20 and is shown in Supporting Figure S1-A. Supporting Figure S1-B and C show the clusters and assays memberships for biochemical and cell-based assays respectively. Assay formats were mapped onto the tree after manual annotation (see below).
Following cluster pre-processing, assays were then manually annotated by assay format, assay technology, and the other BAO categories. The BAO schema with classes, individuals, relationships and their definitions can be downloaded from our website and BAO can also be visualized there.21 For the limited analysis presented here, we focused specifically on assays based on designs to detect luminescence from the luciferase-catalyzed conversion of luciferin substrates22 and assays employing β-lactamase-based technology.23
Luciferase-assays were classified into five sub-categories: reporter-gene, viability, ATP-coupled, luciferin-coupled, and luciferase enzyme activity assays. Briefly, luciferase reporter-gene assays use the luciferase gene downstream of a promoter of interest. The amount of luciferase expressed was quantified by the intensity of light (luminescence) produced in the presence of substrates, ATP and luciferin. Viability assays estimate the proportion of living cells in an assay by measurement of ATP content in a luciferase catalyzed reaction. ATP-coupled assays measure the residual amount of ATP (for example after a kinase reaction) by a coupled luciferase reaction. Luciferin-coupled assays measure the amount of luciferin generated after detoxification by cytochrome P450 enzyme activity. Luciferase enzyme activity assays quantify the luciferase enzyme activity by the amount of light produced in a biochemical reaction. β-lactamase technology is used in either reporter-gene or enzyme activity assays.
PubChem Promiscuity Index (PCIdx)
The PubChem Promiscuity Index (PCIdx) of a substance (by SID) was defined as the number of assays in which this substance is active divided by the number of assays in which it was tested (equation I, where N is the assay count for the substance).
| (I) |
To define “active” we used the PubChem Activity Outcome, which is one of the required fields to be uploaded by the assay depositor. Activity Outcome categorizes tested samples as active, inactive, inconclusive, or unspecified. PubChem does not have rules when to apply the outcome category “active”, which is defined (subjectively) by the depositor. Therefore “active” can have different meanings across different assays. This is clearly not the best way of comparing compounds in a large number of assays and it would be much better to standardize the most important endpoints across all assays. However, currently activity outcome is one of the only two required endpoints (the other one is activity score - also subjectively depositor defined) and therefore the only way to quickly identify “active” compounds.
To compute PCIdx for each compound, all assays in which it was tested and the corresponding activity outcomes were determined by querying the PubChem mirror database above. PCIdx was calculated according to equation I, separately for single concentration assays (PubChem activity outcome method “screening”) and concentration-response assays (activity outcome method “confirmatory”). Only assays of a certain category were considered, for example all luciferase technology assays or a certain subset thereof, such as viability assays or luciferase enzyme inhibition assays.
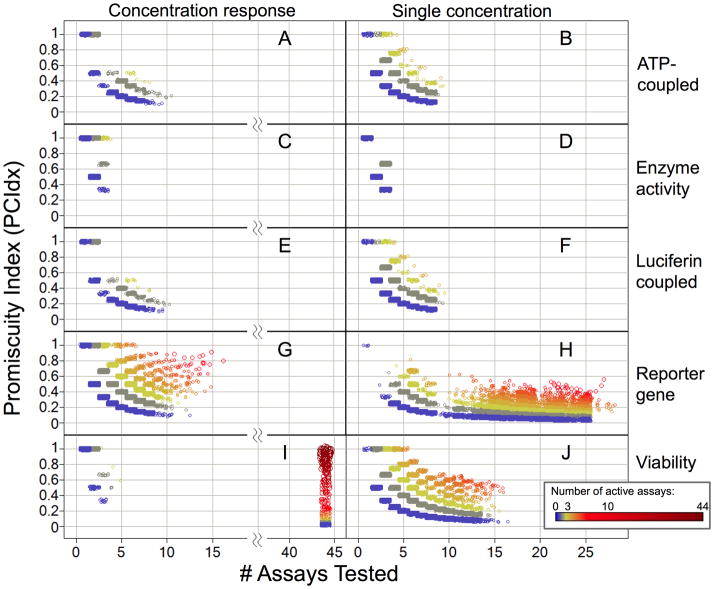
Because the significance of the PCIdx measure increases with more tested assays, we visualized compounds’ promiscuities by plotting PCIdx over the number of assays tested while indicating the number of active assays by a color code (compare Figures 1, S4, S6).
Figure 1.
Compound promiscuity by luciferase assay technologies. For each compound the Promiscuity Index vs. number of tested assays are depicted. A, B: ATP-coupled enzyme activity (e.g. kinase activity, not viability); C, D: luciferase enzyme activity; E, F: luciferin-coupled enzyme activity (e.g. P450); G, H Luciferase reporter gene assays; I, J: cell viability assays (ATP-coupled). A, C, E, G, I: concentration-response assays; B, D, F, H, J: single concentration assays. Color and size indicate the number of assays (of the particular luciferase assay type) in which a compound was active. 87,615 data points with at least one active assay shown: A: 3,457, B: 5,619, C: 2,313, D: 3,646, E: 3,457, F: 5,619, G: 14,200, H: 36,685, I: 1,413, J: 11,206.
Figures 1, S4, S6 and the heat map Figure 3 were created in TIBCO Spotfire DecisionSite.24
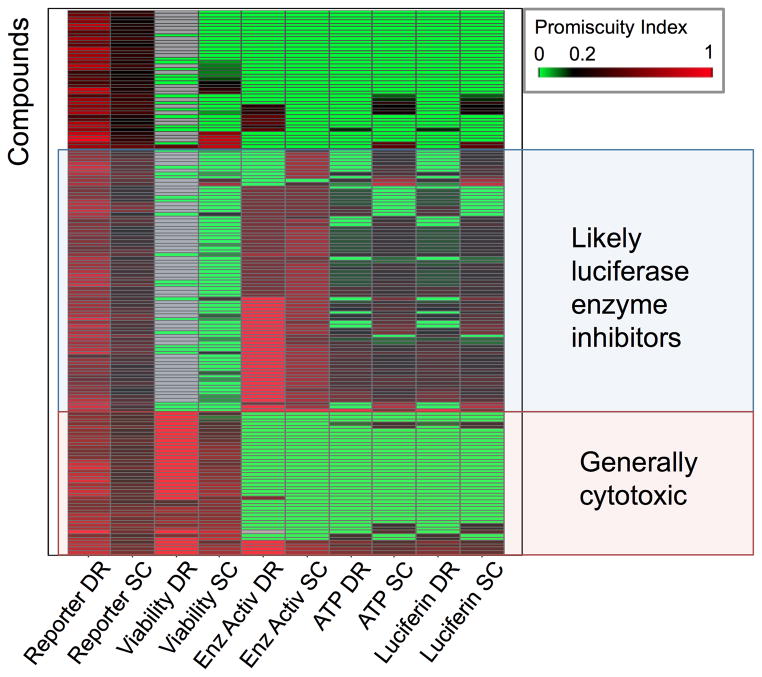
Figure 3.
Heat map of 161 most promiscuous compounds in luciferase reporter gene assays, which are active in at least 5 concentration-response and 5 single concentration (luciferase reporter) assays. DR and SC denote “dose response” and “single concentration”, respectively. Shown are the promiscuity indices of all compounds in the different luciferase assay categories for both concentration-response and single concentration assays, respectively, clustered by their PCIdx profiles. Two groups of promiscuous reporter gene compounds were apparent, suggesting the mechanism for reporter gene assay promiscuity: one in which compounds were also active in viability assays (red shade) and the other where compounds were also active in luciferase enzyme assays (blue shade). Compare Supporting Table S1 for details.
Data Clustering
Data in Figure 3 (corresponding to Table S1) were hierarchically clustered using the unweighted pair group method with arithmetic mean (UPGMA) and PCIdx correlation as similarity measure.
Chemical Structure Clustering
Chemical structures were clustered by maximum common substructure using ChemAxon Library MCS.25
Chemical Structure Similarities
Compound pair-wise similarities and the similarity matrix were computed using extended connectivity atom type fingerprints of length 4 (ECFP4)26 and the Tanimoto metric implemented in Pipeline Pilot 8.19
RESULTS
BioAssay Ontology and Assay Annotations
We have developed a BioAssay Ontology (BAO)21 to facilitate analyses of screening results from large and diverse sets of biological assays spanning multiple technologies and originating from different sources. The BAO project seeks to develop a formal, extensible, knowledge-based description of biological assays by making use of descriptive logic based features of the Web Ontology Language (OWL). Expert curation is an important component of the BAO project, and we have been systematically annotating sets of PubChem BioAssays with BAO terms describing assay concepts. The BAO project will also provide software tools to query and explore data sets in the context of the ontology.
The BioAssay Ontology describes several concepts related to biological screening, including Perturbagen, Target, Format, Assay Design, Detection Technology, and Endpoint, including endpoint data manipulation. Perturbagens deposited in PubChem and the other screening data sources mentioned above are mostly small molecules, but can include various other perturbing agents that are screened in an assay. We refer to targets as “Meta Target” describing not just protein targets, but also pathways, biological processes or events, etc. targeted by the assay. Format describes the biological or chemical features common to each test condition in the assay and includes biochemical, cell-based, organism-based, and variations thereof. Assay Design describes the assay methodology and implementation of how the perturbation of the biological system is translated into a detectable signal. Detection Technology relates to the physical method and technical details to detect and record a signal. Endpoints are the final HTS results as they are usually published (such as IC50, percent inhibition, etc.). Endpoint data manipulation specifies how the raw signal(s) are transformed into reported endpoints (i.e. normalization, correction, etc.). BAO also captures other assay properties such as assay purpose and how assays are related in campaigns. BAO is also designed to handle multiplexed assays. All main BAO components include multiple levels of sub-classes and specification classes, which are linked via object property relationships forming a knowledge representation. The details of the development and description of BAO will be reported elsewhere. The BAO schema with classes, individuals and relationships can be downloaded from our website.21 BAO classes, their subsumption hierarchies and class definitions can also be visualized directly on the BAO website.21
We annotated a set of over 350 PubChem assays and grouped them into related classes by assay technology and detection method. Specifically, we focused on widely used HTS assay technologies that employ luciferase- and β-lactamase-based reporters.22 By analyzing the outcomes of related assays we could readily identify compounds of interest, for example those that were promiscuously active in one or multiple classes of assays. The luciferase assays were annotated and classified into sub-categories that relate to assay design (described in methods). To efficiently annotate assays and to facilitate data analysis across all PubChem assays, we created a local mirror of the PubChem database. This database stores assay descriptions and endpoints in a relational format and can be queried easily using SQL. Mirrored assays were then manually annotated with BAO terms after interpreting the textual descriptions available in PubChem. To aid in the assay annotation process, we clustered the assays based on text fingerprints derived from the free text in assay title, description, protocol, and source (see methods). Supporting Figure S1 illustrates the similarity relationships based on their textual descriptions in PubChem of the 1,498 MLPCN and MLSCN assays and the clusters that were obtained as well as the most important formats (biochemical and cell-based) and the screening center. Figure S1-A shows the minimum spanning tree of the assays (see methods) illustrating that assays from the same center and assays of the same format typically group together locally. Figure S1-B and C show the cluster memberships (see methods). Each cluster contained only assays from one center and most clusters did only contain assays of one format. Clusters of assays also typically were of related designs and biological targets (not shown). In our hands this was an effective method to group similar assays together enabling the sequential annotation of sets of related assays. We found that this greatly reduced errors and accelerated the annotation process compared to random or chronological (by Assay ID: AID) order of annotation.
Analysis of luciferase technology assays
Using the local relational database created from data as of April 2010 we identified a total of 257 assays using a design based on the luciferase-induced conversion of luciferin substrates that results in the emission of light.22 Specifically we annotated the following types of luciferase-technology assays: reporter gene assays (105), cell-viability assays (through detection of ATP, 82), ATP-coupled (other than viability assays, 35), luciferin-coupled assays (23), and enzyme (biochemical) activity assays (12). A histogram of assay types is shown in Supporting Figure S2. We also identified and annotated the assay kits (Supporting Figure S3).
Using the luciferase assay annotations, we computed promiscuity statistics for each compound that was tested in any of the luciferase assays. We developed a Pipeline Pilot (Accelrys Inc.) protocol that queries the relational database to determine how many different assays (of a luciferase-technology category) each compound was tested in and in how many it was found active. This was done separately for single concentration and concentration-response assays. To define active and inactive we used the PubChem activity outcome endpoint. Although this is a subjective, “local” definition (each depositor can define “active” and “inactive” for each assay independently), we found it a useful first approximation. We calculated a PubChem Promiscuity Index (PCIdx) for each category as the quotient of the number of luciferase assays in which a substance was reported as active and the number of assays in which it was tested (see methods, equation I). The larger the ratio of active luciferase assays to assays tested, the higher a compound’s promiscuity PCIdx. However, the significance of this promiscuity measure increases with the number of assays tested. We therefore visualized promiscuity by a scatter plot of PCIdx and the number of assays tested, while also indicating the number of active assays (of each category) by color. Figure 1 shows compound promiscuities for the different luciferase technology categories for single concentration and concentration-response assays (87,615 data points shown overall). It shows a large number of promiscuous compounds identified from viability and reporter gene assays, which we decided to investigate in greater detail. Supporting Figure S4 illustrates promiscuities across all luciferase technology assays taken together.
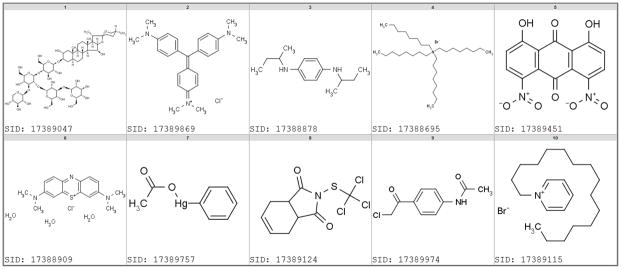
The majority of viability assays were concentration-response series deposited by the National Center for Chemical Genomics (NCGC). Figure 2 shows the most promiscuous cytotoxic compounds (by Substance ID: SID) identified by these assays. We have previously demonstrated that such data can be useful to model acute animal toxicity.27 All of the compounds shown have been tested in 44concentration-response assays and were categorized as active in more than 95 % of them. For example, the toxicity of digitonin (SID 17389047) is related to its lipid (membrane) solubilizing properties. Most of the compounds are chemically reactive, which is likely the cause of their toxicity. Crystal violet (hexamethyl-p-rosaniline chloride, SID 17389869) and methylene blue (SID 17388909) are redox-active and electrophilic dyes, respectively. 17388695, 17389115 are surfactants (phase transfer reagents), 17389451 is a reactive dihydroxyanthraquinone, 17389124 is used as pesticide, and 17389974 is an alkylator.
Figure 2.
Examples of highly promiscuous (cytotoxic) compounds in luciferase viability assays. All compounds have a promiscuity index between 0.95 to 1.0, were tested in 44 assays, and are active in at least 42 assays.
Although luciferase is often used in viability assays, its most common application is in reporter gene assays. To investigate promiscuous compounds in this category, we retrieved all substances that were active in at least 5 single-concentration and 5-concentration-response luciferase reporter gene assays. Figure 3 illustrates the promiscuity indices (PCIdx) of the 161 compounds in each of the luciferase assay categories for dose-response (DR) and single-concentration (SC) assays after hierarchical clustering (see methods). There are two major clusters of compounds. In one group, the compounds were also highly promiscuous across viability assays. This could be expected, since broadly cytotoxic compounds should also show up as actives in luciferase reporter gene assays. Importantly, this pattern was immediately revealed by our analysis method, which utilized activity outcomes across all assays of each category in which a compound had been tested. In the other group, compounds also showed promiscuity in the category of luciferase enzyme inhibition assays. It is therefore likely that the mechanism responsible for their promiscuity across reporter gene assays is the inhibition the luciferase enzyme. Most of those compounds also showed high promiscuity indices in the other categories of luciferase assays. Supporting Table S1 lists all 161 compounds corresponding to Figure 3 (by SID), their PCIdx values for each category and the number of assays in which each compound was active vs. the number of assays in which it was tested.
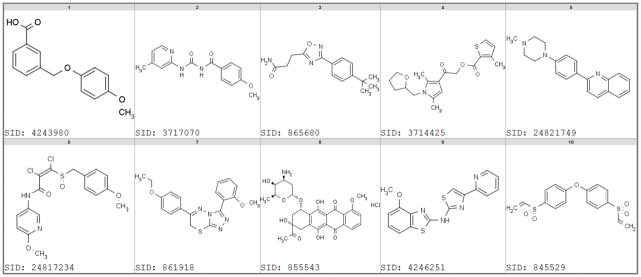
Figure 4 and Table 1 illustrate example chemical structures of both categories of highly promiscuous reporter gene compounds. The first row in Figure 4 and the first 5 entries in Table 1 show selected compounds that likely act via inhibiting luciferase enzyme. They represent 5 different chemical classes including the benzoyl-aryl-urea (SID 3717070), or the 3,5-disubstituted-1,2,4-triazole (SID 865680) scaffolds.16 The second row of Figure 4 and entries 6 to 10 in Table 1 show cytotoxic compounds that were broadly active across cell proliferation assays. They include reactive compounds such as electron-deficient vinyl chloride (SID 24817234) and Michael acceptor (SID 845529), and Daunorubicin (SID 855534), which is a DNA intercalator used as chemotherapeutic.
Figure 4.
Selected examples of promiscuous compounds in luciferase reporter gene assays of two categories. Top row compounds were also active in luciferase enzyme inhibition assays. Bottom row compounds were active in viability assays. Refer to Table 1 for details.
Table 1.
Promiscuity indices (PCIdx) and number of assays tested and found active for selected promiscuous compounds in luciferase reporter gene assays. This table Corresponds to compounds in Figure 4. The top 5 compounds are luciferase enzyme inhibitors, the bottom 5 are cytotoxic.
| SID | Concentration Response | Single Concentration | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reporter Gene Assays | Viability Assays | Enzyme Activ. Assays | Reporter Gene Assays | Viability Assays | Enzyme Activ. Assays | |||||||||||||
| PCIdx | Active | Tested | PCIdx | Active | Tested | PCIdx | Active | Tested | PCIdx | Active | Tested | PCIdx | Active | Tested | PCIdx | Active | Tested | |
| 4243980 | 0.86 | 12 | 14 | 0.50 | 1 | 2 | 0.54 | 7 | 13 | 0.50 | 1 | 2 | ||||||
| 3717070 | 0.75 | 9 | 12 | 1.00 | 1 | 1 | 0.36 | 8 | 22 | 0.50 | 1 | 2 | ||||||
| 865680 | 0.81 | 13 | 16 | 0.33 | 1 | 3 | 0.36 | 8 | 22 | 0.00 | 0 | 2 | ||||||
| 3714425 | 0.69 | 9 | 13 | 1.00 | 2 | 2 | 0.32 | 7 | 22 | 0.67 | 2 | 3 | ||||||
| 24821749 | 0.88 | 7 | 8 | 0.50 | 1 | 2 | 0.44 | 7 | 16 | 0.50 | 1 | 2 | ||||||
| 24817234 | 0.71 | 5 | 7 | 0.50 | 1 | 2 | 0.53 | 8 | 15 | 0.83 | 5 | 6 | ||||||
| 861918 | 0.73 | 8 | 11 | 1.00 | 1 | 1 | 0.36 | 8 | 22 | 0.60 | 6 | 10 | ||||||
| 855543 | 0.75 | 6 | 8 | 1.00 | 1 | 1 | 0.38 | 6 | 16 | 0.40 | 4 | 10 | ||||||
| 4246251 | 0.83 | 10 | 12 | 1.00 | 2 | 2 | 0.23 | 5 | 22 | 0.56 | 5 | 9 | ||||||
| 845529 | 0.71 | 10 | 14 | 0.50 | 2 | 4 | 0.56 | 15 | 27 | 0.55 | 6 | 11 | ||||||
β-lactamase- vs. luciferase-reporter gene assays
Another widely used assay reporter technology relies on β-lactamase.23 Most of the implementations use Fluorescence Resonance Energy Transfer (FRET) substrates resulting in a fluorescence shift upon hydrolysis of the β-lactam.28 As of April 2010, we annotated 92 β-lactamase assays, 74 of which were reporter gene assays (Supporting Figure S5-A). Supporting Figure S5-B shows the assay kits used. To identify small molecule structural classes that were active in a large percentage of the β-lactamase technology assays tested in PubChem, we performed an analysis similar to that for luciferase-based assays. Supporting Figure S6 shows the compounds’ promiscuity plots for β-lactamase enzyme activity and β-lactamase reporter gene assays respectively, and expressed separately for single concentration and concentration-response assays. From Figure S6, several interesting classes of compounds can be identified, including some subtle ones. For example, from quadrant A (biochemical β-lactamase enzyme activity measured by concentration-response assays), a series of 2-alkylsulfonyl-1,3,4-oxadiazoles could be identified, which had previously been demonstrated to covalently modify the enzyme resulting in its inhibition.17 Due to its mechanism, this chemotype shows activity in many other assay types (not shown). However, there are many more compounds that can be identified as highly promiscuous among the β-lactamase reporter gene assays.
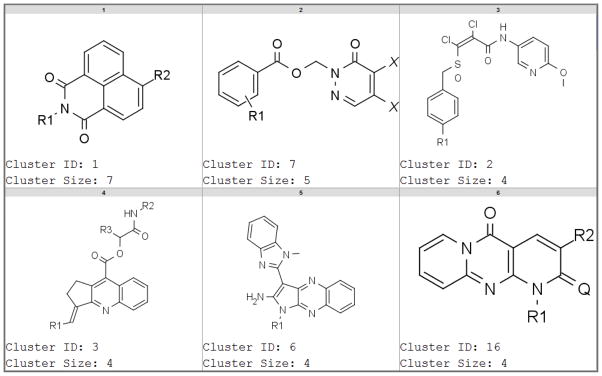
For further analysis, we selected compounds with a promiscuity index (PCIdx) of at least 0.5 and which have been tested in at least 10 reporter gene assays (for single concentration or concentration-response assays). These compounds were clustered by maximum common substructure (MCS). Some of the most promiscuous clusters are shown in Figure 5 by their MCS scaffolds. Supporting Table S2 includes all 97 compounds, their MCS scaffolds, cluster details, PCIdx and the number of active and tested assays. Interestingly, in contrast to the promiscuous luciferase reporter gene compounds (Figure 4, Supporting Table S1), these compounds formed more pronounced (larger) clusters. The mechanism of promiscuity was not immediately obvious from this analysis. However, we hypothesize that compounds in cluster 1 inhibit the β-lactamase enzyme, because these compounds were also promiscuously active in the biochemical β-lactamase inhibition assays. Some of the other series had reactive functional groups, for example cluster 2 (Figure 5) or clusters 7 and 4 (Supporting Table S2), which could therefore be toxic or react chemically with the reporter enzyme or other proteins in the pathways upstream of the promoter.
Figure 5.
Representative chemical scaffolds of the most promiscuous compounds in β-lactamase reporter gene assays (see supporting Table S2 for compounds from all series).
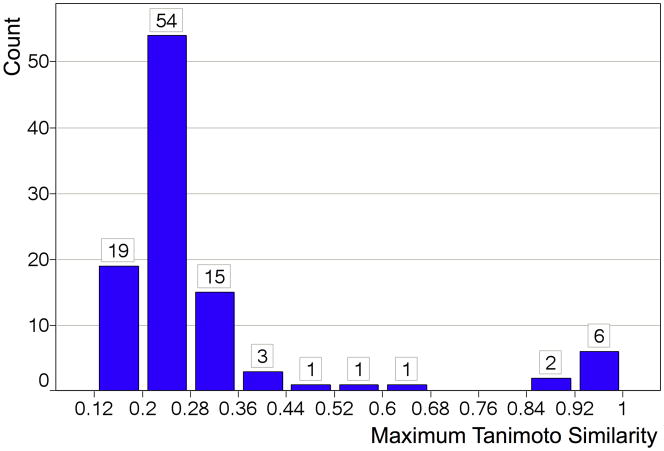
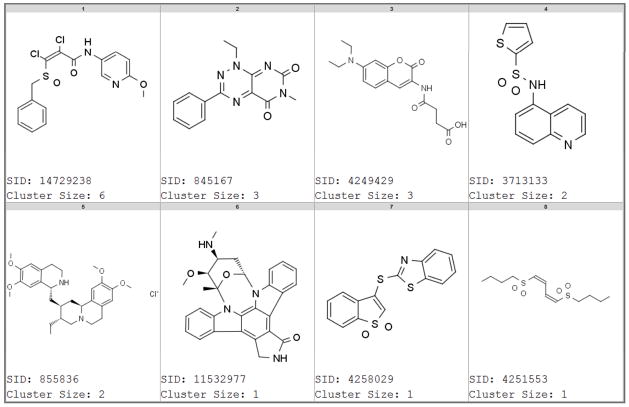
To further investigate how the promiscuity mechanisms were distinct among luciferase and β-lactamase reporter gene assays, we pair-wise compared all highly promiscuous compounds across the two technologies; specifically 102 compounds that were active against the majority of luciferase reporter gene assays vs. 97 compounds active against the majority of β-lactamase reporter gene assays. Compounds were selected with PCIdx ≥ 0.5 and tested in at least 10 assays of their respective reporter technology. Figure 6 shows the similarity histogram of the maximum similar compound among one group for each compound in the other group (see methods). The complete similarity matrix and the histogram of all pair-wise similarities are provided in Supporting Figures S7 and S8. Figure 6 (and Figures S7, S8) indicated that for most of the compounds active against β-lactamase, there was no significantly similar compound active against luciferase. This supports distinct mechanisms of non-specific chemical interferences among luciferase and β-lactamase reporter gene assays. Chemical classes that were promiscuous in both luciferase and β-lactamase reporter gene assays are shown in Figure 7 (all compounds are provided in Supporting Table S3). Their generic mechanisms appeared to include high chemical reactivity such as SID 14729238 or SID 4251553 and general toxicity such as Emetine (SID 855836),27 which is a protein synthesis inhibitor. However, the results suggested that other mechanisms are likely to exist; for example Staurosporine (SID 11532977) is one of the most promiscuous pan-kinase inhibitors.29
Figure 6.
Histogram of the maximum pair-wise Tanimoto similarities of each of the 102 most promiscuous luciferase reporter gene compounds compared against the 97 most promiscuous β-lactamase reporter gene compounds. Tanimoto similarities were computed using ECFP4 fingerprints. Most promiscuous compounds were defined as those with PCIdx ≥ 0.5 and which were tested in at least 10 assays. See Supporting Figure S7 for the full similarity matrix and Figure S8 for the histogram of all pair-wise similarities.
Figure 7.
Compounds representing structural classes that show promiscuous activity across luciferase and β-lactamase reporter gene assays. Supporting Table S3 includes all compounds and their cluster details.
DISCUSSION
The BioAssay Ontology is the first public effort to develop a formal knowledge-based description of HTS assays and screening outcomes.21 The value of large public data repositories such as PubChem will ultimately be determined by how well researchers are able to utilize the information to extract knowledge as a starting point for new research and drug development. Their usefulness will largely be determined by two factors: 1) the content and quality of data in the repository and 2) the ability to retrieve relevant results. The ability to identify, aggregate and analyze data from various assays that are related to a project of interest is particularly important. BAO primarily addresses this second aspect, but it will also help to analyze data quality by identifying redundancies and related data. While developing BAO, we have annotated over 350 PubChem assays to organize them by concepts that are relevant to interpret HTS results. Specifically, we investigated assays based on designs that use the luciferase-catalyzed conversion of luciferin substrates resulting in luminescence and assays detecting β-lactamase via FRET substrates. In contrast to previous reports that focused mostly on individual screening campaigns, BAO has enabled a systematic analysis of many related assays to generate results that could not be obtained from individual screens. Our promiscuity analyses also demonstrated clearly that there is valuable information in the PubChem repository beyond individual screening campaigns, and that the BAO descriptions can facilitate the extraction of new knowledge from large numbers of related data sets.
Among assays employing luciferase technologies, we identified five sub-categories: reporter gene assays, viability assays, ATP-coupled and luciferin-coupled enzyme activity, and biochemical luciferase enzyme activity (Supporting Figure S2). Analyzing compound promiscuity in viability assays revealed the most generally cytotoxic compounds and compound classes. Many of these assays were performed by the NCGC with compounds that were also studied at the Environmental Protection Agency (EPA).30 Toxicity for these highly promiscuous compounds can be mediated by several mechanisms as illustrated by our examples. One common and expected theme that could readily be identified for many of these compounds was that chemical reactivity is related to their cytotoxic effects (Figures 2 and 4).
The majority of the annotated luciferase assays belong to the category of reporter gene assays. We identified the most promiscuous compounds in both single concentration and concentration-response assays, based on the promiscuity index and the number of luciferase reporter gene assays in which a compound was screened. The identified chemotypes are of interest because it is likely that they will be identified in future luciferase reporter gene assays. The fact that many of the most promiscuous luciferase reporter gene compounds have been tested in concentration-response assays indicates that they were selected as interesting hits from primary assays. Based on our observations, researchers would be well advised to exclude these compounds from follow-up studies, because they act via a mechanism that is related to the assay technology and not the biological target of interest. Calculated promiscuities of these compounds in the different sub-types of assays that use luciferase in their design suggested two likely mechanisms of action. One was related to cell viability/toxicity and the other to inhibition of the luciferase enzyme. We have shown specific examples for both cases (Figures 3, 4, Tables 1, S1). The analysis presented here was relatively simple, because it did not take into consideration variations in assay conditions and different luciferase enzymes used. We nevertheless could identify many promiscuous and undesired chemotypes, making this information useful for flagging primary screening hits that should be treated with caution. Our simple analysis that relies on results from many different assays is thus an effective approach to help identifying undesirable compounds and eliminating them before additional resources are spent during hit verification, lead identification and optimization stages. Moreover, this computational analysis could also be used to develop hypotheses on the mechanism of compound promiscuity.
We then performed a similar promiscuity analysis for β-lactamase reporter gene assays to identify chemotypes that were non-specifically active in this category of assays (Figures 5, S6, Table S2). The rationale was the same as for luciferase reporter gene assays: to identify and exclude undesirable hit compounds as early as possible in the discovery and optimization pipeline. In contrast to luciferase reporter gene assays, the β-lactamase promiscuous compounds formed pronounced, larger clusters after maximum common substructure clustering. This could be due to the composition of the library or because the larger number of luciferase (compared to β-lactamase) reporter gene assays selected more diverse highly promiscuous compounds.
Pair-wise comparison of the most promiscuous compounds in luciferase-vs. beta-lactamase reporter gene assays showed that, with a few exceptions, their chemical spaces do not overlap (Figures 6, S7, S8). This suggests distinct mechanisms of promiscuity that are specific to the reporter technology. Although this may be expected, such a quantitative analysis using a large number of assays is relevant for data analysis and it is also directly relevant to HTS assay development. Our analysis demonstrates that the two reporter technologies are orthogonal to one another, because they are prone to distinct chemotypes of artifactual hits. Compounds that were identified as promiscuous in both luciferase and β-lactamase reporter gene assays appear generally cytotoxic due to their chemical reactivity or a mechanism unrelated to the reporter, for example non-selective kinase inhibition (Figure 7, Table S3).
SUMMARY AND CONCLUSIONS
In summary, we have systematically analyzed data from a large number of assays in PubChem to identify compounds that are promiscuously active in specific assay technologies and via distinct mechanisms of action. Such an analysis is only possible with the detailed annotations that we made based upon the BioAssay Ontology, the first reported ontology to formally describe high-throughput screening assays and assay outcomes. There are many advantages of a formal description of bioassays and standardized annotations of data sets such as those in PubChem. Here we demonstrated that analyses across many assays are facilitated by standardized annotations such as those produced by BAO and that the results can provide insights that cannot be obtained by analyzing individual data sets. This is particularly relevant for relatively noisy primary HTS results. Analysis across many assays of the same type can also be expected to be more robust than analyses focused on individual data sets. Although HTS data contain false positives and false negatives, the BAO approach does not rely on each individual result data points, but requires only that the ensemble of results reflect the correct trend, i.e. the fraction of the experiments of a certain category in which a compound is found active.
While undesirable and reactive chemical functionalities that are prone to cause false positives in HTS have been reported in the past 31, the definition of undesired chemical substructures to a large extent depends on the specific assay technologies and biological targets; for example in some applications covalent modifiers may be acceptable or even desired, while in others they have to be excluded. BAO provides a means to identify undesirable chemical substructures in a data driven manner specific to the assay technologies or biological meta-targets that are covered by BAO. With the type of analysis presented here it would thus be possible to identify undesirable chemotypes that are specifically relevant to a given discovery project.
We would not recommend to a-priori remove from a screening library all compounds that show promiscuity, but rather flag them, because such compounds can still be of interest for certain targets and orthogonal assays designs and detection technologies are prone to structurally different artifacts (as we have shown for luciferase and β-lactamase reporter gene assays). By the same token, certain chemotypes may cause artifacts across a large number of assay technologies and biological targets and these could be removed to improve a screening collection. This will require more comprehensive analyses. We are currently annotating more assays from PubChem and will perform similar analyses for various other categories. The curation effort is time-consuming and not an effective long-term strategy to standardize data. While a certain amount of curation will likely be required to consolidate terminology, it would be desirable to add BAO-type annotations at the stage of assay deposition and to make these annotations available in the primary data sources such as PubChem. BAO is available from our website.21
As the number of available data sets increases, the type of analyses presented here would have to be repeated periodically in order to comprehensively and accurately identify promiscuous compounds of a certain category. However this is a straightforward undertaking given standardized assay annotations and endpoints. Utilizing BAO annotations and standardized endpoints we are also currently working on developing predictive classifiers from quantitative outcomes of luciferase assays. Such classifiers could then be used to automatically flag potentially promiscuous compounds.
The BAO software under development21 will facilitate the query, exploration and downloading of curated HTS data by BAO terms and thus will also facilitate the identification of promiscuous compounds for specific assay technologies.
Supplementary Material
Acknowledgments
The work presented here was supported by NIH grant RC2 HG005668. We acknowledge resources from the Center for Computational Science of the University of Miami. Vance Lemmon holds the Walter G. Ross Distinguished Chair in Developmental Neuroscience.
References
- 1.Mayr LM, Bojanic D. Novel trends in high-throughput screening. Curr Opin Pharmacol. 2009;9:580–588. doi: 10.1016/j.coph.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 2.Austin CP, Brady LS, Insel TR, Collins FS. NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 3.Schürer S, Tsinoremas N. Screening Informatics. In: Chen T, editor. A Practical Guide to Assay Development and High-Throughput Screening in Drug Discovery. Taylor and Francis; 2009. [Google Scholar]
- 4.Ling XB. High throughput screening informatics. Comb Chem High Throughput Screen. 2008;11:249–257. doi: 10.2174/138620708783877726. [DOI] [PubMed] [Google Scholar]
- 5.Torr-Brown S. Advances in knowledge management for pharmaceutical research and development. Curr Opin Drug Discov Devel. 2005;8:316–322. [PubMed] [Google Scholar]
- 6.The PubChem Project. Retrieved from http://pubchem.ncbi.nlm.nih.gov/
- 7.Wang Y, Bolton E, Dracheva S, Karapetyan K, Shoemaker BA, Suzek TO, Wang J, Xiao J, Zhang J, Bryant SH. An overview of the PubChem BioAssay resource. Nucleic Acids Res. 2010;38:D255–266. doi: 10.1093/nar/gkp965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.ChEMBL Database. Retrieved from http://www.ebi.ac.uk/chembldb/index.php.
- 9.PDSP Ki Database. Retrieved from http://pdsp.med.unc.edu/kidb.php.
- 10.Jensen NH, Roth BL. Massively parallel screening of the receptorome. Comb Chem High Throughput Screen. 2008;11:420–426. doi: 10.2174/138620708784911483. [DOI] [PubMed] [Google Scholar]
- 11.ChemBank. Retrieved from http://chembank.broad.harvard.edu/
- 12.Seiler KP, George GA, Happ MP, Bodycombe NE, Carrinski HA, Norton S, Brudz S, Sullivan JP, Muhlich J, Serrano M, Ferraiolo P, Tolliday NJ, Schreiber SL, Clemons PA. ChemBank: a small-molecule screening and cheminformatics resource database. Nucleic Acids Res. 2008;36:D351–359. doi: 10.1093/nar/gkm843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Collaborative Drug Discovery. Retrieved from http://www.collaborativedrug.com/
- 14.Hohman M, Gregory K, Chibale K, Smith PJ, Ekins S, Bunin B, Bradford J, Dole K, Spektor A, Blondeau D, Bunin BA. Novel web-based tools combining chemistry informatics, biology and social networks for drug discovery. Drug Discov Today. 2009;14:261–270. doi: 10.1016/j.drudis.2008.11.015. [DOI] [PubMed] [Google Scholar]
- 15.Inglese J, Shamu CE, Guy RK. Reporting data from high-throughput screening of small-molecule libraries. Nat Chem Biol. 2007;3:438–441. doi: 10.1038/nchembio0807-438. [DOI] [PubMed] [Google Scholar]
- 16.Auld DS, Southall NT, Jadhav A, Johnson RL, Diller DJ, Simeonov A, Austin CP, Inglese J. Characterization of chemical libraries for luciferase inhibitory activity. J Med Chem. 2008;51:2372–2386. doi: 10.1021/jm701302v. [DOI] [PubMed] [Google Scholar]
- 17.Babaoglu K, Simeonov A, Irwin JJ, Nelson ME, Feng B, Thomas CJ, Cancian L, Costi MP, Maltby DA, Jadhav A, Inglese J, Austin CP, Shoichet BK. Comprehensive mechanistic analysis of hits from high-throughput and docking screens against beta-lactamase. J Med Chem. 2008;51:2502–2511. doi: 10.1021/jm701500e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Southern M, Griffin P. A Java API for working with PubChem data-sets. Bioinformatics. 2010 doi: 10.1093/bioinformatics/btq715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pipeline Pilot 8.0. San Diego, CA: Accelrys; 2010. [Google Scholar]
- 20.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.BioAssay Ontology. Retrieved from http://www.bioassayontology.org/
- 22.Fan F, Wood KV. Bioluminescent assays for high-throughput screening. Assay Drug Dev Technol. 2007;5:127–136. doi: 10.1089/adt.2006.053. [DOI] [PubMed] [Google Scholar]
- 23.Qureshi SA. Beta-lactamase: an ideal reporter system for monitoring gene expression in live eukaryotic cells. Biotechniques. 2007;42:91–96. doi: 10.2144/000112292. [DOI] [PubMed] [Google Scholar]
- 24.Spotfire DecisionSite 9.0. Palo Alto, CA: TIBCO; 2007. [Google Scholar]
- 25.ChemAxon JChem Software Suite. Retrieved from http://www.chemaxon.com/
- 26.Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 27.Guha R, Schurer SC. Utilizing high throughput screening data for predictive toxicology models: protocols and application to MLSCN assays. J Comput Aided Mol Des. 2008;22:367–384. doi: 10.1007/s10822-008-9192-9. [DOI] [PubMed] [Google Scholar]
- 28.Zlokarnik G, Negulescu PA, Knapp TE, Mere L, Burres N, Feng L, Whitney M, Roemer K, Tsien RY. Quantitation of transcription and clonal selection of single living cells with beta-lactamase as reporter. Science. 1998;279:84–88. doi: 10.1126/science.279.5347.84. [DOI] [PubMed] [Google Scholar]
- 29.Nakano H, Omura S. Chemical biology of natural indolocarbazole products: 30 years since the discovery of staurosporine. J Antibiot (Tokyo) 2009;62:17–26. doi: 10.1038/ja.2008.4. [DOI] [PubMed] [Google Scholar]
- 30.Xia M, Huang R, Witt KL, Southall N, Fostel J, Cho MH, Jadhav A, Smith CS, Inglese J, Portier CJ, Tice RR, Austin CP. Compound cytotoxicity profiling using quantitative high-throughput screening. Environ Health Perspect. 2008;116:284–291. doi: 10.1289/ehp.10727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rishton GM. Reactive compounds and in vitro false positives in HTS. Drug Discovery Today: Technologies. 1997;2:382–384. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.