SUMMARY
Alternative mRNA splicing provides transcript diversity and may contribute to human disease. We demonstrate that expression of several genes regulating RNA processing is decreased in both liver and skeletal muscle of obese humans. We evaluated a representative splicing factor, SFRS10, down-regulated in both obese human liver and muscle and in high fat-fed mice, and determined metabolic impact of reduced expression. SFRS10-specific siRNA induces lipogenesis and lipid accumulation in hepatocytes. Moreover, Sfrs10 heterozygous mice have increased hepatic lipogenic gene expression, VLDL secretion, and plasma triglycerides. We demonstrate that LPIN1, a key regulator of lipid metabolism, is a splicing target of SFRS10; reduced SFRS10 favors the lipogenic β isoform of LPIN1. Importantly, LPIN1β-specific siRNA abolished lipogenic effects of decreased SFRS10 expression. Together, our results indicate that reduced expression of SFRS10, as observed in tissues from obese humans, alters LPIN1 splicing, induces lipogenesis, and therefore contributes to metabolic phenotypes associated with obesity.
INTRODUCTION
Obesity is a global epidemic, with substantial adverse social, economic, and personal health consequences. Foremost among these are the severe metabolic complications of obesity, including insulin resistance, T2D, and increased cardiovascular disease risk. While the precise molecular defects underlying these complications remain unknown, abnormal lipid metabolism is a consistent phenotype. Indeed, both hypertriglyceridemia and “ectopic” lipid accumulation in liver and skeletal muscle are closely linked to both insulin resistance and diabetes risk (Browning and Horton, 2004; Jacob et al., 1999; Kotronen et al., 2007). Thus, we aimed to identify expression signatures of obesity in liver and muscle of human subjects. We now demonstrate results from two independent human cohorts, which both identified decreased expression of genes regulating RNA processing and splicing as the top-ranking expression phenotypes in liver and muscle of obese humans.
RNA processing is a complex cascade including constitutive and alternative splicing, polyadenylation and nuclear export of mature mRNA (Stamm et al., 2005). Alternative splicing occurs for more than 90% of human genes (Wang et al., 2008) in the spliceosome, which consists of pre-mRNA, small ribonucleoproteins (snRNPs) and two major groups of non-snRNP proteins, splicing factors and heterogeneous ribonucleoproteins (HNRNPs). The resulting splice variants have a fundamental role in differentiation and organ development (Bland et al., 2010), and tissue-specific isoforms are commonly observed (Blencowe, 2006; Nilsen and Graveley, 2010). Thus, alternative splicing should be viewed as an important adaptive mechanism used to create protein diversity in response to distinct developmental and metabolic cues (Salomonis et al., 2010; Nilsen and Graveley, 2010). Alternative splicing can be disrupted in several disease states, including cancer (Karni et al., 2007; Venables, 2004) and monogenic human diseases (Faustino and Cooper, 2003). Moreover, several genes linked to obesity and insulin resistance have been shown to be regulated by alternative splicing (Kishore and Stamm, 2006; Sesti et al., 1991; Lefai et al., 2001; Patel et al., 2005; Ghosh et al., 2007; Lee et al., 1996), and defects in RNA processing and nuclear export are associated with lipodystrophy (Agarwal and Garg, 2006).
Our results now indicate that modification of alternative splicing may also contribute to metabolic phenotypes associated with human obesity, as reduced hepatic and muscle expression of a subset of splicing factors is associated obesity, increased hepatic fat content, and hyperinsulinemia. To investigate whether this downregulation could contribute to phenotypes associated with obesity, we further studied a representative splicing factor, SFRS10, which was downregulated in both liver and muscle of obese humans. SFRS10 (official gene name TRA2B), the homolog of Drosophila transformer-2 (Tra2), belongs to the SR-like protein family of splicing factors and is an important modulator of alternative splicing of multiple genes (Nayler et al., 1998). We demonstrate that reduced expression of SFRS10 alters splicing of LPIN1, a key regulator of lipid metabolism (Csaki LS and Reue, 2010; Peterfy et al., 2005; Yao-Borengasser et al., 2006; Ryu et al., 2009; Huang et al., 2011), and contributes to increased hepatic lipogenesis and increased VLDL secretion in mice.
RESULTS
Expression of genes regulating mRNA processing is decreased in obesity
To identify differentially expressed genes in both liver and muscle from insulin resistant humans with obesity, we utilized high-density oligonucleotide arrays in two independent cohorts (Table 1). Up- or downregulated probesets common to both studies are shown in Table S1 (accessions GSE15653 and GSE22435 for liver and muscle data, respectively). GO-based pathway analysis (MAPPFinder) demonstrated that the top-ranking downregulated pathways in both tissues were related to RNA processing and splicing (Figure 1A). For example, 46 of 199 RNA splicing genes were downregulated in liver (Z score 7.5, adjusted p<0.001) and 41 of 199 were downregulated in muscle (Z 11.1, p<0.001). Gene set enrichment analysis (GSEA) also identified the RNA processing gene set as downregulated in obese subjects in liver (nominal p<0.001, FDR 0.039), but not in muscle (nominal p=0.26, FDR 0.87). Expression of 13 genes involved in RNA processing and splicing was decreased in both liver and muscle (Figure 1B). A complete list of genes for each tissue is shown in Tables S2 and S3.
Table 1.
Clinical characteristics of study subjects. Subjects had no known abnormality in glucose metabolism prior to study. Six obese subjects were diagnosed with type 2 diabetes (T2D) during the liver study. Four obese subjects were diagnosed with impaired glucose tolerance (IGT) and three with T2D during the muscle study. Data are mean ± SD.
| Liver study | Skeletal muscle study | |||
|---|---|---|---|---|
| Lean NGT | Obese NGT/T2D | Lean NGT | Obese IGT/T2D | |
| Men / Women | 0 / 5 | 2 / 6 | 0 / 10 | 0 / 7 |
| Age (years) | 36 ± 12 | 43 ± 11 | 59.6 ± 5.0 | 60.0 ± 4.8 |
| Body Mass Index (kg/m2) | 24 ± 5 | 53 ± 7** | 27.4 ± 5.4 | 31.8 ± 6.5* |
| Fasting glucose (mmol/L) | 4.8 ± 0.8 | 6.5 ± 2.0 | 4.5 ± 0.6 | 5.0 ± 0.8 |
| Fasting insulin (pmol/L) | 36.8 ± 33.0 | 148.2 ± 132.6* | 40.3 ± 20.2 | 73.0 ± 23.4* |
p<0.05,
p<0.001 vs. lean of the same study.
Figure 1. RNA processing gene expression is downregulated in obesity.
(A) Top-ranking downregulated pathways in obese humans identified through GO-based pathway analysis (MAPPFinder) of microarray data from liver and muscle. (B) Heatmap of 13 RNA processing genes with decreased gene expression in both tissues. Blue indicates lower and red higher gene expression. NGT, normal glucose tolerance; IGT, impaired glucose tolerance; T2D, type 2 diabetes. (C) Expression of RNA processing genes was determined by RT-PCR from mouse liver and muscle after 4 month of HFD (black bars) compared to chow diet (white bars). Data are mean±SEM. *, p<0.05 vs. chow (n=6). (D) Protein levels of SFRS10, SFPQ and HNRPK were measured by Western blot from liver nuclear extracts. See Fig S1.
We next asked whether decreased expression of RNA processing genes is also present in a mouse model of diet-induced obesity. Indeed, expression of several RNA processing genes was decreased following HFD (4 months) in both liver and muscle (Figure 1C). Reduced expression of SFRS10 and HNRPK was confirmed by Western blot in liver of HFD-fed mice (Figure 1D).
Obesity in both humans and HFD-fed mice is characterized by alterations in both systemic and cellular glucose and lipid metabolism, which could potentially contribute to the observed downregulation of this subset of RNA processing genes. We therefore assessed correlation of the 13 genes downregulated in both tissues with key metabolic phenotypes in humans (Table S4). Of these, liver expression of 9 genes correlated inversely with obesity (BMI), and 5 genes inversely with hepatic lipid content. Skeletal muscle expression of 6 genes correlated inversely with fasting insulin. No correlation between expression and fasting glucose was observed in either tissue.
We chose SFRS10 as a representative splicing factor for further analysis, as it was altered most consistently in both human and animal models. Examination of public databases indicated that hepatic mRNA expression of Sfrs10 was not modulated in mice by (a) insulin deficiency (streptozotocin-induced diabetes) (Yechoor et al., 2002) or (b) experimentally-induced insulin resistance (due to liver-specific deletion of insulin receptor, IRS1, IRS2, or both IRS1 and IRS2) in the absence of obesity (Guo et al., 2009; Biddinger et al., 2008). Similarly, muscle-specific insulin resistance, even with superimposed hyperglycemia, does not alter Sfrs10 expression (Yechoor et al., 2004). Additionally, exposure of HepG2 hepatoma cells and C2C12 myotubes to palmitate or elevated glucose did not affect SFRS10 expression. However, overnight incubation with 10 nM insulin decreased SFRS10 expression (Figure S1), supporting a potential role for chronic insulin exposure in contributing to decreased SFRS10 expression.
Reduced expression of SFRS10 leads to increased lipogenesis in cultured cells
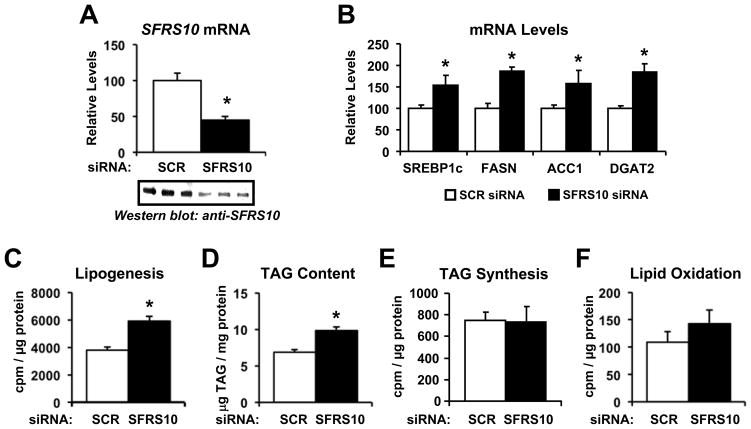
To assess the functional consequences of decreased splicing factor gene expression, we experimentally reduced SFRS10 expression in HepG2 cells. siRNA-mediated knockdown of SFRS10 led to a 50–70% decrease in mRNA and protein levels (Figure 2A). Given that decreased expression of several RNA processing genes was associated with increased hepatic lipid content in human subjects, we examined the effects of reducing SFRS10 on lipid metabolism in hepatic cells. Indeed, SFRS10 knockdown in HepG2 cells induced a 1.5 to 2-fold increase in lipogenic genes, including SREBP1c, FASN, ACC1 and DGAT2 (Figure 2B). While we observed trends for SFRS10 knockdown to increase expression of SREBP1a and PCK1 (PEPCK), expression of other genes influencing lipid or glucose metabolism, including ESR1 (ERRα), NR1H3 (LXRα), NR1H2 (LXRβ), NR1H4 (FXR), PPARA, PPARG, PPARD, PPARGC1A (PGC-1α), and PPARGC1B (PGC-1β), did not differ following SFRS10 knockdown (Table S5). Notably, SFRS10 knockdown-mediated increases in lipogenic gene expression were accompanied by a 1.6-fold increase in lipogenesis, as measured by [14C]-acetate incorporation into the lipid fraction (p<0.05, Figure 2C), and led to a 1.4-fold increase in cellular accumulation of TAG (p<0.05, Figure 2D). TAG synthesis from [14C]-palmitate (Figure 2E) or fatty acid oxidation (Figure 2F) did not differ between control and SFRS10 knockdown, suggesting TAG accumulation was due to enhanced fatty acid synthesis. Similar effects of SFRS10 knockdown were observed in C2C12 myotubes, with induction of SREBP1c and FASN expression, and increased TAG accumulation (Figure S2). These lipogenic effects were specific for SFRS10, as knockdown of the constitutive splicing factor SF3A1 had no effect on lipogenic gene expression or lipid accumulation in either HepG2 or C2C12 cells (Figure S2).
Figure 2. SFRS10 knockdown increases expression of lipogenic genes and leads to TAG accumulation in hepatic cells.
HepG2 cells were transfected with scramble (SCR) or SFRS10 siRNA and analyzed 4 days later. (A) SFRS10 mRNA and protein levels were analyzed by RT-PCR and Western blot. (B) mRNA levels were determined by RT-PCR. (C) Lipogenesis (from 14C-acetate), (D) TAG levels, (E) TAG synthesis, and (F) fatty acid oxidation were measured as described in Methods. Data are mean±SEM of triplicates, representative of 3 independent experiments. *, p<0.05 vs. SCR siRNA. See Fig S2.
To determine whether increased Sfrs10 expression could also modulate lipid metabolism, Hepa1c hepatoma cells were transfected with a DNA construct expressing Sfrs10. As seen in Figure S3, experimental overexpression of Sfrs10 in Hepa1c cells significantly decreased expression of the lipogenic genes Fasn, Agpat2, and Dgat2 (p<0.05 for SFRS10 vs. GFP, panel B).
Sfrs10 +/- mice have increased lipogenic gene expression, VLDL secretion, and plasma triglycerides
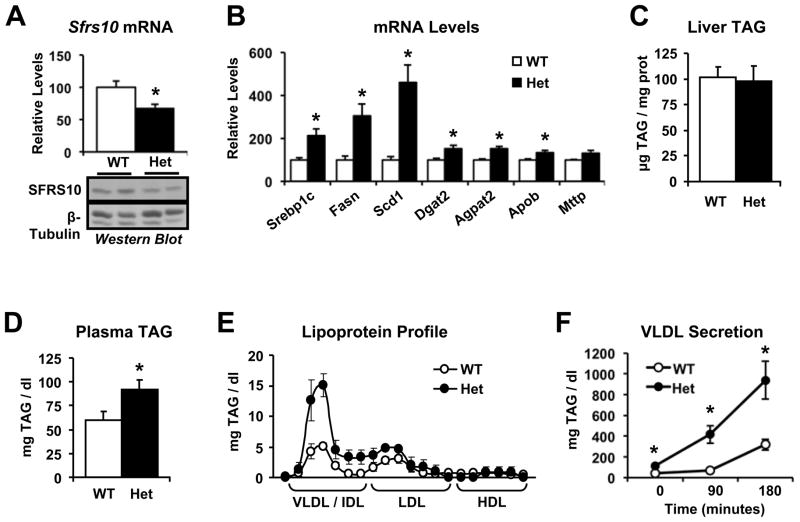
We next studied whether decreased expression of Sfrs10 induces hepatic lipogenesis in a mouse model in vivo. For this purpose, the Sfrs10 locus was disrupted as described in Methods. No Sfrs10 -/- pups were detected, confirming that genetic disruption of the Sfrs10 gene is lethal during embryonic life (Mende et al., 2010). However, heterozygous Sfrs10 +/- mice were viable, had normal growth and body weight and showed no differences in blood glucose and insulin levels compared to wild-type animals (Figure S4). As shown in Figure 3A, hepatic Sfrs10 mRNA levels were decreased by 30% in heterozygous mice compared to wild-type, a magnitude similar to the decrease in obese humans. Sfrs10 protein levels were only slightly decreased in the liver of heterozygous mice (Figure 3A), likely due to the known autoregulation of Sfrs10 protein mediated by a negative feedback loop (Stoilov et al., 2004). When present at high levels, Sfrs10 binds to its own exon 2, activating its inclusion, and generating the Tra2b4 isoform that is not translated into protein. Conversely, low Sfrs10 protein levels induce exon 2 skipping, favoring the Tra2b1 isoform, the main functional isoform. In accord with this autoregulation, we observed an 80% decrease in Tra2b4 mRNA levels in the liver of Sfrs10 heterozygous mice, with a much smaller 25% decrease in Tra2b1 (Figure S4). These data indicate that lower Sfrs10 expression in the heterozygous mice promotes skipping of its own exon 2 in an attempt to restore protein levels.
Figure 3. Sfrs10 heterozygous (Het) mice show increased lipogenic gene expression and hypertriglyceridemia.
Wild-type (WT) and Sfrs10 Het mice were fasted for 16 hours and then refed for 10 hours before sacrifice. (A) Liver Sfrs10 mRNA and protein levels were determined by RT-PCR and Western blot. (B) Liver mRNA was quantified by RT-PCR. (C) Liver TAG and (D) plasma TAG were measured as in Methods. (E) Plasma lipoprotein profile was determined by FPLC. (F) VLDL secretion was calculated by quantifying plasma TAG after Tyloxapol administration. Data are mean±SEM of at least 5 mice/group and are representative of 2 independent cohorts. *, p<0.05 vs. WT. See Fig S4.
Despite this autoregulation, and in agreement with the in vitro data, Sfrs10 heterozygous mice showed increased hepatic expression of lipogenic and TAG synthesis genes, including Srebp1c, Fasn, Scd1, Dgat2 and Agpat2 in the postprandial state (1.5 to 4.6-fold compared to wild-type, p<0.05, Figure 3B). While liver TAG accumulation remained unchanged (Figure 3C), plasma TAG levels were 52% higher in the heterozygous mice (p<0.05, Figure 3D). In particular, we observed a marked increase in plasma levels of the TAG-enriched VLDL fraction (3-fold, p<0.05, Figure 3E), indicating hepatic origin of the higher plasma TAG. To determine if this pattern was indeed due to increased hepatic VLDL secretion, we measured plasma TAG following administration of tyloxapol, an inhibitor of TAG clearance. Plasma TAG were 2–3 fold higher in heterozygous mice, confirming increased secretion (p<0.05 at all time points, Figure 3F). Together, these data indicate that decreased expression of Sfrs10 is sufficient to increase hepatic lipogenic gene expression, increase VLDL secretion, and induce hypertriglyceridemia in a mouse model in vivo.
Lpin1 splicing is regulated by Sfrs10
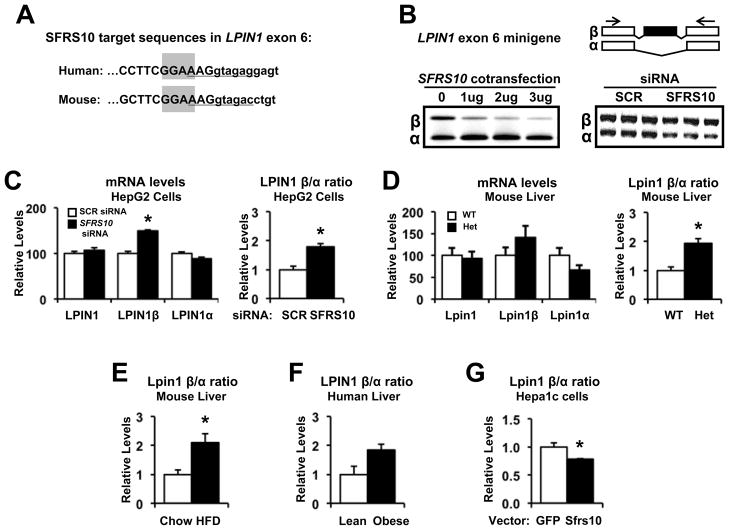
Our data suggest that specific targets of Sfrs10 upstream of Srebp1c and Fasn may mediate effects on lipid accumulation. It has been shown that Sfrs10 binds to RNA with the sequence NGAA. The protein binds to AGAA with an affinity of 2.25 μM and to GGAA with 4.5 μM (Clery et al., 2011). Therefore, we considered known genes with alternatively spliced isoforms, particularly those regulating lipogenesis. Of these, Lpin1 is known to regulate lipid metabolism (Csaki LS and Reue, 2010; Bou et al., 2010) and has two major alternatively spliced isoforms, α and β, and a third variant, γ, recently identified in brain (Han and Carman, 2010). The β isoform, generated by inclusion of exon 6, is associated with increased expression of lipogenic genes (Peterfy et al., 2005; Csaki LS and Reue, 2010). Interestingly, the alternatively spliced exon 6 of Lpin1 contains a GGAA sequence motif that binds to SFRS10, and these sequences are highly conserved between human LPIN1 and mouse Lpin1 genes (Figure 4A). The location of the Sfrs10 site is unusual, as it partially overlaps with the U1 binding sites at the 5’ splice site. To determine if alterations in SFRS10 expression could indeed modulate splicing of LPIN1, we used a minigene construct containing the alternative exon 6 of the human LPIN1 gene. Cotransfection of the minigene construct with a plasmid expressing SFRS10 caused exon 6 skipping (Figure 4B, left). Conversely, SFRS10 siRNA increased inclusion of exon 6 (Figure 4B, right). These findings suggest that SFRS10 competes with U1 snRNA for binding at the 5’ splice site. With higher SFRS10 expression, U1 snRNA could be replaced by SFRS10, leading to exon skipping; with lower SFRS10 expression, U1 snRNA could bind the 5’ splice site and thus initiate exon inclusion.
Figure 4. SFRS10 regulates LPIN1 splicing.
(A) The putative binding site of SFRS10, GGAA, is highlighted in gray within alternatively spliced exon 6 sequence (Ensembl release 61) of human and mouse LPIN1. The U1 snRNA binding site at the 5’ splice site is underlined. (B) SFRS10 cotransfection increases exclusion of LPIN1 exon 6 in a minigene system (left), while SFRS10 siRNA increases inclusion (right). PCR primers are shown as arrows. (C–D) Expression of total LPIN1, LPIN1β and LPIN1α isoforms was determined (RT-PCR) in: (C) HepG2 cells after SCR (white bars) or SFRS10 siRNA (black bars), and (D) liver samples from WT (white, n=7) and Sfrs10 heterozygous (black, n=5) mice. (E–G) Expression of Lpin1β relative to Lpin1α was measured by RT-PCR in liver from: (E) HFD (black, n=6) and chow (white, n=6) mice, (F) lean (white, n=6) or obese (black, n=14) humans, and (G) Hepa1c cells after GFP (white, n=5) or SFRS10 (black, n=5) overexpression. Data are mean±SEM. *, p<0.05 vs. control. See Fig S2.
We next assessed whether SFRS10 knockdown affected splicing of endogenous LPIN1 in HepG2 cells. Notably, SFRS10 siRNA did not change total LPIN1 expression (Figure S5), but altered the generation of LPIN1 splice isoforms, yielding an increase in the β isoform in parallel with a modest decrease in the α isoform (Figure 4C, left). As a result, the ratio of the endogenous β isoform in relation to the α isoform (LPIN1β/α ratio) increased in response to SFRS10 knockdown (Figure 4C, right), confirming that modified SFRS10 expression leads to altered LPIN1 splicing. A similar increase in Lpin1β/α ratio was observed in response to Sfrs10 knockdown in C2C12 myotubes (Figure S2). This effect was specific to SFRS10, as knockdown of the constitutive splicing factor SF3A1 did not alter LPIN1 splicing, as indicated by unchanged LPIN1β/α ratio (Figure S2). Similar results were observed in liver of Sfrs10 heterozygous mice: no change in total Lpin1 expression (Figure 4D, left), but an increase in the Lpin1β/α ratio (Figure 4D, right). Consistent with this effect, and in parallel with reduced SFRS10 levels, we also observed a significant increase in the Lpin1β/α ratio in liver of HFD fed mice (Figure 4E), and a similar trend in liver of obese humans (Figure 4F). Conversely, overexpression of Sfrs10 in Hepa1c cells reduced the Lpin1β/α ratio as compared to GFP control (Figure 4G). Together, these results indicate that SFRS10 levels can modulate LPIN1 splicing, and thus ratios between the β and α isoforms of LPIN1.
Lipin1 protein was initially identified as a type 1 phosphatidic acid phosphatase (PAP-1) acting in the TAG synthesis pathway (Han et al., 2006; Donkor et al., 2007), but also subsequently recognized as a transcriptional regulator of lipid metabolism (Finck et al., 2006). LPIN1β induces lipogenic gene expression in adipocytes (Peterfy et al., 2005). Indeed, Sfrs10 downregulation leads to increased expression of lipogenic genes in hepatoma cells (Figure 2B), C2C12 myotubes (Figure S2, panel A) and in Sfrs10 heterozygous mice (Figure 3B). However, we found no differences in hepatic PAP-1 activity (Figure S4) in heterozygous mice as compared to wild-type mice or in TAG synthesis from palmitate in cells (Figure 2E).
To investigate if the effects of SFRS10 knockdown on lipogenic gene expression were mediated by increases in the LPIN1β isoform, we developed distinct siRNA oligonucleotides directed against either total LPIN1 or specifically targeting exon 6, the LPIN1β -specific exon (Table S6). While a total LPIN1 siRNA decreased both LPIN1α and β isoforms, the LPIN1β-targeted siRNA decreased LPIN1β expression by 68% without affecting LPIN1α expression, demonstrating specificity of the LPIN1β siRNA (Figure 5A and Figure S5). Isolated knockdown of either total or the β-specific isoform of LPIN1 had no significant effects on FASN expression. However, when SFRS10 expression was also reduced (via cotransfection with SFRS10 siRNA), the LPIN1β-specific siRNA abolished SFRS10-mediated increases in genes regulating fatty acid synthesis and TAG synthesis (Figure 5A). Importantly, LPINβ knockdown also prevented the SFRS10 siRNA-induced increase in lipogenesis (Figure 5B) and TAG accumulation (Figure 5C), and prevented increases in lysophosphatidic acid, an intermediate in the TAG synthesis pathway (Figure 5D). These findings indicate that effects of reduced SFRS10 on LPIN1 splicing, favoring the LPIN1β isoform, are sufficient to increase expression of lipogenic genes and activate lipogenesis.
Figure 5. Increased expression of lipogenic genes and lipogenesis in response to SFRS10 siRNA is reversed with LPIN1β knockdown.
HepG2 cells were transfected with the indicated siRNA and analyzed 4 days later. (A) mRNA levels were determined by RT-PCR. (B) Lipogenesis, (C) TAG accumulation and (D) lysophosphatidic acid levels were measured as in Methods. Data are mean±SEM of triplicates, representative of 3 independent experiments. *, p<0.05 vs. SCR siRNA. #, p<0.05 vs. SFRS10 siRNA. See Fig S5.
DISCUSSION
In the current study, we demonstrate downregulation of a subset of RNA processing genes in liver and skeletal muscle of obese humans. Expression of several splicing factors was inversely related to BMI, hepatic lipid accumulation, and hyperinsulinemia in humans. Moreover, diet-induced obesity reduced expression of several RNA processing genes in mice, and exposure of cultured hepatoma cells to insulin reduced expression of SFRS10, suggesting a potential role for obesity-linked insulin resistance and/or chronic hyperinsulinemia in vivo. Additional factors, including genetic variation, could potentially contribute to obesity-associated differences in expression regulation. In particular, a polymorphism close to the SFRS10 locus has been associated with obesity in several populations (Thorleifsson et al., 2009; Cheung et al., 2010).
To examine whether decreased expression of these splicing factors was merely a consequence of, or could directly contribute to obesity-related metabolic phenotypes, we modulated expression of a representative alternative splicing factor, SFRS10, selected due to its consistent downregulation in both human and rodent tissues. Using both siRNA in cultured cells, and in vivo using a mouse model of Sfrs10 heterozygosity induced by gene targeting, we observed that experimental reduction in Sfrs10 expression led to increased lipogenesis, likely mediated by upregulation of key lipogenic genes such as Fasn and Srebp1c. Moreover, we show that heterozygosity for Sfrs10 increases VLDL secretion, resulting in marked hypertriglyceridemia in vivo – thus indicating that downregulation of SFRS10 can indeed contribute to disordered lipid metabolism.
In this study, we identify Lpin1 as a key splicing target of Sfrs10 which mediates its effects on lipogenesis. Originally, a Lpin1 null mutation was discovered as a cause for lipodystrophy in mice (Peterfy et al., 2001); mutations in LPIN1 in humans have been linked to abnormal lipid accumulation in muscle (Zeharia et al., 2008). Subsequent studies identified Lpin1 as both a phosphatidate phosphatase in the TAG synthesis pathway and also a transcriptional coactivator of PPARα (Finck et al., 2006; Csaki LS and Reue, 2010). Conversely, overexpression of Lpin1 in either fat or muscle leads to obesity, with additional metabolic effects depending on tissue site of overexpression (Phan and Reue, 2005). In humans, transcriptional regulation of Lpin1 alone is unlikely to explain hepatic lipid accumulation in obesity, as total Lpin1 expression is reduced in parallel with insulin resistance in adipose tissue and liver (Croce et al., 2007; Donkor et al., 2008; Suviolahti et al., 2006; Yao-Borengasser et al., 2006) .
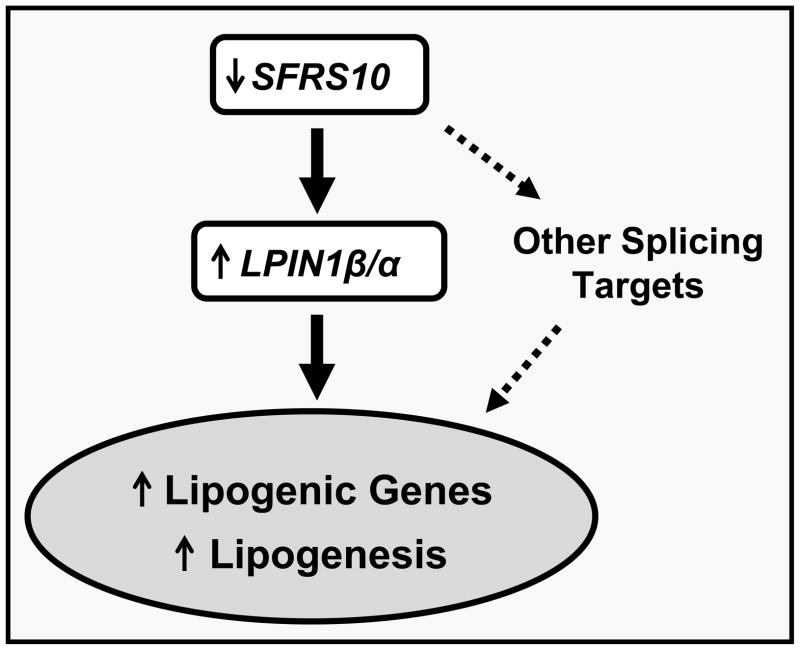
Alternatively spliced isoforms of LPIN1, including α and β, have distinct patterns of cellular localization and regulation (Peterfy et al., 2005; Khalil et al., 2009; Han and Carman, 2010). For example, hepatic LPIN1α and β expression in humans does not correlate in individual subjects, indicating independent isoform regulation (Croce et al., 2007). The β isoform is associated with increased expression of lipogenic genes in adipose tissue (Peterfy et al., 2005) and has more prominent effects than the α isoform to promote TAG secretion in hepatoma cells (Khalil et al., 2009). By contrast, adenoviral expression of Lpin1β in primary hepatocytes modestly reduces TAG secretion (Chen et al., 2008). Furthermore, Lpin1 isoforms have been shown to form homo- and hetero-oligomers (Liu et al., 2010). Thus, regulation of Lpin1 expression, isoform splicing, and function is complex, and precise molecular mechanisms mediating these effects may be tissue-specific (Phan and Reue, 2005), differ in fed/fasting transitions and between cellular and animal models. However, our studies using the Lpin1 exon 6 minigene indicate that both knockdown and overexpression of Sfrs10 alter Lpin1 splicing, independent of Lpin1 transcriptional regulation. Moreover, effects of reduced Sfrs10 expression to promote lipogenesis are dependent upon the Lpin1β isoform. Thus, we propose a model in which reduced expression and splicing activity of Sfrs10 alters Lpin1 splicing, favoring generation of the Lpin1β isoform, contributing to lipid synthesis and hypertriglyceridemia (Figure 6).
Figure 6. Human obesity is associated with decreased expression of RNA processing genes and can influence metabolic phenotypes.
Reduced expression of the splicing factor SFRS10 alters splicing of LPIN1, leading to dysregulation of lipogenic pathways and contributing to hypertriglyceridemia. Other alterations in RNA processing in human obesity should be identified (dashed arrows).
We have considered several possible mechanisms by which Lpin1β-dependent effects of reduced Sfrs10 expression could enhance lipogenesis and VLDL secretion. Firstly, lipin1 has PAP-1 activity, yielding DAG and thus promoting TAG synthesis (Donkor et al., 2007). However, we observed no change in TAG synthesis from labeled palmitate in HepG2 cells with Sfrs10 knockdown, and there were no differences in PAP-1 activity, nor in phosphatidic acid or DAG content (not shown), in liver from Sfrs10 heterozygous vs. wild type mice. These data likely reflect the normal mRNA expression of Lpin1, 2, and 3 in the setting of reduced Sfrs10 expression. While we recognize that activity assays and static measures of PA and DAG do not assess potential increases in flux through the PA-DAG pathway, these data indicate that effects of Sfrs10 to promote lipogenesis and hepatic VLDL secretion are unlikely to be mediated via increased lipin1-dependent PAP-1 activity.
A second mechanism potentially contributing to lipin1β-dependent effects on lipogenesis is its role as transcriptional coactivator. Lpin1 overexpression in liver and adipose increases expression of genes modulating lipid oxidation (Finck et al., 2006; Donkor et al., 2008), potentially via interactions with PGC-1α and PPARα. In the context of Sfrs10 reductions, Lpin1β effects on lipid oxidation are not observed. Rather, increased lipogenesis appears to be the dominant mechanism mediating TAG accumulation in HepG2 cells, as evidenced by LPIN1β-dependent increases in lipogenic gene expression, and increased synthesis of TAG from acetate in response to SFRS10 knockdown. We did not observe increased TAG secretion into the medium in HepG2 cells (data not shown), possibly reflecting the impaired capacity of HepG2 cells to secrete TAG compared to hepatocytes in vivo (Gibbons et al., 1994). However, the prominent effects of Sfrs10 on lipogenesis were confirmed in vivo, as demonstrated by the striking increases in lipogenic gene expression and parallel increases in TAG secretion and plasma VLDL-TAG in Sfrs10 heterozygous mice. Our data support a role for differential transcriptional effects of the β vs. α splice variants. While we do not yet fully understand mechanisms responsible for these effects, these splice variants have differential cellular localization (Peterfy et al., 2005); the Lpin1β polybasic motif may play a particularly important role in nuclear localization and regulation of gene expression, again independently of PAP-1 activity (Ren et al., 2010). Emerging data highlight the complexity of this system, as subcellular localization of Lipin1β in turn may be regulated by local phospholipase activity (Huang et al., 2011).
We recognize that general dysregulation of RNA processing gene expression, as observed in our obese and insulin resistant human cohorts, may have an impact on a broad range of cellular pathways, and that we have focused solely on effects of SFRS10 as a representative gene (Zhong et al., 2009). Furthermore, LPIN1 is most likely just one of multiple potential splicing targets of SFRS10. In fact, numerous exons have the NGAA sequence that can serve as an SFRS10 binding site (Clery et al., 2011). Identification of additional alternative splicing events mediated by SFRS10 and, more broadly, in human obesity, may provide targets that contribute to obesity or insulin resistance-associated phenotypes.
Additional mechanisms may also contribute to dysregulated expression and function of genes regulating mRNA splicing/processing in human obesity. For example, insulin may also regulate SFRS10 expression, as overexpression of constitutively active Fox01 (resistant to nuclear exclusion by insulin) in liver increases expression of Sfrs10 (Zhang et al., 2006). Furthermore, activation of Cdc2-like kinase family proteins (clk) (Jiang et al., 2009) by insulin may alter phosphorylation and activity of splicing factors, including SFRS10 (Stoilov et al., 2004). Future studies are warranted to explore the potential interactions between insulin signaling, regulation of Clk kinases, and SFRS10.
In summary, we have now identified RNA processing genes as a group of genes linked to human obesity. Using the SFRS10-LPIN1 cascade as an example, we demonstrate that altered expression and splicing function of SFRS10 may modulate metabolic pathways critical for obesity and related metabolic phenotypes. These findings have several implications. First, genes and molecules regulating mRNA processing should be investigated as potential candidates in obesity and insulin resistant states. Secondly, alternatively spliced isoforms of known metabolic genes should be identified and characterized, as their alternative splicing may serve as an important regulatory step. A recent publication suggests sequence polymorphisms associated with obesity may also facilitate alternative splicing of obesity genes (Goren et al., 2008). Moreover, identification of pathways regulating alternative splicing in obesity may have implications for other chronic diseases linking with insulin resistance, as suggested for SFRS10 in Alzheimer’s disease (Glatz et al., 2006). Finally, our study suggests that modulation of splicing factors and alternative splicing may be a potential therapeutic target for obesity-associated tissue lipid accumulation and consequent metabolic complications.
EXPERIMENTAL PROCEDURES
Human subjects and tissue biopsies
Biopsies were obtained from two independent human cohorts: Boston (liver) and Finland (muscle). For the liver cohort, intraoperative biopsies were obtained from lean nondiabetic control subjects undergoing cholecystectomy (n=5) and from obese subjects undergoing gastric bypass surgery (n=8). Although these subjects had no known history of IGT or T2D, four of the obese subjects were diagnosed with T2D within 1 week prior to gastric bypass on the basis of oral glucose tolerance testing (National Diabetes Data Group criteria (1979)). For LPIN1β/α ratio determination (Figure 4F) additional liver samples were obtained from subjects with similar metabolic profiles (Pihlajamaki et al., 2009). The muscle cohort consisted of 17 postmenopausal Caucasian women of Finnish ancestry who required cholecystectomy. Muscle biopsies (rectus abdominis) were obtained during abdominal surgery from lean postmenopausal women with normal glucose tolerance (NGT, n=10) or obese women, who were diagnosed with IGT (n=4) or T2D (n=3) during the study. All subjects had normal liver, kidney and thyroid function, no history of excessive alcohol intake, and no major chronic illness. All samples were washed with phosphate-buffered saline, immediately frozen in liquid nitrogen, and stored at −80°C.
Informed consent was obtained from all subjects after the purpose and potential risks of the study were explained. All human studies were performed in accord with the Helsinki Declaration and were approved by local institutional review, including the Institutional Review Boards of the Joslin Diabetes Center and Beth Israel Deaconess Medical Center and Ethics Committee of the Kuopio University Hospital.
Analytical methods in human studies
Plasma glucose was measured using the glucose oxidase method (2300 Stat Plus, Yellow Springs Instrument Co Inc, Yellow Springs, Ohio). Plasma insulin concentration was determined by radioimmunoassay (RIA, Diagnostic Systems Laboratories, Webster, TX) or commercial double-antibody solid-phase radioimmunoassay (Insulin RIA 100, Pharmacia Diagnostics AB, Uppsala, Sweden). Liver fat content was quantified from hematoxylin and eosin-stained sections (Pihlajamaki et al., 2009).
Animal care and treatment
All protocols were approved by the Joslin Institutional Animal Care and Use Committee. Mice were housed 4 per cage in an OLAW-certified animal facility, with 12h light cycle. For HF diet experiments, six week old ICR mice were placed on chow (17% calories from fat) or HF diet (42% calories from milkfat, Harlan Teklad) for 4 months.
Sfrs10 mouse generation and analysis
Mice with heterozygous gene trap insertion at the Sfrs10 locus (Tra2bGt(P142D08)Wrst) were generated using a mouse embryonic stem cell line (ID 3SP142D08 http://www.genetrap.org/cgi-bin/annotation.py?cellline=3SP142D08) containing a Rosabetageo vector inserted in intron 1 of the Sfrs10 gene. Mice were generated by blastocyst injection and transferred to pseudopregnant recipient dams. Sv129S2-derived offspring were intercrossed with C57BL/6J mice. Genotyping was performed by PCR using tail DNA and two different primer pairs, one flanking the insertion site and the other recognizing the neo gene in the inserted sequence (available upon request). For plasma and tissue analysis, 8–10 week old males were fasted overnight and refed ad libitum for 10 hours. Blood glucose was measured using a Bayer Contour glucometer and insulin levels determined by ELISA (Crystal Chem Inc.). Mice were anesthetized with pentobarbital prior to tissue harvest.
Cell culture
Human HepG2 cells, mouse Hepa1c cells and mouse C2C12 myoblasts were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% (HepG2 and Hepa1c) or 20% fetal bovine serum (C2C12). To initiate differentiation, confluent C2C12 cells were incubated in DMEM containing 2% horse serum (Invitrogen). Full differentiation was observed by day 5. For fatty acid incubations, palmitate (Alltech) was complexed with BSA, yielding a final stock of 5 mM.
RNA isolation and expression analysis
Total RNA was isolated from human and mouse tissues with Trizol (Invitrogen) and from HepG2 and C2C12 cells using RNeasy with DNase I treatment (Qiagen). cDNA was synthesized and analyzed by RT-PCR (ABI Prism 7000 or 7700 Sequence Detection System, Applied Biosystems), using SYBR Green (cell/mouse) or Taqman pre-designed Assays-on-Demand (human). Primer sequences are available upon request.
For microarray analysis, 15 μg of cRNA were hybridized to human Affymetrix GeneChip HG-U133A arrays. Target preparation, hybridization and scanning were performed in Joslin Diabetes Center Genomics Core (liver) or Turku Centre for Biotechnology (muscle). Signal intensities were quantitated using GeneChip Operating Software (GCOS). Global scaling was used to standardize signal intensities. MAPPFinder (www.genmapp.org) and GSEA (www.broad.mit.edu/gsea) were used to identify differentially expressed pathways and gene sets (Subramanian et al., 2005). For correlation analyses, Pearson correlations and associated p values for each probe set were calculated by comparison with permuted data (Peter Park, R version 2.0.1).
Protein isolation and Western blotting
Mouse liver or cultured cells were homogenized in buffer containing protease inhibitors (Sigma), sodium fluoride (100 mM), sodium orthovanadate (2 mM) and 1% Triton X-100. Protein concentrations were determined using BCA assay (Pierce, Rockford, IL). Proteins were separated by SDS-PAGE for subsequent Western blotting. Anti-SFRS10 antibody (S4070) was obtained from Sigma, and anti-SFPQ (ab11825) and anti-HNRPK antibodies (ab18195) from Abcam (Cambridge, MA).
siRNA of SFRS10, SF3A1, LPIN1 and LPIN1β isoform
HepG2 cells were transfected at 20–40% confluency and C2C12 myotubes were transfected on differentiation day 3 with 100 nM SFRS10, SF3A1, or LPIN1 SmartPool siRNA or siCONTROL non-targeting siRNA. Dharmafect 3 (C2C12) or 4 (HepG2) transfection reagent was used (Dharmacon, Lafayette, Colorado). We designed 3 siRNAs specifically targeting exon 6 of LPIN1 (Ensembl release 61, February 2011). The most specific and efficient siRNA targeting sequence, AAGAACUAGACAGACCUCCUU, was used for subsequent studies. To study siRNA knockdown effects on lipogenesis and lipid accumulation, cells were treated overnight with 1% BSA or 1% BSA/500 μM palmitate. Efficiency of transfection (>70–80%, not shown) was determined using siGLO-RISC free non-targeting siRNA (Dharmacon).
DNA constructs and transfection
The mouse Sfrs10 cDNA (ATCC, Manassas, VA) was cloned into the pAdTrack-CMV vector, containing GFP as tracer. Hepa1c cells were transiently transfected using PolyFect (Qiagen, ratio 3:1 DNA/Polyfect) with either pAdTrack-Sfrs10 or pAdTrack-CMV (control) plasmids. To enrich for Sfrs10-expressing cells, GFP-positive cells were selected through FACS (Flow Cytometry Core, Joslin) and mRNA analyzed from >100,000 GFP-positive cells.
In vivo cellular splicing assay
The minigene was constructed using a fragment of human LPIN1 exon 6 flanked by 500 nt intronic regions, which was cloned into an exon trap vector (Stoss et al., 1999). Effects of SFRS10 on inclusion of LPIN1 exon 6 were investigated by transfecting 1 ug of LPIN1 exon 6 minigene into 2 cell models: (a) HepG2 cells (Mirus transfection reagent, Madison, WI) 2 days after SFRS10 siRNA transfection (as above), and (b) HEK293 cells, together with SFRS10, using calcium phosphate. Cells were harvested next day and RNA extracted (Qiagen, Valencia, CA). cDNA was created using plasmid-specific primers, and PCR done using minigene-specific primers flanking LPIN1 exon 6.
Lipogenesis, TAG synthesis and accumulation, palmitate oxidation, PAP-1 assay, plasma lipoprotein profile and VLDL secretion
Labeled [14C]-acetate (13 mM, 1μCi/6 well plate, lipogenesis) or [14C]-palmitate (50 nM, 5μCi/6 well plate, TAG synthesis) was added to the media for 2 hours at day 4 after siRNA transfection. Cells were lysed as described above, and lipids extracted with chloroform/methanol. Lipogenesis was assessed by measuring radioactivity in the lipid fraction. Thin layer chromatography (TLC) with Silica Gel G plates (Analtech, Newark, DE) and hexane/diethyl ether/acetic acid (70/29/1) as a solvent was used to separate fractions. Incorporation of [14C]-palmitate into TAG fraction of conditioned medium was performed by lipid extraction and TLC separation of lipid classes (see above). To assess palmitate oxidation, cells were pretreated for 4 hours with 125 μM palmitate, followed by addition of [14C]-palmitate (100 μM, 1 μCi/12 well plate); released CO2 was trapped in 0.1N KOH for 2 hours. Mg+2-dependent PAP-1 activity was assessed in Triton X-100 extracts from wild type or SFRS10 heterozygous mice after an overnight fast and refeeding for 10 hours (Ren et al., 2010).
To measure TAG content from cells or liver tissue, lipids were extracted with chloroform/methanol, total TAG content determined using Triglyceride Assay Kit (Sigma). LPA levels were measured from flash-frozen cell pellets by mass spectrometry (Xiao et al., 2000). Plasma lipoprotein profile was determined by FPLC (Vanderbilt DRTC lipid core).
To measure VLDL secretion, mice were fasted overnight and refed for 2 hours. Mice were then fasted for 4 hours before intraperitoneal administration of Tyloxapol (Sigma) at a dose of 500 mg/kg. Tail vein blood samples were obtained before injection (time 0) and at 90 and 180 minutes for measurement of TAG (Sigma).
Statistical analysis
Data analysis was performed with the SPSS/Win programs (version 10.0, SPSS Inc, Chicago, Illinois, USA). A p-value<0.05 was considered statistically significant. Data are presented as mean ± SEM.
Supplementary Material
HIGHLIGHTS.
Expression of splicing factors is decreased in liver and muscle of obese humans
Sfrs10 downregulation increases lipogenesis, VLDL secretion and plasma TAG
SFRS10 directly regulates splicing of LPIN1, a key regulator of lipid metabolism
Modulation of LPIN1 splicing is required for SFRS10-mediated effects on lipogenesis
Acknowledgments
We gratefully acknowledge support from NIH DK062948 (MEP) and DK060837 (MEP and ABG), DK70648 (ABG), M01 RR001032 (GCRC), D36836 (Joslin DERC), Lilly Foundation (MEP), Graetz Fund (MEP), Academy of Finland (ML and JP), Finnish Diabetes Research Foundation (JP and ML), EVO-fund of Kuopio University Hospital (5167, ML), European Union (EUGENE2 LSHM-CT-2004-512013, ML), and EURASNET (SS). JP also received support from Sigrid Juselius Foundation, Maud Kuistila Foundation, Northern Savo Cultural Foundation and Viipuri Tuberculosis Foundation. AM is supported by RO1GM50388 and P20RR021964. HR is the recipient of an American Heart Association Post Doctoral Fellowship. SS was supporWe appreciate the assistance of Chris Burge and Xinshu Xiao, Massachusetts Institute of Technology, with computational identification of splicing targets, and thank Amit Khanna for help with minigene construction. We thank Joyclyn Yee and Martha Vokes for assistance with microarray analysis.
Abbreviations
- BMI
body mass index
- T2D
type 2 diabetes
- NGT
normal glucose tolerance
- IGT
impaired glucose tolerance
- TAG
triglycerides
- LPA
lysophosphatidic acid
- RT-PCR
real-time polymerase chain reaction
Footnotes
Supplemental data include 5 Supplemental Tables and 5 Supplemental Figures.
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal AK, Garg A. Genetic disorders of adipose tissue development, differentiation, and death. Annu Rev Genomics Hum Genet. 2006;7:175–99. doi: 10.1146/annurev.genom.7.080505.115715. [DOI] [PubMed] [Google Scholar]
- Biddinger SB, Hernandez-Ono A, Rask-Madsen C, Haas JT, Aleman JO, Suzuki R, Scapa EF, Agarwal C, Carey MC, Stephanopoulos G, et al. Hepatic insulin resistance is sufficient to produce dyslipidemia and susceptibility to atherosclerosis. Cell Metab. 2008;7:125–134. doi: 10.1016/j.cmet.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland CS, Wang ET, Vu A, David MP, Castle JC, Johnson JM, Burge CB, Cooper TA. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res. 2010;38:7651–7664. doi: 10.1093/nar/gkq614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Bou KM, Blais A, Figeys D, Yao Z. Lipin - The bridge between hepatic glycerolipid biosynthesis and lipoprotein metabolism. Biochim Biophys Acta. 2010;1801:1249–1259. doi: 10.1016/j.bbalip.2010.07.008. [DOI] [PubMed] [Google Scholar]
- Browning JD, Horton JD. Molecular mediators of hepatic steatosis and liver injury. J Clin Invest. 2004;114:147–52. doi: 10.1172/JCI22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Gropler MC, Norris J, Lawrence JC, Jr, Harris TE, Finck BN. Alterations in hepatic metabolism in fld mice reveal a role for lipin 1 in regulating VLDL-triacylglyceride secretion. Arterioscler Thromb Vasc Biol. 2008;28:1738–1744. doi: 10.1161/ATVBAHA.108.171538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung CY, Tso AW, Cheung BM, Xu A, Ong KL, Fong CH, Wat NM, Janus ED, Sham PC, Lam KS. Obesity susceptibility genetic variants identified from recent genome-wide association studies: implications in a chinese population. J Clin Endocrinol Metab. 2010;95:1395–1403. doi: 10.1210/jc.2009-1465. [DOI] [PubMed] [Google Scholar]
- Clery A, Jayne S, Benderska N, Dominguez C, Stamm S, Allain FH. Molecular basis of purine-rich RNA recognition by the human SR-like protein Tra2-beta1. Nat Struct Mol Biol. 2011;18:443–450. doi: 10.1038/nsmb.2001. [DOI] [PubMed] [Google Scholar]
- Croce MA, Eagon JC, LaRiviere LL, Korenblat KM, Klein S, Finck BN. Hepatic lipin 1beta expression is diminished in insulin-resistant obese subjects and is reactivated by marked weight loss. Diabetes. 2007;56:2395–2399. doi: 10.2337/db07-0480. [DOI] [PubMed] [Google Scholar]
- Csaki LS, Reue K. Lipins: multifunctional lipid metabolism proteins. Annu Rev Nutr. 2010;30:257–272. doi: 10.1146/annurev.nutr.012809.104729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donkor J, Sariahmetoglu M, Dewald J, Brindley DN, Reue K. Three mammalian lipins act as phosphatidate phosphatases with distinct tissue expression patterns. J Biol Chem. 2007;282:3450–3457. doi: 10.1074/jbc.M610745200. [DOI] [PubMed] [Google Scholar]
- Donkor J, Sparks LM, Xie H, Smith SR, Reue K. Adipose tissue lipin-1 expression is correlated with peroxisome proliferator-activated receptor alpha gene expression and insulin sensitivity in healthy young men. J Clin Endocrinol Metab. 2008;93:233–239. doi: 10.1210/jc.2007-1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–37. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Finck BN, Gropler MC, Chen Z, Leone TC, Croce MA, Harris TE, Lawrence JC, Jr, Kelly DP. Lipin 1 is an inducible amplifier of the hepatic PGC-1alpha/PPARalpha regulatory pathway. Cell Metab. 2006;4:199–210. doi: 10.1016/j.cmet.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Ghosh N, Patel N, Jiang K, Watson JE, Cheng J, Chalfant CE, Cooper DR. Ceramide-activated protein phosphatase involvement in insulin resistance via Akt, serine/arginine-rich protein 40, and ribonucleic acid splicing in L6 skeletal muscle cells. Endocrinology. 2007;148:1359–1366. doi: 10.1210/en.2006-0750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons GF, Khurana R, Odwell A, Seelaender MC. Lipid balance in HepG2 cells: active synthesis and impaired mobilization. J Lipid Res. 1994;35:1801–1808. [PubMed] [Google Scholar]
- Glatz DC, Rujescu D, Tang Y, Berendt FJ, Hartmann AM, Faltraco F, Rosenberg C, Hulette C, Jellinger K, Hampel H, et al. The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2-BETA1 changes in sporadic Alzheimer's disease. J Neurochem. 2006;96:635–44. doi: 10.1111/j.1471-4159.2005.03552.x. [DOI] [PubMed] [Google Scholar]
- Goren A, Kim E, Amit M, Bochner R, Lev-Maor G, Ahituv N, Ast G. Alternative approach to a heavy weight problem. Genome Res. 2008;18:214–220. doi: 10.1101/gr.6661308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Copps KD, Dong X, Park S, Cheng Z, Pocai A, Rossetti L, Sajan M, Farese RV, White MF. The Irs1 branch of the insulin signaling cascade plays a dominant role in hepatic nutrient homeostasis. Mol Cell Biol. 2009;29:5070–5083. doi: 10.1128/MCB.00138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, Carman GM. Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J Biol Chem. 2010;285:14628–14638. doi: 10.1074/jbc.M110.117747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han GS, Wu WI, Carman GM. The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J Biol Chem. 2006;281:9210–9218. doi: 10.1074/jbc.M600425200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA. piRNA-Associated Germline Nuage Formation and Spermatogenesis Require MitoPLD Profusogenic Mitochondrial-Surface Lipid Signaling. Dev Cell. 2011;20:376–387. doi: 10.1016/j.devcel.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob S, Machann J, Rett K, Brechtel K, Volk A, Renn W, Maerker E, Matthaei S, Schick F, Claussen CD, et al. Association of increased intramyocellular lipid content with insulin resistance in lean nondiabetic offspring of type 2 diabetic subjects. Diabetes. 1999;48:1113–1119. doi: 10.2337/diabetes.48.5.1113. [DOI] [PubMed] [Google Scholar]
- Jiang K, Patel NA, Watson JE, Apostolatos H, Kleiman E, Hanson O, Hagiwara M, Cooper DR. Akt2 regulation of Cdc2-like kinases (Clk/Sty), serine/arginine-rich (SR) protein phosphorylation, and insulin-induced alternative splicing of PKCbetaII messenger ribonucleic acid. Endocrinology. 2009;150:2087–2097. doi: 10.1210/en.2008-0818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karni R, de Stanchina E, Lowe SW, Sinha R, Mu D, Krainer AR. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nat Struct Mol Biol. 2007;14:185–93. doi: 10.1038/nsmb1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil M, Sundaram M, Zhang H, Links P, Raven J, Manmontri B, Sariahmetoglu M, Tran K, Reue K, Brindley D, et al. The levels and compartmentalization of phosphatidate phoshatase-1 (lipin-1) control the assembly and secretion of hepatic VLDL. Lipid Res. 2009;50:47–58. doi: 10.1194/jlr.M800204-JLR200. [DOI] [PubMed] [Google Scholar]
- Kishore S, Stamm S. The snoRNA HBII-52 regulates alternative splicing of the serotonin receptor 2C. Science. 2006;311:230–2. doi: 10.1126/science.1118265. [DOI] [PubMed] [Google Scholar]
- Kotronen A, Westerbacka J, Bergholm R, Pietilainen KH, Yki-Jarvinen H. Liver fat in the metabolic syndrome. J Clin Endocrinol Metab. 2007;92:3490–7. doi: 10.1210/jc.2007-0482. [DOI] [PubMed] [Google Scholar]
- Lee GH, Proenca R, Montez JM, Carroll KM, Darvishzadeh JG, Lee JI, Friedman JM. Abnormal splicing of the leptin receptor in diabetic mice. Nature. 1996;379:632–635. doi: 10.1038/379632a0. [DOI] [PubMed] [Google Scholar]
- Lefai E, Roques M, Vega N, Laville M, Vidal H. Expression of the splice variants of the p85alpha regulatory subunit of phosphoinositide 3-kinase in muscle and adipose tissue of healthy subjects and type 2 diabetic patients. Biochem J. 2001;360:117–26. doi: 10.1042/0264-6021:3600117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GH, Qu J, Carmack AE, Kim HB, Chen C, Ren H, Morris AJ, Finck BN, Harris TE. Lipin proteins form homo- and hetero-oligomers. Biochem J. 2010;432:65–76. doi: 10.1042/BJ20100584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mende Y, Jakubik M, Riessland M, Schoenen F, Rossbach K, Kleinridders A, Kohler C, Buch T, Wirth B. Deficiency of the splicing factor Sfrs10 results in early embryonic lethality in mice and has no impact on full-length SMN/Smn splicing. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq094. [DOI] [PubMed] [Google Scholar]
- Nayler O, Cap C, Stamm S. Human transformer-2-beta gene (SFRS10): complete nucleotide sequence, chromosomal localization, and generation of a tissue-specific isoform. Genomics. 1998;53:191–202. doi: 10.1006/geno.1998.5471. [DOI] [PubMed] [Google Scholar]
- Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NA, Kaneko S, Apostolatos HS, Bae SS, Watson JE, Davidowitz K, Chappell DS, Birnbaum MJ, Cheng JQ, Cooper DR. Molecular and genetic studies imply Akt-mediated signaling promotes protein kinase CbetaII alternative splicing via phosphorylation of serine/arginine-rich splicing factor SRp40. J Biol Chem. 2005;280:14302–9. doi: 10.1074/jbc.M411485200. [DOI] [PubMed] [Google Scholar]
- Peterfy M, Phan J, Reue K. Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J Biol Chem. 2005;280:32883–9. doi: 10.1074/jbc.M503885200. [DOI] [PubMed] [Google Scholar]
- Peterfy M, Phan J, Xu P, Reue K. Lipodystrophy in the fld mouse results from mutation of a new gene encoding a nuclear protein, lipin. Nat Genet. 2001;27:121–4. doi: 10.1038/83685. [DOI] [PubMed] [Google Scholar]
- Phan J, Reue K. Lipin, a lipodystrophy and obesity gene. Cell Metab. 2005;1:73–83. doi: 10.1016/j.cmet.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Pihlajamaki J, Boes T, Kim EY, Dearie F, Kim BW, Schroeder J, Mun E, Nasser I, Park PJ, Bianco AC, et al. Thyroid Hormone-Related Regulation of Gene Expression in Human Fatty Liver. J Clin Endocrinol Metab. 2009 doi: 10.1210/jc.2009-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Federico L, Huang H, Sunkara M, Drennan T, Frohman MA, Smyth SS, Morris AJ. A phosphatidic acid binding/nuclear localization motif determines lipin1 function in lipid metabolism and adipogenesis. Mol Biol Cell. 2010;21:3171–3181. doi: 10.1091/mbc.E10-01-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu D, Oh KJ, Jo HY, Hedrick S, Kim YN, Hwang YJ, Park TS, Han JS, Choi CS, Montminy M, et al. TORC2 regulates hepatic insulin signaling via a mammalian phosphatidic acid phosphatase, LIPIN1. Cell Metab. 2009;9:240–251. doi: 10.1016/j.cmet.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Salomonis N, Schlieve CR, Pereira L, Wahlquist C, Colas A, Zambon AC, Vranizan K, Spindler MJ, Pico AR, Cline MS, et al. Alternative splicing regulates mouse embryonic stem cell pluripotency and differentiation. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0912260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesti G, Marini MA, Tullio AN, Montemurro A, Borboni P, Fusco A, Accili D, Lauro R. Altered expression of the two naturally occurring human insulin receptor variants in isolated adipocytes of non-insulin-dependent diabetes mellitus patients. Biochem Biophys Res Commun. 1991;181:1419–1424. doi: 10.1016/0006-291x(91)92097-4. [DOI] [PubMed] [Google Scholar]
- Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- Stoilov P, Daoud R, Nayler O, Stamm S. Human tra2-beta1 autoregulates its protein concentration by influencing alternative splicing of its pre-mRNA. Hum Mol Genet. 2004;13:509–524. doi: 10.1093/hmg/ddh051. [DOI] [PubMed] [Google Scholar]
- Stoss O, Stoilov P, Hartmann AM, Nayler O, Stamm S. The in vivo minigene approach to analyze tissue-specific splicing. Brain Res Brain Res Protoc. 1999;4:383–394. doi: 10.1016/s1385-299x(99)00043-4. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suviolahti E, Reue K, Cantor RM, Phan J, Gentile M, Naukkarinen J, Soro- Paavonen A, Oksanen L, Kaprio J, Rissanen A, et al. Cross-species analyses implicate Lipin 1 involvement in human glucose metabolism. Hum Mol Genet. 2006;15:377–86. doi: 10.1093/hmg/ddi448. [DOI] [PubMed] [Google Scholar]
- Thorleifsson G, Walters GB, Gudbjartsson DF, Steinthorsdottir V, Sulem P, Helgadottir A, Styrkarsdottir U, Gretarsdottir S, Thorlacius S, Jonsdottir I, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41:18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–54. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao-Borengasser A, Rasouli N, Varma V, Miles LM, Phanavanh B, Starks TN, Phan J, Spencer HJ3, McGehee RE, Jr, Reue K, et al. Lipin expression is attenuated in adipose tissue of insulin-resistant human subjects and increases with peroxisome proliferator-activated receptor gamma activation. Diabetes. 2006;55:2811–8. doi: 10.2337/db05-1688. [DOI] [PubMed] [Google Scholar]
- Yechoor VK, Patti ME, Saccone R, Kahn CR. Coordinated patterns of gene expression for substrate and energy metabolism in skeletal muscle of diabetic mice. Proc Natl Acad Sci U S A. 2002;99:10587–10592. doi: 10.1073/pnas.142301999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yechoor VK, Patti ME, Ueki K, Laustsen PG, Saccone R, Rauniyar R, Kahn CR. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: An in vivo analysis in MIRKO mice. Proc Natl Acad Sci U S A. 2004 doi: 10.1073/pnas.0407574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeharia A, Shaag A, Houtkooper RH, Hindi T, de Lonlay P, Erez G, Hubert L, Saada A, de Keyzer Y, Eshel G, et al. Mutations in LPIN1 cause recurrent acute myoglobinuria in childhood. Am J Hum Genet. 2008;83:489–494. doi: 10.1016/j.ajhg.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Patil S, Chauhan B, Guo S, Powell DR, Le J, Klotsas A, Matika R, Xiao X, Franks R, et al. FoxO1 regulates multiple metabolic pathways in the liver: effects on gluconeogenic, glycolytic, and lipogenic gene expression. J Biol Chem. 2006;281:10105–17. doi: 10.1074/jbc.M600272200. [DOI] [PubMed] [Google Scholar]
- Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.