Summary
Bmi1 is required for the self-renewal of stem cells in many tissues including the lung epithelial stem cells, Bronchioalveolar Stem Cells (BASCs). Imprinted genes, which exhibit expression from only the maternally- or paternally-inherited allele, are known to regulate developmental processes but their role in adult cells remains a fundamental question. Many imprinted genes were de-repressed in Bmi1 knockout mice, and knockdown of Cdkn1c (p57) and other imprinted genes partially rescued the self-renewal defect of Bmi1 mutant lung cells. Expression of p57 and other imprinted genes was required for lung cell self-renewal in culture and correlated with repair of lung epithelial cell injury in vivo. Our data suggest that Bmi1-dependent regulation of expressed alleles at imprinted loci, distinct from imprinting per se, is required for control of lung stem cells. We anticipate that the regulation and function of imprinted genes is crucial for self-renewal in diverse adult tissue-specific stem cells.
Introduction
Many adult tissues including the lung maintain homeostasis or achieve injury repair via stem cell populations. In the distal murine lung, Clara cells, the bronchiolar non-ciliated columnar epithelial cells, and alveolar type II cells (AT2) cells, the secretory epithelial cells in the alveolar space, have long been proposed to function as stem or progenitor cells. Clara cells are a self-maintaining cell population that gives rise to new Clara cells and ciliated cells during steady state lung homeostasis, demonstrating their role as adult progenitor cells.(Rawlins et al., 2009) AT2 cells, similarly, are thought to function during development and after injury in adults as progenitors for the alveolar type I (AT1) cells that perform gas exchange. BASCs are an adult lung stem cell population that proliferates in response to distal lung cell injury when either Clara cell or AT1 cell damage occurs. BASCs may uniquely have bronchiolar and alveolar lineage potential as demonstrated by their ability to give rise to Clara and AT2 cells in culture, yet this activity remains to be shown in vivo.(Kim et al., 2005) Ciliated cells undergo morphological changes after Clara cell injury in vivo, yet they do not directly contribute to lung repair and may be considered differentiated cells of the distal lung.(Rawlins et al., 2007)
Bmi1, a member of the Polycomb Repressive Complex 1 (PRC1), is required for the self-renewal of adult stem cells including BASCs.(Dovey et al., 2008; Kim et al., 2005; Park et al., 2004; Sauvageau and Sauvageau, 2010). Serial plating of BASCs serves as an assay for measuring the self-renewal capacity of lung stem cells, and Bmi1-deficient BASCs exhibited little or no self-renewal. Furthermore, Bmi1 knockout mice exhibited an impaired ability to repair Clara cell injury that was associated with failure of BASC expansion in vivo.(Dovey et al., 2008) In the lung and other tissues, suppression of the Cdkn2a locus encoding p16/p19 is an important function of Bmi1 that is required for stem cell self-renewal, yet this activity cannot account for the full range of Bmi1 functions. Reducing levels of p16/p19 in Bmi1 mutants in vivo or by knockdown in culture only partially rescued the BASC defects,(Dovey et al., 2008) suggesting that other Bmi1 target genes are important in controlling their self-renewal.
Results
Imprinted gene de-repression in Bmi1-deficient lung cells
To test our hypothesis that additional targets of Bmi1 are required for the self-renewal of lung stem cells, we compared gene expression profiles of FACS-purified cell populations from Bmi1 wild-type and mutant lungs. As expected, multiple homeodomain genes were de-repressed in Bmi1 mutant lung cells, as were Cdkn2a (p16/p19) and Cdkn2b (p15), (Figure 1A, Table S1). Gene expression differences were validated by quantitative RT-PCR (qPCR) for 25 out of 30 genes examined (Figure 1B; Table S1). Other INK4 or CIP/KIP CDK inhibitor genes, including Cdkn1a (encoding p21) and Cdkn1b (encoding p27), were not differentially expressed (Fig 1B), even though p21 is a Bmi1 target in neural stem cells.(Fasano et al., 2007) However, a different CIP/KIP family member, Cdkn1c, encoding p57 (referred to hereafter as p57 to designate gene or protein), was highly up-regulated in Bmi1 mutant lung cells (Figure 1A,B). p57 levels were 6.8- and 21.5-fold higher in Bmi1 mutant cells compared to wild-type cells by microarray and qPCR, respectively (p= 2.83E-4 and 3.38E-13, respectively).
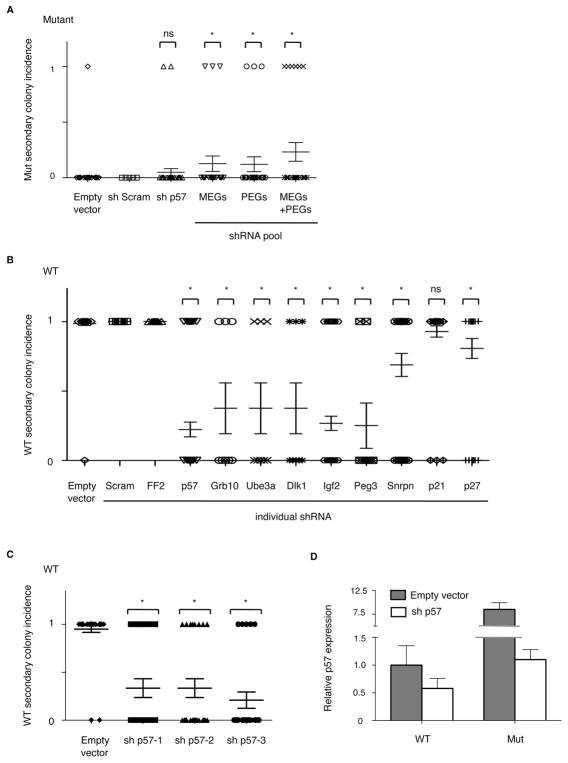
Figure 1.
De-repression of imprinted genes in Bmi1 mutant lung cells. (A) Gene expression differences of homeobox (hox) genes, paternally expressed genes (PEGs), and maternally expressed genes (MEGs) from three samples each of Bmi1 wild-type (WT) and mutant lung cells, as assessed by Affymetrix Mouse 430 2.0 gene expression microarray. Red indicates up-regulated expression, green is down-regulated expression, and differences with FDR (False Discovery Rate) <0.1 are marked on left of heatmap (in red, up-regulated; in green, down-regulated). (B) Validation of differential expression of homeodomain genes and CDK inhibitors in Bmi1 wild-type and mutant lung cells by qPCR. Expression from mutant samples (grey bars) shown as fold change relative to wild-type samples (black bars) set to 1. (C) Table showing the 10 gene sets revealed to be most overrepresented in Bmi1 mutant samples using GSEA. Red, upregulated gene sets; green, downregulated gene sets. (D) qPCR analysis of expression of imprinted gene network members in lung cells from wild-type (black bars) and mutant (grey bars) mice, normalized as in (b). The differential expression of all genes shown was statistically significant. (E) Comparison of p57 expression in wild-type (black bars) and mutant (gray bars) ciliated cells (Epcam-positive, CD24-high), BASCs (Epcam-positive, Sca-1-low, CD24-low) and AT2 cells (Epcam-positive, Sca-1-neg, CD24-pos/neg). Asterisk (*) indicates P≤0.05. Error bars, standard deviation. See also Table S1, Table S2, Fig S1.
p57 belongs to another set of genes, previously known to be regulated by imprinting, that also demonstrated significant de-repression in the Bmi1 mutant cells. Importantly, imprinted genes were amongst the most highly dysregulated gene sets identified in the mutant cells using gene set enrichment analysis (GSEA Enrichment Score = 0.67, FDR q-val < 0.001) (Figure 1C; Table S2). Of the 84 imprinted genes (MRC Harwell, mousebook.org) queried by microarray or qPCR, 33 were significantly dysregulated in Bmi1 mutant lung cells (Figure 1B–D, Table S1). Both maternally expressed genes (MEGs) and paternally expressed genes (PEGs) were significantly upregulated in Bmi1 mutants (Figures 1A–D).
A particularly interesting subset of imprinted genes, an imprinted gene network (IGN) that is transcriptionally down-regulated during postnatal growth in several tissues including the lung and implicated in somatic growth control,(Finkielstain et al., 2009; Gabory et al., 2009; Lui et al., 2008; Varrault et al., 2006) was de-repressed in Bmi1 mutant lung cells. For example, the IGN members H19, Dlk1, and Igf2 were 5.8-, 3.4-, 5.1-fold upregulated in mutant cells by array, respectively, and increased expression was validated by qPCR (Figure 1D, Table S1). All of the IGN members we examined were significantly de-repressed in the lung in the absence of Bmi1, including p57, H19, Dlk1, Igf2, Plagl1, Grb10, Gtl2 (Meg3), Mest, Ndn, and Peg3 (Figure 1D, Table S1).
We next determined the specificity of de-repression of p57 and other imprinted genes in the Bmi1-deficient lung stem cell compartment. There was no significant difference in the number of BASCs in wildtype and Bmi1-deficient lungs (data not shown and (Dovey et al., 2008)), ruling out gene expression differences caused by different BASC abundance. The CD31-negative, CD45-negative Sca-1-positive lung cell population typically used in self-renewal experiments contained BASCs and ciliated cells.(Kim et al., 2005) We used an improved cell sorting strategy to further enrich each cell type on the basis of Sca-1, Epcam and CD24 abundance and further examine imprinted gene expression patterns. The CD31-, CD45-negative Epcam-positive Sca-1-low, CD24-low distal lung cell population contains epithelial lung stem/progenitor cells and allows for further BASC enrichment, whereas the Epcam-positive cells with high levels of CD24 are ciliated cells and the Epcam-positive Sca-1-negative cells are AT2 cells (McQualter et al., 2010)(Figure S1, unpublished data, Kim Lab). The IGN members p57, Igf2 and Dlk1 were elevated in the more purified Bmi1-deficient BASCs (Figure 1E and data not shown). Importantly, p57, the most differentially expressed imprinted gene on our microarray list (Table S1), exhibited BASC-specific de-repression; Bmi1-mutant BASCs had 1.7-fold more p57 than their wild-type counterparts (p = 0.05) whereas there was no difference in the abundance of p57 in ciliated cells or AT2 cells (relative to wild-type BASC levels set to 1, 2.4-fold mutant ciliated vs. 3.9-fold wild-type ciliated, p = 0.50 and 0.30-fold mutant AT2 vs. 0.43-fold wild-type AT2, p = 0.50,)(Figure 1E).
Functional analysis of imprinted genes in lung cell self-renewal
We used knockdown to determine that elevated imprinted gene expression levels contributed to the self-renewal defect of Bmi1 mutant lung cells. Uninfected Bmi1 mutant lung cells or those infected with negative control retrovirus rarely formed secondary colonies (Figure 2A, Dovey et al., 2008). Knockdown of p57 in Bmi1-mutant cells significantly reduced p57 levels (Fig 2D, S2) and increased secondary colony formation of Bmi1-mutant cells (4.8% vs 0.7% from shEmpty control; p = 0.09; Figure 2A), although the difference was not statistically significant. To test the idea that simultaneous dysregulation of imprinted genes caused self-renewal defects, we used a pooled shRNA approach to more substantially minimize imprinted gene over-expression in Bmi1 mutant cells. Interestingly, small pools of shRNAs directed against MEGs (p57, Grb10, Ube3a) or PEGs (Igf2, Snrpn, Peg3, Dlk1) confirmed for knockdown activity (Figure S2) further improved secondary epithelial colony formation to 13% and 12%, respectively (p=0.008 and p=0.001). The most significant increase in Bmi1 mutant secondary colonies was observed with a combined pool of MEG and PEG shRNAs (23%; p=2E-7; Figure 2A, Fig S2), consistent with the hypothesis that coordinated, Bmi1-dependent regulation of imprinted genes is required for proper lung stem cell self-renewal.
Figure 2.
Lung cell self-renewal in culture is dependent on precise regulation of imprinted gene expression. Formation of secondary colonies following knockdown of various imprinted genes in Bmi1 mutant (A) or WT (B,C) lung cells by retroviral short hairpin RNA (shRNA) expression is shown. Each data point represents an individual well of primary colony cells plated for secondary colony formation, with 0 plotted for wells that lacked secondary colonies and 1 for wells with 1 or more secondary colonies. Infection with empty retroviral vector, sh-Scrambled (Scram), and sh-Firefly luciferase (FF2) served as negative controls. Additional controls, Snrpn, p21, p27. (A) Secondary colony formation in mutant cells infected with empty, Scram shRNA, p57 (sh2) shRNA, a MEG pool that included p57 (sh3), Grb10, and Ube3a shRNAs, a PEG pool that included Igf2, Snrpn, Peg3, and Dlk1 shRNAs, or a combined pool with all shRNAs (MEG + PEG) is shown. (B,C) Secondary colony formation in wild-type cells after infection with individual shRNA as indicated or 3 different shRNAs directed against p57. Results in A,B,C are data from four, three and three experiments, respectively. Asterisk (*) indicates P<0.05. D. qPCR analysis of knockdown efficiency of p57 by sh3 in wild-type or Bmi1 mutant lung cells; p57 expression was measured relative to empty retrovirus control-infected wild-type cells. Error bars, standard error (A–C) or standard deviation (D). See also Fig S2.
Several of the imprinted genes for which combined knockdown rescued Bmi1 mutant cells were also required for wild-type lung cell self-renewal. Virus expressing individual shRNAs for six different imprinted genes negatively impacted wild-type lung cell self-renewal (Figure 2B) (p57, Grb10, Ube3a, Dlk1, Igf2, Peg3; p values compared to shEmpty, 2.9E-29, 1.6E-12, 1.6E-12, 1.6E-12, 3.4E-26, and 3.4E-17, respectively); p57 and Igf2 knockdown had the most significant impact on lung stem cell function. We confirmed a requirement for p57 in the self-renewal of wild-type lung cells using three different p57 shRNAs (Figure 2C), which all effectively reduced p57 expression levels (Fig S2). Individual shRNA for Snrpn, an imprinted gene that was not differentially expressed in Bmi1-deficient lung cells (Table S1), impaired self-renewal (67% secondary colony formation compared to 99% shEmpty, p=2.6E-6)(Figure 2B), although to a lesser extent than knockdown of p57 and other IGN members (e.g. 22% secondary colony formation shp57). Similar results were seen with shRNA against p27 (81% secondary colony formation, p=0.00089), a non-imprinted p57 family member, whereas shRNA for the third non-imprinted p57 family member, p21, did not impair secondary colony formation (p=0.12)(Figure 2B).
We directly compared p57 gene expression levels after knockdown in mutant and wild-type cells (Figure 2D). Importantly, mutant lung cells infected with empty vector virus exhibited elevated levels of p57 compared with equally treated wild-type cells, confirming the de-repression of p57 even in cultured, infected mutant cells. p57 shRNA in mutant cells restored p57 expression to wild-type control levels. Notably, Bmi1-deficient lung cells have elevated levels of many other imprinted genes, which also likely contributed to inability to completely rescue secondary colony forming-ability solely with p57 knockdown. p57 shRNA in wild-type lung cells further reduced p57 expression levels by 50% below normal wild-type levels, correlating with a reduction in secondary colony formation. Thus, increased p57 levels are incompatible with self-renewal, as are insufficient p57 levels. Together these data were consistent with the hypothesis that lung cell self-renewal relies on exquisite control of imprinted genes, perhaps particularly by function of the IGN.(Varrault et al., 2006) Since p57 exhibited more specific differential expression in BASCs than other imprinted genes, we chose to further examine p57 regulation in vivo.
p57 dynamic expression in the lung injury response
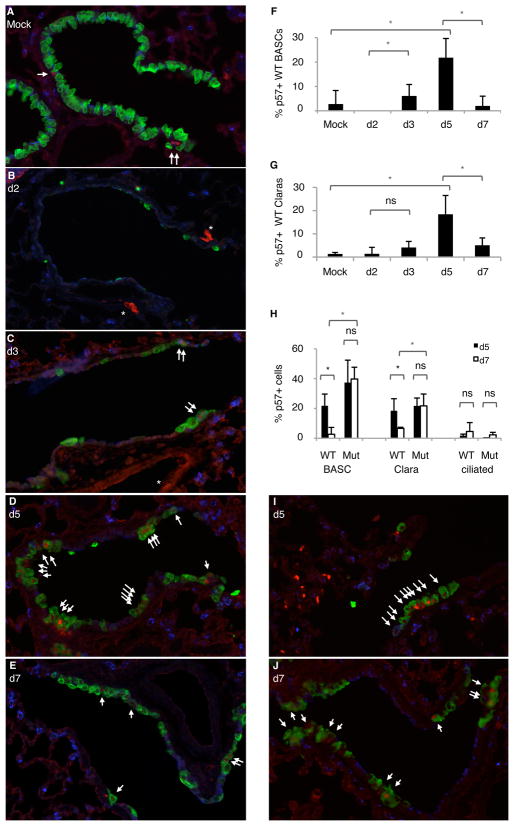
To further test the importance of p57 in lung stem cell function, we evaluated the normal expression patterns of p57 in vivo following lung injury with naphthalene, which is toxic to Clara cells but not to BASCs.(Kim et al., 2005) We performed four-color IF to score the incidence of Clara cells (CCSP-positive, SPC-negative), and BASCs (CCSP-positive, SPC-positive) with positive p57 staining at key time points during the course of lung injury repair (Figure 3A–E, Figure S3A). The expression of p57 in lungs from mock treated adult mice was relatively scarce, with expression restricted to BASCs and Clara cells (2.8 +/− 5.6% and 1.3 +/−0.62%, of p57+ cells, respectively) and most frequently found in ciliated cells (14.3 +/− 3.9% of p57+ cells) (Figure 3F,G, data not shown). As expected, two days following lung injury, most Clara cells were cleared from the airways, with only ciliated cells and BASCs persisting (Figure 3B; data not shown and (Kim et al., 2005)). The number of BASCs positive for p57 was lowest at this time point (none were detected), and there was a significant increase in p57+ BASCs three days after naphthalene (6.1%, p=0.04) that peaked after five days (21.8%, p=0.01) (Figure 3F). Following the known pattern of BASC expansion and decline after appreciable repair of Clara cell injury,(Kim et al., 2005) there was a rapid decline in p57+ BASCs by day 7 after injury (2%, p=0.01) (Figure 3F). The percentage of Clara cells with p57 similarly peaked at day 5 (18.5%, p=0.006) and declined at day 7 after injury (5.1%, p=0.02)(Figure 3D,E,G). In contrast to BASCs and Clara cells, the incidence of ciliated cells (assessed with acetylated tubulin staining) with p57 expression was not different at days 5 and 7 after injury (Figure 3H). Thus, a dynamic pattern of p57 expression correlated with lung stem cell activity in vivo.
Fig. 3.
p57 is dynamically regulated during bronchiolar cell repair in wild-type lung but not Bmi1 mutants. Sections were stained with antisera for p57 (red), SP-C (blue), and CCSP (green) along with DAPI (not shown) and four color imaging was performed to score BASCs (CCSP+ SPC+) and Clara cells (CCSP+ SPC-). Ciliated cells were assessed by acetylated tubulin staining. (A–E) Sections from wild-type lungs, isolated from vehicle-treated control mice (A, mock) or mice 2 (B), 3 (C), 5 (D), or 7 (E) days following treatment with naphthalene. For simplicity, only p57, CCSP and SPC staining are shown. Arrows, p57 positive Clara cells. (*), Background red signal in red blood cells/blood vessels (*). (F,G) Fluctuation in p57 levels in wild-type lungs at different time points after naphthalene treatment in BASCS (F) and Clara cells (G) was determined by four color staining as described above. (H) Quantification of BASCs, Clara cells or ciliated cells positive for p57 in Bmi1 mutant and wild-type lung cells at day 5 (d5, black bars) and day 7 (d7, white bars) following naphthalene-induced lung injury. Wild-type data are same as shown in F,G. Asterisk (*) indicates P<0.05; ns = non-significant difference. Error bars, standard deviation. (I,J) Bmi1 mutant lungs 5 days (I) or 7 days (J) after naphthalene treatment were stained as in A–E. See also Fig S3.
Bmi1 deficiency abrogated the normal dynamic pattern of p57 expression in lung stem cells after injury in vivo. The percentage of p57+ BASCs and Clara cells in mutant and wild-type lungs was comparable five days after naphthalene (BASCS: 37.4% mutant vs. 21.8% wild-type, p=0.13; Claras: 21.7 mutant vs. 18.5% wild-type, p=0.57) (Figure 3H). However, p57+ stem cell populations did not decline after naphthalene in Bmi1 mutants (37.4% day 5 vs. 39.77% day 7, p =0.82 for mutant BASCs and 21.7% day 5 vs. 21.8% day 7, p = 0.99 for mutant Claras) (Figure 3J), and retained a significant increase relative to wild-type populations at day 7 (BASCs: 39.8% mutant vs. 2% wild-type, p=0.0004; Claras: 21.8% mutant vs. 5.1% wild-type, p=0.01)(Figure 3H). Importantly, the aberrant expression pattern of p57 was correlated with a significant deficiency in BASC expansion and Clara cell repair in Bmi1 mutant lungs detectable at day 7 that persisted even 30 days after naphthalene treatment (Figure S3B, data not shown and (Dovey et al., 2008)). p57 expression in ciliated cells, the differentiated cells of the distal lung, was similar in wild-type and Bmi1-deficient lungs after naphthalene treatment (Figure 3H). These data suggested that Bmi1 regulation of p57 is crucial in stem cells during lung injury repair in vivo.
Bmi1-dependent regulation of imprinted loci
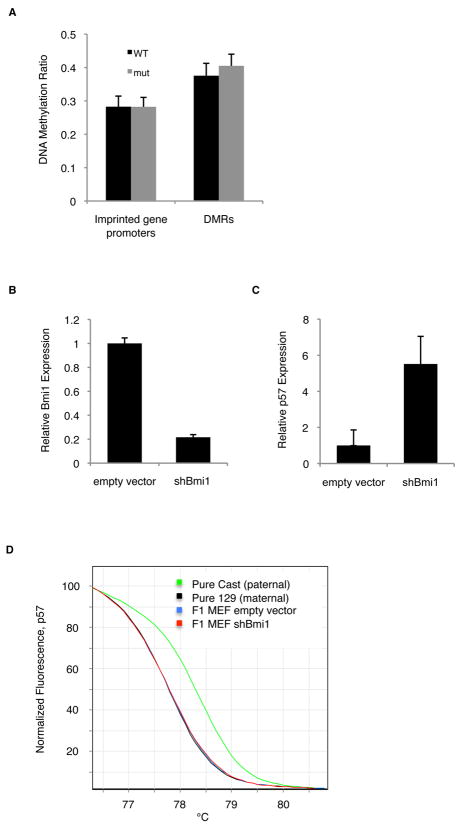
We next wished to determine the molecular nature of imprinted gene deregulation in Bmi1 mutant cells. The mono-allelic expression of imprinted genes is regulated by DNA methylation at maternal or paternal Differentially Methylated Regions (DMRs), many of which regulate the expression of imprinted genes in clusters.(Edwards and Ferguson-Smith, 2007) We confirmed that p57 and Gtl2 (Meg3) showed mono-allelic, imprinted expression in wild-type BASCs (Figure S4A). To evaluate whether the de-repressed expression of imprinted genes in Bmi1 mutant lungs was due to loss of DNA methylation at DMRs, we performed RRBS (reduced representation bisulfite sequencing) (Gu et al.) of Bmi1 mutant and wild-type BASCs. A high degree of sequence coverage was achieved genome-wide in all samples, including the promoter regions of known imprinted genes and germline DMRs (not shown). DNA methylation levels at all DMRs and imprinted gene promoters from wild-type and Bmi1 mutant cells were comparable (37.6% wild-type vs 40.5% mutant; p=0.57 for DMRs and 28.3% vs 28.2%, p=0.99 for promoters; Figure 4A). Methylation at individual imprinted gene germline DMRs examined was also similar, irrespective of whether most of the genes in the cluster or only some genes in the cluster showed de-repression in mutants (Figure S4B).
Fig. 4.
Bmi1-dependent regulation of imprinted loci. (A) The average DNA methylation ratio was assessed in 4 independent biological replicates each of Bmi1 wild-type and mutant BASCs by RRBS. The DNA methylation ratio was averaged across all wild-type (black bars) or Bmi1 mutant (gray bars) imprinted gene promoter regions and DMRs; error bars represent standard errors for each pair. (B–D) Modulation of Bmi1 levels and high resolution melt curve analysis (HRM) to determine allele-specific expression in F1 Cast/129 MEFs. Levels of Bmi1 (B) and p57 (C) after infection with lentivirus expressing shBmi1 are shown, relative to empty vector virus. HRM results (D) for p57 showing expression from pure Cast cDNA (green, paternal imprinted allele control), pure 129 cDNA (black, maternal expressed allele control), empty virus infected F1 MEFs (blue), and shBmi1 F1 MEFs (red). Values shown are the average of three replicates from a representative experiment. See also Fig S4.
We performed chromatin immunoprecipitation (ChIP) to assess the binding and activity of PRC1 components at imprinted loci. ChIP for the PRC1 component Ring1b enriched for p57, Dlk1 and Igf2 loci in wild-type lung cells similar to the positive control Bmi1 target p16 (Figure S4C). In Bmi1 mutant lung cells, there was a modest decrease in Ring1b enrichment for p16, p57 and Dlk1 loci, but this change was only significant at p57 (p=0.05). Likewise, ChIP for the PRC1 catalytic mark, ubiquitinated histone H2A, enriched for all loci examined in wild-type lung samples, but enrichment in Bmi1 mutant cells was only slightly decreased and did not change at p57 (Figure S4C).
Finally, to evaluate whether Bmi1-dependent regulation of imprinted loci occurs in a bi- or mono-allelic fashion, we modulated Bmi1 levels in an SV40-transformed clonal mouse embryonic fibroblast (MEF) line derived from the F1 progeny of a Castaneus male and a 129 female.(Gimelbrant et al., 2005) Using lentivirus to knockdown Bmi1 levels,(Fasano et al., 2007) we observed an 80% reduction in Bmi1 levels in shBmi1 MEFs compared to MEFs transduced with empty vector (Figure 4B). Importantly, the shBmi1 cells exhibited a 5.5-fold and a 4.5-fold increase in p57 and Igf2 expression, respectively (p = 0.02 and p = 2e-5 respectively)(Figure 4C, Figure S4D), further confirming our findings in lung cells that Bmi1 regulates imprinted gene expression. qPCR coupled with high resolution melt curve analysis (HRM) was used to differentiate between expression products derived from the maternal and paternal alleles with differential SNPs. The p57 expression in F1 MEFs was primarily from the maternal p57 allele, as expected for a MEG (Figure 4D, Fig S4D). In the shBmi1 cells, no shifts were observed in the HRM, indicating that imprinted gene expression remained derived from the maternal, expressed allele despite changes in overall p57 expression levels (Figure 4D, Fig S4C). Gtl2 (Meg3) exhibited a similar pattern of expression from the materal, expressed allele in control and shBmi1 cells (not shown). Therefore, the increased expression of p57 and other imprinted genes that occurs as a consequence of Bmi1 deficiency is unlikely to be from loss of imprinting per se, and rather is a consequence of the dysregulation of the expressed allele at imprinted loci.
Discussion
Our work shows that imprinted genes are important Bmi1 targets in adult lung stem cells. Defects in cultured Bmi1 mutant lung cells were attributable in part to de-repression of imprinted genes, whereas insufficient levels of p57 were also incompatible with wild-type lung stem cell self-renewal. Precise, dynamic control of p57 levels was observed during a time course of lung injury and repair in vivo. There was no evidence that Bmi1 deficiency generally altered DNA methylation of imprinted loci, but instead Bmi1 is implicated by our studies in the regulation of the expressed allele at these loci. Thus, an important mechanism for stem cell self-renewal is the control of expression levels of imprinted genes, providing an extra layer on top of typical imprinting mechanisms to regulate these critical loci.
Our results show for the first time that the PRC1 functions in imprinted gene regulation in adult stem cells. The Polycomb Repressive Complex 2 (PRC2) regulates imprinted gene repression in the placenta and early embryo at a small number of imprinted loci.(Mager et al., 2003) ln our studies, a much broader set of imprinted loci were overexpressed than observed in PRC2 or PRC1 mutant embryos,(Mager et al., 2003; Terranova et al., 2008; Umlauf et al., 2004). DNA methylation was intact in Bmi1 mutant adult lung cells, suggesting that imprinting mechanisms per se were not the cause of de-repression of imprinted genes. Importantly, non-germline DMRs are associated with regulation of expression from imprinted loci (e.g., for Igf2, Feil et al., 1994 and Moore et al., 1997; for p57, Bhogal et al., 2004). We observed an increase in methylation at a silencing non-germline DMR in Bmi1 mutants (Fig. S4b), suggesting that Bmi1-dependent changes in p57 expression do not rely on altered methylation. The non-germline regions regulating Igf2 were not sufficiently covered in our RRBS data set. Therefore, we cannot rule out the possibility that non-germline DMRs or other differentially methylated sites were aberrant in Bmi1 mutant lung cells and were partially responsible for changes in gene expression. On the other hand, our allelic expression analysis supports a role for Bmi1 in regulation of the expressed allele at imprinted loci. Since we have not yet been able to ascertain direct binding of Bmi1 at imprinted loci, the action of Bmi1 in this context may be indirect. Alternatively, other direct modes of PRC1-dependent gene regulation, including chromatin compaction (Eskeland et al., 2010), may be essential for full repression of these loci. We suggest that the Bmi1-dependent regulation of imprinted loci that is tied mechanistically to adult stem cell functions is distinct from the typical DNA methylation imprinting mechanisms that occur in development, adding another level of regulation to fine tune control of the expressed allele at imprinted gene loci.
Our results demonstrate that a delicate, dynamic balance of expression from imprinted gene loci is needed for adult lung stem cell function. We have not yet confirmed that all of the imprinted genes we studied exhibit imprinted expression in BASCs with the exception of p57 and Gtl2. Slc22a3 and Ube3a, known to show tissue-specific imprinting, were predominantly biallelically expressed in BASCs (Fig S4a), reinforcing the idea that Bmi1 modifies the expression of this unique subset of genes independently of classic imprinting mechanisms. We focused on the effects of imprinted gene dysregulation in Bmi1-deficient BASCs, yet it is likely that other adult lung stem cell populations are impacted. Supporting this idea, we also observed dynamic expression of p57 during the naphthalene injury response in Clara cells, another lung stem/progenitor cell population. p57 levels fluctuated in stem cells during a time course of injury repair and high levels of p57 expression were incompatible with self-renewal whereas insufficient levels of p57 also prevented self-renewal. Therefore, the in vitro and in vivo data support a model in which there are important threshold levels of imprinted loci expression for stem cell function.
Bmi1 regulation of both MEGs and PEGs, many of which are thought to operate in an imprinted gene network (IGN) and which are also known to have growth-inhibiting and growth-promoting roles during development, respectively, (Feil, 2009; Kelsey, 2007) was required for lung stem cell self-renewal. Findings related to the IGN are consistent with our results: the IGN similarly includes MEGs and PEGs,(Varrault et al., 2006) manipulation of expression levels of individual IGN members has additive effects on expression levels of other IGN members,(Gabory et al., 2009; Varrault et al., 2006) and changes in expression of the IGN with age in the lung and other tissues is not due to differential methylation.(Lui et al., 2008) The IGN may function to provide an equilibrium between genes of opposing roles in order to facilitate growth control and development (Varrault et al., 2006) and we now link the IGN to stem cell function.
This work may implicate imprinted genes as self-renewal factors in diverse tissue-specific stem cells. Supporting this hypothesis, in a meta-analysis of public microarray datasets, the IGN was overrepresented in Bmi1-knockout lung cells, MEFs, cerebellum, and HSCs yet not in Bmi1-deficient ES cells (Supplemental). p57 is a suggested mediator of HSC quiescence,(Yamazaki et al., 2006) yet its function in stem/progenitor cells has only been shown during embryonic and early postnatal development.(Bilodeau et al., 2009; Dugas et al., 2007; Dyer and Cepko, 2000; Georgia et al., 2006; Joseph et al., 2009; Mascarenhas et al., 2009; Park et al., 2005; Umemoto et al., 2006) PRC1 complex activity appears to be dispensable for self-renewal of embryonic stem (ES) cells, yet is required for silencing target genes important for cell fate decisions in ES cell differentiation.(Bernstein et al., 2007; Leeb et al., 2010) Interestingly, studies comparing ES and iPS cells further implicated an imprinted locus in the differentiation capacity of pluripotent cells.(Liu et al.; Stadtfeld et al.) Therefore, the PRC complexes have separable functions in pluripotent, embryonic, and adult stem cells, with regulation of imprinted loci being a particularly crucial role of PRC1 in adult stem cell self-renewal.
Experimental Procedures
Mice, tissues and cells
Lung injury was performed as described (Dovey et al., 2008). Immunostaining was done with antisera for p57 (EP2515Y, Epitomics or Ab-3, Thermo Scientific), CCSP for Clara cells and BASCs (sc-25555, Santa Cruz), SP-C for AT2 cells and BASCs (sc-7705, Santa Cruz), acetylated tubulin for ciliated cells (6-11B-1, Sigma-Aldrich). BASCs were scored as cells with individual nuclei positive for CCSP and SPC. Primary lung cells were isolated from 6 week – 4 month old Bmi1 wild-type or mutant mice and cultured as described (Dovey et al., 2008).
Retroviral transduction of shRNA
shRNA sequences were cloned into MSCV-based retroviral vectors. Virus-containing media was concentrated, primary cells were infected with polybrene four days after plating, and resorted by FACS three days after infection, using GFP expression to select for infected cells..
Gene expression analysis
RNA was purified with Absolutely RNA purification kits (Agilent Technologies), amplified using the WT-Ovation Pico RNA Amplification System (Nugen), and analyzed using Affymetrix Mouse Genome 430 2.0 microarrays. qPCR was with TaqMan assays (Applied Biosystems) and a BioRad iQ5 Thermal Cycler with the housekeeping gene GAPDH for normalization.
Statistical analysis
GraphPad Prism (GraphPad Software, Inc) and Excel (Microsoft, Inc) were used for graphing and statistical functions. P-values were calculated using Student’s T-test, with values < 0.05 considered statistically significant..
Supplementary Material
Highlights.
An imprinted gene network is dysregulated in the absence of Bmi1
Precise regulation of imprinted loci is required for lung stem cell function
Imprinted genes may be important stem cell regulators in diverse tissues
Acknowledgments
We thank Kim Lab members for discussions; S. Temple, C. Dulac, C. Gregg, D. Tenen, C. Hetherington, S. Elledge, J. Luo, N. Solimini, M. Hemann, C. Meacham, B. Wilson for reagents and discussions; DFCI and CHB HemOnc FACS facilities; CHB Molecular Genetics Core facility; R. Bronson for histology; L. Zon, S. Orkin, G. Daley for critical reading; M. Goodell for sharing unpublished data. This work was supported by the Ladies Auxiliary to the Veterans of Foreign Wars (CMF), the National Defense Science & Engineering Graduate Fellowship (ANL), RO1 HL090136, U01 HL100402, American Cancer Society Research Scholar Grant #RSG-08-082-01-MGO, the V Foundation for Cancer Research, a Basil O’Conner March of Dimes Starter Award, and the Harvard Stem Cell Institute (CFK).
Footnotes
Supplemental Information is included.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Bhogal B, Arnaudo A, Dymkowski A, Best A, Davis TL. Methylation at mouse Cdkn1c is acquired during postimplantation development and functions to maintain imprinted expression. Genomics. 2004;84(6):961–970. doi: 10.1016/j.ygeno.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Bilodeau S, Roussel-Gervais A, Drouin J. Distinct developmental roles of cell cycle inhibitors p57Kip2 and p27Kip1 distinguish pituitary progenitor cell cycle exit from cell cycle reentry of differentiated cells. Mol Cell Biol. 2009;29:1895–1908. doi: 10.1128/MCB.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovey JS, Zacharek SJ, Kim CF, Lees JA. Bmi1 is critical for lung tumorigenesis and bronchioalveolar stem cell expansion. Proc Natl Acad Sci U S A. 2008;105:11857–11862. doi: 10.1073/pnas.0803574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Ibrahim A, Barres BA. A crucial role for p57(Kip2) in the intracellular timer that controls oligodendrocyte differentiation. J Neurosci. 2007;27:6185–6196. doi: 10.1523/JNEUROSCI.0628-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. p57(Kip2) regulates progenitor cell proliferation and amacrine interneuron development in the mouse retina. Development. 2000;127:3593–3605. doi: 10.1242/dev.127.16.3593. [DOI] [PubMed] [Google Scholar]
- Edwards CA, Ferguson-Smith AC. Mechanisms regulating imprinted genes in clusters. Curr Opin Cell Biol. 2007;19:281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi Al, Wutz A, Bickmore WA. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Molecular Cell. 2010;38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano CA, Dimos JT, Ivanova NB, Lowry N, Lemischka IR, Temple S. shRNA knockdown of Bmi-1 reveals a critical role for p21-Rb pathway in NSC self-renewal during development. Cell Stem Cell. 2007;1:87–99. doi: 10.1016/j.stem.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Feil R, Walter J, Allen ND, Reik W. Developmental control of allelic methylation in the imprinted mouse Igf2 and H19 genes. Development. 1994;120:2933–2943. doi: 10.1242/dev.120.10.2933. [DOI] [PubMed] [Google Scholar]
- Feil R. Epigenetic asymmetry in the zygote and mammalian development. Int J Dev Biol. 2009;53:191–201. doi: 10.1387/ijdb.082654rf. [DOI] [PubMed] [Google Scholar]
- Finkielstain GP, Forcinito P, Lui JC, Barnes KM, Marino R, Makaroun S, Nguyen V, Lazarus JE, Nilsson O, Baron J. An extensive genetic program occurring during postnatal growth in multiple tissues. Endocrinology. 2009;150:1791–1800. doi: 10.1210/en.2008-0868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabory A, Ripoche MA, Le Digarcher A, Watrin F, Ziyyat A, Forne T, Jammes H, Ainscough JF, Surani MA, Journot L, et al. H19 acts as a trans regulator of the imprinted gene network controlling growth in mice. Development. 2009;136:3413–3421. doi: 10.1242/dev.036061. [DOI] [PubMed] [Google Scholar]
- Georgia S, Soliz R, Li M, Zhang P, Bhushan A. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Dev Biol. 2006;298:22–31. doi: 10.1016/j.ydbio.2006.05.036. [DOI] [PubMed] [Google Scholar]
- Gimelbrant AA, Ensminger AW, Qi P, Zucker J, Chess A. Monoallelic expression and asynchronous replication of p120 catenin in mouse and human cells. J Biol Chem. 2005;280:1354–1359. doi: 10.1074/jbc.M411283200. [DOI] [PubMed] [Google Scholar]
- Gu H, Bock C, Mikkelsen TS, Jager N, Smith ZD, Tomazou E, Gnirke A, Lander ES, Meissner A. Genome-scale DNA methylation mapping of clinical samples at single-nucleotide resolution. Nat Methods. 2010;7:133–136. doi: 10.1038/nmeth.1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Andersson ER, Vlachos P, Sodersten E, Liu L, Teixeira AI, Hermanson O. p57Kip2 is a repressor of Mash1 activity and neuronal differentiation in neural stem cells. Cell Death Differ. 2009;16:1256–1265. doi: 10.1038/cdd.2009.72. [DOI] [PubMed] [Google Scholar]
- Kim CF, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, Jacks T. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–835. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes & development. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Luo GZ, Yang W, Zhao X, Zheng Q, Lv Z, Li W, Wu HJ, Wang L, Wang XJ, et al. Activation of the imprinted Dlk1-Dio3 region correlates with pluripotency levels of mouse stem cells. J Biol Chem. 2010;285:19483–19490. doi: 10.1074/jbc.M110.131995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui JC, Finkielstain GP, Barnes KM, Baron J. An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am J Physiol Regul Integr Comp Physiol. 2008;295:R189–196. doi: 10.1152/ajpregu.00182.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager J, Montgomery ND, de Villena FP, Magnuson T. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nature genetics. 2003;33:502–507. doi: 10.1038/ng1125. [DOI] [PubMed] [Google Scholar]
- Mascarenhas MI, Parker A, Dzierzak E, Ottersbach K. Identification of novel regulators of hematopoietic stem cell development through refinement of stem cell localization and expression profiling. Blood. 2009;114:4645–4653. doi: 10.1182/blood-2009-06-230037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQualter JL, Yuen K, Williams B, Bertoncello I. Evidence of an epithelial stem/progenitor cell hierarchy in the adult mouse lung. Proc Natl Acad Sci U S A. 2010;107:1414–1419. doi: 10.1073/pnas.0909207107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Constancia M, Zubair M, Bailleul B, Feil R, Sasaki H, Reik W. Multiple imprinted sense and antisense transcripts, differential methylation and tandem repeats in a putative imprinting control region upstream of mouse Igf2. Proc Natl Acad Sci U S A. 1997;94:12509–12514. doi: 10.1073/pnas.94.23.12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HC, Boyce J, Shin J, Appel B. Oligodendrocyte specification in zebrafish requires notch-regulated cyclin-dependent kinase inhibitor function. J Neurosci. 2005;25:6836–6844. doi: 10.1523/JNEUROSCI.0981-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park IK, Morrison SJ, Clarke MF. Bmi1, stem cells, and senescence regulation. J Clin Invest. 2004;113:175–179. doi: 10.1172/JCI20800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, Hogan BL. The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell. 2009;4:525–534. doi: 10.1016/j.stem.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Ostrowski LE, Randell SH, Hogan BL. Lung development and repair: contribution of the ciliated lineage. Proc Natl Acad Sci U S A. 2007;104:410–417. doi: 10.1073/pnas.0610770104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell. 2010;7:299–313. doi: 10.1016/j.stem.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ZD, Gu H, Bock C, Gnirke A, Meissner A. High-throughput bisulfitesequencing in mammalian genomes. Methods. 2009;48:226–232. doi: 10.1016/j.ymeth.2009.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtfeld M, Apostolou E, Akutsu H, Fukuda A, Follett P, Natesan S, Kono T, Shioda T, Hochedlinger K. Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature. 2010;465:175–181. doi: 10.1038/nature09017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell. 2008;15:668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- Umemoto T, Yamato M, Nishida K, Yang J, Tano Y, Okano T. Limbal epithelial side-population cells have stem cell-like properties, including quiescent state. Stem Cells. 2006;24:86–94. doi: 10.1634/stemcells.2005-0064. [DOI] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Cao R, Cerqueira F, Wagschal A, Zhang Y, Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nature genetics. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Varrault A, Gueydan C, Delalbre A, Bellmann A, Houssami S, Aknin C, Severac D, Chotard L, Kahli M, Le Digarcher A, et al. Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev Cell. 2006;11:711–722. doi: 10.1016/j.devcel.2006.09.003. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Iwama A, Takayanagi S, Morita Y, Eto K, Ema H, Nakauchi H. Cytokine signals modulated via lipid rafts mimic niche signals and induce hibernation in hematopoietic stem cells. Embo J. 2006;25:3515–3523. doi: 10.1038/sj.emboj.7601236. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.