Abstract
Pax6, a transcription factor containing the bipartite paired DNA-binding domain, has critical roles in development of the eye, nose, pancreas, and central nervous system. The 2.5 Å structure of the human Pax6 paired domain with its optimal 26-bp site reveals extensive DNA contacts from the amino-terminal subdomain, the linker region, and the carboxy-terminal subdomain. The Pax6 structure not only confirms the docking arrangement of the amino-terminal subdomain as seen in cocrystals of the Drosophila Prd Pax protein, but also reveals some interesting differences in this region and helps explain the sequence specificity of paired domain–DNA recognition. In addition, this structure gives the first detailed information about how the paired linker region and carboxy-terminal subdomain contact DNA. The extended linker makes minor groove contacts over an 8-bp region, and the carboxy-terminal helix–turn–helix unit makes base contacts in the major groove. The structure and docking arrangement of the carboxy-terminal subdomain of Pax6 is remarkably similar to that of the amino-terminal subdomain, and there is an approximate twofold symmetry axis relating the polypeptide backbones of these two helix–turn–helix units. Our structure of the Pax6 paired domain–DNA complex provides a framework for understanding paired domain–DNA interactions, for analyzing mutations that map in the linker and carboxy-terminal regions of the paired domain, and for modeling protein–protein interactions of the Pax family proteins.
Keywords: Pax6, paired domain, protein–DNA, helix–turn–helix
Pax proteins, which contain a conserved 128-amino-acid DNA-binding ‘paired’ domain, named after the prototypical Drosophila paired gene, have critical roles in mammalian development and oncogenesis (for review, see Noll 1993; Strachan and Read 1994; Stuart et al. 1994; Read 1995; Mansouri and Gruss 1996; Dahl et al. 1997). Missense mutations within the paired domains of Pax genes produce a number of mouse and human developmental disorders (Baldwin et al. 1995; Prosser and van Heyningen 1998), whereas chromosomal translocations of the human PAX3 and PAX7 genes are associated with alveolar rhabdomyosarcoma, a pediatric cancer of muscle (for review, see Barr 1997). These results underscore the importance of the Pax paired domain in protein–DNA recognition and in the regulation of gene expression.
One Pax gene that provides a particularly useful paradigm for studies investigating the developmental function of this gene family is Pax6 . Pax6 is expressed in the developing eye, nose, pancreas, and central nervous system (CNS) (Walther and Gruss 1991; Turque et al. 1994; Grindley et al. 1995; Davis and Reed 1996; Koroma et al. 1997). In humans and mice, Pax6 haploinsufficiency results in the aniridia and Small eye (Sey) ocular phenotypes, whereas homozygous Pax6 mutants result in a complete failure of eye development along with CNS and pancreatic defects (Hogan et al. 1986; Schmahl et al. 1993; Glaser et al. 1994; Quinn et al. 1996; Caric et al. 1997; Ericson et al. 1997; Sander et al. 1997; St-Onge et al. 1997; Warren and Price 1997). In transgenic mice, overexpression of human PAX6 also produces ocular developmental defects (Schedl et al. 1996). Moreover, mutations in a homologous Drosophila Pax6 gene result in the eyeless (ey) phenotype, and Pax6 misexpression in Drosophila results in ectopic eye formation (Quiring et al. 1994; Halder et al. 1995). Most recently, several additional genes in the Drosophila eye-forming regulatory hierarchy have been identified and their functional inter-relationships have been determined (Chen et al. 1997; Pignoni et al. 1997; for review, see Desplan 1997). While confirming that Pax6 is a key regulator of eye development, these results have focused attention on the identity of Pax6 target genes and on the mechanism by which Pax6 recognizes DNA.
The mammalian Pax gene family consists of nine members that can be organized into groups based upon sequence similarity, structural features, and genomic organization. The four groups include Pax1 and Pax9; Pax2, Pax5, and Pax8; Pax3 and Pax7; and Pax4 and Pax6 (for review, see Stuart et al. 1994). However, some similarities extend across multiple groups or throughout the entire Pax family. Previous studies have shown that the paired domains of the Pax2, Pax3, Pax5, and Pax6 proteins can recognize similar DNA sequences (Czerny et al. 1993; Epstein et al. 1994a, 1996; Chalepakis and Gruss 1995; Czerny and Busslinger 1995). Biochemical and crystallographic studies have shown that the paired domain actually consists of independent amino-terminal and carboxy-terminal subdomains (hereafter referred to as the ‘N subdomain’ and the ‘C subdomain’) (Czerny et al. 1993; Epstein et al. 1994b; Xu et al. 1995). The crystal structure of a complex containing the Drosophila paired (Prd) paired domain and a DNA-binding site revealed the folding arrangement of the N and C subdomains and provided a model for the docking of the N subdomain (Xu et al. 1995). However, the arrangement of the C subdomain in the Prd–DNA cocrystal leaves open several important questions about paired domain–DNA interactions.
The Prd structure shows that the C subdomain contains three α helices and folds like a homeodomain, but the C subdomain does not make any DNA contacts in the Prd–DNA cocrystals. In Drosophila, it has been possible to rescue the paired phenotype with constructs lacking the C subdomain, suggesting that it may be dispensable in this context (Cai et al. 1994). However, for other paired domains, genetic and biochemical evidence shows that the C subdomain has important functions and can make DNA contacts. This domain is well conserved among Pax6 homologs, and a missense mutation in the C subdomain of human Pax6 results in foveal hypoplasia (Azuma et al. 1996). In addition, selected optimal binding sites for the Pax6 paired domain show conserved bases over a 20-bp region, and DNA footprinting experiments show that both subdomains are required to protect this site: Deletion of the C subdomain contracts the footprint to 16 bp (Epstein et al. 1994b). Pax6 binding sites identified in lens crystallin genes (for review, see Cvekl and Piatigorsky 1996) and in the promoter for a neural cell adhesion molecule (Chalepakis et al. 1994) have sequences similar to that of the optimized site, further supporting the physiological significance of these extended binding sites and the role of the C subdomain of Pax6 in DNA recognition.
Studies of other Pax proteins also highlight the importance of the C subdomain in DNA recognition. For Pax5, one set of extended DNA sites found in promoters of Pax5-regulated genes requires both the N and C subdomains for efficient binding. DNA footprinting experiments confirm that these extended sites are protected by the intact Pax5 paired domain, but not by the isolated N subdomain (Czerny et al. 1993). Pax3 and Pax7 (which normally have one more residue in the linker than PAX6) have alternative splice forms with linkers identical in length to the Pax6 linker. These isoforms can recognize the extended sites identified for Pax5 and Pax6, and optimal binding to these extended sites also requires the intact C subdomains of Pax3 and Pax7 (Vogan et al. 1996). There are also alternative splice forms of Pax6 (known as Pax6-5a) and Pax8 that contain insertions that disrupt the N subdomain and therefore bind DNA exclusively via their C subdomains (Epstein et al. 1994b; Kozmik et al. 1997). These results highlight structural and functional similarities in many members of the Pax family and emphasize the importance of understanding how the linker region and the C subdomain contact DNA.
To better understand paired domain–DNA interactions and the function of the C subdomain in particular, we have determined the 2.5-Å resolution crystal structure of a complex containing the human Pax6 paired domain with its optimal DNA-binding site. This cocrystal structure reveals specific DNA contacts made by the N subdomain, the extended linker, and the C subdomain. It provides a general model for understanding Pax mutations, the relationship of Pax subfamilies, and the protein–protein and protein–DNA interactions that are relevant for the biological function of the paired domains.
Results
Overall structure of the Pax6 paired domain–DNA complex
The Pax6 paired domain was crystallized with a 26-bp DNA duplex containing the optimal Pax6 binding site (sequences shown in Fig. 1A–C). The Pax6 paired domain, like the Prd paired domain (Xu et al. 1995), contains two globular subdomains (Fig. 2) linked by an extended polypeptide chain (residues 61–76). The N subdomain (residues 1–60) contains a short β motif (an antiparallel β hairpin, followed by a type II β turn) and also includes three α helices that fold like a homeodomain. The C subdomain (77–133) contains three α helices with a related homeodomain-like fold. There are no protein–protein contacts between the N and C subdomains.
Figure 1.
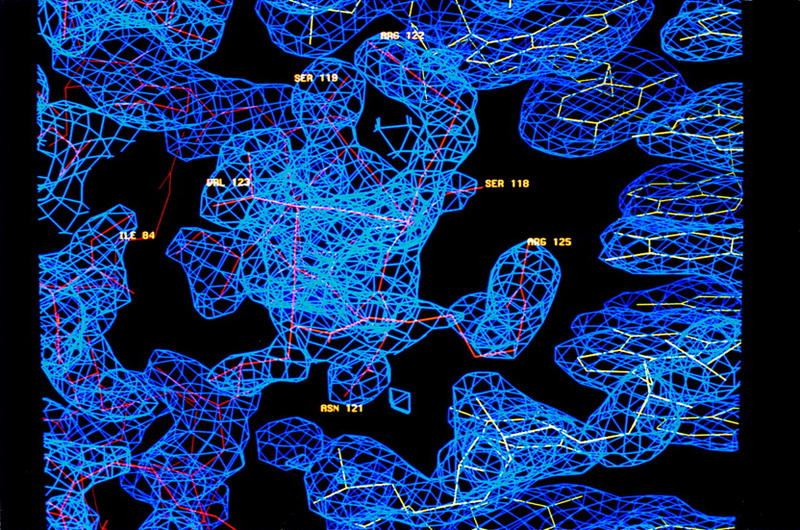
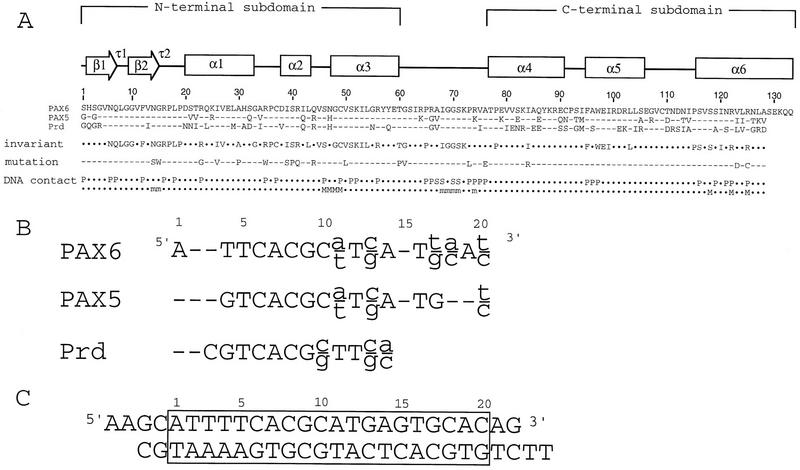
Sequences of paired domains and binding sites. (A) The sequence and secondary structure of the Pax6 paired domain, with sequences of paired domains from Pax5 and the Drosophila Prd protein, are shown (Treisman et al. 1991; Adams et al. 1992; Glaser et al. 1992). The protein contains the (conventional) 128-residue paired domain and five subsequent residues (SEKQQ) from Pax6. Lines below these sequences indicate residues that are conserved in almost all paired domains and also show missense mutations in the Pax6 paired domain (Azuma et al. 1996, 1998; Tang et al. 1997; Prosser and van Heyningen 1998 and http://www.mrc.hgu.ac.uk/Softdata/Pax6/ cited therein; Wolf et al. 1998; Grønskov et al. 1999; Hanson et al. 1999; T. Glaser, pers. comm.). Note that both the N17S and I29V missense mutations were identified in the same allele along with a 12-bp insertion in intron 5; hence, the functional significance of each charge by itself is uncertain. DNA contacts from the Pax6 crystals are summarized in the last two lines: One line indicates contacts with the sugar (S) phosphate (P) backbone; the other indicates base contacts [(M) major groove; (m) minor groove]. Note that the Pax6 nomenclature differs by three residues from that used in this paper because the Pax6 paired domain begins at residue 4 of the Pax6 protein. (B) DNA-binding sites for the Pax6 and Pax5 paired domains are longer than that for the Prd paired domain. Consensus binding sites for the Prd and Pax6 paired domains were determined from in vitro selections (Epstein et al. 1994a; Jun and Desplan 1996); the binding site for Pax5 paired domain was deduced from a combination of in vitro selection (Czerny and Busslinger 1995) and alignments of functional promoter sequences (Czerny et al. 1993). The extended sites recognized by Pax6 and Pax5 reflect binding of the C subdomain. (C) DNA oligonucleotide used in cocrystallization, with a box marking the Pax6 binding site. (D) The density-modified MI map shows clear electron density for the protein and the DNA. The map is contoured at 2.0 ς; this section shows the interface of the carboxy-terminal HTH motif (red) with the DNA (yellow). Several residues are labeled.
Figure 2.
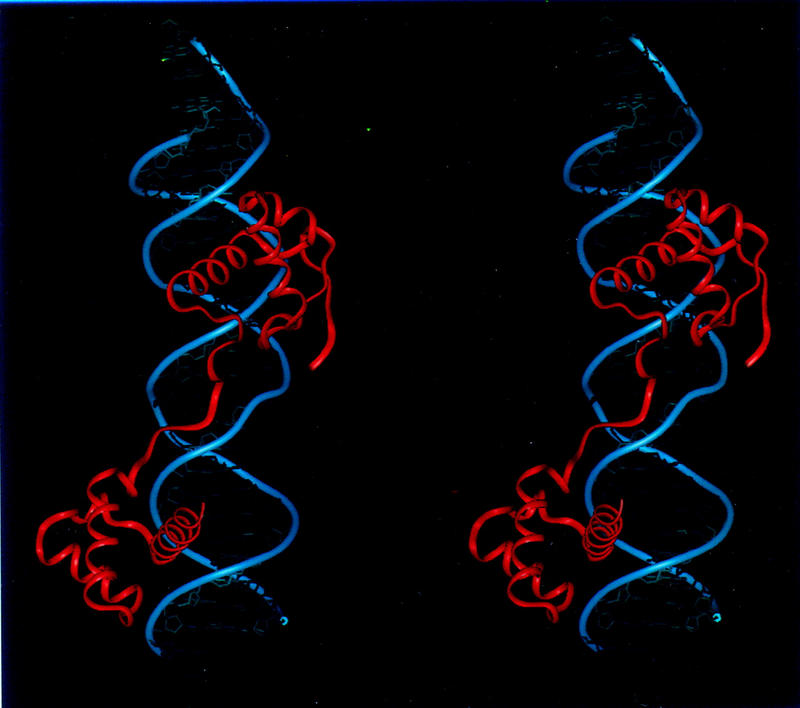
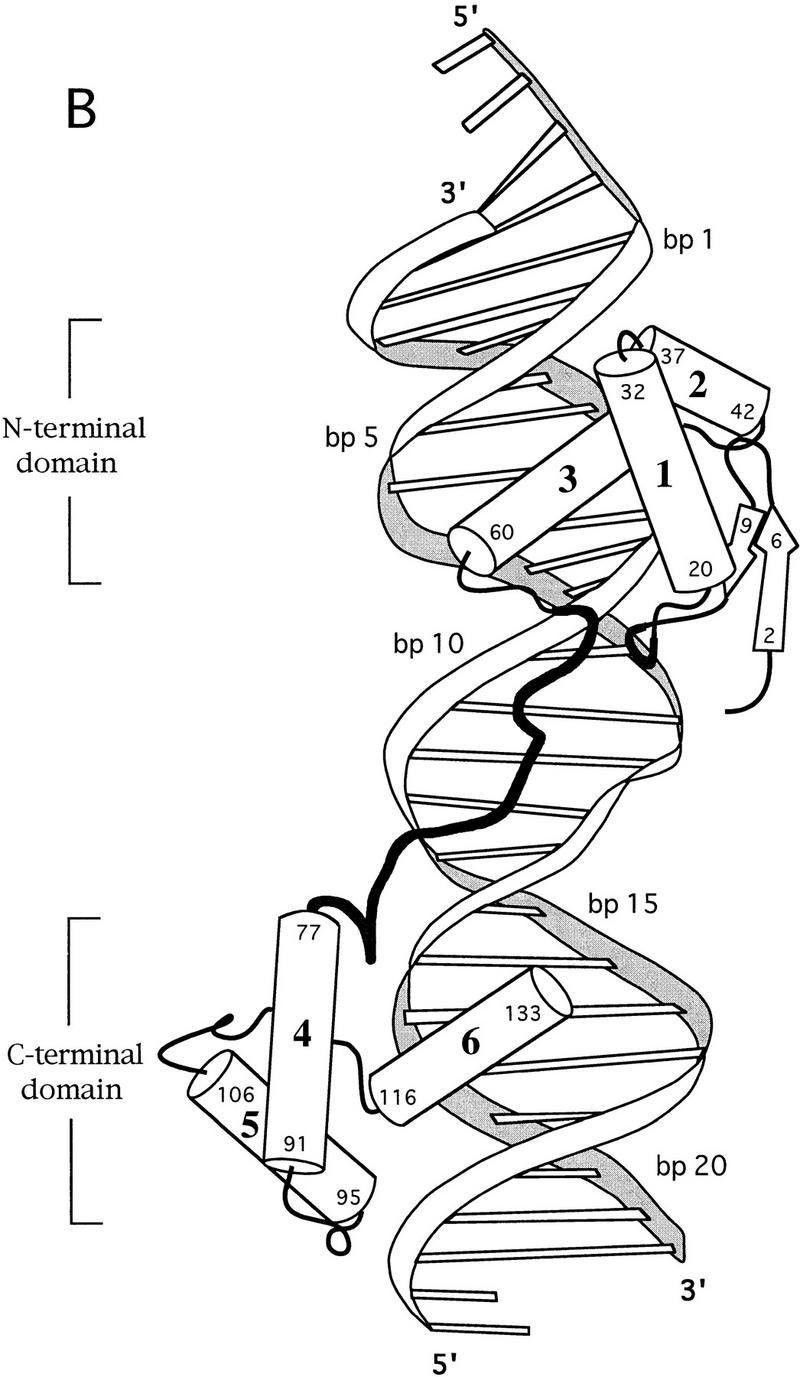
Overview of the Pax6 paired domain–DNA complex. (A) Stereo view with ribbons drawn through the Cα atoms of the protein (red) and through the phosphate atoms of the DNA backbone (blue). The N subdomain is at the top. (B) Sketch of the Pax6 paired domain–DNA complex in a similar orientation. Cylinders represent α helices; arrows represent β strands. Helices 1–6 are labeled; residue numbers indicate termini of the corresponding secondary structure elements.
Sequence comparisons show that the N subdomain is relatively well conserved among Pax proteins, and this part of the Pax6 structure is very similar to Prd. The first few residues of the N subdomain form a β hairpin that spans the minor groove of the DNA and contacts the sugar phosphate backbone of both DNA strands. This β hairpin is followed by a β turn (residues 13–16) that makes important base contacts in the minor groove. The β hairpin and β turn pack against the subsequent helical portion of this subdomain, which contains three α helices (helices 1–3 of the paired domain, residues 20–60, Fig. 2A,B). This N subdomain uses a helix–turn–helix (HTH) unit to dock against the major groove at one end of the binding site. The extended linker, which contains residues 61–76 and connects the two subdomains, binds in the minor groove near the center of the site. The linker makes numerous contacts with the sugar phosphate backbone and the DNA bases over an 8-bp region. The C subdomain contains three α helices (helices 4, 5, and 6 of the paired domain, Fig. 2A,B) and uses a HTH motif to dock against the major groove in the distal portion of the Pax6 binding site. Helix 6 (the ‘recognition helix’ of the C subdomain) fits directly into the major groove. Docking of this subdomain also is stabilized by the phosphate contacts from the amino-terminal portion of helix 5 and from the carboxy-terminal portion of the linker. Because the role of the C subdomain in the Pax6 complex is dramatically different than in the Prd complex (Xu et al. 1995), we begin by discussing this region in more detail.
Major groove contacts made by the carboxy-terminal HTH unit
The overall folding arrangement of the Pax6 C subdomain is very similar to that seen with Prd [root mean square (rms) distance of 1.23 Å when superimposing Cα atoms of residues 80–124], but each helix of the Pax6 C subdomain is slightly longer than the corresponding helix of Prd, and, most significantly, no DNA contacts were observed with the Prd C subdomain.
Helices 5 (residues 95–106) and 6 (residues 116–133) form a HTH unit, and contacts with DNA bases are mediated by the amino-terminal portion of helix 6. Base contacts from this helix include (1) van der Waals contacts between Arg-122 and the methyl group of thymine 16; (2) van der Waals contacts between Arg-125 and the methyl group of thymine 19; (3) a water-mediated contact between Ser-118 and the N7 of guanine 17; and (4) a water-mediated contact between the Oδ of Asn-121 and the N7 of guanine 20 (Figs. 3 and 4). These observed base contacts are fully corroborated by data from biochemical studies (Czerny et al. 1993, 1995; Epstein et al. 1994a). During site selection studies, thymines were highly preferred at positions 16 and 19, whereas guanine or adenine (which both have the N7 hydrogen-bond acceptor) were preferred in positions 17 and 20. Our results also are consistent with methylation protection studies showing that the N7 positions of guanines 17 and 20 are fully protected by binding of Pax5 or Pax6.
Figure 3.
Stereo view of the interface between the C domain and the DNA. The orientation of the complex is similar to that in Fig. 2, A and B. DNA is represented by solid sticks; the protein backbone is represented with open sticks. Side chains of key residues that contact the DNA are shown (Phe-95 and Trp-97 with open sticks; Ser-118, Ser-119, Asn-121, Arg-122, and Arg-125 with solid sticks). (●) Water molecules; (broken lines) hydrogen bonds. Corresponding superpositions between the C subdomain (residues 80–128) and the three helices of Engrailed (residues 10–58) give an rms distance of 1.71 Å; superpositions with the three helices of the Hin recombinase (residues 148–180) give rms distances of 1.86 Å.
Figure 4.
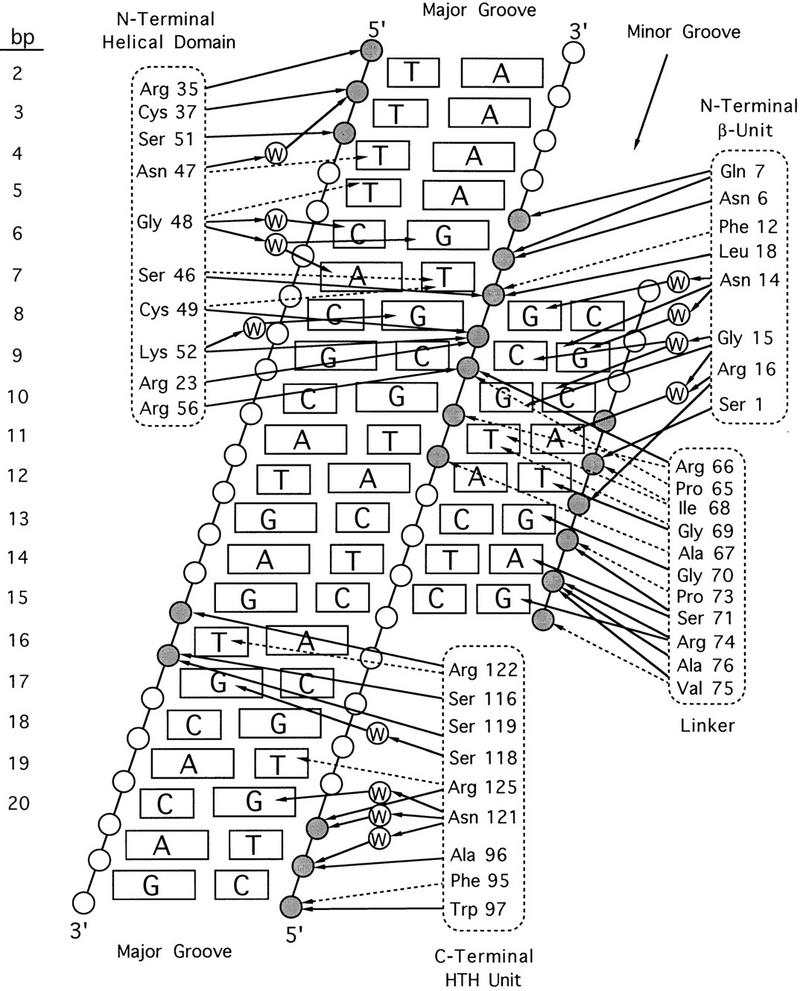
Diagram of DNA contacts in the Pax6 paired domain–DNA complex. DNA is represented as a cylindrical projection. Circles labeled W denote water molecules; other circles represent phosphates; shaded circles mark sites where Pax6 contacts the DNA backbone. All contacts made by Pax6 are indicated with arrows. (Solid arrows) Hydrogen bonds; (broken arrows) van der Waals contacts.
The C subdomain of Pax6 also makes contacts with flanking phosphates on both sides of the major groove (Figs. 3 and 4). Contacts with one strand of the DNA involve serines 116 and 119 (from the amino terminus of helix 6) and Arg-122 (Fig. 4). Contacts with the other DNA strand involve Asn-121 and Arg-125 from helix 6 and Phe-95, Ala-96, and Trp-97 from the amino terminus of helix 5. Finally, we also note that docking of the C subdomain may also be constrained by phosphate contacts (discussed below) from the carboxy-terminal portion of the linker region.
Minor groove contacts by the linker
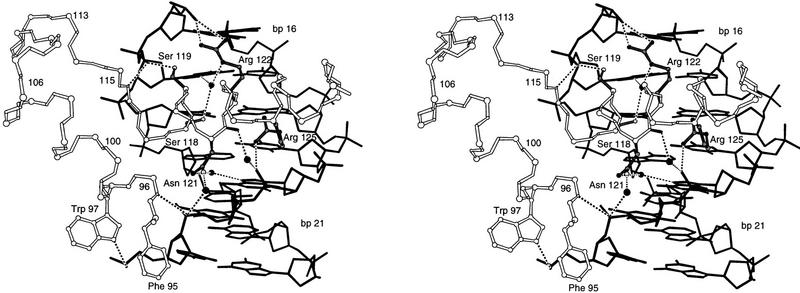
The extended polypeptide linker (residues 61–76) lies in the minor groove and makes extensive contacts over an 8-bp region of the DNA (Figs. 2, 4, and 5). The conformation of the amino-terminal region of the linker is quite similar to that seen with the Prd paired domain, but the Pax6 linker is much better ordered and makes many more contacts with the DNA. Residues 65–67 make several contacts with the DNA backbone, and there are extensive base and phosphate contacts from the residues that follow. Ile-68, which is an invariant residue among paired family proteins, fits directly into the minor groove and makes van der Waals contacts with thymines 11 and 12 and with the sugar of guanine 10. The main chain NH of Gly-69 hydrogen bonds with the O2 of thymine 11, whereas the NH and carbonyl groups of Gly-70 hydrogen bond, respectively, with the N7 and the N2 of guanine 13. The Ser-71 side chain contacts the N3 of adenine 14. Pro-73 appears to play an especially important role in DNA recognition: The side chain packs against the sugar of guanine 15, and this proline also changes the direction of the polypeptide main chain, allowing the carbonyl oxygen of residue 72 to hydrogen bond with the Nε of Arg-74. This, in turn, allows the Arg-74 side chain to reach back and to make both a direct and a water-mediated contact with guanine 15. The main chain NH groups of residues 74–76 form an interesting loop around the phosphate of thymine 16 and make an extensive set of chelating contacts with this phosphate.
Figure 5.
Stereo view of the interface between the linker and the DNA. The orientation of the complex is similar to that in Figs. 2, A and B. DNA is represented by solid sticks; the protein backbone is represented by open sticks. Side chains of key residues (Ile-68, Ser-71, Pro-73, Arg-74, and Val-75) that contact DNA are in black. (●) Water molecules; (broken lines) hydrogen bonds.
The conformation of the linker appears to be stabilized by a set of protein–protein interactions with the N and C subdomains. These interactions are particularly extensive in the amino-terminal portion of the linker: (1) Gly-61 packs against the Tyr-57 side chain (which is in the hydrophobic core of the N subdomain); (2) Ile-63 makes hydrophobic contacts with the Arg-23 and Gln-24 side chains; (3) Arg-64 makes a salt bridge with Asp-20; (4) Pro-65 and Ile-68 interact with Arg-16 and Pro-17 of the β turn motif. Residues 62, 63, and 64 of the linker form a half-circle loop that is stabilized by a hydrogen bond between the Ser-62 side chain and the main chain NH of Arg-64. There also are several stabilizing contacts in the carboxy-terminal portion of the linker: The Val-75 side chain packs against the ring of Pro-115, the residue immediately preceding the DNA recognition helix, and Ala-76 interacts with the Val-123 side chain. Essentially, Val-75 and Ala-76 serve to cover and complete one section of the hydrophobic core of the C subdomain.
Contacts by the N subdomain
Comparisons with the Prd paired domain show that the amino acid sequence is highly conserved in this region (Prd and Pax6 have 68% identity for residues 1–60). Structural comparisons of these proteins also show that the folding, docking, and DNA contacts are exceedingly similar in this region (Fig. 2D). Superimposing residues 2–60 gives an rms distance of 0.45 Å for corresponding Cα atoms.
Although the overall structures of the Pax6 and Prd N subdomains are very similar, there are important differences in their binding site sequences and base contacts. Asn-47 of Pax6, which is the first residue of the recognition helix (helix 3), replaces a histidine that occurs at this position in Prd. This change helps explain a key difference in binding site specificity of various paired domains. In Prd, residue 47 is a histidine, which hydrogen bonds with a guanine at position 4. In contrast, Asn-47 of Pax6 recognizes an AT base pair by making a van der Waals contact with the methyl group of thymine 4 (Fig. 6A). This arrangement is further stabilized by a water-mediated interaction between the Asn-47 side chain and the phosphate of thymine 2. This hydrophobic contact between Asn-47 and thymine 4 explains the observed sequence preference and reveals a novel structural basis for interaction between an AT base pair and asparagine. In many other protein–DNA complexes, aspargine makes a pair of hydrogen bonds with adenine. In the Pax6 N subdomain, the position and the orientation of the polypeptide backbone preclude Asn-47 from making this typical set of hydrogen bonds with the AT base pair. Additional, more subtle differences in the base contacts of the Pax and Prd N subdomains involve water-mediated contacts from Gly-48 and Lys-52 (Fig. 4).
Figure 6.
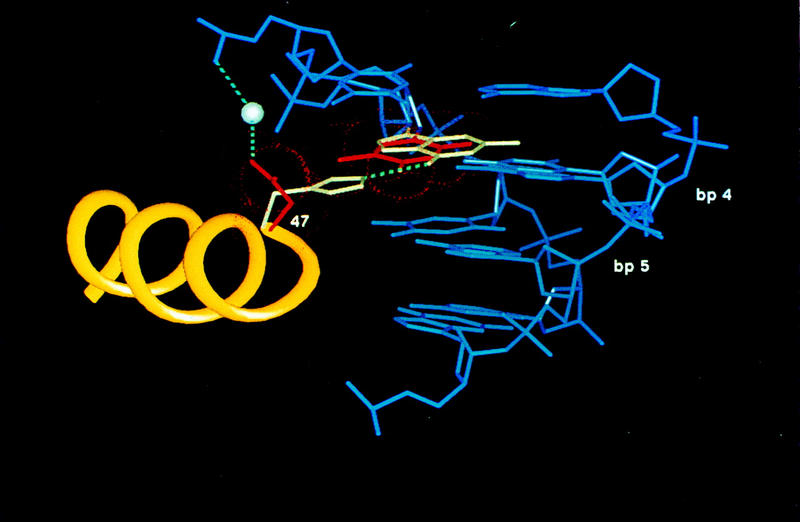
Key differences in DNA contacts made by the Pax6 and Prd N subdomains. (A) Comparison of the role of residue 47 in Pax6 and Prd. Complexes were aligned by superimposing the amino-terminal HTH motifs of Prd and Pax6. Helix 3 is yellow; neighboring regions of the DNA are blue. His-47 of Prd (white) makes a hydrogen bond (broken line) with the guanine (white) at base pair 4 of the Prd site; Asn-47 of Pax6 (shown in red) makes van der Waals contacts (dotted red spheres) with the thymine (red) at base pair 4 of the Pax6 site and makes a water-mediated contact with a phosphate. (B) Stereo view of contacts made by Gly-15 and Arg-16 where the β turn of Pax6 fits into the minor groove. (Broken lines) Hydrogen bonds with the Pax6 site (bases shown in black); (●) critical water molecule. Bases from the corresponding region of paired are shown with open lines. (Complexes were superimposed by superimposing the β turns.) In Pax6, the carbonyl oxygen of Gly-15 contacts the N2 of a guanine at base pair 10; Prd has a contact at essentially the same position in space but it involves the N2 of a guanine on the opposite strand of the DNA. In Pax6, the critical water molecule contacts the N3 of the adenine at base pair 11; Prd has a water molecule at essentially the same position in space, but it contacts the O2 of a thymine, which occurs at base pair 11 of the Prd site.
Another interesting set of differences involve the minor groove contacts made by the β turn units. In Pax6, the side chain Oδ of Asn-14 makes a hydrogen bond with the N2 of guanine 9 and makes a water-mediated hydrogen bond with the same guanine. The carbonyl oxygen of Gly-15 hydrogen bonds with the N2 position of guanine 10. Gly-15 also makes van der Waals contact with base pair 10 and makes water-mediated contacts with the O2 of cytosine 9. Gly-15 and Arg-16 together make a water-mediated contact with the N3 of adenine 11. Although the overall fold and docking of the Pax6 β turn unit (residues 13–16) is very similar to that of Prd, there are significant differences in the DNA sequences of the binding sites in this region and corresponding differences in the base contacts (Figs. 1, 4, and 6B).
Comparing this β turn unit with that of Prd provides a striking example of ambiguities involved in minor groove recognition. Thus, the carbonyl oxygen of Gly-15 contacts the N2 position of guanine in each complex, but the N2 position is right in the center of the minor groove (Seeman et al. 1976), and these guanines are on opposite strands in the two different complexes. Similar ambiguities occur with the water-mediated contact involving Gly-15 and Arg-16. This water contacts the N3 of adenine 11 in the Pax6 complex, but in Prd it makes an essentially isosteric contact with the O2 of a thymine that occurs at a corresponding position in the minor groove. In comparing the amino-terminal regions of Prd and Pax6, we also note that residues Ser-1 and His-2 of Pax6 make a few contacts with the DNA backbone. Corresponding residues of Prd were unstructured, and these new DNA contacts may help to stabilize the overall docking of the β turn.
DNA conformation
The Pax6 binding site has a relatively standard B-DNA conformation in the crystals, and the DNA duplexes stack to form a pseudocontinuous helix. In the Pax6 cocrystals, the DNA within the 20-bp Pax6 binding site has an average helical twist of 34.7° (10.4 bp/turn) and an average rise of 3.36 Å/base pair, as determined with the CURVES program (Lavery and Sklenar 1988). However, there are significant local deformations where the β turn and the linker bind in the minor groove. Thus, the helical twist between base pair 11 and 12 is only 15°, and this correlates with penetration of the Ile-68 side chain of the linker into this region of the minor groove. There is overwinding at the neighboring position, with a helical twist of 48° between base pairs 12 and 13. This region also has a 27° bend that opens the minor groove in the region where the β turn makes base contacts. This bend may be a common characteristic of paired domain–DNA complexes, because the Prd complex has a similar (20°) bend at this site (Xu et al. 1995).
Discussion
Basis for DNA recognition by the Pax C subdomain
The structure of the Pax6 paired domain–DNA complex helps explain the roles of the linker region and the C subdomain in paired domain–DNA interactions. It provides a plausible model for other paired domain–DNA complexes that contact extended sites and also helps explain why the C subdomain of Prd does not bind DNA.
Sequence comparisons suggest that the overall fold of the C subdomain is conserved throughout the Pax family. In all nine members of the family, this region shows a high degree (>50%) of homology. No insertions or deletions are seen in the alignment, and hydrophobic core residues are especially well conserved. We therefore presume that all paired C subdomains contain a similar HTH fold. Five of six side chains that contact DNA are also conserved throughout the Pax family. The only variation occurs at position 121, where Pax6 has an asparagine, but Pax3 and about half of the paired domains have a serine. However, these residues could readily make similar contacts. The side chain carbonyl of residue 121 makes a water-mediated contact to base 19. The Ser-121 side chain may also make a similar contact as it has been shown that the C subdomain of Pax3 has DNA selectivity similar to that of Pax6 (Vogan and Gros 1997). Given the conservation of the C subdomain and the similar DNA-binding specificities of many paired domains, the Pax6 structure may provide a good basis for modeling DNA contacts by the C subdomain in other Pax proteins. The paired domain has some homology with the DNA-binding domains of Tc1 transposases and these seem to use similar docking arrangements (Franz et al. 1994; Ivics et al. 1996; van Pouderoyen et al. 1997).
Although the C subdomain is involved in recognizing the extended intact site, it appears that the N subdomain plays a dominant role in DNA binding of the intact paired domain. The binding site for the N subdomain shows a clear consensus sequence, the crystal structure shows more contacts in this region, and the isolated N subdomain still binds DNA strongly. There are situations in which the primary contacts come from the C subdomain, but it is possible that these involve other docking arrangements. An alternative splice form of Pax6, with binding of the N subdomain disrupted by an insertion of 14 amino acids between helices 2 and 3, can recognize DNA (site 5aCON) exclusively via the C subdomain (Epstein et al. 1994b). Similarly, a Pax8 alternative splice form exists that contains an additional serine in helix 3 of the N subdomain (Kozmik et al. 1997). This form is also unable to bind to an N subdomain recognition sequence but recognizes a DNA sequence identical to the Pax6 5aCON site. Interestingly, several sequences in the 5aCON site that are selected by the C subdomain are not strongly selected by the intact Pax6 paired domain. It is not obvious how to align the 5aCON site with the binding site of the intact PAX6 paired domain, and it is possible that the isolated C subdomain has a distinctive docking arrangement. However, in the context of the intact protein, the DNA-binding ability of the C subdomain may be overshadowed by the greater affinity and specificity of the N subdomain. Considering the extensive contacts by the N subdomain and the additional contacts from the linker (discussed in the next section), binding of the N subdomain and the linker may constrain the docking modes accessible for the C subdomain.
Selection studies with Prd (Jun and Desplan 1996) give a shorter binding site than for Pax6, and our previous crystallographic studies revealed that the C subdomain of Prd does not contact this site. Sequence comparison between Pax6 and Prd reveals two differences among the six DNA-contacting residues of the C subdomain (Fig. 1A). First, at position 119, Pax6 has a serine and its side chain hydroxyl makes a strong hydrogen bond with the phosphate oxygen of guanine 17. Prd has an alanine at this position and thus would not only lose a critical contact but also place a hydrophobic group near the phosphate. Second, at position 121, Pax6 has an asparagine and Prd has a serine. This difference may be less critical as Pax3 and Pax7 also have serines at this position and yet their C subdomains are able to contact DNA. The inability of the Prd C subdomain to bind DNA may thus result from the difference of a single residue at position 119. Given the relatively weak binding of the C subdomain (at least in the context of the full-length paired domain), losing one strong hydrogen bond from residue 119 could readily explain why DNA binding was not observed for the Prd C subdomain.
A unique role for the paired domain linker
The Pax6 linker that connects the N and C subdomains is well ordered (unlike the corresponding region of the Prd complex) and makes extensive base contacts in the minor groove. Selections show that binding site sequence is well conserved in this region, and minor groove contacts from the linker explain the recognition specificity. Contacts in the Pax6 complex rationalize the observed specificity, and the energetic significance of the linker–DNA interactions is also highlighted by the two Pax missense mutations that occur in this region, G66D (Baldwin et al. 1995) and P73L (T. Glaser, pers. comm.). In addition, we note that the amino acid sequence of the linker is highly conserved in all paired domains and that all the base-contacting residues are invariant (with the exception of the Arg-74/Lys-74 difference noted above). Binding site selections show very similar preferred sequences from base pair 11 to 15 for the Prd, Pax2, Pax5, and Pax6 paired domains. The observations suggest that the Pax6 linker should provide a good model for other Pax proteins.
DNA binding by covalently linked modules has been observed in several other systems, but Pax6 reveals a novel paradigm for the role of a linker region. It is interesting to contrast the role of the linker in the Pax structure with (1) the role of the linker in the POU domains, where the flexible linkers seen in the Oct-1 (Klemm et al. 1994) and Pit-1 structures (Jacobson et al. 1997) primarily serve to tether the N and C subdomains, and (2) the role of the linkers in the zinc fingers (Pavletich and Pabo 1991; Elrod-Erickson et al. 1996) where relatively short well-ordered linkers make water-mediated phosphate contacts from the outer edge of the major groove. Pax6 provides an impressive example of how an extended polypeptide chain can be used to trace along and contact DNA bases in the minor groove. Minor groove contacts by an extended polypeptide chain have been seen in other complexes, such as the homeodomain with an extended amino-terminal arm (Kissinger et al. 1990; Wolberger et al. 1991) or the Hin recombinase, with amino-terminal and carboxy-terminal arms that bind in the minor groove (Feng et al. 1994). However, compared with these amino- and carboxy-terminal arms, the Pax6 linker is much better ordered and makes more numerous DNA contacts. Having the linker tethered on both sides by the N and C subdomains may help stabilize the overall structure of the linker region, and protein–protein interactions between the ends of the linker and the adjacent subdomains also presumably constrain the linker conformation and enhance specificity.
Approximate twofold symmetry axis relates the N and C subdomains
The overall fold and docking arrangement of the C subdomain is similar to that of the N subdomain, and there is an approximate twofold symmetry axis (through the center of the extended binding site) that relates the polypeptide backbones of these two subdomains. (In Fig. 2B, this approximate twofold axis would be perpendicular to the page and go through the minor groove near base pair 12.) However, the detailed interactions at the protein–DNA interface are almost entirely different for these two subdomains: There are no recognizable similarities in the amino acid sequences of these domains, in the DNA sequences of their binding sites, or even in the relative position of residues from the HTH units that make critical base and phosphate contacts. However, the overall similarity in the folding and docking arrangements is quite striking. We infer that the paired domain may have arisen by gene duplication of a three-helix unit and that detailed similarities in the amino acid sequences of the domains or in their DNA contacts were lost during subsequent divergent evolution. [A conceptually similar internal twofold axis occurs in the TBP/TATA-box complex (Kim et al. 1993a,b), and it has been proposed that TBP evolved via ancient gene duplications.]
Correlation with Pax developmental mutants
Missense mutations that produce murine and human developmental disorders can readily be explained from our structure. Of the 18 Pax6 paired domain missense mutations known to us (Hanson et al. 1994, 1999; Azuma et al. 1996, 1998; Tang et al. 1997; Prosser and van Heyningen 1998 and http://www.mrc.hgu.ac.uk/Softdata/Pax6/ cited therein; Wolf et al. 1998; Grønskov et al. 1999; T. Glaser, pers. comm.), 8 mutations involve residues that directly contact DNA. These are distributed throughout the N and C subdomains and the intervening linker. Mutations affecting residues that lie at the DNA–protein interface include N14S and G15W in the β turn region; R23G and R35W in the N subdomain; P73L and A76E in the linker; and R125C in the C subdomain. (Our numbering scheme refers to the isolated paired domain as shown in Fig. 1: The Pax6 protein has three additional amino-terminal residues.)
Several other PAX6 missense mutants may affect folding or stability of the proteins. The mutants A30P, S40P, and T60P introduce potentially disruptive prolines into α helical regions. Mutations in the hydrophobic core of the N subdomain (I39S and V50L) or in the hydrophobic core of the C subdomain (I84R and V123D) may disrupt the folding and stability of the protein. The R41Q mutation changes an invariant residue in an α helical region of the N subdomain. It is not clear from the structure that the Q44R missense mutation would be disruptive, and Arg occurs at this position in other Pax domains. However, this mutation also alters the nucleotide sequence within a suboptimal PAX6 splice donor and is thought to interfere with RNA splicing (I. Hanson and V. van Heyningen, pers. comm.).
As more PAX6 missense mutations are analyzed, it may become possible to correlate the position of a mutant, and the relative effect of the mutation on DNA binding, with the observed developmental defects. There are intriguing trends in the current data. Thus, mutations that are expected to completely abolish N subdomain function (A30P, S40P, V50L, and T60P) all result in aniridia. Other missense mutations (such as R23G, R35W, and P73L) retain partial DNA-binding activity and less severe phenotypic effects (Tang et al. 1997; T. Glaser, pers. comm.). It also is important to recognize that C subdomain mutants could exert their biological effects by altering binding by the 5a isoform, which binds exclusively via the C subdomain (Epstein et al. 1994b).
Protein–protein contacts of paired domains
Paired domains can bind DNA cooperatively by interacting with other DNA-binding domains such as the homeodomain (Underhill et al. 1995; Jun and Desplan 1996; Sheng et al. 1997; Underhill and Gros 1997; Fortin et al. 1997) and the Ets domain (Fitzsimmons et al. 1996). Like Pax3, Pax4, and Pax7 proteins, the intact Pax6 protein contains a paired-type homeodomain, which is located about 80 residues downstream of the paired domain. Although further data are needed to clarify the respective roles of these domains in gene regulation, some sites are cooperatively recognized by the paired domain and the homeodomain. For example, DNA binding to the adhesion molecule L1 promoter requires both the Pax6 paired domain and homeodomain, and footprinting experiments reveal that the homeodomain protects the DNA immediately adjacent to the binding site for the Pax6 N subdomain (Chalepakis et al. 1994). Modeling of the homeodomain and the Pax6 paired domain with this spacing shows that the homeodomain and the Pax6 amino-terminal HTH unit can both dock in the major groove, contacting opposite sides of the double helix. The first β turn and the loop between helices 2 and 3 of the paired domain are closest to the homeodomain, which has an amino-terminal arm reaching the paired domain from the minor groove.
Recently, it has been shown that the Pax5 paired domain can recruit Ets DNA-binding domains to the Pax5 C subdomain DNA-binding site to form ternary complexes on a B-cell-specific promoter (Fitzsimmons et al. 1996). The Pax6 paired domain also exhibits overlapping DNA-binding specificity with Ets family members and could also potentially interact (Plaza et al. 1994). The structure and docking of Pax5 should be nearly identical to Pax6: The C subdomains are 75% identical and all of the DNA-contacting residues are conserved. In the B-cell promoter, the Ets binding site is adjacent to the binding site of the Pax5 C subdomain, and our structure provides a plausible basis for modeling the relevant protein–protein interactions (Fig. 7). Modeling the Fli-1 Ets domain (Liang et al. 1994) with the docking arrangement of the PU.1 Ets domain (Kodandapani et al. 1996) indicates that residues of the Ets recognition helix can pack against the second and third helices of the Pax6 C subdomain in neighboring portions of the major groove. Tyrosine 341 of Ets would pack against Val-117 of Pax5, tyrosine 343 packs against Trp-97, and Asp-344 would form a charge interaction with Arg-100. These proposed contacts are consistent with the pattern of conserved residues and the effects of mutations at this interface. The striking conservation of these residues also raises the possibility that cooperative interactions with Ets domains may occur with other paired domains.
Figure 7.
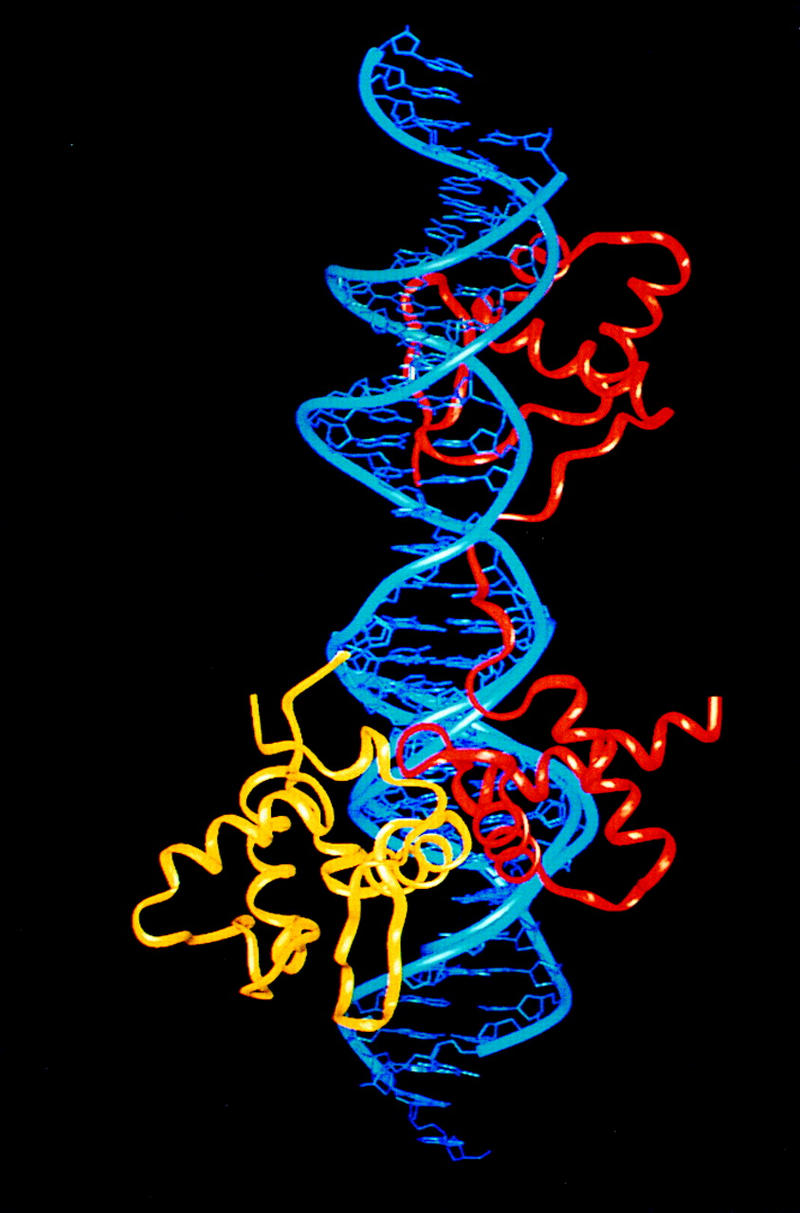
Proposed model for cooperative DNA binding of Pax6 (red) with the Ets domain (yellow). In this model, protein–protein interactions are mediated by the second and third helices of the Pax6 C subdomain with the third helix and the following hairpin region of Ets domain. The model was generated by superimposing phosphates of the Pax and Ets (Liang et al. 1994; Kodandapani et al. 1996) complexes in a way that reflects the relative spacing of the binding sites (Fitzsimmons et al. 1996). (The N subdomain is at the top, but the complex has been rotated, relative to Fig. 2, around a vertical axis so that the proposed contacts between Pax6 and Ets are easier to see.)
Materials and methods
Protein and DNA preparation
A DNA fragment encoding residues 4–136 of the human Pax6 protein was expressed from the T7 promoter of the PET29b vector (Novagen). As indicated in Figure 1A, this region includes the 128-residue paired domain and 5 subsequent residues (just beyond the carboxy-terminal end of the conventional 128-residue domain) that tend to be conserved in the Pax6 proteins (Loosli et al. 1996). E. coli BL21(DE3) cells with this expression vector and a pLys S plasmid were grown at 37°C, and induced, after reaching OD600 = 0.8, with 0.5 mm IPTG for 3 hr.. Cells were harvested and resuspended (150 ml/10-liter culture) in buffer A (40 mm HEPES at pH 7.5, 5 mm DTT, 1 mm EDTA) with 200 mm NaCl, 1 μg/ml DNase I, and 1 μg/ml each of the protease inhibitors pepstatin, aprotinin, benzamidine, and PMSF. The resuspended cells were frozen at −80°C and lysed by thawing at room temperature for 30 min. The lysate was centrifuged at 30,000g for 30 min and the supernatant was diluted with an equal volume of buffer A. The crude extract was precipitated by adding polyethyleneamine (at 4°C with vigorous stirring) to a final concentration of 0.25% (wt/vol), and centrifuged 40 min later (30,000g, 15 min). The supernatant was loaded onto an S-Sepharose column and eluted with a gradient from 100 to 350 mm NaCl in buffer A. The paired domain eluted between 200 and 250 mm NaCl. These fractions were pooled, diluted with 4 volumes of buffer A, and loaded onto a 20 ml calf thymus (double-stranded) DNA–cellulose column. The column was washed with 100 ml of buffer A plus 100 mm NaCl. The paired domain was eluted from this nonspecific DNA column with a step of buffer A plus 200 mm NaCl and then was loaded directly onto a 10-ml agarose column that contained about 10 mg of biotinylated Pax6 DNA bound to streptavidin beads. This column was washed with 50 ml of buffer A plus 200 mm NaCl, and the paired domain was eluted with 50 ml buffer A plus 1000 mm NaCl. At this stage, the affinity-purified protein gave a single band on an overloaded SDS gel. To remove any DNA that might be present in these samples, fractions containing the paired domain were diluted with 4 volumes of buffer A, loaded onto a heparin column, and eluted with a gradient from 200 to 600 mm NaCl in buffer A. The final sample was then dialyzed against buffer A, concentrated to 20 mg/ml, and stored at −80°C.
DNA oligonucleotides used for crystallization were purified with two rounds of reverse phase HPLC on C4 columns (trityl-on and trityl-off) before annealing (Klemm et al. 1994). For iodinated oligonucleotides, the second HPLC column was replaced with a mono-Q column (Kim et al. 1993a), and the DNA was eluted in a buffer containing 50 mm triethylammonium acetate (pH 8.0) with a gradient of 500–700 mm NaCl.
Crystallization
Crystals were grown at room temperature with the hanging drop vapor diffusion method using ammonium acetate as a volatile salt. When initially set up, the drops contained (1) 1 μl of 0.5 mm protein–DNA complex and (2) 1 μl of the well buffer (40 mm HEPES at pH 7.5, 10 mm spermine, 10 mm DTT, 5 mm EDTA, and 20% PEG-200) supplemented with 200 mm ammonium acetate. The Pax6–DNA complex becomes less soluble at lower ionic strength, and crystals grow (in ∼1 week) as ammonium acetate diffuses out of the drop and into the well. Crystals that diffracted beyond 2.5 Å were obtained with the DNA duplex shown in Figure 1C. A series of iodinated derivatives were prepared by making DNA oligonucleotides in which different sets of thymines had been replaced with iodo-uracil (Fig. 1C). For data collection, crystals were transiently mixed with three volumes of 30% PEG-200, then flash cooled in a stream of nitrogen gas at about −160°C.
Structure determination and refinement
The crystals form in space group P212121, with a = 33.84 Å, b = 61.68 Å, and c = 171.11 Å. Data were collected on a Rigaku R-Axis image plate and were reduced, scaled, and merged with Denzo and Scalepack (Otwinowski and Minor 1997). Derivatives were prepared by substituting iodouracil for thymine at specific positions in the binding site (Table 1), and data sets from these cocrystals were local-scaled to the native data using Maxscale (M.A. Rould, unpubl.). An initial set of phases had been obtained by molecular replacement methods (using the Prd N subdomain and 10 bp of DNA as a model). The positions of heavy atoms were determined by difference Fourier methods, and heavy atom parameters were refined with the MLPHARE program of CCP4 (Collaborative Computational Project 1994). The initial MIR map had a mean figure of merit of 0.79, and this MIR map was further improved with solvent flattening and histogram matching as implemented in the DM program of CCP4. The density-modified MIR map (Fig. 1D) showed clear density for every DNA base and for almost every sidechain of the protein. Model building was done with TOM FRODO (M. Israel, A.J. Chirino, and C.M. Cambillau, pers. comm.) and was facilitated by using the conserved regions of the Prd–DNA complex as an initial starting point. Refinement was done with X-PLOR (Brünger 1992a), repeatedly using positional refinement with tightly restrained individual B-factor refinement, and using simulated annealing OMIT maps to guide rebuilding. The free R factor was used to monitor the overall progress of refinement, and we found that a bulk solvent correction (Brünger 1992a) significantly improved both the free and working R factors. Before the last cycle of refinement, local scaling of the observed and calculated structure factors with Maxscale (M.A. Rould, pers. comm.) was used to correct for absorption errors and anisotropic diffraction. The final model includes 84 water molecules, and each is in a position that allows at least one hydrogen bond with the protein or the DNA. The final model has an R factor of 23.3% and a free R of 25.6% (with excellent stereochemistry) for all data from 20–2.5 Å resolution. All residues are in allowed regions of the Ramachandran plot.
Table 1.
MIR phasing and refinement statistics (20.0–2.5 Å)
|
|
Native
|
Derivatives
|
||||
|---|---|---|---|---|---|---|
|
1
|
2
|
3
|
4
|
5
|
||
| Base pairs with iodouracil | 12 + 14 | 2 + 4 + 12 + 14 | 1 + 2 + 12 + 14 | 1 + 2 + 12 | 1 + 12 + 14 | |
| Measured reflections | 252,170 | 244,376 | 198,940 | 194,410 | 192,455 | 377,231 |
| Unique reflections | 13,002 | 12,806 | 12,332 | 11,011 | 12,353 | 12,969 |
| Completeness (%) | 99.3 | 97.2 | 93.1 | 86.1 | 94.4 | 99.0 |
| Rsym (%) | 3.9 | 5.5 | 4.7 | 5.1 | 4.9 | 4.9 |
| RCullis | 0.55 | 0.59 | 0.67 | 0.51 | 0.52 | |
| Refinement: | |||||
|---|---|---|---|---|---|
| All data
|
F > 2ς
|
Nonhydrogen atoms at complex: 2077 | |||
| R | 23.3% | 22.6% | |||
| Rfree | 25.6% | 24.1% | Water molecules in model: 84 | ||
| Protein | DNA | rms ΔB between bonded atoms: 3.3 Å2 | |||
| rms bond length (Å) | 0.009 | 0.009 | |||
| rms bond angles (°) | 1.344 | 1.627 |
(Rsym) ΣhΣi|Ih,i − Ih|/ΣhΣiIh,i where Ih is the mean intensity of the i observations of reflection h.
(Cullis R-factor) Σ|FPH ± FP|−FH,calc|/Σ|FPH ± FP| (centric reflections only).
(Phasing power) √[ΣFh,calc2/Σ(FPH,obs − FPH,calc)2]
(Rfree) Σ|Fobs| − |Fcalc|/Σ|Fobs|, for a 10% subset of all reflections that were never used in crystallographic refinement (Brünger 1992b).
R-factor uses the same equation as Rfree but is calculated for those reflections (90%) used in crystallographic refinement (Brünger 1992b).
Ideal stereochemical parameters for protein refinement are from Engh and Huber (1991); ideal parameters for DNA are from PARNDBX.DNA from the Nucleic Acid Database.
Acknowledgments
Crystallographic studies were supported by National Institutes of Health (NIH) grant GM31471 (C.O.P.) and by the Howard Hughes Medical Institute (HHMI) and used equipment purchased with support from the PEW Charitable Trusts; R.L.M. and J.A.E. were supported by NIH grant R01 EY10123. We thank Claude Desplan for many helpful comments on the manuscript. C.O.P. is an investigator of HHMI, and H.E.X. and J.A.E. were HHMI postdoctoral fellows. Coordinates have been deposited with the Brookhaven Data Bank (PDB Code 6pax).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pabo@mit.edu; FAX (617) 253-8728.
References
- Adams B, Dörfler P, Aguzzi A, Kozmik Z, Urbánek P, Maurer-Fogy I, Busslinger M. Pax-5 encodes the transcription factor BSAP and is expressed in B lymphocytes, the developing CNS, and adult testis. Genes & Dev. 1992;6:1589–1607. doi: 10.1101/gad.6.9.1589. [DOI] [PubMed] [Google Scholar]
- Azuma N, Nishina S, Yanagisawa H, Okuyama T, Yamada M. PAX6 missense mutation in isolated foveal hypoplasia. Nat Genet. 1996;18:141–142. doi: 10.1038/ng0696-141. [DOI] [PubMed] [Google Scholar]
- Azuma N, Hotta Y, Tanaka H, Yamada M. Missense mutations in the PAX6 gene in Aniridia. Invest Ophthalmol Vis Sci. 1998;39:2524–2528. [PubMed] [Google Scholar]
- Baldwin CT, Hoth CF, Macina RA, Milunsky A. Mutations in Pax3 that cause Waardenburg syndrome type I: Ten new mutations and review of the literature. Am J Med Genet. 1995;58:115–122. doi: 10.1002/ajmg.1320580205. [DOI] [PubMed] [Google Scholar]
- Barr FG. Chromosomal translocations involving paired box transcription factors in human cancer. Int J Biochem Cell Biol. 1997;29:1449–1461. doi: 10.1016/s1357-2725(97)00095-2. [DOI] [PubMed] [Google Scholar]
- Bertuccioli C, Fasano L, Jun S, Wang S, Sheng G, Desplan C. In vivo requirement for the paired domain and homeodomain of the paired segmentation gene product. Development. 1996;122:2673–2685. doi: 10.1242/dev.122.9.2673. [DOI] [PubMed] [Google Scholar]
- Brünger AT. X-PLOR Manual version 3.0. New Haven, CT: Yale University Press; 1992a. [Google Scholar]
- ————— The free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992b;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- Cai J, Lan Y, Appel LF, Weir M. Dissection of the Drosophila paired protein: Functional requirements for conserved motifs. Mech Dev. 1994;47:139–150. doi: 10.1016/0925-4773(94)90086-8. [DOI] [PubMed] [Google Scholar]
- Caric D, Gooday D, Hill RE, McConnell SK, Price DJ. Determination of the migratory capacity of embryonic cortical cells lacking the transcription factor Pax-6. Development. 1997;124:5087–5096. doi: 10.1242/dev.124.24.5087. [DOI] [PubMed] [Google Scholar]
- Chalepakis G, Gruss P. Identification of DNA recognition sequences for the Pax3 paired domain. Gene. 1995;162:267–270. doi: 10.1016/0378-1119(95)00345-7. [DOI] [PubMed] [Google Scholar]
- Chalepakis G, Wijnholds J, Giese P, Schachner M, Gruss P. Characterization of Pax-6 and Hoxa-1 binding to the promoter region of the neural cell adhesion molecule L1. DNA & Cell Biol. 1994;13:891–900. doi: 10.1089/dna.1994.13.891. [DOI] [PubMed] [Google Scholar]
- Chen R, Amoui M, Zhang Z, Mardon G. Dachshund and eyes absent proteins form a complex and function synergistically to induce ectopic eye development in Drosophila. Cell. 1997;91:893–903. doi: 10.1016/s0092-8674(00)80481-x. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr Sect D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Cvekl A, Piatigorsky J. Lens development and crystallin gene expression: Many roles for Pax-6. BioEssays. 1996;18:621–630. doi: 10.1002/bies.950180805. [DOI] [PubMed] [Google Scholar]
- Czerny T, Busslinger M. DNA-binding and transactivation properties of Pax-6: Three amino acids in the paired domain are responsible for the different sequence recognition of Pax-6 and BSAP (Pax-5) Mol Cell Biol. 1995;15:2858–2871. doi: 10.1128/mcb.15.5.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerny T, Schaffner G, Busslinger M. DNA sequence recognition by Pax proteins: Bipartite structure of the paired domain and its binding site. Genes & Dev. 1993;7:2048–2061. doi: 10.1101/gad.7.10.2048. [DOI] [PubMed] [Google Scholar]
- Dahl E, Koseki H, Balling R. Pax genes and organogenesis. BioEssays. 1997;19:755–765. doi: 10.1002/bies.950190905. [DOI] [PubMed] [Google Scholar]
- Davis JA, Reed RR. Role of Olf-1 and Pax-6 transcription factors in neurodevelopment. J Neurosci. 1996;16:5082–5094. doi: 10.1523/JNEUROSCI.16-16-05082.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desplan C. Eye development: Governed by a dictator or a junta? Cell. 1997;91:861–864. doi: 10.1016/s0092-8674(00)80475-4. [DOI] [PubMed] [Google Scholar]
- Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 Å: A model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- Engh RR, Huber R. Accurate bond and angle parameters for x-ray protein-structure refinement. Acta Crystallogr. 1991;A47:392–400. [Google Scholar]
- Epstein JA, Cai J, Glaser T, Jepeal L, Maas RL. Identification of a Pax paired domain recognition sequence and evidence for DNA-dependent conformational changes. J Biol Chem. 1994a;269:8355–8361. [PubMed] [Google Scholar]
- Epstein JA, Glaser T, Cai J, Jepeal L, Walton DS, Maas RL. Two independent and interactive DNA-binding subdomains of the Pax6 paired domain are regulated by alternative splicing. Genes & Dev. 1994b;18:2022–2034. doi: 10.1101/gad.8.17.2022. [DOI] [PubMed] [Google Scholar]
- Epstein JA, Shapiro DN, Cheng J, Lam PY, Maas RL. Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc Natl Acad Sci. 1996;93:4213–4218. doi: 10.1073/pnas.93.9.4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson J, Rasbass P, Schedl A, Brenner-Moroton S, Kawakami A, van Heyningen V, Jessell TM, Briscoe J. Pax6 controls progenitor cell identity and neuronal fate in response to graded Shh signaling. Cell. 1997;90:169–180. doi: 10.1016/s0092-8674(00)80323-2. [DOI] [PubMed] [Google Scholar]
- Feng J, Johnson RC, Dickerson RE. Hin recombinase bound to DNA: The origin of specificity in major and minor groove interactions. Science. 1994;263:348–355. doi: 10.1126/science.8278807. [DOI] [PubMed] [Google Scholar]
- Fitzsimmons D, Hodsdon W, Wheat W, Maira SM, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes & Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- Fortin AS, Underhill DA, Gros P. Reciprocal effect of Waardenburg syndrome mutations on DNA binding by the Pax-3 paired domain and homeodomain. Hum Mol Genet. 1997;6:1781–1790. doi: 10.1093/hmg/6.11.1781. [DOI] [PubMed] [Google Scholar]
- Franz G, Loukeris TG, Dialektaki G, Thompson CRL, Savakis C. Mobile Minos elements from Drosophila hydei encode a two-exon transposase with similarity to the paired DNA-binding domain. Proc Natl Acad Sci. 1994;91:4746–4750. doi: 10.1073/pnas.91.11.4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser T, Walton DS, Maas RL. Genomic structure, evolutionary conservation and aniridia mutations in the human Pax6 gene. Nat Genet. 1992;2:232–239. doi: 10.1038/ng1192-232. [DOI] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- Grindley J, Davidson DR, Hill RE. The role of Pax6 in eye and nasal development. Development. 1995;121:1433–1442. doi: 10.1242/dev.121.5.1433. [DOI] [PubMed] [Google Scholar]
- Grønskov, K., T. Rosenberg, A. Sand, and K. Brøndum-Nielsen. 1999. Mutational analysis of PAX6: 16 novel mutations including 5 missense mutations with a mild aniridia phenotype. Eur. J. Hum. Genet. (in press). [DOI] [PubMed]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila . Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hanson I, Fletcher JM, Jordan T, Brown A, Taylor D, Adams RJ, Punnett HH, van Heyningen V. Mutations at the Pax6 locus are found in heterogeneous anterior segment malformations including Peter’s anomaly. Nat Genet. 1994;6:168–173. doi: 10.1038/ng0294-168. [DOI] [PubMed] [Google Scholar]
- Hanson I, Churchill A, Love J, Axton R, Moore T, Clarke M, Meire F, van Heyningen V. Missense mutations in the most ancient residues of the PAX6 paired domain underlie a spectrum of human congenital eye malformations. Hum Mol Genet. 1999;8:165–172. doi: 10.1093/hmg/8.2.165. [DOI] [PubMed] [Google Scholar]
- Hogan BL, Horsburgh G, Cohen J, Hetherington CM, Fisher G, Lyon MF. Small eyes (Sey): A homozygous lethal mutation on chromosome 2 which affects the differentiation of both lens and nasal placodes in the mouse. J Embryol Exp Morphol. 1986;97:95–110. [PubMed] [Google Scholar]
- Ivics Z, Izsvak Z, Minter A, Hackett PB. Identification of functional domains and evolution of Tc1-like transposable elements. Proc Natl Acad Sci. 1996;93:5008–5013. doi: 10.1073/pnas.93.10.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson EM, Li P, Leon-del-Rio A, Rosenfeld MG, Aggarwal AK. Structure of Pit-1 POU domain bound to DNA as a dimer: Unexpected arrangement and flexibility. Genes & Dev. 1997;11:198–212. doi: 10.1101/gad.11.2.198. [DOI] [PubMed] [Google Scholar]
- Jun S, Desplan C. Cooperative interactions between paired domain and homeodomain. Development. 1996;122:2639–2650. doi: 10.1242/dev.122.9.2639. [DOI] [PubMed] [Google Scholar]
- Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993a;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993b;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- Kissinger CR, Liu B, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 Å resolution: A framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- Klemm JD, Rould MA, Aurora R, Herr W, Pabo CO. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S, Maki RA, Ely KR. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature. 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- Koroma BM, Yang JM, Sundin OH. The Pax-6 homeobox gene is expressed throughout the corneal and conjunctival epithelia. Invest Ophthalmol Vis Sci. 1997;38:108–120. [PubMed] [Google Scholar]
- Kozmik Z, Czerny T, Busslinger M. Alternatively spliced insertions in the paired domain restrict the DNA sequence specificity of Pax6 and Pax8. EMBO J. 1997;16:6793–6803. doi: 10.1093/emboj/16.22.6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavery R, Sklenar H. Definition of generalized helicoidal parameters and an axis of curvature for irregular nucleic acids. J Biomol Struct Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- Liang H, Mao X, Olejniczak ET, Nettesheim DG, Yu L, Meadows RP, Thompson CB, Fesik SW. Solution structure of the ets domain of Fli-1 when bound to DNA. Nat Struct Biol. 1994;1:871–875. doi: 10.1038/nsb1294-871. [DOI] [PubMed] [Google Scholar]
- Loosli F, Kmita-Cunisse M, Gehring WJ. Isolation of a Pax-6 homolog from the ribbonworm Lineus sanguineus . Proc Natl Acad Sci. 1996;93:2658–2663. doi: 10.1073/pnas.93.7.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansouri A, Hallonet M, Gruss P. Pax genes and their roles in cell differentiation and development. Curr Opin Cell Biol. 1996;8:851–857. doi: 10.1016/s0955-0674(96)80087-1. [DOI] [PubMed] [Google Scholar]
- Noll M. Evolution and role of Pax genes. Curr Opin Genet Dev. 1993;3:595–605. doi: 10.1016/0959-437x(93)90095-7. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO. Zinc finger-DNA recognition: Crystal structure of a Zif268-DNA complex at 2.1 Å. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- Pignoni F, Hu B, Zavitz KH, Xiao J, Garrity PA, Zipursky SL. The eye specification proteins So and Eya form a complex and regulate multiple steps in Drosophila eye development. Cell. 1997;91:881–891. doi: 10.1016/s0092-8674(00)80480-8. [DOI] [PubMed] [Google Scholar]
- Plaza S, Grevin D, MacLeod K, Stehelin D, Saule S. Pax-QNR/Pax-6, a paired- and homeobox-containing protein, recognizes Ets binding sites and can alter the transactivating properties of Ets transcription factors. Gene Expr. 1994;4:43–52. [PMC free article] [PubMed] [Google Scholar]
- Prosser J, van Heyningen V. PAX6 mutations reviewed. Hum Mut. 1998;11:93–108. doi: 10.1002/(SICI)1098-1004(1998)11:2<93::AID-HUMU1>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- Quinn JC, West JD, Hill RE. Multiple functions for Pax6 in mouse eye and nasal development. Genes & Dev. 1996;10:435–446. doi: 10.1101/gad.10.4.435. [DOI] [PubMed] [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring W. Homology of the eyeless gene of Drosophila to the Small eye gene in mice and humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Read AP. Pax genes—Paired feet in three camps. Nat Genet. 1995;9:333–334. doi: 10.1038/ng0495-333. [DOI] [PubMed] [Google Scholar]
- Sander M, Neubuser A, Kalamaras J, Ee HC, Martin GR, German MS. Genetic analysis reveals that PAX6 is required for normal transcription of pancreatic hormone genes and islet development. Genes & Dev. 1997;11:1662–1673. doi: 10.1101/gad.11.13.1662. [DOI] [PubMed] [Google Scholar]
- Schedl A, Ross A, Lee M, Engelkamp D, Rashbass P, van Heyningen V, Hastie ND. Influence of Pax6 gene dosage on development: Overexpression causes severe eye abnormalities. Cell. 1996;86:71–82. doi: 10.1016/s0092-8674(00)80078-1. [DOI] [PubMed] [Google Scholar]
- Schmahl W, Knoedlseder M, Favor J, Davidson D. Defects of neuronal migration and the pathogenesis of cortical malformations are associated with small eye (Sey) in the mouse. Acta Neuropathol. 1993;86:126–135. doi: 10.1007/BF00334879. [DOI] [PubMed] [Google Scholar]
- Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng G, Harris E, Bertuccioli C, Desplan C. Modular organization of Pax/homeodomain proteins in transcriptional regulation. Biol Chem. 1997;378:863–872. doi: 10.1515/bchm.1997.378.8.863. [DOI] [PubMed] [Google Scholar]
- St-Onge L, Sosa-Pineda B, Chowdhury K, Mansouri A, Gruss P. Pax6 is required for differentiation of glucagon producing alpha cells in mouse pancreas. Nature. 1997;387:406–409. doi: 10.1038/387406a0. [DOI] [PubMed] [Google Scholar]
- Strachan T, Read AP. Pax genes. Curr Opin Genet Dev. 1994;4:427–438. doi: 10.1016/0959-437x(94)90032-9. [DOI] [PubMed] [Google Scholar]
- Stuart ET, Kioussi C, Gruss P. Mammalian Pax genes. Annu Rev Genet. 1994;28:219–236. doi: 10.1146/annurev.ge.28.120194.001251. [DOI] [PubMed] [Google Scholar]
- Tang HK, Chao L-Y, Saunders GF. Functional analysis of paired box missense mutations in the Pax6 gene. Hum Mol Genet. 1997;6:381–386. doi: 10.1093/hmg/6.3.381. [DOI] [PubMed] [Google Scholar]
- Treisman J, Harris E, Desplan C. The paired box encodes a second DNA-binding domain in the paired homeodomain protein. Genes & Dev. 1991;5:594–604. doi: 10.1101/gad.5.4.594. [DOI] [PubMed] [Google Scholar]
- Turque N, Plaza S, Radvanyi F, Carriere C, Saule S. Pax-QNR/Pax-6, a paired box- and homeobox-containing gene expressed in neurons, is also expressed in pancreatic endocrine cells. Mol Endocrinol. 1994;8:929–938. doi: 10.1210/mend.8.7.7984154. [DOI] [PubMed] [Google Scholar]
- Underhill DA, Gros P. The paired-domain regulates DNA binding by the homeodomain within the intact Pax-3 protein. J Biol Chem. 1997;272:14175–14182. doi: 10.1074/jbc.272.22.14175. [DOI] [PubMed] [Google Scholar]
- Underhill DA, Vogan KJ, Gros P. Analysis of the mouse Splotch-delayed mutation indicates that the Pax-3 paired domain can influence homeodomain DNA-binding activity. Proc Natl Acad Sci. 1995;92:3692–3696. doi: 10.1073/pnas.92.9.3692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Pouderoyen G, Ketting RF, Perrakis A, Plasterk RHA, Sixma TK. Crystal structure of the specific DNA-binding domain of Tc3 transposase of C. elegans in complex with transposon DNA. EMBO J. 1997;16:6044–6054. doi: 10.1093/emboj/16.19.6044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogan KJ, Gros P. The C subdomain makes an important contribution to the DNA binding activity of the Pax-3 paired domain. J Biol Chem. 1997;272:28289–28295. doi: 10.1074/jbc.272.45.28289. [DOI] [PubMed] [Google Scholar]
- Vogan KJ, Underhill DA, Gros P. An alternative splicing event in the Pax-3 paired domain identifies the linker region as a key determinant of paired domain DNA-binding activity. Mol Cell Biol. 1996;16:6677–6686. doi: 10.1128/mcb.16.12.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther C, Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- Warren N, Price DJ. Roles of Pax-6 in murine diencephalic development. Development. 1997;124:1573–1582. doi: 10.1242/dev.124.8.1573. [DOI] [PubMed] [Google Scholar]
- Wolberger C, Vershon AK, Liu B, Johnson AD, Pabo CO. Crystal structure of a MATα2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991;67:517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- Wolf MTF, Lorenz B, Winterpacht A, Drechsler M, Schumacher V, Royer-Pokora B, Blankenagel A, Zabel B, Wildhardt G. Ten novel mutations found in aniridia. Hum Mutat. 1998;12:304–313. doi: 10.1002/(SICI)1098-1004(1998)12:5<304::AID-HUMU3>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Xu W, Rould MA, Jun S, Desplan C, Pabo CO. Crystal structure of a paired domain-DNA complex at 2.5 Å resolution reveals structural basis for Pax developmental mutations. Cell. 1995;80:639–650. doi: 10.1016/0092-8674(95)90518-9. [DOI] [PubMed] [Google Scholar]