Abstract
RNA polymerase II (RNAP II) is responsible for transcription of mRNA precursors in eukaryotic cells. Recent studies, however, have suggested that RNAP II also participates in subsequent RNA processing reactions through interactions between the carboxy-terminal domain (CTD) of the RNAP II largest subunit and processing factors. Using reconstituted in vitro splicing assays, we show that RNAP II functions directly in pre-mRNA splicing by influencing very early steps in assembly of the spliceosome. We demonstrate that the phosphorylation status of the CTD dramatically affects activity: Hyperphosphorylated RNAP IIO strongly activates splicing, whereas hypophosphorylated RNAP IIA can inhibit the reaction.
Keywords: RNAP II isoforms, RNA processing, in vitro slicing, phosphorylation
Splicing of mammalian pre-mRNA is a nuclear process in which introns are removed from primary transcripts synthesized by RNA polymerase II (RNAP II). Splicing takes place in a large macromolecular complex called the spliceosome, which is composed of small nuclear ribonucleoprotein (snRNP) particles and non-snRNP proteins including members of the serine/arginine-rich (SR) protein family (for review, see Moore et al. 1993; Kramer 1996). Although cytological studies have suggested that splicing can occur cotranscriptionally (e.g., Beyer et al. 1988; Bauren and Wieslander 1994) and factors required for splicing are found localized at sites of active transcription (e.g., Zhang et al. 1994), functional coupling between transcription and splicing does not seem obligatory because splicing can be reconstituted in vitro with pretranscribed RNA and splicing-competent cell extracts.
Recent studies, however, have provided evidence indicating that the carboxy-terminal domain (CTD) of the largest subunit of RNAP II links transcription with pre-mRNA processing (for reviews, see Corden and Patturajan 1997; Neugebauer and Roth 1997; Steinmetz 1997). The CTD is comprised of multiple repeats of the consensus sequence YSPTSPS, which is highly conserved throughout evolution (for review, see Corden 1990) and subject to reversible phosphorylation during the transcription cycle (for review, see Dahmus 1996). RNAP II with a hypophosphorylated CTD (RNAP IIA) is preferentially included in the preinitiation complex at the promoter, whereas RNAP II with a hyperphosphorylated CTD (RNAP IIO) is associated with elongation complexes.
Biochemical studies have shown that RNAP II, via the CTD, can physically interact with capping enzymes (Cho et al. 1997; McCracken et al. 1997a; Yue et al. 1997), polyadenylation factors (McCracken et al. 1997b), and splicing factors, including both snRNPs and SR-like proteins (Chabot et al. 1995; Yuryev et al. 1996; Mortillaro et al. 1996; Kim et al. 1997). Notably, only RNAP IIO has been found to associate with capping and splicing factors, and this isoform has also been detected in active spliceosomes (Mortillaro et al. 1996). In addition, in vivo studies using mammalian cultured cells have demonstrated that RNAs transcribed by RNAP II with a shortened CTD undergo inefficient capping, splicing, and polyadenylation (McCracken et al. 1997a,b) and that overexpression of phosphorylated CTD peptides inhibits splicing (Du and Warren 1997). Antibodies directed against the CTD and CTD peptides have also been shown to inhibit splicing in vitro (Yuryev et al. 1996). These observations have supported the view that the phosphorylated CTD of elongating RNAP II may serve as a platform on which processing factors bind, thus helping to promote efficient splicing by recruiting necessary factors to the vicinity of the nascent transcript. But an important, unaddressed issue is whether RNAP II plays a direct, active role in the splicing reaction. Perhaps the simplest view has been that RNAP II functions only as an ‘escort,’ helping to deliver factors to sites of processing, but does not participate in the actual reaction. However, our recent finding that RNAP II, and specifically the CTD, is necessary for polyadenylation in vitro in the absence of transcription (Hirose and Manley 1998) led us to consider the possibility that RNAP II might also function directly in the more complex pre-mRNA splicing reaction.
Here, using in vitro splicing assays, we show that RNAP II participates directly in splicing in the absence of transcription. Purified RNAP IIO was found to strongly activate splicing of several different premRNAs, whereas RNAP IIA was capable of inhibiting the reaction. RNAP IIO functions very early to accelerate one of the first steps in spliceosome assembly, working in a manner distinct from, but complementary to, that of SR proteins. We discuss these results with respect to the coupling of mRNA transcription and processing.
Results and Discussion
That RNAP II plays a direct role in polyadenylation (Hirose and Manley 1998) led us to consider the possibility that the enzyme might also function in splicing. Arguing against this, S100 extracts of HeLa cells lack RNAP II (Weil et al. 1979), yet can be activated for splicing of many pre-mRNAs by addition of SR protein splicing factors (for review, see Fu 1995; Manley and Tacke 1996; Valcárcel and Green 1996). In the case of polyadenylation, the requirement for RNAP II had escaped attention because high concentrations of another factor, the small molecule creatine phosphate (CP), can substitute for it (Hirose and Manley 1998). Perhaps CP, or some other factor, has masked an RNAP II requirement.
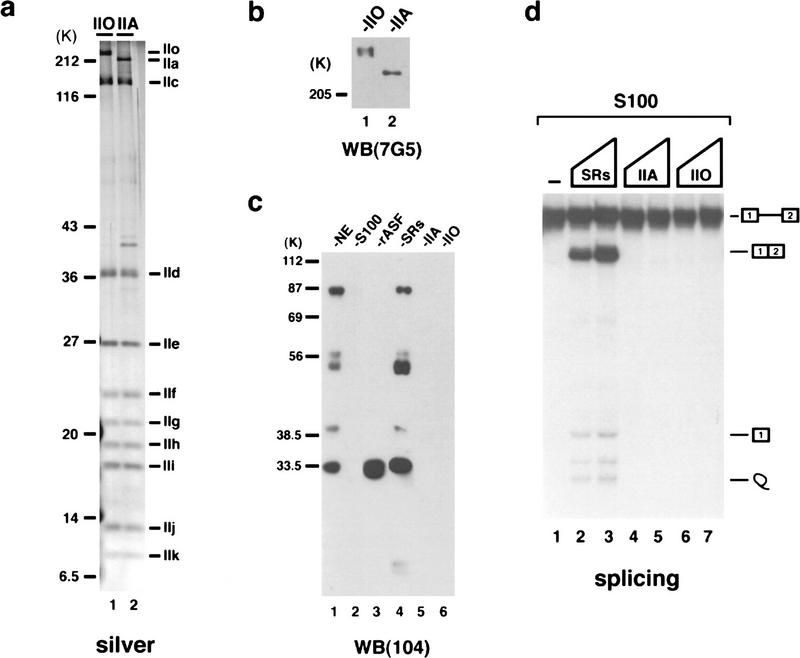
To examine the possibility that RNAP II functions directly in splicing, we first purified RNAP II from HeLa cells, following the procedure of Lu et al. (1991). Given that some studies suggested that only the RNAP IIO isoform is able to interact with splicing factors, this procedure was especially appropriate because it allowed separation of RNAP IIO from RNAP IIA. Aliquots of the two purified preparations were resolved by SDS-PAGE and visualized by silver staining (Fig. 1a), which revealed that both were essentially homogeneous. Western blotting with an antibody that recognizes the CTD (Besse et al. 1995) confirmed that neither isoform was detectably contaminated with the other (Fig. 1b). Given the known ability of SR proteins to activate splicing, we wished to provide additional evidence that the RNAP preparations were not contaminated with SR proteins. Figure 1c presents a Western blot utilizing an antibody that recognizes an epitope shared in all SR proteins (Roth et al. 1990). SR proteins were not detectable in either RNAP preparation (lanes 5,6) nor in an S100 extract used in the experiments described below (lane 2). For comparison, nuclear extract (NE; lane 1) and a preparation of purified SR proteins (lane 4) are also shown. The SR proteins and RNAP II preparations were then tested in standard S100 complementation splicing assays, using a β-globin-derived pre-mRNA (Fig. 1d). As expected, the SR proteins strongly activated splicing (lanes 2,3), and RNAP IIA and IIO were both inactive (lanes 4–7). Addition of either RNAP to reaction mixtures containing SR proteins but lacking CP failed to activate splicing (results not shown), indicating that the roles of RNAP II in splicing, if any, and of CP are distinct from their function in polyadenylation.
Figure 1.
Purification and characterization of the RNAP IIO and IIA. (a) Silver-stained 10% SDS–polyacrylamide gel containing 80 ng of purified hyperphosphorylated (IIO, lane 1) and hypophosphorylated (IIA, lane 2) RNAP II. The positions of size marker proteins are indicated at left; individual RNAP II subunits are indicated at right. The band of ∼40 kD in the IIA sample reflects a breakdown product of a larger subunit, as it was not observed reproducibly in separate analyses of the same preparation. (b) Western blot analysis of 40 ng of purified IIO (lane 1) and IIA (lane 2) probed with mAb 7G5. (c) Western blot analysis of 2 μg of HeLa NE (lane 1), 2 μg of HeLa cell S100 (lane 2), 100 ng of baculovirus recombinant ASF/SF2 (lane 3), 0.5 μg of purified SR proteins (lane 4), and 400 ng each of IIO (lane 5) or IIA (lane 6) probed with mAb 104. (d) In vitro splicing of β-globin pre-mRNA in S100 extract supplemented with buffer (lane 1), 250 (lane 2) and 500 ng (lane 3) of SR proteins, 40 (lane 4) and 80 ng (lane 5) RNAP IIA, and 40 (lane 6) and 80 ng (lane 7) of RNAP IIO. Reaction conditions were as described. Positions of the pre-mRNA, intermediates, and products of splicing are indicated schematically.
Next we set out to determine whether RNAP II might be capable of activating splicing under conditions where the efficiency of the reaction was reduced. In Figure 2a, splicing of the β-globin pre-mRNA was again assayed in S100 extract plus SR proteins, but the efficiency was lowered (lane 2) simply by a slight alteration in the concentrations of monovalent salts (see Materials and Methods). Strikingly, addition of RNAP IIO to reaction mixtures resulted in strong concentration-dependent activation of splicing, ∼15-fold at the highest amount tested (120 ng; lane 5). In sharp contrast, identical amounts of RNAP IIA not only failed to activate splicing but actually inhibited it, such that splicing became undetectable as the concentration of RNAP IIA was increased (lanes 6–8). Equivalent amounts of control proteins, for example, glutathione S-transferase (GST; lanes 9–11), BSA, and heat-inactived RNAP II (data not shown) were without effect. To corroborate these results, we repeated the experiment except substituting a single recombinant SR protein, ASF/SF2 for the total SR protein preparation. The results (Fig. 2a, lanes 12–20) were essentially identical: RNAP IIO strongly stimulated splicing and RNAP IIA inhibited it.
Figure 2.
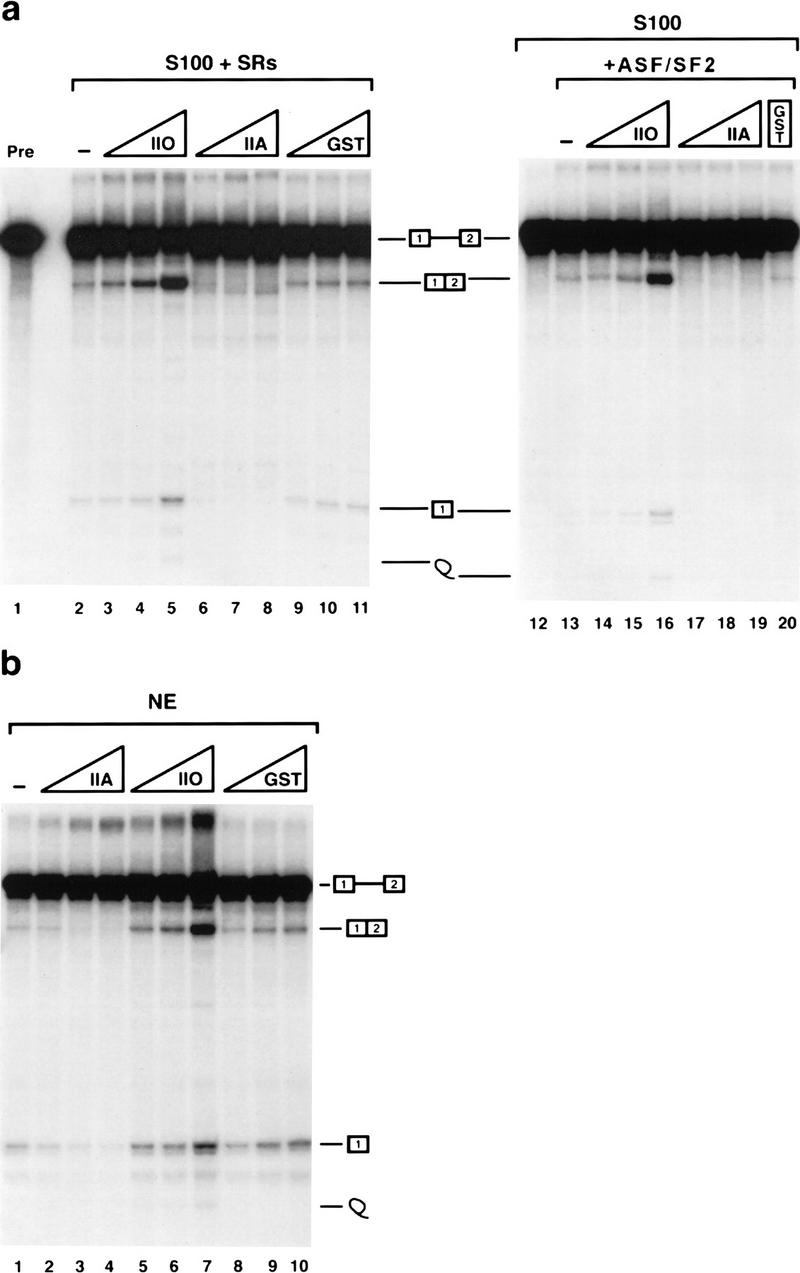
RNAP IIO activates and IIA inhibits splicing of β-globin pre-mRNA. (a) In vitro splicing of β-globin pre-mRNA in S100 supplemented with 250 ng of SR proteins (left) or 80 ng of ASF/SF2 (right) in the presence of buffer (lanes 2,13) or increasing amounts (40, 80, or 120 ng) of RNAP IIO (lanes 3–5,14–16), IIA (lanes 6–8,17–19) or GST (lanes 9–11,20). (Lanes 1,12) The precursor RNA and reaction products in S100 without SR proteins or ASF/SF2, respectively. (b) In vitro splicing of β-globin pre-mRNA in NE in the presence of buffer (lane 1), increasing amounts (40, 80, or 120 ng) of RNAP IIO (lanes 2–4), IIA (lanes 5–7), or GST (lanes 8–10).
We also wished to determine whether the RNAP II isoforms might be capable of influencing splicing when added to NE, which contains both RNAP II and SR proteins. In this case, splicing efficiency was reduced by using a lower amount of NE, a strategy employed previously to show that splicing could be activated by certain SR proteins (Yeakley et al. 1996) or by the splicing regulator Tra2 (Tacke et al. 1998). The results (Fig. 2b) show that the two RNAP II isoforms had the same effects on splicing of the β-globin pre-mRNA in NE as in S100: RNAP IIA inhibited splicing (lanes 2–4) and RNAP IIO activated it (lanes 5–7). Therefore, under these conditions RNAP IIO can be a limiting factor for splicing, whereas the IIA isoform can have a dominant-negative effect.
How does RNAP II actually function to influence splicing? One possibility is that the enzyme, specifically the CTD, simply mimics SR proteins. To test this we compared the effect of adding additional SR proteins with that of adding RNAP IIO. The results in Figure 3a show that splicing reactions reconstituted with S100 and 250 ng of SR proteins (as in Fig. 2a) were not affected significantly by an additional 120 ng of SR proteins, whereas the same mass of RNAP IIA or IIO again had large effects. (These amounts constitute nearly a 20-fold molar excess of additional SR proteins relative to the added RNAP II.) Essentially identical results (not shown) were obtained with a range of SR protein concentrations and with another pre-mRNA (IgM; see below). Given the difference in the behavior of the two RNAP II isoforms, it seemed possible that the CTD alone might be necessary and/or sufficient for the effects observed. Figure 3b, lanes 1–8, shows, however, that the CTD was not sufficient, as neither phosphorylated nor unphosphorylated recombinant GST–CTD (both of which could activate polyadenylation; results not shown) had a significant, reproducible effect on splicing. This distinguishes the effect of RNAP IIO on splicing from its previously described function in 3′-end formation, where the CTD was shown to be sufficient to activate 3′ cleavage (Hirose and Manley 1998). Not, unexpectedly, however the CTD was necessary, as RNAP IIB, a proteolytic form lacking the CTD, was without significant effect, positive or negative, on splicing (Fig. 3b, lanes 9–15). We also examined a time course of the RNAP IIO-supplemented S100 splicing reaction to determine when activation of splicing could first be detected. As shown in Figure 3c, RNAP IIO enhanced splicing very early in the reaction, at or before the first catalytic step, as judged by the significant increase in reaction intermediates (5′ exon and lariat–3′ exon) observed as early as 10 min and clearly by the 20-min time point.
Figure 3.
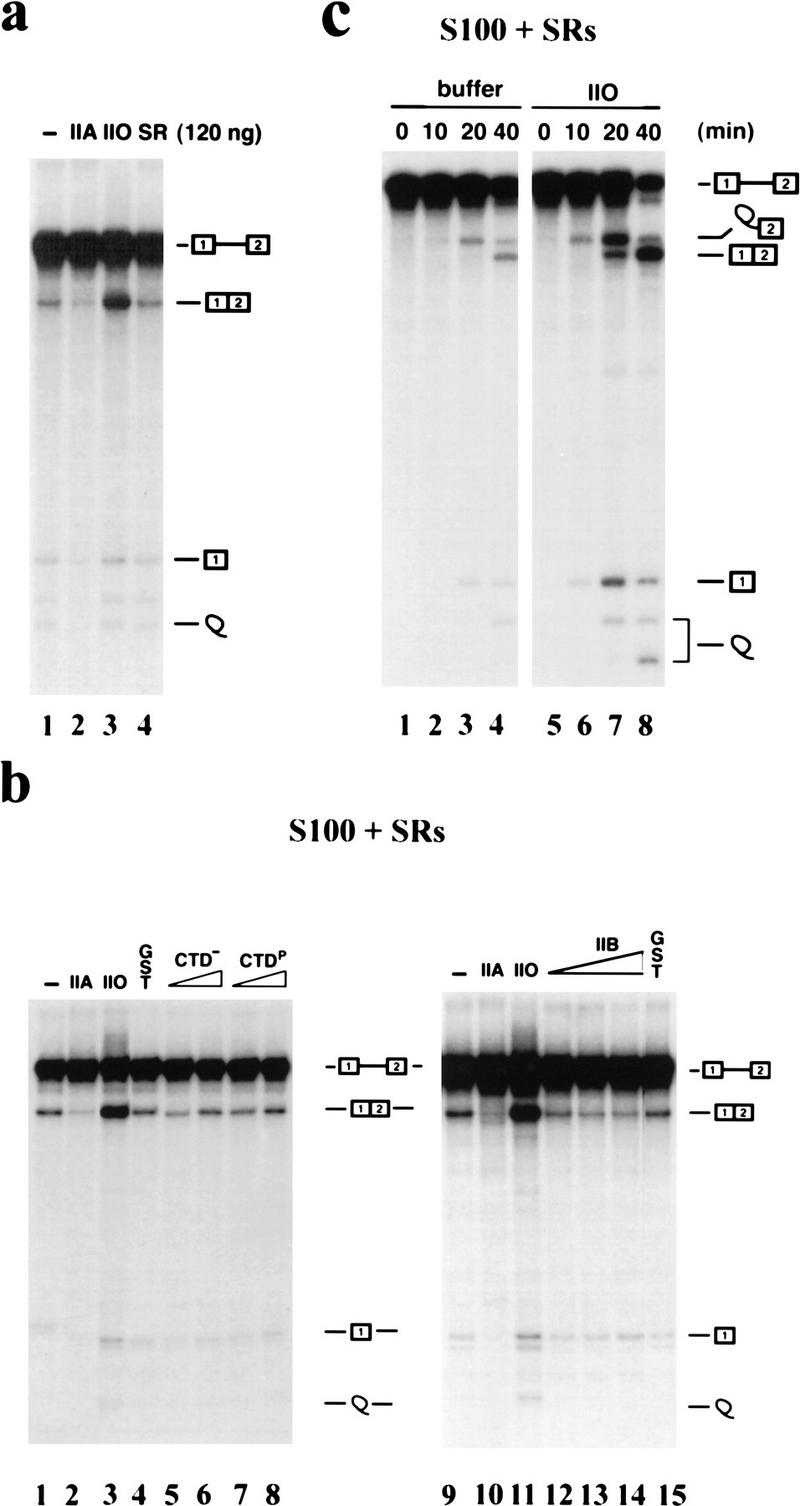
Characterization of RNAP II’s effects on splicing. Splicing reactions with β-globin pre-mRNA were reconstituted with S100 extract plus 250 ng of SR proteins and the indicated additional components. (a) Extra SR proteins do not affect splicing. Reaction mixtures were supplemented with 120 ng of RNAP IIA (lane 2), RNAP IIO (lane 3), or 120 ng of additional HeLa SR proteins (lane 4). (b) The CTD is necessary but not sufficient. Reactions mixtures were supplemented with 100 ng of RNAP IIA (lanes 2,10), 100 ng of RNAP IIO (lanes 3,11), 60 ng of GST (lanes 4,15), 30 or 60 ng of GST–CTD (lanes 5,6), 30 or 60 ng of phosphorylated GST–CTD (lanes 7,8), or 30, 60, or 120 ng of RNAP IIB (lanes 12–14). (c) Time course of splicing. Reaction mixtures were supplemented with 100 ng of RNAP IIO (lanes 5–8) and incubated at 30°C for the indicated times.
We also tested whether effects of RNAP IIO and IIA on splicing could be observed with additional pre-mRNAs. Splicing of the IgM M1 and M2 exons has been well studied, known, for example, to require an SR protein-dependent exonic splicing enhancer (e.g., Watakabe et al. 1993). Figure 4a shows that this pre-mRNA responded to addition of both RNAP IIO and IIA almost exactly as did the β-globin pre-mRNA. RNAP IIO strongly activated IgM splicing (∼10-fold) in both S100 and NE, whereas RNAP IIA nearly abolished it. We also examined splicing of HIV tat pre-mRNA, which behaves distinctively from the above pre-mRNAs. tat RNA is spliced very poorly in NE, except when supplemented with additional ASF/SF2 (Krainer et al. 1990), and it can also be committed to splicing specifically by ASF/SF2 (Fu 1993). In vitro splicing of the tat pre-mRNA was performed with S100 plus SR proteins or with NE, in the presence of increasing amounts of both isoforms of RNAP II (Fig. 4b). RNAP IIO again activated splicing in a concentration-dependent manner in the S100-reconstituted system (lanes 6–8). However, unlike with the other pre-mRNAs, RNAP IIA did not significantly affect tat splicing (lanes 3–5). As expected, the tat pre-mRNA was not spliced efficiently in NE (lane 10). Strikingly, RNAP IIO (lanes 13,14), but not IIA (lanes 11,12), was able to activate splicing, without supplementation with exogenous ASF/SF2. Together, these results suggest that RNAP IIO can function as a general activator of splicing, whereas the function of RNAP IIA as a splicing inhibitor may be more substrate specific.
Figure 4.
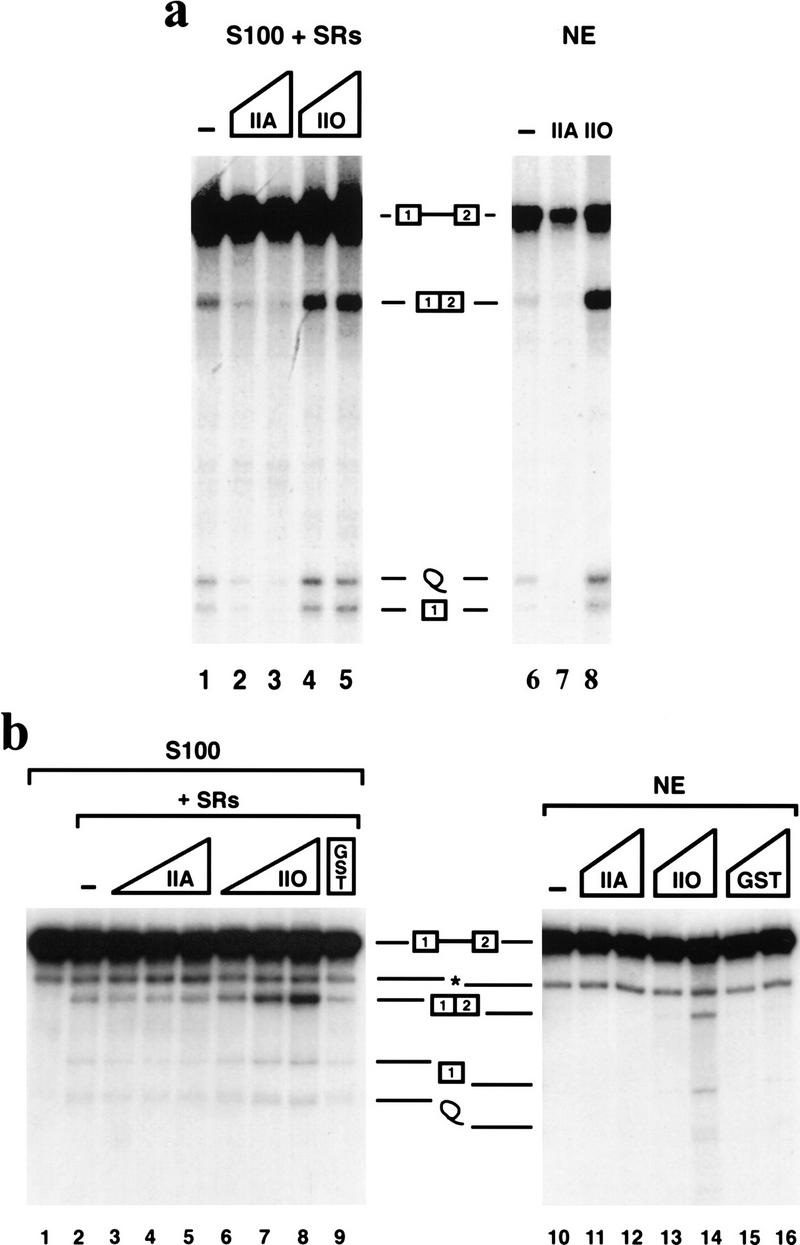
RNAP II affects splicing of two other pre-mRNAs. IgM M1–M2 (a) and HIV tat (b) pre-mRNAs were incubated in S100 plus SR proteins (left) or in NE (right) as in Fig. 2. Reaction mixtures were supplemented with the indicated proteins as follows: (a) IgM M1–M2 splicing; 80 (lane 2) or 120 ng (lanes 3,7) of RNAP IIA, or 80 (lane 4) or 120 ng (lanes 5,8) of RNAP IIO. (b) HIV tat splicing; 40, 80, or 120 ng of RNAP IIA (lanes 3–5), RNAP IIO (lanes 6–8) or 120 ng of GST (lane 9), or 60 or 120 ng of RNAP IIA (lanes 11,12), RNAP IIO (lanes 13,14), or GST (lanes 15,16). (*) The band is a truncated pre-mRNA resulting from artifactual cleavage (Krainer et al. 1990).
Pre-mRNA splicing requires the assembly of a series of spliceosomal complexes on the substrate RNA preceding the chemical steps of the reaction (for review, see Moore et al. 1993). Upon incubation of pre-mRNA in splicing extract, three specific complexes, which can be resolved on nondenaturing polyacrylamide gels, are formed: prespliceosomal complex A, early spliceosomal complex B, and catalytically active late spliceosomal complex C. To investigate whether either form of RNAP II affects spliceosome assembly, we performed nondenaturing gel analysis of spliceosomes formed on β-globin pre-mRNA with S100 plus SR proteins and RNAP II. Figure 5 shows a time course of spliceosome assembly in the presence of buffer (lanes 1–6), 80 ng of RNAP IIO (lanes 7–12), or 80 ng of RNAP IIA (lanes 13–18). Addition of RNAP IIO resulted in substantial increases in all three complexes. Most notable were the significant increases in A complex at very early times of the reaction. Complex formation was markedly enhanced even at the earliest time point tested (2 min; cf. lanes 2 and 8), indicating that RNAP IIO accelerates the rate of one of the first steps in spliceosome assembly. In contrast, RNAP IIA inhibited appearance of all complexes at all time points, with one significant exception: There was no reduction in A complex formation at the 2-min time point (cf. lanes 2 and 14). This finding leads to the conclusions that the step(s) enhanced by RNAP IIO is distinct from the step(s) inhibited by RNAP IIA and that RNAP IIA can intervene in spliceosome assembly subsequent to initial recognition of the pre-mRNA by splicing factors.
Figure 5.
RNAP IIO facilitates, but IIA inhibits, spliceosome formation. Standard splicing reaction mixtures with β-globin pre-mRNA and S100 plus 250 ng of SR proteins in the presence of buffer (lanes 1–5), 80 ng of RNAP IIO (lanes 7–12) or 80 ng of RNAP IIA (lanes 13–18) were incubated for the time indicated above each lane, and splicing complexes were processed and fractionated by native gel electrophoresis as described. The positions of the free precursor RNA, nonspecific complex H, prespliceosomal complex A, early spliceosomal complex B, and catalytically active late spliceosomal complex C are indicated at right.
We have presented evidence that RNAP II participates directly in pre-mRNA splicing and that the phosphorylation state of the CTD strongly affects its activity. These findings extend previous in vitro and in vivo studies that suggested functional interactions between RNAP II and splicing factors, that is, inhibition of splicing by antibodies directed against the CTD (Chabot et al. 1995; Yuryev et al. 1996), by peptides corresponding to CTD heptapeptide repeats (Yuryev et al. 1996; Du and Warren 1997) and by truncation of the CTD of actively transcribing RNAP II (McCracken et al. 1997b). The CTD has been shown to interact directly with SR-like proteins (Yuryev et al. 1996), and RNAP IIO can be detected associated with known splicing factors, including snRNPs (Chabot et al. 1995; Mortillaro et al. 1996). Because RNAP IIO appears to be the elongating form of the enzyme (Dahmus 1996) and functions very early in spliceosome assembly, an attractive model is that the enzyme, via the CTD and likely associated proteins (e.g., SCAFs; Patturajan et al. 1998b), facilitates binding of U1 and/or U2 snRNP particles to the pre-mRNA 5′ splice site and/or branch site, respectively. These RNAP II–snRNP interactions are likely transient and serve to help commit the nascent RNA to splicing and assure proper pairing of splice sites. By this view, the final steps in spliceosome assembly, and certainly splicing catalysis, could come after RNAP IIO has dissociated from the splicing complex, perhaps already functioning to nucleate complex assembly at downstream splicing signals. This is consistent with electron micrographic visualization of actively transcribed genes, which indicates the presence of possible splicing complexes (i.e., snRNPs) on putative splice sites of nascent RNAs (Beyer et al. 1988).
RNAP IIA, on the other hand, can inhibit splicing, apparently by disrupting early pre-splicing complexes. This effect may be related to findings of Yuryev et al. (1996), who reported that an unphosphorylated CTD peptide containing eight heptapeptide repeats strongly inhibited splicing in vitro, whereas mutant derivatives did not. However, inhibition required at least a 103-fold higher molar concentration of peptide relative to the levels of RNAP IIA used here, suggesting either a different mechanism or that intact RNAP IIA functions to inhibit splicing much more efficiently than does a CTD peptide. Considerable evidence suggests that RNAP IIA assembles in the preinitiation complex but is rapidly phosphorylated upon initiation of transcription (Dahmus 1996). How then could this isoform play a role in splicing? Although RNAP IIO is the principal elongating form, IIA has been implicated in elongation on a small number of genes. Remarkably, where identified, these tend to be intronless genes, such as those encoding histones or heat shock proteins (Weeks et al. 1993; O’Brien et al. 1994). RNAP IIA’s negative effect on splicing in vitro may reflect a proofreading mechanism that prevents inaccurate splicing by disrupting inappropriate prespliceosomal complexes. In any case, especially given that RNAP II appears to exist in multiple partly phosphorylated forms (e.g., Patturajan et al. 1998a), our results not only indicate that RNAP II is a direct participant in splicing but also suggest that differential CTD phosphorylation has the potential to play an important role in splicing regulation.
Materials and methods
Proteins
NE and cytoplasmic S100 for splicing assays were prepared from HeLa cells essentially as described (Tacke and Manley 1995; Tacke et al. 1998). SR proteins were purified from HeLa cells by the method of Zahler et al. (1992). Recombinant baculovirus-encoded ASF/SF2 was expressed in SF9 cells and purified as described (Tacke and Manley 1995). Purification and separation of the RNAP IIA and IIO from HeLa cell nuclear extract pellets was performed as described (Lu et al. 1991; Hirose and Manley 1998). Recombinant GST–CTD was purified from Escherichia coli, phosphorylated or mock phosphorylated, and repurified as described (Hirose and Manley 1998). Purified calf thymus RNAP IIB was a gift of X. Sun and D. Reinberg (Rutgers University) (see also Hirose and Manley 1998). Western blots were performed as described previously (Tacke and Manley 1995).
In vitro splicing
32P-Labeled pre-mRNA substrates were prepared as described (Tacke et al. 1998). Unless stated otherwise in the figure legends, splicing reactions were carried out at 30°C for 80 min in 15 μl containing 4 μl of NE or S100 supplemented with the amounts of purified SR proteins, recombinant ASF/SF2, GST–CTD, and/or RNAP IIO, IIA, or IIB indicated in the figure legends. The final concentrations of buffer components in all splicing reactions were 13 mm HEPES (pH 7.9), 0.13 mm EDTA, 13% glycerol, 1 mm DTT, 2 mm MgCl2, 1 mm ATP, 10 mm CP (disodium salt), 0.16 units of RNasin (Promega), 1–2 ng of labeled substrate RNA, and 2% (wt/vol) polyvinyl alcohol. With respect to monovalent salts, in the experiment in Figure 1d, reaction mixtures also contained 60 mm KCl and 8 mm (NH4)2SO4, whereas in Figures 2–5, they contained 27 mm KCl and 20 mm (NH4)2SO4. The former concentrations were optimal for SR protein-dependent splicing in S100, whereas the latter reproducibly allowed significant response to RNAP II. We do not know if this was due to the slight reduction in monovalent cations (76 vs. 67 mm) or the changed ratio of KCl to (NH4)2SO4. Splicing products were deproteinized and analyzed on 6% polyacrylamide gels containing 8 m urea. Splicing efficiencies were determined by PhosphorImager (Molecular Dynamics).
Spliceosome formation assays
Reaction mixtures were exactly as above, except that 2% polyvinyl alcohol was replaced with 7% glycerol. β-Globin pre-mRNA was incubated in S100 extract plus 250 ng of purified SR proteins in the absence or presence of 80 ng of RNAP IIA or IIO at 30°C for the times indicated in Figure 5. Heparin (0.5 mg/ml) was then added, and reaction mixtures were incubated for an additional 5 min at 30°C. Five microliters of each reaction mixture was resolved on a nondenaturing 4% polyacrylamide gel.
Acknowledgments
We thank C. Kedinger and M. Vigneron for mAb 7G5; X. Sun and D. Reinberg for purified RNAP IIB; S.H. Xiao and J. Prasad for plasmids; and X.H. Shi and Y. Sun for assistance in preparation of HeLa cell extracts. We are grateful to S.H. Xiao and J. Prasad for advice and discussions. This work was supported by grant R37GM 48259 from the National Institutes of Health. Y.H. was partly supported by the Japanese Ministry of Education, Science, and Culture.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jlm2@columbia.edu; FAX (212) 865-8246.
References
- Bauren G, Wieslander L. Splicing of Balbiani ring 1 gene pre-mRNA occurs simultaneously with transcription. Cell. 1994;76:183–192. doi: 10.1016/0092-8674(94)90182-1. [DOI] [PubMed] [Google Scholar]
- Besse S, Vigneron M, Pichard E, Puvion-Dutilleul F. Synthesis and maturation of viral transcripts in herpes simplex virus type 1 infected HeLa cells: The role of interchromatin granules. Gene Expr. 1995;4:143–161. [PMC free article] [PubMed] [Google Scholar]
- Beyer AL, Yvonne N, Osheim YN. Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes & Dev. 1988;2:754–765. doi: 10.1101/gad.2.6.754. [DOI] [PubMed] [Google Scholar]
- Chabot B, Bisotto S, Vincent M. The nuclear matrix phosphoprotein p255 associates with splicing complexes as part of the [U4/U6.U5] tri-snRNP particle. Nucleic Acids Res. 1995;23:3206–3213. doi: 10.1093/nar/23.16.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EJ, Takagi T, Moore CR, Buratowski S. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes & Dev. 1997;11:3319–3326. doi: 10.1101/gad.11.24.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 1990;10:383–387. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- Corden JL, Patturajan M. A CTD function linking transcription to splicing. Trends Biochem Sci. 1997;22:413–416. doi: 10.1016/s0968-0004(97)01125-0. [DOI] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Du L, Warren SL. A functional interaction between the carboxy-terminal domain of RNA polymerase II and pre-mRNA splicing. J Cell Biol. 1997;136:5–18. doi: 10.1083/jcb.136.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu XD. Specific commitment of different pre-mRNAs to splicing by single SR proteins. Nature. 1993;365:82–85. doi: 10.1038/365082a0. [DOI] [PubMed] [Google Scholar]
- ————— The superfamily of arginine/serine-rich splicing factors. RNA. 1995;1:663–680. [PMC free article] [PubMed] [Google Scholar]
- Hirose Y, Manley JL. RNA polymerase II is an essential mRNA polyadenylation factor. Nature. 1998;395:93–96. doi: 10.1038/25786. [DOI] [PubMed] [Google Scholar]
- Kim E, Du L, Bregman DB, Warren SL. Splicing factors associate with hyperphosphorylated RNA polymerase II in the absence of pre-mRNA. J Cell Biol. 1997;136:19–28. doi: 10.1083/jcb.136.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krainer AR, Conway GC, Kozak D. Purification and characterization of pre-mRNA splicing factor SF2 from HeLa cells. Genes & Dev. 1990;4:1158–1171. doi: 10.1101/gad.4.7.1158. [DOI] [PubMed] [Google Scholar]
- Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley JL, Tacke R. SR proteins and splicing control. Genes & Dev. 1996;10:1569–1579. doi: 10.1101/gad.10.13.1569. [DOI] [PubMed] [Google Scholar]
- McCracken S, Fong N, Rosonina E, Yankulov K, Brothers G, Siderovski D, Hessel A, Foster S, Shuman S, Bentley DL Amgen EST Program. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 1997a;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken S, Fong N, Yankulov K, Ballantyne S, Pan G, Greenblatt J, Patterson SD, Wickens M, Bentley DL. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997b;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA. Splicing O9F precursors to mRNAs by the spliceosome. In: Gesteland RF, Atkins JF, editors. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 303–358. [Google Scholar]
- Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer KM, Roth MB. Transcription units as RNA processing units. Genes & Dev. 1997;11:3279–3285. doi: 10.1101/gad.11.24.3279. [DOI] [PubMed] [Google Scholar]
- O’Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL. Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem. 1998a;273:4689–4694. doi: 10.1074/jbc.273.8.4689. [DOI] [PubMed] [Google Scholar]
- Patturajan M, Wei X, Berezney R, Corden JL. A nuclear matrix protein interacts with the phosphorylated C-terminal domain of RNA polymerase II. Mol Cell Biol. 1998b;18:2406–2415. doi: 10.1128/mcb.18.4.2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth MB, Murphy C, Gall JG. A monoclonal antibody that recognizes a phophorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz EJ. Pre-mRNA processing and the CTD of RNA polymerase II: The tail that wags the dog? Cell. 1997;89:491–494. doi: 10.1016/s0092-8674(00)80230-5. [DOI] [PubMed] [Google Scholar]
- Tacke R, Manley JL. The human splicing factors ASF/SF2 and SC35 possess distinct, functionally significant RNA binding specificities. EMBO J. 1995;14:3540–3551. doi: 10.1002/j.1460-2075.1995.tb07360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacke R, Tohyama M, Ogawa S, Manley JL. Human Tra2 proteins are sequence-specific activators of pre-mRNA splicing. Cell. 1998;93:139–148. doi: 10.1016/s0092-8674(00)81153-8. [DOI] [PubMed] [Google Scholar]
- Valcárcel J, Green MR. The SR protein family: Pleiotropic functions in pre-mRNA splicing. Trends Biochem Sci. 1996;21:296–301. [PubMed] [Google Scholar]
- Watakabe A, Tanaka K, Shimura Y. The role of exon sequences in splice site selection. Genes & Dev. 1993;7:407–418. doi: 10.1101/gad.7.3.407. [DOI] [PubMed] [Google Scholar]
- Weeks JR, Hardin SE, Shen J, Lee JM, Greenleaf AL. Locus-specific variation in phosphorylation state of RNA polymerase II in vivo: Correlations with gene activity and transcript processing. Genes & Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- Weil PA, Luse DS, Segall J, Roeder RG. Selective and accurate initiation of transcription at the Ad2 major late promoter in a soluble system dependent on purified RNA polymerase II and DNA. Cell. 1979;18:469–484. doi: 10.1016/0092-8674(79)90065-5. [DOI] [PubMed] [Google Scholar]
- Yeakley JM, Morfin JP, Rosenfeld MG, Fu XD. A complex of nuclear proteins mediates SR protein binding to a purine-rich splicing enhancer. Proc Natl Acad Sci. 1996;93:7582–7587. doi: 10.1073/pnas.93.15.7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue Z, Maldonado E, Pillutla R, Cho H, Reinberg D, Shatkin AJ. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc Natl Acad Sci. 1997;94:12898–12903. doi: 10.1073/pnas.94.24.12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. The C-terminal domain of the largest subunit of RNA polymerase II interacts with a novel set of serine/arginine-rich proteins. Proc Natl Acad Sci. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahler AM, Lane WS, Stolk JA, Roth MB. SR proteins: A conserved family of pre-mRNA splicing factors. Genes & Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- Zhang G, Taneja KL, Singer RH, Green MR. Localization of pre-mRNA splicing in mammalian nuclei. Nature. 1994;372:809–812. doi: 10.1038/372809a0. [DOI] [PubMed] [Google Scholar]