Abstract
Somitogenesis has been linked both to a molecular clock that controls the oscillation of gene expression in the presomitic mesoderm (PSM) and to Notch pathway signaling. The oscillator, or clock, is thought to create a prepattern of stripes of gene expression that regulates the activity of the Notch pathway that subsequently directs somite border formation. Here, we report that the zebrafish gene after eight (aei) that is required for both somitogenesis and neurogenesis encodes the Notch ligand DeltaD. Additional analysis revealed that stripes of her1 expression oscillate within the PSM and that aei/DeltaD signaling is required for this oscillation. aei/DeltaD expression does not oscillate, indicating that the activity of the Notch pathway upstream of her1 may function within the oscillator itself. Moreover, we found that her1 stripes are expressed in the anlage of consecutive somites, indicating that its expression pattern is not pair-rule. Analysis of her1 expression in aei/DeltaD, fused somites (fss), and aei;fss embryos uncovered a wave-front activity that is capable of continually inducing her1 expression de novo in the anterior PSM in the absence of the oscillation of her1. The wave-front activity, in reference to the clock and wave-front model, is defined as such because it interacts with the oscillator-derived pattern in the anterior PSM and is required for somite morphogenesis. This wave-front activity is blocked in embryos mutant for fss but not aei/DeltaD. Thus, our analysis indicates that the smooth sequence of formation, refinement, and fading of her1 stripes in the PSM is governed by two separate activities.
Keywords: Somitogenesis, Delta, her1, oscillator, wave front, zebrafish
Somites are reiterated, epithelial structures within the paraxial mesoderm of the vertebrate embryo that give rise to the vertebrae and muscle of the trunk and tail. They are derived from the unsegmented, mesenchymal, presomitic mesoderm (PSM) flanking the notochord and form in an anterior to posterior sequence as clusters of cells undergo furrow formation and epithelialization. The somites are patterned by the adjacent notochord, neural tube, lateral plate mesoderm, and surface ectoderm giving rise to the appropriate, innervated muscle types and vertebrae. In turn, the somites influence the pattern of the neural tube along the anterior–posterior axis and provide the appropriate signals to guide ventrally migrating neural crest cells (Stern et al. 1991; Itasaki et al. 1996; for review, see Bronner-Fraser 1999).
Embryological experiments had suggested that during somitogenesis, segmentation of the paraxial mesoderm takes place in the PSM before morphological signs of segments are evident (Elsdale et al. 1976; Kimmel et al. 1991). Furthermore, it has been shown in the chick that anterior–posterior polarity of the somite is established early and is maintained independently of its orientation with respect to the environment (Aoyama and Asamoto 1988). Additional grafting experiments suggest that somite borders form only when anterior and posterior somite compartments are juxtapposed to each other (Stern and Keynes 1987). More recently, identification of genes expressed in a striped pattern within the PSM and subsequent gene knockout experiments in the mouse have given clear evidence for a molecular prepattern that is linked to somite formation (summarized in del Barco Barrantes et al. 1999).
The clock and wave front and the Meinhardt models describe mechanisms that could regulate somite formation (Cooke and Zeeman 1976; Meinhardt 1982, 1986; Cooke 1998). Both models predicted the existence of an oscillator within the cells of the PSM that produces a spatial pattern that governs somite formation. The existence of an oscillator was revealed by the analysis of c-hairy1, a chick homolog of the Drosophila pair-rule gene hairy, that is expressed in a single stripe that progresses through the cells of the PSM in a posterior to anterior direction. A single stripe traverses the entire PSM in 180 min (two somite cycles) and fades in the anterior PSM as the next somite border is formed (Palmeirim et al. 1997; Forsberg et al. 1998). This wave of expression repeats each somite cycle and does not appear to be directed by cell displacement or by an intercellular signal. It was postulated that these waves of expression are created by an oscillator within each cell that coordinates expression of this gene (Palmeirim et al. 1997). In the clock and wave-front model, the oscillator imposes a stepwise, and thus segmental, progression to the wave front by controlling competence of the cells to respond to (the morphogenic signals of) the wave front that moves from anterior to posterior. In the original version of the model, the wave front is an independent entity, and the stepwise interaction between the wave front and the clock is what leads to regulated somite furrow formation (Cooke and Zeeman 1976; Cooke 1998). A modification of this model was made such that the wave front is a direct output of the clock. In this version, cells would “count” the number of cycles that they had been through; upon reaching the appropriate count, they would form a somite furrow that then must be integrated with the existing spatial pattern (the last formed somite border) (Cooke 1998). The Meinhardt model proposes that the wave front and the oscillator are derived from a common underlying mechanism. In this model, cells in the PSM oscillate between signals responsible for anterior and posterior specification of half-somites. A single cell cannot express both the anterior and the posterior signals. However, an anterior-expressing cell will instruct its neighbors to express posterior signals, and posterior-expressing cells instruct its neighbors to express anterior signals. In computer simulations, these interactions create oscillations that spread in a wave-like fashion from posterior to anterior and refine into bands that come to rest at the appropriate distance because they are stabilized by anterior–posterior signals (the wave front) from the last formed somite. Upon stabilization, the somite furrow would form (Meinhardt 1982, 1986). Although molecular evidence for the clock or oscillator exists, none has been found for either an oscillator-dependent or -independent wave front.
The oscillator has been linked to the Notch pathway in several ways. lfng expression oscillates in both the mouse and chick PSM (Forsberg et al. 1998; McGrew et al. 1998; Aulehla and Johnson 1999). This oscillation, in contrast to c-hairy oscillation, is dependent on protein synthesis, initially suggesting that the Notch pathway acts downstream of the clock (Palmeirim et al. 1997; McGrew et al. 1998). Mouse knockouts or mutants of several components of the Notch pathway, including Notch1, Delta-like 1 (Dll1), Delta-like 3 (Dll3), RBP-Jκ (suppressor of hairless), presenilin, and lunatic fringe (lfng), produce embryos with defects in segmentation and/or anterior–posterior patterning of the somites (Conlon et al. 1995; Oka et al. 1995; Hrabé Angelis et al. 1997; Wong et al. 1997; Evrard et al. 1998; Kusumi et al. 1998; Zhang and Gridley 1998). Further analysis indicated that Dll1, Notch1, and RBP-Jκ are required for proper lfng expression in the mouse, suggesting that the Notch pathway is required for at least some readouts of the clock (del Barco Barrantes et al. 1999).
Somitogenesis in the zebrafish embryo commences at 10.5 hr post-fertilization with a somite pair being created approximately every 30 min until 26–30 somite pairs are formed (for review, see Holley and Nüsslein-Volhard 1999). In the zebrafish, the Notch homologs Notch1a, Notch1b, Notch5, and Notch6 have complex expression patterns that include the PSM and/or the developing somites (Bierkamp and Campos-Ortega 1993; Westin and Lardelli 1997). The Notch ligand homologs, DeltaD and DeltaC, are expressed in the tailbud and in two stripes in the anterior PSM. After somite formation, DeltaD is expressed in the anterior and DeltaC is expressed in the posterior of each somite (Dornseifer et al. 1997; Haddon et al. 1998). Similar to data from Xenopus experiments, misexpression of DeltaD or DeltaC via mRNA injection leads to defects in somite formation (Dornseifer et al. 1997; Jen et al. 1997, 1999; Takke and Campos-Ortega 1999).
her1, a hairy homolog in the zebrafish, is expressed in the tailbud and in two or three stripes in the more anterior PSM. The most anterior stripe fades just before morphological somites can be distinguished, and, at the same time, new stripes of expression emerge from the tailbud. Cell labeling experiments suggested that the stripes of her1 expression correspond to the odd number somites beginning with the fifth somite, whereas the intervening nonexpressing stripes correspond to the even number somites (Müller et al. 1996).
In the zebrafish, the fss-type mutants are defective in both the segmentation and anterior–posterior patterning of the somites. fss is required for the formation of all somites, whereas in beamter (bea) embryos, the first three to four somites form but the remainder do not. In after eight (aei), deadly seven (des), and white tail (wit) embryos, the first seven to nine somites form but the more posterior somites do not. In each of these mutants, in the unsegmented regions, the segmental expression of genes such as MyoD is lost, and expression is seen throughout the somitic mesoderm (Jiang et al. 1996; van Eeden et al. 1996). Moreover, each of these mutants display defects in her1 expression within the PSM (van Eeden et al. 1998).
Here, we show that the fss-type gene aei encodes the zebrafish DeltaD protein. In addition to the previously described somite phenotype, we found that aei/DeltaD embryos exhibit a neuronal hyperplasia. We undertook an extensive analysis of her1 expression and found that, like c-hairy and lfng in the chick and in the mouse or chick, respectively, its expression oscillates within the PSM. Moreover, we found that her1 stripes are expressed in the anlage of consecutive somites, indicating that her1 is not expressed in a pair-rule pattern. aei/DeltaD activity is required for the oscillation of her1 expression, whereas aei/DeltaD expression itself does not oscillate, indicating that the Notch pathway functions upstream of her1 mRNA oscillation, possibly within the clock itself. Analysis of her1 expression in aei, fss, and aei/fss embryos uncovered a wave-front activity that is blocked in embryos mutant for fss but not aei/DeltaD. This analysis indicates that her1 expression has two phases. The first phase of expression within the posterior and intermediate PSM involves periodic oscillations of stripes of her1 expression and requires aei/DeltaD signaling. This establishes a molecular prepattern of her1 stripes within the PSM that appears to be stabilized during the second phase of expression by an anterior wave-front activity acting through fss. This wave-front activity then appears to be required to transform the molecular prepattern into regularly spaced and patterned somites. Finally, cell transplantation experiments demonstrate that fss is required cell-autonomously to propagate this molecular and morphogenic wave-front activity.
Results
aei is DeltaD
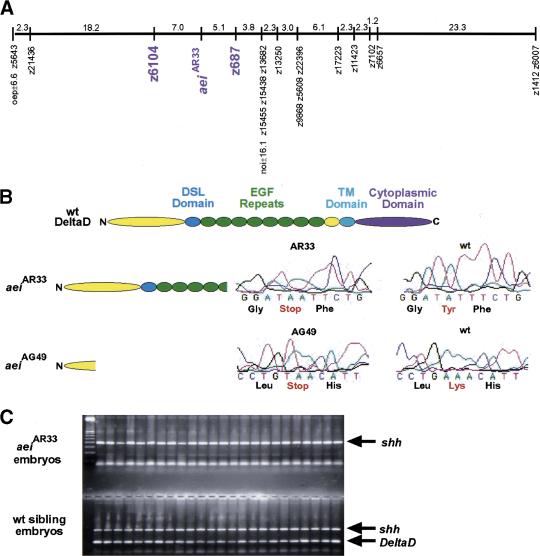
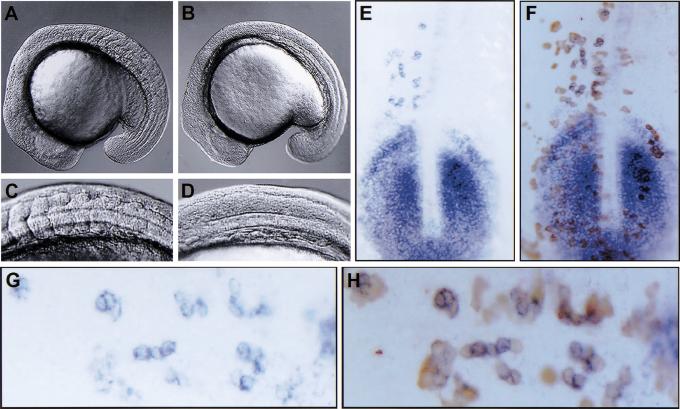
Radiation hybrid mapping placed DeltaD within a 5- to 10-cM region to the right of z6104 on linkage group 13, whereas aei was mapped within the interval between z6104 and z687 (Fig. 1A; Geisler et al. 1999). No additional markers were available within this interval to map DeltaD more accurately with respect to aei. Thus, DeltaD was cloned from aeiAR33 (previously aeitr233) via RT–PCR and sequenced in the hope of uncovering a polymorphism that could be used for mapping via PCR. A T → A substitution was found that created a stop codon within the fifth EGF repeat of DeltaD (Fig. 1B). PCR primers were designed such that the 3′ base of one primer matched the wild-type sequence but not the mutant sequence (allele-specific PCR). To further destabilize primer annealing, the penultimate base of the primer was altered such that it did not match either sequence. The second primer matched both wild-type and mutant sequences perfectly. As an internal control, primers specific to sonic hedgehog (shh) that maps to linkage group 7 were included in each reaction. PCR was performed on DNA preparations from individual mutant aeiAR33 and wild-type sibling embryos. Whereas the sibling embryos gave both the shh and DeltaD PCR products, 0 out of 175 aeiAR33 embryos gave the DeltaD product (Fig. 1C). Genetically, this maps DeltaD within 0.3 cM of aei. Sequencing of a second allele, aeiAG49 (previously aeitg249) revealed a stop codon 63 amino acids from the amino terminus, well before the Delta:Serrate:Lag-2 (DSL) domain that is thought to mediate ligand–receptor interactions, producing what should be a null allele (Fig. 1B; for review, see Artavanis-Tsakonas et al. 1999). The analysis of Islet-1-expressing neurons in the trunk of 13 somite stage aei embryos revealed a hyperplasia of Rohon-Beard neurons (Fig. 2A–C). In each aei allele, the number of Islet-1-expressing neurons is roughly doubled. This neuronal hyperplasia phenotype is analogous to the classic neurogenic phenotype observed for Notch pathway mutants in Drosophila. Additionally, the fact that the two alleles exhibit the same severity of phenotype suggests that despite having the DSL domain, aeiAR33 is also a null allele.
Figure 1.
aei is DeltaD. (A) The genetic map of linkage group 13 is shown with aei and the markers to which it was linked (purple). Numbers indicate genetic distance in cM. (B) Schematics are shown of a wild-type DeltaD protein and of the two predicted protein products produced in aeiAR33 and aeiAG49. Sequencing trace profiles of the region altered in each mutant allele are presented along with the corresponding wild-type sequences. (C) The products of mapping PCR reactions performed on 24 aeiAR33 embryos and 24 wild-type embryos are displayed. Although all embryos produce the shh control product, only the sibling embryos produce the DeltaD product.
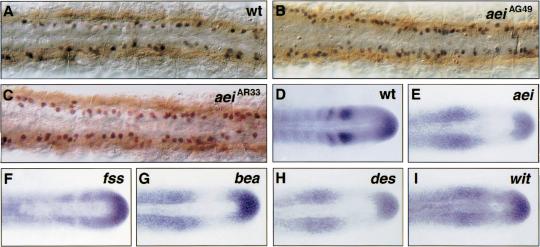
Figure 2.
aei/DeltaD is required for both neurogenesis and patterning of aei/DeltaD expression in the PSM. (A–C) Examination of Islet-1-expressing neurons (black/blue) in the trunk of aei embryos revealed a neuronal hyperplasia. Using myosin (brown) as a reference for somite position, Islet-1-expressing neurons anterior to the posterior border of somite 5 were counted in wild-type (A) and aei embryos (B,C). Wild-type embryos (n = 20) averaged 33 Islet-1-expressing neurons in this region, whereas aeiAG49 (n = 19) and aeiAR33 (n = 20) averaged 58 and 61 such neurons, respectively. All embryos are at about the 13-somite stage. Photos show dorsal views of the trunk region. (D–I) Expression of aei/DeltaD is mispatterned in each of the fss-type mutants. Note, in fss embryos (F), 10%–15% show some evidence of aei/DeltaD stripe formation. Most embryos, however, exhibit expression patterns as seen in aei/DeltaD and the other fss-type mutants. All embryos are at about the 15-somite stage. Photos show dorsal views of the tailbud and posterior trunk. In all panels, anterior is to the left and posterior to the right.
aei/DeltaD regulates its own expression in the PSM
During somitogenesis, aei/DeltaD is expressed in the tailbud and in two stripes in the anterior PSM. The anterior-most stripe refines and persists after furrow formation within the anterior of each somite (Fig. 2D; Dornseifer et al. 1997). In each of the fss-type mutants, the striped expression in the anterior PSM is lost and aei/DeltaD is expressed in all cells of this region (Fig. 2E–I). Thus, in each of the mutants in which somite formation is affected, aei/DeltaD expression is perturbed. The expression of DeltaD in fss embryos is sometimes different from that observed in the other mutants (Fig. 2F). This is perhaps significant and is discussed later within the context of additional data. Finally, Figure 2E indicates that aei/DeltaD activity is required for proper regulation of aei/DeltaD expression.
Examination of her1 expression
her1 is misexpressed in each of the fss-type mutants, but unlike aei/DeltaD expression that is affected in basically the same way in each of the fss-type mutants, the her1 expression pattern differentiates the mutants into three groups (van Eeden et al. 1998). fss embryos form stripes of her1 expression but always lack the anterior-most stripe. In bea, no her1 stripes are formed, and her1 is expressed uniformly throughout the PSM. In aei, des, and wit, her1 stripes do not form, and only a disorganized expression domain is seen in the anterior PSM. To better comprehend the significance of these phenotypic differences, a better understanding of her1 expression in wild-type embryos was required. We staged embryos at the 7-, 8-, 12-, and 15-somite stages and examined her1 expression using MyoD as a reference. MyoD is the most reliable, robust, segmentally expressed in situ marker for the zebrafish somitic mesoderm. MyoD labels the posterior of each somite, and whereas the anterior border of the MyoD expression domain changes as the somite matures, the posterior border of expression remains at the posterior border of each somite. In Figures 3D and 5A, a distinct row of cells can be seen along the posterior of most of the MyoD stripes. These cells are morphologically distinct in that they form the epithelium of the somite and express MyoD less strongly than the cells to their immediate anterior (Weinberg et al. 1996). Measurements were made in reference to the most posterior cells in each MyoD stripe (the posterior border of the somite) and the anterior border of each her1 stripe.
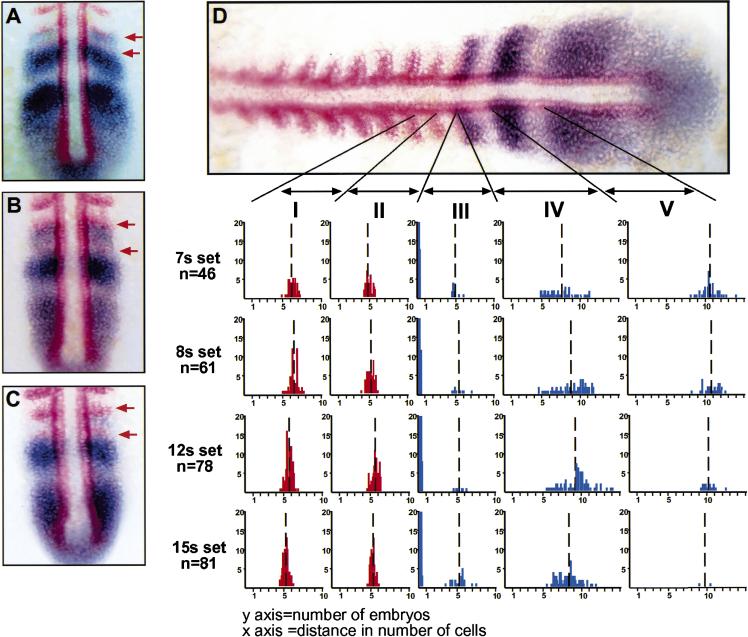
Figure 3.
her1 is expressed in the anlage of consecutive somites. Embryos (7-, 8-, 12-, and 15-somite stage) were double-stained for her1 (blue) and MyoD (red). Some embryos from each data set were found to have her1 stripes immediately posterior to consecutive MyoD stripes: (A) 7 somites; (B) 12 somites; (C) 15 somites. The last two MyoD stripes are indicated by red arrows. (D) A summary of all of the measurement data is shown. The distance between the posterior of the third to last and penultimate MyoD stripes (interval I), between the posterior of the penultimate and the last MyoD stripes (interval II), between the posterior of the last MyoD stripe and the anterior of the first her1 stripe (interval III), between the anterior of the first and the anterior of the second her1 stripe (interval IV), and between the anterior of the second and the anterior of the third her1 stripe (interval V) were made. Vertical broken lines represent mean values. Note that in most cases, including the embryo shown in D, the last MyoD stripe and the first her1 stripe are directly juxtaposed giving a value of 0 for that measurement. This is indicated by the large number of data points at 0 in interval III. Adobe Photoshop 5.0 was used to analyze the expression domains of the digitally photographed in situ images. Distances between expression domains were measured in pixels, and the number of pixels per cell (seven) was calibrated using the in situ as well as DAPI stained embryos.
Figure 5.
aei/DeltaD signaling is required for oscillation of her1 expression. (A–D) Anterior is left and posterior is right. Expression of her1 (blue) and MyoD (red) in wild-type (A), aei (B), fss (C), and aei;fss (D) embryos. (E–H) A hypothetical time course illustrating aei/DeltaD (blue) stripe formation relative to MyoD expression (red) is depicted. Anterior is top and posterior is bottom. In E, one aei/DeltaD stripe is seen immediately posterior to the most posterior MyoD stripe. Posterior to this aei/DeltaD stripe is a broad, weaker domain of DeltaD expression. This weaker domain of expression appears to refine such that a region of nonexpression begins to form immediately posterior to the strong aei/DeltaD stripe (F,G). As this intervening region not expressing aei/DeltaD becomes more refined, the posterior stripe of expression increases in intensity (H). (I–L) A hypothetical time course comparing aei/DeltaD expression (blue) with her1 expression (red) is depicted. aei/DeltaD expression goes through the same process of refinement as shown in E–H, whereas the her1 stripes proceed anteriorly. Although in J and K the anterior limit of the posterior her1 and aei/DeltaD stripes are not aligned as the anterior limits of the more anterior stripes are, ultimately, these more posterior stripes do align (L). Measurements of the distance between the anterior border of the two aei/DeltaD stripes suggest that the posterior aei/DeltaD stripe may progress anteriorly over one to two cell diameters.
During somitogenesis, MyoD is expressed in the adaxial cells that form longitudinal stripes on each side of the notochord and in lateral stripes in the posterior of each somite. Formation of these lateral stripes precedes somite border formation by 1–2 somites, that is, an embryo with 12 somites may have 13–14 MyoD stripes (Weinberg et al. 1996). Thus, whereas MyoD expression arises just before somite boundary formation, her1 expression fades before boundary formation. In 11% of the examined embryos, a MyoD stripe arose before the her1 stripe just anterior to it had faded. Some embryos from each of the data sets in Figures 3 and 4 (40 of 341 total) exhibited her1 stripes immediately posterior to consecutive stripes of MyoD expression indicating that her1 is expressed in the anlage of consecutive somites (Fig. 3A–C). Moreover, the distance between the posterior-most MyoD stripe and the anterior-most her1 stripe was never the size of two somites. These two stripes were either directly adjacent to each other (in most cases) or were separated by a distance of roughly one somite (Fig. 3D, III). These results suggest that her1 is not expressed in a pair-rule pattern and are in contradiction with previous studies (Müller et al. 1996).
Figure 4.
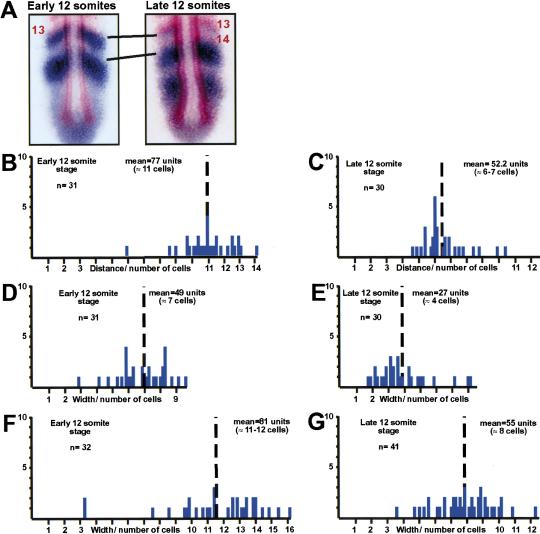
The stripes of her1 expression progress anteriorly during somitogenesis. (A) Early 12-somite embryos and late 12-somite embryos were stained for her1 (blue) and MyoD (red). Measurements were made between the anterior of the her1 stripe immediately posterior to the thirteenth MyoD stripe and the anterior of the next, posterior her1 stripe. In many of the late 12-somite embryos (as pictured), the fourteenth MyoD stripe already had formed, but the her1 stripe immediately posterior to the thirteenth MyoD stripe had not faded. (B–G) Plots of individual measurements are shown with the y-axis representing the number of embryos and the x-axis representing distance from anterior (left) to posterior (right). The distance between the two her1 stripes decreases with time (cf. B to C). Likewise, the width of the two her1 stripes along the anterior–posterior axis decreases with time at the same rate (cf. D to E and F to G). Mean values are indicated by the broken vertical line. Means were compared by two-sample t-test. The difference between the means in B and C is 29 pixels with a 95% C.I. from 19.5 to 30.7 pixels. The difference between the means in D and E is 21.6 pixels with a 95% C.I. from 15.9 to 27.4 pixels. The difference between the means in F and G is 25.7 pixels with a 95% C.I. from 17.8 to 33.7 pixels. Thus, the differences between the means in each comparison is statistically significant.
Analysis of the graphs suggests that the distances between the more posterior her1 stripes (Fig. 3D, IV) are more variable than the distances between MyoD stripes (Fig. 3D, I and II). For instance, the distance between the anterior of the first and second her1 stripe varied continuously from one to two somites in length (Fig. 3D, IV). The distances between the anterior of the second and third her1 stripes also appear more variable than the distances between MyoD stripes. However, the sample size for this measurement is significantly smaller given that not all embryos at the seven- or eight-somite stage have three her1 stripes, and, as somitogenesis proceeds and the tailbud becomes more compact, usually only one or two her1 stripes are observed (cf. graphs in Fig. 3D, V).
Could the variation in spacing between posterior her1 stripes be indicative of a directionally refining expression pattern? Given that the spacing between consecutive MyoD stripes is one somite in length, that the distance between the posterior-most MyoD stripe and the anterior-most her1 stripe is either 0 or one somite in length, and that her1 is expressed in the anlage of consecutive somites, the distances between the first and second and second and third her1 stripes must decrease during time as the PSM matures. Concomitantly, the width of each her1 stripe along the anterior–posterior axis also should diminish. For instance, Figure 3, B and C, shows embryos in which the last MyoD stripe directly abuts a her1 stripe that is between 6 and 12 cells in width. This her1 stripe must diminish in width as the PSM matures. Although the interface between the MyoD stripe and this her1 stripe refines as indicated by the variability observed in the 79% of embryos that have this interface, this border is maintained because the only measurable distance between these stripes is one somite in length: The anterior border does not gradually fade away (posteriorly) from the MyoD stripe. Moreover, the variability seen in interval II of Figure 3D that has a range of three cells means that the MyoD stripe does not move posteriorly to maintain the interface with the her1 stripe as it fades from the anterior: This would require that the range of variability in interval II be >12 cells. Thus, the data in Figure 3 indicate that the fading of each her1 stripe along the anterior–posterior axis must occur directionally from the posterior.
The question remains as to whether the variabilities seen in intervals IV and V of Figure 3D are due to the distances between the anterior borders of the her1 stripes decreasing as the PSM matures. To test this hypothesis, a more precise staging experiment was performed. Embryos were examined under a dissecting microscope as they developed and fixed at either the early or late 12-somite stage. Embryos that just had begun to form the thirteenth furrow were classified as early 12 somites, whereas those with the thirteenth furrow well formed on both sides were called late 12 somites. These embryos were stained for both her1 and MyoD, and the distances between the anterior of the her1 stripe immediately posterior to the thirteenth MyoD stripe and the anterior of the next, posterior her1 stripe were measured (Fig. 4A). This distance averaged almost two somites in length in the early 12-somite embryos (Fig. 4B), whereas in the late 12-somite embryos, the average distance was closer to 1 somite in length (Fig. 4C). Thus, the distance between these two her1 stripes decreases with time. Analysis of the width of these two her1 stripes along the anterior–posterior axis indicates that both the anterior (Fig. 4D,E) and posterior (Fig. 4F,G) her1 stripes fade from the posterior at equal rates as time progresses. The differences between the means compared are all statistically significant. Time-lapse analysis of somitogenesis indicates that these decreases in distances are due to neither cell migration nor cell compaction toward the anterior (supplemental information). This latter point is demonstrated again by nuclear staining (DAPI) (data not shown). Finally, this decrease in distance is not due to patterned programmed cell death within the tailbud as demonstrated by TUNEL (data not shown).
Cumulatively, these observations suggest that her1 expression oscillates within the zebrafish PSM such that stripes of her1 expression progress anteriorly through the cells of the PSM as somitogenesis proceeds. The anterior border of a her1 stripe would have to progress anteriorly at a rate of six to eight cells per somite cycle (30 min), until it is a one-somite distance posterior of where the her1 stripe anterior to it had stopped and faded.
aei/DeltaD signaling is required for her1 oscillation
As described previously, each of the fss-type genes is required for proper regulation of her1 expression (van Eeden et al. 1998). The observation that her1 oscillates within the PSM now allows a better understanding of how somitogenesis is affected in the fss-type mutants. As shown in Figure 5B, aei/DeltaD is required for her1 stripe formation. Although stripes of her1 expression are sometimes seen in aei/DeltaD embryos before the five- to six-somite stage, we have not observed her1 stripes in aei/DeltaD embryos past this stage (van Eeden et al. 1998). In aei embryos, her1 is expressed constantly in the anterior PSM just posterior to the MyoD expression domain. There is little variability to this expression pattern, and no stripes of expression are ever seen separating from the tailbud. Thus, there is no oscillation of her1 expression. This means that cells in the posterior PSM do not express her1 at all but do so once they mature and are just posterior to the MyoD expression domain. This anterior expression is de novo, independent of the oscillation of her1, and must be constantly induced or propagated to always flank the MyoD expression domain posteriorly.
If her1 expression oscillates and aei/DeltaD signaling is required for this oscillation, then the question arises as to whether aei/DeltaD expression also oscillates. If aei/DeltaD expression does oscillate, then this would indicate that it, along with her1, is a readout of the clock. If, however, aei/DeltaD does not oscillate, then this would suggest that aei/DeltaD signaling functions within the clock itself. aei/DeltaD is expressed in one or two stripes in the anterior PSM. The anterior-most stripe is always immediately posterior to a MyoD stripe. Measurements of the distance between the anterior borders of the two aei/DeltaD stripes indicate that the anterior border of the posterior stripe is not more than approximately eight cells (<1.5 somites) away from the posterior-most MyoD stripe (n = 64). Thus, although a aei/DeltaD stripe may progress anteriorly over roughly two cell diameters to be within a one-somite distance from the stripe anterior to it, this refinement in expression of aei/DeltaD cannot account for the oscillation of her1 that cycles within the more posterior region of the PSM. The oscillation of her1 is probably dependent on the protein product of aei/DeltaD that is derived from mRNA that is transcribed within the posterior tailbud. These observations indicate that aei/DeltaD signaling, but not oscillation of aei/DeltaD expression, is required for the oscillation of her1 expression. This is supported by the fact that although aei/DeltaD stripes do not form completely in fss embryos (Fig. 2F), her1 expression nevertheless oscillates (Fig. 5C). Moreover, misexpression of aei/DeltaD via mRNA injection perturbs somite border formation but not her1 transcription (Takke and Campos-Ortega 1999), indicating that regulated expression of aei/DeltaD is not required for her1 oscillation but is required for regular somite border formation.
Although aei/DeltaD stripes may progress in an anterior direction to a limited extent, examination of aei/DeltaD expression relative to MyoD and her1 expression reveals more about the formation of aei/DeltaD stripes. Formation of each aei/DeltaD stripe appears to result from a refining of a diffuse band of expression immediately posterior to a mature aei/DeltaD stripe (Fig. 5E). This band of expression refines both anteriorly and posteriorly (Fig. 5F,G). While the borders of the aei/DeltaD stripes refine, the level of expression concomitantly increases (Fig. 5H). This refinement occurs while stripes of her1 expression progress anteriorly until the anterior border of the her1 stripe aligns with the crisp anterior border of an aei/DeltaD stripe (Fig. 5I–L). The two stripes then continue to refine and fade together (see the anterior-most stripes in Fig. 5I–L). This analysis also shows that consecutive her1 stripes overlap with consecutive aei/DeltaD stripes that refine and are ultimately expressed in the anterior half of every somite.
fss is required for propagation of a wave-front activity
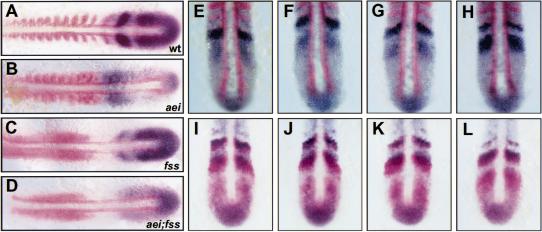
As reported previously, fss, which has the strongest morphological phenotype of the fss-type mutants, has the most subtle effect on her1 expression (van Eeden et al. 1998). In fss embryos, her1 stripes emerge from the tailbud region, but the anterior-most stripe is always absent (Fig. 5C). There are no stripes of MyoD expression that could be used as a reliable reference point in fss embryos; so we are not able to do the same measuring experiments as were performed in wild-type embryos. However, by analogy with the wild-type embryos, we interpret the formation of stripes and variability (observed by eye) of her1 expression in fss embryos as indicating that oscillation of her1 expression occurs. Thus, the activity of fss is required for the maintenance or refinement of the her1 pattern established by the oscillator. In aei;fss double mutant embryos, no her1 stripes are formed as in aei/DeltaD embryos, and the anterior expression that is constant in aei/DeltaD embryos is lost (Fig. 5D). Therefore, fss is required for this anterior activity that in the absence of her1 oscillation is sufficient to induce or propagate her1 expression in the anterior PSM. This anterior activity interacts with the oscillator-derived pattern to produce the smooth changes seen in her1 expression. This fss-dependent activity has some characteristic of the “wave front” in that it interacts with the oscillator-derived pattern in the anterior PSM and is required for the formation of the somites. However, this observed activity is not identical to the wave front; so we refer to it as a “wave-front activity” to make this distinction. aei/DeltaD, on the other hand, is required for the oscillation of her1 and does not affect propagation of the wave-front activity. This difference also is seen in the morphology of the somitic mesoderm in the two mutants. In aei/DeltaD, the posterior somites fail to form, but unpatterned or irregular furrows are seen in the posterior somitic mesoderm (Fig. 6A,C). In contrast, the somitic mesoderm in fss embryos remains smooth with no evidence of furrow formation (Fig. 6B,D). In time-lapse movies of wild-type somitogenesis, a “morphogenic wave front” can be seen as cells in the PSM undergo the furrow formation and cell shape changes that constitute epithelial somite formation in a single anterior to posterior wave. Accordingly, in time-lapse analysis, this morphogenic wave front is preserved in aei/DeltaD embryos (albeit unpatterned), but none is observed in fss embryos (supplemental information; http://www.eb.tuebingen.mpg.de/abt.3/research_interests/somitogenesis.html).
Figure 6.
fss activity is required cell autonomously to propagate a molecular and morphogenic wave-front activity. (A–D) Approximately 17-somite stage embryos are shown. C and D are higher magnification views of the embryos in A and B, respectively. aei/DeltaD embryos (A,C) have a qualitatively different phenotype than fss embryos (B,D). The “unsegmented” posterior somitic mesoderm in aei/DeltaD embryos is convoluted with irregular somite borders (A,C), whereas no border morphogenesis is observed in fss embryos (B,D). (E–H) her1 expression (blue) in a fss embryo that has received wild-type cells (brown) via transplantation is shown. G and H are higher magnification views of E and F, respectively, with anterior to the left. Embryos were first stained for her1 expression, dissected, mounted, and photographed. Then, the dissected tailbuds were stained for the biotin–dextran-labeled donor cells. All her1-expressing cells in the anterior domain are wild-type donor cells (cf. G to H). This result was observed in a total of 61 embryos from five independent experiments.
fss function is required cell automomously
her1 expression, therefore, has two phases: the initial oscillating phase that requires aei/DeltaD signaling and a second phase in the anterior, mature PSM that is under control of a wave-front activity acting through fss. The characteristics of the wave-front activity remain unknown. To address how fss functions and, thus, to try to characterize the wave-front activity, cell transplantation experiments were performed. Wild-type cells within the anterior PSM, where her1 is normally not expressed in fss embryos, are able to express her1 but not able to induce expression in host cells (61 embryos from five experiments; Fig. 6E–H). This indicates that fss functions cell autonomously. However, this cell-autonomous requirement is not absolute because in reciprocal transplantations, fss cells in a wild-type anterior PSM environment will express her1 (49 embryos from three experiments; data not shown). Thus, a community effect is able to overcome the cell-autonomous requirement for fss. This same phenomenon is observed with floating head that encodes a homeobox gene (Talbot et al. 1995). Although this gene encodes for a transcription factor that is required for notochord development, small clones of floating head mutant cells in a wild-type environment can become notochord (Amacher and Kimmel 1998). These experiments show that mutations in fss do not directly block the propagation of an anteriorly derived extracellular signal that controls her1 expression and subsequent somite morphogenesis.
Although this data indicates that fss function is required cell autonomously, it also suggests that fss has cell nonautonomous effects. Transplanted wild-type cells are capable of expressing her1 in the fss host. However, the images in Figure 6, E–H, also show that these wild-type cells do not turn off her1 as they should. If the transplanted wild-type cells were able to regulate her1 expression in a completely wild-type manner, then a “mosaic stripe” pattern should be observed. Although fss appears to be required cell autonomously, it is likely that these cells are required to communicate with their neighbors to consummate the patterning events initiated by the oscillator. The observation that aei/DeltaD is sometimes differently expressed in fss than the other mutants may reflect the distinct role fss plays in this process. For example, in aei/DeltaD embryos, the oscillator readout is perturbed, and thus, no prepattern is established, and aei/DeltaD is expressed consistently in a broad disorganized domain. In fss, the oscillator is functioning and the prepattern is established but not maintained or refined. Thus, in some fss embryos aei/DeltaD stripe formation, which appears to be dependent on her1 (Takke and Campos-Ortega 1999), is initiated but not completed because the wave-front activity that is required to interact with the oscillator pattern at this point is compromised. The reason that only ∼15% of fss embryos appear as in Figure 2F may be because the oscillator-derived pattern is present in the anterior PSM during only a small interval within the somite cycle. In summary, fss appears to function cell autonomously to promote her1 expression and somite formation by propagation of an anterior wave-front activity. However, because of intercellular interactions involving the cells requiring fss activity, fss has indirect cell-nonautonomous effects.
Discussion
Using MyoD as a reference, we performed a detailed study of her1 expression within the PSM. We provide evidence that her1 expression oscillates within the cells of the PSM such that stripes of expression emerge from the tailbud and progress through the PSM in a posterior to anterior direction. The anterior border of a stripe progresses six to eight cell diameters each somite cycle (∼30 min), and a given cell will go through multiple cycles of expression. As somitogenesis proceeds and the tailbud decreases in size, cells of the PSM would go through fewer cycles of her1 expression: A cell in the posterior PSM at the 7- or 8-somite stage would go through eight or so cycles, whereas a cell at the same position in a 12- or 15-somite stage embryo would go through six to seven or five cycles, respectively.
Here, we have demonstrated that the fss-type gene aei is the Notch ligand DeltaD. Additionally, we show that aei/DeltaD signaling is required for her1 oscillation throughout the PSM but that aei/DeltaD stripes appear to form and refine only within the anterior PSM. During gastrulation, both aei/DeltaD and her1 are expressed in the marginal zone, and it is the continuation of this expression that is seen in the posterior tailbud (Müller et al. 1996; Dornseifer et al. 1997). The transient expression of aei/DeltaD in the posterior tailbud (and/or earlier expression) is the likely source of aei/DeltaD protein whose signaling activity is required for the oscillation of her1 expression. The fact that her1 oscillates in fss embryos in the absence of aei/DeltaD stripe formation again supports the conclusion that oscillation of aei/DeltaD signaling but not transcription is required for her1 oscillation. Accordingly, ubiquitous misexpression of aei/DeltaD or an activated form of Notch via mRNA injection perturbs somite formation, but only the activated Notch affects her1 stripe formation (Takke and Campos-Ortega 1999), again indicating that regulated Notch activity but not regulated DeltaD expression is required for the oscillation of her1 expression. Cumulatively, these findings indicate that Notch signaling is required within the oscillator or at least for some readouts of the oscillator.
By examining her1/MyoD expression in aei, fss, and aei/fss mutant embryos, a wave-front activity was uncovered that requires fss but not aei/DeltaD activity. Accordingly, in time-lapse analysis, the morphogenic wave front is preserved in aei/DeltaD but not fss embryos. This wave-front activity is capable of continually inducing her1 expression in the anterior PSM just posterior to the MyoD expression domain independently of her1 oscillation. This de novo expression, however, is unpatterned in that her1 is expressed in a broad domain in the anterior PSM. Thus, although neither aei/DeltaD signaling nor her1 oscillation appears to be required for the anterior induction of her1 by the wave front, they are required for restraining the inductive activity. The oscillator thus appears to provide pattern, whereas the wave-front activity appears to provide a spatial–temporal signal that stabilizes and translates this pattern into regularly patterned somites.
Recent research had suggested that the oscillator acts to regulate Notch signaling that then directs somite border formation, possibly via Eph:Ephrin interactions (for review, see Durbin et al. 1998; Pourquié 1999). This relationship was largely inferred from the observation that although c-hairy oscillation was independent of protein synthesis, oscillation of the Notch pathway gene lfng was dependent on translation (Palmeirim et al. 1997; Forsberg et al. 1998; McGrew et al. 1998; Aulehla and Johnson 1999). However, the observation that lfng expression within the PSM is affected in Dll1 and RBP-Jκ mutant mice even though neither of these genes shows such a dynamic expression pattern suggested that Notch signaling may act upstream of oscillating gene expression in the PSM. Here, we provide clear evidence that Notch pathway signaling is required for her1 oscillation. This regulation of her1 activity then appears to be required for the patterned expression of aei/DeltaD in the anterior PSM as aei/DeltaD stripe formation is perturbed in mutant embryos in which her1 oscillation is lost (aei/DeltaD, bea, des, and wit mutant embryos). The failure to maintain and refine her1 expression in fss embryos also correlates with defect in aei/DeltaD stripe formation. Moreover, misexpression of her1 via mRNA injection perturbs aei/DeltaD stripe formation and somite formation (Takke and Campos-Ortega 1999). Thus, genetic data indicate that aei/DeltaD acts upstream of her1 to regulate oscillation of her1 expression, and the combination of genetic and misexpression data shows that her1 functions upstream of aei/DeltaD stripe formation that is then required for proper somite border formation. The relative changes in expression of these two genes within the PSM during somite formation are depicted in Figure 7.
Figure 7.
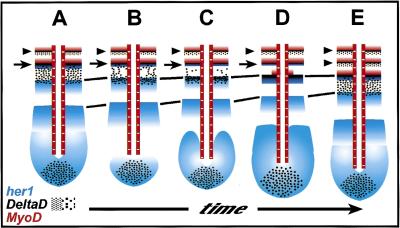
A model for the concomitant changes in MyoD, her1, and aei/DeltaD gene expression during zebrafish somite formation. Anterior is top and posterior is bottom. (A–E) A time series during one cycle of somite formation is represented. Somite furrows are indicated by arrowheads. MyoD is expressed in the posterior of each somite, whereas aei/DeltaD is weakly expressed in the anterior of each somite. The anterior-most stripe of her1 overlaps with a strong stripe of aei/DeltaD (arrows). These two stripes refine and fade together (A–D) leaving only a faint stripe of aei/DeltaD expression (E). Around this time, the next, posterior stripe of MyoD emerges (D), and somite furrow formation occurs (second arrowhead in E). During this time period, the more posterior stripes of her1 expression progress anteriorly (A–E) (equivalent stripes are indicated by connecting lines). Meanwhile, the weak domain of aei/DeltaD expression immediately posterior to the strong stripe of expression (arrow) in the anterior PSM (A) begins to refine (B,C). Ultimately, this process results in a strong stripe of expression that aligns anteriorly with a stripe of her1 expression (D). Posterior to these overlapping stripes, a broad, weak domain of expression of aei/DeltaD forms (E) as was originally present in A. In the more posterior PSM and tailbud, an additional stripe of her1 expression emerges (B–E). The expression of aei/DeltaD in the posterior tailbud remains relatively constant (A–E).
In both the mouse and in the zebrafish, many of the mutants in which somite segmentation is perturbed show an anterior to posterior polarity in phenotype in that the anterior regions are less severely affected than the posterior regions. In the zebrafish, aei/DeltaD, des, and wit form the anterior-most seven to nine somites but not the more posterior somites. Interestingly, the development of these anterior somites exhibits several differences from the development of the more posterior somites. The first six somites form more rapidly (three pairs per hour) than the posterior somites (two pairs per hour) (Westerfield 1995). These anterior somites also display a more synchronous development. For example, in the first five somites, the adaxial cells undergo shape change and rearrangement simultaneously, whereas within the posterior somites the adaxial cells rearrange in an anterior to posterior progression as each somite matures (van Eeden et al. 1996). The refinement of snail expression, the initiation of engrailed expression, and the formation of the lateral MyoD stripes occur simultaneously in the anterior five to seven somites but sequentially in the posterior somites (Kimmel et al. 1991; Hammerschmidt and Nüsslein-Volhard 1993; Weinberg et al. 1996). The first her1 stripe was originally fate mapped to the anlage of the fifth somite (Müller et al. 1996). However, our data indicates that her1 oscillates through the PSM, which brings the fate mapping data into question. It is possible that if Müller et al. (1996) very carefully staged and positioned their embryos, they could have labeled the first and second her1 stripes and also labeled the anlage of the fifth and seventh somite. In other words, by carefully staging the embryos, probably within at most a 10-min interval, and labeling in the same region that happened to correspond to the anlage of somite 5 and 7, they also could have caught her1 expression just as the two stripes were passing through this region. Because the her1 stripes would continue to move anteriorly, they ultimately could stop and fade in the anterior of the anlage of somite 3 or 4, for example. Thus, it is not clear when the oscillator output of her1 stripes begins to function during somitogenesis. Within this context, it is interesting to note that aei/DeltaD activity is not required for anterior somite formation, indicating that segmentation of these somites occurs independently of aei/DeltaD-dependent oscillations. This raises the possibility that anterior somite formation occurs independently of any oscillator. Conversely, fss activity is necessary for formation and anterior–posterior patterning of these somites, indicating that the wave-front activity or some additional function of fss is required.
How do the results presented here affect our understanding of the conservation of segmentation mechanisms in higher animals? Much excitement arose from the initial finding that her1 was expressed in a pair-rule pattern as in the Drosophila embryo (Müller et al. 1996). In conjunction with the finding that the amphioxus engrailed gene (AmphiEn) is expressed in the posterior of the eight anterior-most somites, many suggested a possible conservation of segmentation mechanisms between protostomes and deuterostomes (Holland et al. 1997). Although there is precedent for such conservation in the roles that the hox genes play in defining segment identity and that the dpp;Bmp/sog;chordin patterning system plays in establishing embryonic dorsal–ventral polarity, it seems less likely now that such conservation exists for segmentation mechanisms because here we find that her1 is not expressed in a pair-rule pattern. It seems more likely that the conservation of hairy function during segmentation reflects a conserved use of the Notch pathway during somitogenesis. That Notch function and a molecular oscillator are required for somitogenesis in all vertebrates seems likely at this point. However, differences in how this regulatory network is organized may exist. For example, which Notch pathway genes are direct outputs of the oscillator may vary. Although lfng expression oscillates in both the chick and mouse, the one known lfng homolog in zebrafish does not oscillate (Forsberg et al. 1998; McGrew et al. 1998; Aulehla and Johnson 1999; S. Holley and T. Vogt, unpubl.). One intriguing possibility concerning conservation of segmentation mechanisms between protostomes and deuterostomes concerns the development of the anterior somites. In amphioxus where AmphiEn is expressed in a metameric pattern, the anterior somites form via a mechanism distinct from the posterior somites (Holland et al. 1997). Given that anterior somitogenesis appears distinct in both mouse and zebrafish, it is possible that the as yet unidentified genetic mechanism that controls this process exhibits some homology to protostome segmentation mechanisms.
Materials and methods
Fish work
Fish were raised as described by Westerfield (1995). Eggs were derived from natural crosses. aei;fss double mutant embryos were derived from a cross between double heterozygous parents. Morphologically, the fss phenotype is dominant to aei. One quarter of the morphologically fss embryos displayed the composite (fss + aei) her1 expression phenotype. These were classified as double mutant embryos.
Mapping
Radiation hybrid mapping was performed as described by Geisler et al. (1999). For the allele-specific PCR, 5-day-old embryos were chilled on ice for 30 min. Single embryos were placed in 96-well microtiter plates in 25 μl of 70 mg/ml proteinase K in TE and incubated at 70°C for 4 hr and at 75°C for 30 min. Each preparation was then diluted with 50 μl ddH2O, and 5 μl was used for each PCR reaction. For the PCR reactions, primers AR33 MAP-3 (5′-gttgcactgatcttgaaaacacctacagct-3′), AR33 MAP-2 (5′-ccagcataacccatcgggcactggcagaca-3′), shh 7 (5′-ccggagatccgcgcctcgac-3′), and shh 10 (5′-ctgtgtcatgagcctgtccgctc-3′) were used at a final concentration of 0.225 μm. Additional final PCR conditions were 10 mm Tris-HCl (pH 8.3), 50 mm KCl, 1.5 mm MgCl2, 0.01% gelatin, and 0.5 units of Taq polymerase. PCR cycling conditions were as follows: 94°C for 2 min; 35 cycles of 94°C for 30 sec, 67°C for 30 sec, 73°C for 1 min; and 73°C for 5 min. Reactions were analyzed by electrophoresis in 2% agarose gels. Three 96-well microtiter dishes containing 72 mutant and 24 sibling embryos were assayed twice, and only samples that produced the same result were counted. Sixty-one of seventy-two (84.7%) sibling embryos gave both the shh and DeltaD product. Two hundred and six aeiAR33 embryos gave the shh product but not the DeltaD product.
Cloning and sequencing
Total RNA was isolated from mutant embryos using TriStar reagent (Angewandte Gentechnologie Systeme GmbH) according to kit protocol. Poly(A) RNA was subsequently isolated using Qiagen Oligotex. RT–PCR was performed using the SuperScript kit (GIBCO BRL). Three independently derived PCR products were either cloned into Bluescript or into PCR2.1 using the TA-cloning kit (Invitrogen). All clones were sequenced using the Thermo Sequenase fluorescent labeling primer cycle sequencing kit (Amersham), run on an A.L.F. sequencer (Pharmacia) and analyzed with the Lasergene software package.
In situ and antibody stainings
In situ hybridizations were performed according to standard protocols. Briefly, embryos were hybridized simultaneously with digoxigenin- and fluorescein-labeled RNA probes. Antibody incubation and NBT/BCIP staining for the digoxigenin-labeled probe were performed, followed by fixation, 2 × 15-min incubations in 100 mm glycine (pH 2.2), through a methanol series to 100% methanol for 1 hr, through a methanol series to PBST, washing, preincubation, and incubation with the anti-fluorescein antibody. The second staining reaction used the alkaline phosphatase substrate Fast Red (Roche). Embryos were cleared in 75% glycerol in PBST.
For antibody stainings, embryos were fixed in 4% PFA, blocked, incubated with the α-Islet-1 antibody (39.4D5 monoclonal, 1:500 dilution; Developmental Studies Hybridoma Bank), washed, incubated with 2% biotinylated anti-mouse (Vector), washed, and stained according to standard procedures. The first staining reaction, using the Elite ABC peroxidase kit (Vector), was performed in the presence of 0.03% CoCl2 to give the black/gray stain. The second incubation with the α-myosin antibody (A4.1025 monoclonal, 1:50 dilution; Developmental Studies Hybridoma Bank) was performed as above, and the second staining reaction was performed as above but without CoCl2 to give the brown stain. Embryos were then fixed, washed, and cleared.
Cell transplantations
Cell transplantations were performed essentially as described by Westerfield (1995). Donor embryos were labeled with a fresh mixture of 1.25% biotin–dextran, MW10,000 and 1.25% tetramethyl-rhodamine–dextran MW10,000 (Molecular Probes). Roughly 20 cells were transplanted to host embryos. After in situ hybridizations were performed, donor cells were visualized using the Elite ABC peroxidase kit (Vector). Briefly, after the standard NBT/BCIP staining reaction, embryos were fixed for 20 min in 4% PFA, washed, put through a methanol series, incubated in methanol for 1 hr, through a methanol series to PBST. Tailbuds then were dissected, flat-mounted, and photographed. The tailbuds were recovered and incubated in 2% Blocking Reagent (Roche) for 1 hr. Meanwhile, 1 μl each of solutions A and B from the Elite ABC kit were mixed in 1 ml of blocking solution and preincubated for 45 min to 1 hr. The AB mixture was added to the embryos and incubated for 15 min. Embryos were washed three times for 30 min in PBST and preincubated in DAB (20 μg/ml) for 15 min. The staining reaction was initiated by adding 2 μl 0.3% H2O2/ml. Embryos were then fixed, washed, and cleared in 75% glycerol.
Photomicroscopy and graphics work
All embryos were digitally photographed (400 dpi) and analyzed using Adobe Photoshop 5.0. Figures were compiled using Freehand 8.0.
Acknowledgments
We are grateful to Dörthe Jülich and Silke Geiger-Rudolph for technical assistance. We are particularly thankful to Prof. Klaus Dietz, Department of Medical Biometry (University of Tübingen, Germany) for help with the statistical analysis of our data. We thank Dr. Jose Campos-Ortega for the gift of the her1 and DeltaD constructs and Heike Schauerte for the shh primers. We thank Barbara Conrad for advice on TUNEL. We thank Hans Meinhardt, Ralf Sommer, Carl Neumann, Elke Ober, Marcus Dekens, Florian Maderspacher, Henry Roehl, and Holger Knaut for critical comments on the manuscript. S.A.H. was supported by a postdoctoral fellowship from the Damon Runyon-Walter Winchell Cancer Research Foundation (DRG-1435).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL holley@bio.tuebingen.mpg.de; FAX 49 7071 601484.
References
- Amacher SL, Kimmel CB. Promoting notochord fate and repressing muscle development in zebrafish. Development. 1998;125:1397–1406. doi: 10.1242/dev.125.8.1397. [DOI] [PubMed] [Google Scholar]
- Aoyama H, Asamoto K. Determination of somite cells: Independence of cell differentiation and morphogenesis. Development. 1988;104:15–28. doi: 10.1242/dev.104.1.15. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Aulehla A, Johnson RL. Dynamic expression of lunatic fringe suggests a link between notch signaling and an automomous cellular oscillator driving somite segmentation. Dev Biol. 1999;207:49–61. doi: 10.1006/dbio.1998.9164. [DOI] [PubMed] [Google Scholar]
- Bierkamp C, Campos-Ortega JA. A zebrafish homologue of the Drosophila neurogenic gene Notch and its pattern of transcription during early embryogenesis. Mech Dev. 1993;43:87–100. doi: 10.1016/0925-4773(93)90027-u. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M. Rostrocaudal differences within the somites confer segmental pattern to trunk neural crest migration. In: Ordahl C, editor. Somitogenesis. San Diego, CA: Academic Press; 1999. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- Cooke J. A gene that resuscitates a theory-somitogenesis and a molecular oscillator. Trends Genet. 1998;14:85–88. doi: 10.1016/s0168-9525(98)01396-1. [DOI] [PubMed] [Google Scholar]
- Cooke J, Zeeman EC. A clock and wavefront model for control of the number of repeated structures during animal morphognesis. J Theor Biol. 1976;58:455–476. doi: 10.1016/s0022-5193(76)80131-2. [DOI] [PubMed] [Google Scholar]
- del Barco Barrantes I, Elia A, Wunnch K, Hrabde De Angelis M, Mak T, Rossant J, Conlon R, Gossler A, Luis de la Pompa J. Interaction between Notch signaling and Lunatic Fringe during somite boundary formation in the mouse. Curr Biol. 1999;9:470–480. doi: 10.1016/s0960-9822(99)80212-7. [DOI] [PubMed] [Google Scholar]
- Dornseifer P, Takke C, Campos-Ortega JA. Overexpression of a zebrafish homologue of the Drosophila neurogenic gene Delta perturbs differentiation of primary neurons and somite development. Mech Dev. 1997;63:159–171. doi: 10.1016/s0925-4773(97)00037-3. [DOI] [PubMed] [Google Scholar]
- Durbin L, Brennan C, Shiomi K, Cooke J, Barrios A, Shanmugalingam S, Guthrie B, Lindberg R, Holder N. Eph signaling is required for segmentation and differentiation of the somites. Genes & Dev. 1998;12:3096–3109. doi: 10.1101/gad.12.19.3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsdale T, Pearson M, Whitehead M. Abnormalities in somite segmentation following heat shock to Xenopus embryos. J Embyol Exp Morphol. 1976;35:625–635. [PubMed] [Google Scholar]
- Evrard YA, Lun Y, Aulehla A, Gan L, Johnson RL. lunatic fringe is an essential mediator of somite segmentation and patterning. Nature. 1998;394:377–381. doi: 10.1038/28632. [DOI] [PubMed] [Google Scholar]
- Forsberg H, Crozet F, Brown NA. Waves of mouse Lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. Curr Biol. 1998;8:1027–1030. doi: 10.1016/s0960-9822(07)00424-1. [DOI] [PubMed] [Google Scholar]
- Geisler R, Rauch G-J, Baier H, van Bebber F, Brobeta L, Dekens MP, Finger K, Fricke C, Gates MA, Geiger H, et al. A radiation hybrid map of the zebrafish genome. Nat Genet. 1999;23:86–89. doi: 10.1038/12692. [DOI] [PubMed] [Google Scholar]
- Haddon C, Smithers L, Schneider-Maunoury S, Coche T, Henrique D, Lewis J. Multiple delta genes and lateral inhibition in zebrafish primary neurogenesis. Development. 1998;125:359–370. doi: 10.1242/dev.125.3.359. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt M, Nüsslein-Volhard C. The expression of a zebrafish gene homologous to Drosophila snail suggests a conserved function in invertebrate and vertebrate gastrulation. Development. 1993;119:1107–1118. doi: 10.1242/dev.119.4.1107. [DOI] [PubMed] [Google Scholar]
- Holland LZ, Kene M, Williams NA, Holland ND. Sequence and embryonic expression of the amphioxus engrailed gene (AmphiEn): The metameric pattern of transcription resembles that of its segment-polarity homolog in Drosophila. Development. 1997;124:1723–1732. doi: 10.1242/dev.124.9.1723. [DOI] [PubMed] [Google Scholar]
- Holley SA, Nüsslein-Volhard C. Somitogenesis in zebrafish. In: Ordahl C, editor. Somitogenesis. San Diego, CA: Academic Press; 1999. pp. 97–127. [Google Scholar]
- Hrabé Angelis M, McIntyre J, Gossler A. Maintenance of somite borders in mice requires the Delta homologue Dll1. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- Itasaki N, Sharpe J, Morrison A, Krumlauf R. Reprogaramming Hox expression in the vertebrate hindbrain: Influence of paraxial mesoderm and rhombomere transposition. Neuron. 1996;16:487–500. doi: 10.1016/s0896-6273(00)80069-0. [DOI] [PubMed] [Google Scholar]
- Jen W-C, Wettstein D, Turner D, Chitnis A, Kintner C. The Notch ligand, X-Delta-2, mediates segmentation of the paraxial mesoderm in Xenopus embryos. Development. 1997;124:1169–1178. doi: 10.1242/dev.124.6.1169. [DOI] [PubMed] [Google Scholar]
- Jen W-C, Gawantka V, Pollet N, Niehrs C, Kintner C. Periodic repression of Notch pathway genes governs the segmentation of Xenopus embryos. Genes & Dev. 1999;13:1486–1499. doi: 10.1101/gad.13.11.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y-J, Brand M, Heisenberg C-P, Beuchle D, Furutani-Seiki M, Kelsh RN, Warga RM, Granato M, Haffter P, Hammerschmidt M, et al. Mutations affecting neurogenesis and brain morphology in the zebrafish, Danio rerio. Development. 1996;123:205–216. doi: 10.1242/dev.123.1.205. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Schilling TF, Hatta K. Patterning of body segments of the zebrafish embryo. Curr Top Dev. 1991;25:77–110. doi: 10.1016/s0070-2153(08)60412-3. [DOI] [PubMed] [Google Scholar]
- Kusumi K, Sun ES, Kerrebrock AW, Bronson RT, Chi DC, Bulotsky MS, Spencer JB, Birren BW, Frankel WN, Lander ES. The mouse pudgy mutation disrupts Delta homogue Dll3 and intiation of early somite boundaries. Nat Genet. 1998;19:274–278. doi: 10.1038/961. [DOI] [PubMed] [Google Scholar]
- McGrew MJ, Dale JK, Fraboulet S, Pourquié O. The Lunatic Fringe gene is a target of the molecular clock linked to segmentation in avian embryos. Curr Biol. 1998;8:979–982. doi: 10.1016/s0960-9822(98)70401-4. [DOI] [PubMed] [Google Scholar]
- Meinhardt H. Models of biological pattern formation. London, UK: Academic Press; 1982. [Google Scholar]
- ————— . Models of segmentation. In: Bellairs R, Ede DA, Lash JW, editors. Somites in developing embryos. 1986. . NATO ASI series A, vol 118. Plenum Press, New York, NY. [Google Scholar]
- Müller M, Weiszäcker E, Campos-Ortega JA. Expression domains of a zebrafish homologue of the Drosophila pair-rule gene hairy correspond to primordia of alternating somites. Development. 1996;122:2071–2078. doi: 10.1242/dev.122.7.2071. [DOI] [PubMed] [Google Scholar]
- Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, Honjo T. Disruption of the mouse RBP-J Kappa results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquié O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segementation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Pourquié O. Notch around the clock. Curr Opin Genet Dev. 1999;9:559–565. doi: 10.1016/s0959-437x(99)00011-8. [DOI] [PubMed] [Google Scholar]
- Stern CD, Keynes RJ. Interactions between somite cells: The formation and maintenance of segment boundaries in the chick embryo. Development. 1987;99:261–272. doi: 10.1242/dev.99.2.261. [DOI] [PubMed] [Google Scholar]
- Stern CD, Jaques KF, Lim T, Fraser SE, Keynes RJ. Segmental lineage restrictions in the chick embryo spinal cord depend on the adjacent somites. Development. 1991;113:239–244. doi: 10.1242/dev.113.1.239. [DOI] [PubMed] [Google Scholar]
- Takke C, Campos-Ortega JA. her1, a zebrafish pair-rule gene, acts downstream of notch signaling to control somite development. Development. 1999;126:3005–3014. doi: 10.1242/dev.126.13.3005. [DOI] [PubMed] [Google Scholar]
- Talbot WS, Trevarrow B, Halpern ME, Melby AE, Farr G, Postlethwait JH, Jowett T, Kimmel CB, Kimelman D. A homeobox gene essential for zebrafish notochord development. Nature. 1995;378:150–157. doi: 10.1038/378150a0. [DOI] [PubMed] [Google Scholar]
- van Eeden FJM, Granato M, Schach U, Brand M, Furutani-Seiki M, Haffter P, Hammerschmidt M, Heisenberg C-P, Jiang Y-J, Kane DA, et al. Mutations affecting somite formation and patterning in the zebrafish Danio rerio. Development. 1996;123:153–164. doi: 10.1242/dev.123.1.153. [DOI] [PubMed] [Google Scholar]
- van Eeden FJM, Holley SA, Haffter P, Campos-Ortega J, Nüsslein-Volhard C. Zebrafish segmentation and pair-rule patterning. Dev Genet. 1998;23:65–76. doi: 10.1002/(SICI)1520-6408(1998)23:1<65::AID-DVG7>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Anderman P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Westerfield M. The zebrafish book. Eugene, OR: University of Oregon Press; 1995. [Google Scholar]
- Westin J, Lardelli M. Three novel Notch genes in zebrafish: Implications for vertebrate Notch gene evolution and function. Dev Genes Evol. 1997;207:51–63. doi: 10.1007/s004270050091. [DOI] [PubMed] [Google Scholar]
- Wong PC, Zheng H, Chen H, Becher MW, Sirinathsinghji DJS, Trumbauer ME, Chen HY, Price DL, Van der Ploeg LHT, Sisodia SS. Presenilin 1 is required for Notch1 and Dll1 expression in the paraxial mesoderm. Nature. 1997;387:288–292. doi: 10.1038/387288a0. [DOI] [PubMed] [Google Scholar]
- Zhang N, Gridley T. Defects in somite formation in lunatic fringe deficient mice. Nature. 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]