Abstract
Purpose
This prospective study aimed to develop a robust and clinically-applicable method to identify high-risk early stage lung cancer patients and then to validate this method for use in future translational studies.
Patients and Methods
Three published Affymetrix microarray data sets representing 680 primary tumors were used in the survival-related gene selection procedure using clustering, Cox model and random survival forest (RSF) analysis. A final set of 91 genes was selected and tested as a predictor of survival using a qRT-PCR-based assay utilizing an independent cohort of 101 lung adenocarcinomas.
Results
The RSF model built from 91 genes in the training set predicted patient survival in an independent cohort of 101 lung adenocarcinomas, with a prediction error rate of 26.6%. The mortality risk index (MRI) was significantly related to survival (Cox model p < 0.00001) and separated all patients into low, medium, and high-risk groups (HR = 1.00, 2.82, 4.42). The MRI was also related to survival in stage 1 patients (Cox model p = 0.001), separating patients into low, medium, and high-risk groups (HR = 1.00, 3.29, 3.77).
Conclusions
The development and validation of this robust qRT-PCR platform allows prediction of patient survival with early stage lung cancer. Utilization will now allow investigators to evaluate it prospectively by incorporation into new clinical trials with the goal of personalized treatment of lung cancer patients and improving patient survival.
Keywords: Lung cancer, qRT-PCR, Prognosis
Lung cancer is the leading cause of cancer-related death, and non-small cell lung cancer (NSCLC) accounts for almost 80 percent of deaths from lung cancer. 1 Surgery is the major treatment option for patients with early stage (stage IA and IB) NSCLC, yet as many as 35–50% of these patients will relapse within five years, indicating that a “high-risk” subgroup of these patients might benefit from adjuvant chemotherapy if properly identified. 2 While recent studies have demonstrated significantly improve survival using adjuvant chemotherapy in patients undergoing pulmonary resection for stage IB, II or IIIA NSCLC, these patients are subject to treatment-related toxicity. 2–5 It remains a critical and unsolved challenge to estimate more precisely the risk for survival or recurrence in individual patients in order to provide adjuvant therapy to high risk patients and avoid providing adjuvant therapy to low risk patients.
The emerging use of gene expression signatures may enable clinicians to make treatment decisions based on specific characteristics of individual patients and their tumor. Many microarray-based gene profiles with gene numbers varying from dozens to thousands have been reported to predict patient survival in lung cancer. 6–14 Due to the need for specialized laboratory facilities, the large number of contributing genes and complex statistical analyses, microarray-based gene expression profiles are not very practical for clinical utilization. Alternatively, a quantitative real-time PCR (qRT-PCR) method may be utilized more efficiently. Several qRT-PCR profiles have been reported, 15–22 but none of these gene sets have been refined or tested adequately for clinical use.
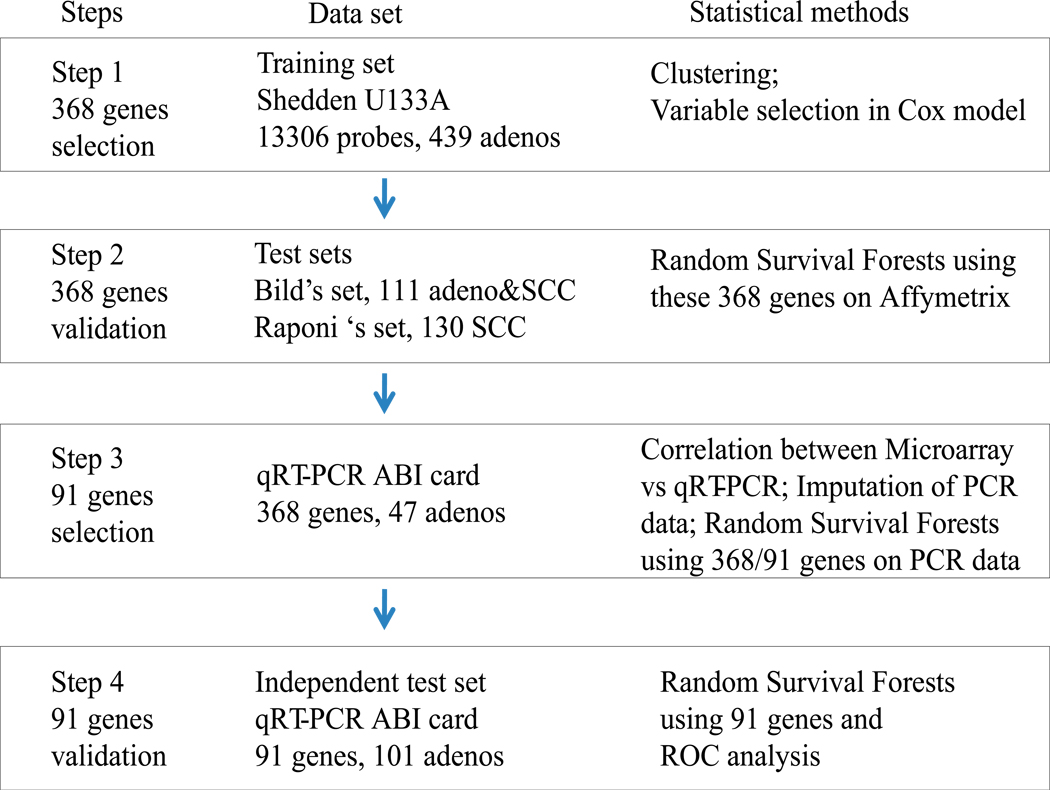
In this study, we have combined microarray gene profiles of a large NSCLC data set 8, 12, 13 and a qRT-PCR-based approach, utilizing a novel gene selection method. A novel aspect of the design was to include genes representing diverse biological processes and then to measure gene expression using qRT-PCR. Finally, we developed and then verified a 91-gene qRT-PCR card-based platform survival classifier using an independent cohort of 101 lung adenocarcinomas. The strategy used in this study is shown in Figure 1. This strategy was prospectively defined and executed as planned.
FIGURE 1.
Strategy for the development and validation of the 91-gene qRT-PCR classifier for lung cancer prognosis.
Patients and Methods
The methods are briefly described in the text of the paper, with full descriptions in the Supplementary Methods.
Published microarray data collection
Three published Affymetrix microarray data sets representing 680 primary tumors were used in the survival-related gene selection procedure. The primary training data set included 439 lung adenocarcinomas from a consortia study of four centers, 13 and a combined 111 lung adenocarcinomas and squamous carcinoma (SCC) data set represented test set one 8 and a 130 lung SCC data set was used as test set two. 12 The clinical information for these three data sets is provided in Table 1. Other microarray data sets 7, 9, 10, 14 were not chosen due to platform differences, sample sizes less 100 or not having survival information. Our primary outcome was overall survival for all datasets, censored at 5 years. The information concerning adjuvant chemotherapy or radiation therapy was provided in the original papers and also summarized in Table 1.
TABLE 1.
Clinical Characteristics of Samples Used in this Study.
| Data set | Training set | Bild test set | Raponi test set | Validation set | |
|---|---|---|---|---|---|
| Platform | U133A | U133 plus2.0 | U133A | qRT-PCR | |
| Sample number | 439 | 111 | 130 | 101 | |
| Type of cancer | AD | 58 AD/53 SCC | SCC | AD | |
| Age average (SD) | 64.4 (10.1) | 64.8 (9.6) | 67.5 (9.9) | 67.0 (9.6) | |
| Gender | |||||
| Female | 218 (49.7%) | 48 (43.2%) | 48(36.9%) | 53(52.5%) | |
| Male | 221 | 63 | 82 | 48 | |
| Stage | |||||
| Stage I | 276(62.9%) | 67(63.2%) | 73(56.2%) | 59(58.4%) | |
| Stage II | 104 | 18 | 34 | 16 | |
| Stage III | 59 | 21 | 23 | 26 | |
| Differentiation | |||||
| Well | 60 | NA | 15 | 28 | |
| Moderate | 208 | NA | 76 | 38 | |
| Poor | 166(38.3%) | NA | 39(30%) | 34(33.7%) | |
| Dead (5 year) | 186(42.4%) | 58(52.3%) | 52(40%) | 44(43.6%) | |
| Alive | 253 | 53 | 78 | 57 | |
| Median survival (m) | 47 | 31.1 | 34.5 | 28.8 | |
| Adjuvant therapy | 108 | unknown | 48 | 42 | |
| No adjuvant therapy | 329 | unknown | 69 | 58 | |
Abbreviation: AD, adenocarcinoma; SCC, squamous cell cancer. Adjuvant therapy includes chemo- and/or radio-therapy.
Patients and tissue specimens for qRT-PCR measurements
A subset of 47 of the 439 patients had qRT-PCR measured. In addition we identified an independent validation set of 101 lung adenocarcinomas procured from patients having pulmonary resection for cancer between February 1992 and November 2007 at the University of Michigan (a total 120 samples were examined using qRT-PCR, including 12 paired normal lung and tumor tissues and 7 duplicate tumor samples representing different portion of these tumors). This study was approved by the Institutional Review Board of University of Michigan. None of the patients received preoperative chemotherapy or radiation therapy. A total of 68 patients were stage 1b or above 1b, and 38 of these 68 patients (56%, 38/68) received adjuvant chemotherapy or radiation therapy. No adjuvant therapy was provided to the 29 of 33 stage 1a patients. The clinical information for this cohort is presented in Table 1.
Custom TaqMan low density arrays and quantitative RT-PCR
Regions containing a minimum of 70% tumor cellularity were utilized for RNA isolation. RNA quality was analyzed by 2100 Bioanalyzer (Agilent Santa Clara, CA). Custom TaqMan Low Density Arrays (384-well micro fluidic cards) were obtained from Applied Biosystems Inc. (ABI). The primers of survival related genes including an endogenous loading control gene (18S RNA, beta-actin and GAPDH) and blank controls were pre-applied to the cards. The preparation and running of the micro-fluidic cards (qRT-PCR) followed the guidelines of the product protocols. Cycle threshold (Ct) values were generated for each card by automatic selection of a threshold. The technical performance and repeatability measures were tested (Supplementary Figure S1, S2 and S3) before performing experiments using large numbers of samples.
Statistical analysis
Initial microarray data processing and filtering
The preprocessing and filtering steps were identical to those described in Shedden et al 13 (Supplementary Methods). After pre-screening, 13,306 probes were left for further analysis. All genes in training and testing datasets were median-centered and the median absolute deviation (MAD) scaled before use in subsequent analyses.
Pre-selection of survival-related clusters and genes
The first step in the strategy was to select approximately 370 promising genes from the Affymetrix data.
Using the 439 training samples, genes were first separated into 300 groups using K-means clustering. A two-stage selection procedure was then implemented; first, selection of clusters and second, selection of genes within each of the selected clusters. The top 73 clusters whose cluster mean was most associated with survival were selected using backward elimination and stepwise regression using a Cox proportional hazard model. Within each of the selected clusters, the second selection identified a subset of genes prognostic for survival based on a combination of various criteria which were descripted in Supplementary Methods. This approach led to a set of 73 clusters and a total of 368 genes from the selected clusters considered relevant to patient survival of lung cancer. We also tested the survival predictability of these 368 genes using two independent datasets (Bild et al., 110 samples and Raponi et al., 130 samples) with Random Survival Forests (RSF) (Supplementary Results).
Normalization and imputation of qRT-PCR values
The second step in the strategy was to measure the pre-selected 368 genes using qRT-PCR technology on a subset of the 439 training samples. The qRT-PCR measurements on the 47 samples were standardized using the control gene 18S RNA. The qRT-PCR measurements on the remaining 392 patients were treated as missing data and a multiple imputation strategy was used to make full use of all 439 patients to build a prediction model. The multiple imputation (MI) was performed using IVEware.23 The details of multiple imputation and alternative strategies of normalizing the microarray data for the training set of 439 samples are described in the Supplementary Methods. The Spearman’s correlations between qRT-PCR and Affymetrix-based measurements were calculated.
Random Survival Forests for survival analysis and prediction
The third step of the strategy was to further refine the selected genes down to approximately 91 genes for final evaluation and validation using qRT-PCR measurements. The details of RSF are described in the Supplementary Methods.
To assess the statistical significance of the predictions on the validation dataset, the mortality risk index (MRI) was included as a continuous variable in a univariate Cox model both for all 101 tumors and for stage 1 tumors only. Kaplan-Meier survival analyses are shown using the MRI to separate patients into three risk tertiles (high, medium, and low-risk, 1/3rd in each group).
Results
Identification of a survival-related 91 gene subset
In order to identify a 91-gene qRT-PCR platform-based classifier obtained from a subset of the 368 genes selected in the Affymetrix platform, three major criteria were considered: correlation between Affymetrix and qRT-PCR, association of gene with survival and representation of a broad spectrum of biological processes.
First, we defined genes whose qRT-PCR measurement showed high correlation with Affymetrix microarray measurements based on the same 47 samples used in the training set. There were 301 out of 368 (301/368, 82%) genes which had a significantly high correlation value larger than 0.5 (p < 0.001, Table 2 and Supplementary Figure S5).
TABLE 2.
Spearman Correlation Between qRT-PCR and Affymetrix Microarray Data.
| Spearman correlation |
Number of genes |
|---|---|
| > 0.9 | 67 |
| 0.8–0.9 | 92 |
| 0.7–0.8 | 71 |
| 0.6–0.7 | 46 |
| 0.5–0.6 | 25 |
| 0.4– 0.5 | 23 |
| 0.3–0.4 | 15 |
| 0.2–0.3 | 13 |
| < 0.2 | 16 |
Second, based on the Affymetrix expression values and the measured qRT-PCR expression values, we imputed the qRT-PCR values for the remaining patients in the training dataset. We performed a RSF using 1000 trees and repeated it 10 times on each of the 20 imputed training data sets. We selected genes that had either: (a) a p values from the Cox model adjusted for stage and age on the imputed PCR data that was less than 0.05, or (b) average variable importance measure (VIMP) from the RSF (mean of 10 VIMPs per dataset) was larger than the “noise” VIMP average from RSF.
The final step was a subjective one of reducing the number of genes to 91, while retaining representation from each cluster if possible and selecting multiple genes from the largest clusters if the cluster and the gene appeared to be strongly associated with survival. A set of 91 genes from 53 clusters were selected.
In order to compare the relative prediction capability of the 91-gene classifier to the 368-gene classifier based on the Affymetrix data, we performed the similar RSF prediction analysis as done with the 368-gene signature described in Supplementary Results. The 91-gene signature gave a similar prediction result as compared to using 368 genes with both of the two test sets. The prediction error rates were 40.7% (33.9% for adenocarcinomas and 43.9% for SCC) and 36.3%, respectively for the Bild and Raponi test sets (Supplementary Table S2). This indicated that the 91-gene signature was comparable to the 368-gene signature in predicting patient survival in lung cancer.
The annotation of the 91 genes is provided in Supplementary Table S3, and the main biological categories are indicated in Supplementary Figure S6. Among these, signal transduction, transcription regulation, cell cycle, cell adhesion, and proliferation are the major biological processes.
Validation of the 91-gene classifier in an independent test set
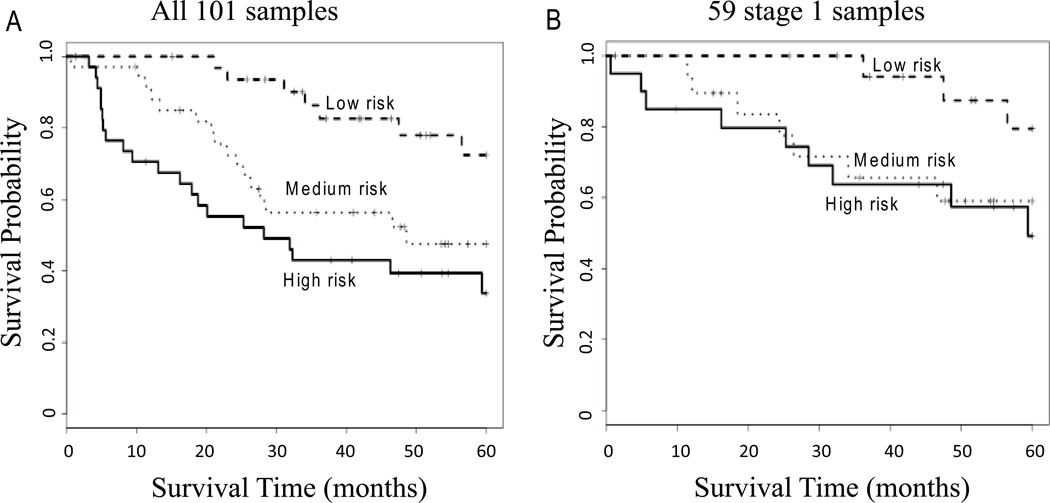
In order to validate the 91gene classifier for lung cancer prognosis, we utilized the qRT-PCR card-based platform with a completely independent cohort of 101 lung adenocarcinomas. The qRT-PCR data was normalized as described above. The RSF prediction model was built with the 91 genes, tumor stage and patient age information using the average of 20 imputed training sets of 439 tumors. The qRT-PCR data obtained from the 101 tumors was then tested with the RSF prediction model. The prediction error rate for the 101 test cohort was 26.6% (Table 3). We then tested the usefulness of predictors used to build the RSF using a univariate Cox model with the MRI as a continuous covariate. The RSF prediction was significant for the 101 patient cohort (likelihood ratio test (LRT) p < 0.00001). Using the MRI produced from the RSF, three risk groups were also identified, with patient 5-year survival being significantly different between low, medium, and high-risk groups (HR = 1.00, 2.82, 4.42, p = 0.0008; Figure 2A and Table 3). For stage I tumors only, this MRI was also significantly related to survival (Cox model LRT, p = 0.001) and separated patients into low, medium, and high risk groups (HR = 1.00, 3.29, 3.776, p = 0.04; Figure 2B and Table 3). The area under the curve (AUC)s from receiver operating characteristic (ROC) analyses were both 0.77 for all patients and for stage 1 only (Supplementary Figure S7). A notable feature of the validation shown in Figure 2 is the large separation between the curves in the first two years of follow-up, with almost no patients dying in the first two years for the low-risk group, but with significant number of deaths in the first two years for the high-risk group.
TABLE 3.
Prediction Results of the 91-gene qRT-PCR Signature in the Validation set (n = 101).
| RSF* | Cox model** | log-rank test*** | |||
|---|---|---|---|---|---|
| Test error rate | P | HR | 95%CI | P | |
| 26.6% | 5.21e-09 | ||||
| Low-risk | 1 | 0.0008 | |||
| Medium-risk | 2.82 | 1.16 – 6.88 | |||
| High-risk | 4.42 | 1.88 – 10.42 | |||
RSF prediction model built from 439 training set including 91 genes, stage and age;
Mortality risk index (MRI) as continuous value, likelihood ratio test (LRT) was used, univariate Cox model;
MRI separated test patients to 3 risk groups (low, medium and high-risk, 1/3rd in each group)
FIGURE 2.
Prediction results of the 91-gene qRT-PCR signature in the 101 samples of validation set. A, Kaplan-Meier survival curve using patient mortality risk index (MRI) from the RSF prediction model built from training set including 91 genes, stage and age. This predictor could significantly separate high, medium and low-risk groups (1/3rd in each group, HR = 1.00, 2.82, 4.42, p = 0.0008) among all 101 patients (A), as well as among the 59 stage 1 patient (1/3rd in each group, HR = 1.00, 3.29, 3.776) (B).
In order to confirm the multiple imputation strategy we developed produces results comparable to other methods. We build a RSF prediction model directly from the Affymetrix microarray data of training set of 439 patients (median-centered and MAD-scaled) and applied it to the 101 qRT-PCR validation set (similarly MAD-scaled). We found the RSF survival prediction results on the validation set of 101 qRT-PCR patients are similar for using both multiple imputation (Table 3 and Figure 2A) and MAD-scaled microarray data (Supplementary Table S4 and Supplementary Figure S8) from the training sets of 439 patients.
To evaluate whether the 91 gene set improves the prediction compared to clinical variables, age and stage in the validation set, we compared two Cox models via LRT; a model with age and stage versus a model with age, stage and the mortality index. We found that the set of 91 genes improves the prediction capability as compared to age and stage only (LRT p < 0.0001) using all 101 patients.
We also compared the effect of adjuvant therapy for these 3 risk groups defined by the MRI shown in Figure 2A. We didn’t observe any benefit from adjuvant therapy in the high and medium risk groups (p = 0.8 and 0.5, respectively). A reduced survival for low risk patients was observed if adjuvant therapy was given (p = 0.01, Supplementary Fig. S10). More detailed results and discussion regarded the effect of adjuvant therapy are provided in Supplementary Results.
Discussion
Lung cancer is a heterogeneous disease, and it is often difficult to accurately predict patient survival using tumor pathological characteristics or staging information only. Several groups, including ours, have postulated that improved estimation of an individual patient’s potential risk for recurrent disease can be achieved by a combination of clinical information and certain molecular markers including gene mutations (e.g., EGFR and KRAS mutations), DNA copy number change (e.g., MET and IGF1R), as well as gene or microRNA expression. 25–32 Studies based on gene expression signatures from microarrays or using qRT-PCR assays have been reported as predictive of patient survival in lung cancer, 6–19, 33–36 but there is sparse overlap of survival-related genes from different studies 18. The small amount of gene overlap may reflect sample collection methods, processing protocols, single-institutional subject cohorts, statistical methods, small sample sizes, different analysis platforms and different probes utilized. 13, 18, 37, 38 Ein-Dor 39 and coworkers have suggested that because of biological heterogeneity it may require thousands of samples to identify robust and reproducible gene subsets for most tumor types. Boutros et al demonstrated that thousands of different 6-gene signatures can predict patient survival if only 6 genes were used. 15 The largest microarray based study of lung adenocarcinomas showed that combining a cluster and Cox model based method for gene selection plus using clinical covariates provided the best overall survival predictive ability. 13 Because the number of genes in each cluster varied from dozens to hundreds however it would be very difficult to apply some microarray-based gene classifiers in the clinical setting. These studies indicate that for an optimal survival classifier, a large sample size, including more genes in the signature, and appropriate statistical methods incorporating accurate clinical information are needed. As an approach, qRT-PCR is a more reproducible, simple, efficient and clinical practicable assay. In this study, we attempted to combine these major criteria to reduce the number of survival-related genes to a reasonable, but not necessarily a very small number. Then, we developed and validated a qRT-PCR card-based 91-gene survival classifier, using the four major procedures shown in Figure 1, for the purpose of developing a clinically-practicable qRT-PCR based assay.
Of the 91 genes in this study, the functional analysis showed that more than 20 different biological processions were involved. Most of these processes were cancer-related and most of these genes have been reported by others as being involved in cancer development or used for cancer diagnosis or prognosis (Supplementary Table S3). We have compared our 91-gene list with 20 other qRT-PCR-based or microarray-based studies for lung cancer prognosis. Eleven genes were reported also by other 7 studies 6, 16, 19, 20, 34, 41, 42(Supplementary Table S5). No overlapping genes to this study were reported by the other 13 studies (Supplementary Table S6). Several genes including DUSP6 and ERBB3 were also used in Chen et al 5-gene signature, 16 and ERBB3 was used in Raz et al 4-gene model. 19 SLC2A1 and MEF2C were presented in Lu et al 64-gene profile. 34 Interestingly, five genes were also reported by Beer et al6 in 2002.
Gene cluster analysis and a risk index created from Cox models have been successfully utilized before as the statistical approach for gene expression profile-based survival prediction. 13 Genes in the same cluster which are coordinately expressed in a dataset often represent similar biological functions or define similar pathological features. In our approach we specifically chose genes representative of as many clusters as possible to aid in prediction performance in the likely case of lung adenocarcinoma tumor heterogeneity. We used both Cox models and RSF to aid in the identification of genes and development of the classifier. In general, performances of RSF and Cox model were similar, with RSF being complementary to the Cox model providing genes important for survival prediction based on the VIMP value. 24, 43 To our knowledge, this is the first study to combine clustering, Cox model and RSF prediction models for survival-related gene selection and then to predict survival from qRT-PCR data in lung cancer. This is also the first study to impute a large microarray data to a mimic qRT-PCR value in the training set in the prediction study and we found a similar performance (Table 3 and Figure 2) as using Affymetrix-microarray based values (MAD-scaled) (Supplementary Table S4 and Supplementary Figure S8).
The strength of this study is that it was prospectively planned and executed as described in Figure 1. The planned strategy was of incrementally refining and reducing the number of genes as the study transitioned from an Affymetrix platform to a qRT-PCR platform. Another advantage of this study is the large sample size used in the training set, with the data being from one uniform study although measured at four centers. Utilizing the power of imputation we were able to make use of the survival data from all 439 subjects in the training data rather than just the subset of those who had both Affymetrix and qRT-PCR measurements. We find the prediction results using Affymetrix-based measurements using RSF and the 368 genes were similar to the imputation method thus supporting its use. Further, the successful survival prediction for the 91-gene qRT-PCR platform also included stage 1 cancers. Interestingly, these 91 genes also can predict patient survival with SCC indicating that some of the same biological processes may be shared with lung SCC 44 and suggesting a greater utility of the prognostic gene set. The use of prospective clinical trials to test the prediction of benefit from chemotherapy for patient groups defined by the MRI are needed in order to broadly use this classifier for treatment selection in lung cancer.
Supplementary Material
Acknowledgments
Supported by MICHR/CSTA and MIIE UL1RR024986 (DG Beer), and CTSADB (S Kim).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure: The authors declare no conflicts of interest.
References
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Strauss GM, Herndon JE, 2nd, Maddaus MA, et al. Adjuvant paclitaxel plus carboplatin compared with observation in stage IB non-small-cell lung cancer: CALGB 9633 with the Cancer and Leukemia Group B, Radiation Therapy Oncology Group, and North Central Cancer Treatment Group Study Groups. J Clin Oncol. 2008;26:5043–5051. doi: 10.1200/JCO.2008.16.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bezjak A, Lee CW, Ding K, et al. Quality-of-life outcomes for adjuvant chemotherapy in early-stage non-small-cell lung cancer: results from a randomized trial, JBR.10. J Clin Oncol. 2008;26:5052–5059. doi: 10.1200/JCO.2007.12.6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douillard JY, Rosell R, De Lena M, et al. Adjuvant vinorelbine plus cisplatin versus observation in patients with completely resected stage IB-IIIA non-small-cell lung cancer (Adjuvant Navelbine International Trialist Association [ANITA]): a randomised controlled trial. Lancet Oncol. 2006;7:719–727. doi: 10.1016/S1470-2045(06)70804-X. [DOI] [PubMed] [Google Scholar]
- 5.Arriagada R, Bergman B, Dunant A, et al. Cisplatin-based adjuvant chemotherapy in patients with completely resected non-small-cell lung cancer. N Engl J Med. 2004;350:351–360. doi: 10.1056/NEJMoa031644. [DOI] [PubMed] [Google Scholar]
- 6.Beer DG, Kardia SL, Huang CC, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 7.Bhattacharjee A, Richards WG, Staunton J, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci U S A. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 9.Garber ME, Troyanskaya OG, Schluens K, et al. Diversity of gene expression in adenocarcinoma of the lung. Proc Natl Acad Sci U S A. 2001;98:13784–13789. doi: 10.1073/pnas.241500798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen JE, Pavey SJ, Passmore LH, et al. Expression profiling defines a recurrence signature in lung squamous cell carcinoma. Carcinogenesis. 2007;28:760–766. doi: 10.1093/carcin/bgl207. [DOI] [PubMed] [Google Scholar]
- 11.Larsen JE, Pavey SJ, Passmore LH, et al. Gene expression signature predicts recurrence in lung adenocarcinoma. Clin Cancer Res. 2007;13:2946–2954. doi: 10.1158/1078-0432.CCR-06-2525. [DOI] [PubMed] [Google Scholar]
- 12.Raponi M, Zhang Y, Yu J, et al. Gene expression signatures for predicting prognosis of squamous cell and adenocarcinomas of the lung. Cancer Res. 2006;66:7466–7472. doi: 10.1158/0008-5472.CAN-06-1191. [DOI] [PubMed] [Google Scholar]
- 13.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomida S, Koshikawa K, Yatabe Y, et al. Gene expression-based, individualized outcome prediction for surgically treated lung cancer patients. Oncogene. 2004;23:5360–5370. doi: 10.1038/sj.onc.1207697. [DOI] [PubMed] [Google Scholar]
- 15.Boutros PC, Lau SK, Pintilie M, et al. Prognostic gene signatures for non-small-cell lung cancer. Proc Natl Acad Sci U S A. 2009;106:2824–2828. doi: 10.1073/pnas.0809444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen HY, Yu SL, Chen CH, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 17.Endoh H, Tomida S, Yatabe Y, et al. Prognostic model of pulmonary adenocarcinoma by expression profiling of eight genes as determined by quantitative real-time reverse transcriptase polymerase chain reaction. J Clin Oncol. 2004;22:811–819. doi: 10.1200/JCO.2004.04.109. [DOI] [PubMed] [Google Scholar]
- 18.Lau SK, Boutros PC, Pintilie M, et al. Three-gene prognostic classifier for early-stage non small-cell lung cancer. J Clin Oncol. 2007;25:5562–5569. doi: 10.1200/JCO.2007.12.0352. [DOI] [PubMed] [Google Scholar]
- 19.Raz DJ, Ray MR, Kim JY, et al. A multigene assay is prognostic of survival in patients with early-stage lung adenocarcinoma. Clin Cancer Res. 2008;14:5565–5570. doi: 10.1158/1078-0432.CCR-08-0544. [DOI] [PubMed] [Google Scholar]
- 20.Bianchi F, Nuciforo P, Vecchi M, et al. Survival prediction of stage I lung adenocarcinomas by expression of 10 genes. J Clin Invest. 2007;117:3436–3444. doi: 10.1172/JCI32007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu CQ, Ding K, Strumpf D, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. J Clin Oncol. 2010;28:4417–4424. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu CQ, Strumpf D, Li CY, et al. Prognostic gene expression signature for squamous cell carcinoma of lung. Clin Cancer Res. 2010;16:5038–5047. doi: 10.1158/1078-0432.CCR-10-0612. [DOI] [PubMed] [Google Scholar]
- 23.Raghunathan TE, Lepkowski JM, Van Hoewyk J, et al. A multivariate technique for multiply imputing missing values using a sequence of regression models. Surv Methodol. 2001;27:11. [Google Scholar]
- 24.Ishwaran H, Kogalur UB, Blackstone EH, et al. Random Survival Forests. Ann Appl Stat. 2008;2:841–860. [Google Scholar]
- 25.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature genetics. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cappuzzo F, Marchetti A, Skokan M, et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:1667–1674. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raponi M, Dossey L, Jatkoe T, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–5783. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 29.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen G, Gharib TG, Huang CC, et al. Proteomic analysis of lung adenocarcinoma: identification of a highly expressed set of proteins in tumors. Clin Cancer Res. 2002;8:2298–2305. [PubMed] [Google Scholar]
- 31.Chen G, Gharib TG, Wang H, et al. Protein profiles associated with survival in lung adenocarcinoma. Proc Natl Acad Sci U S A. 2003;100:13537–13542. doi: 10.1073/pnas.2233850100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen G, Wang X, Yu J, et al. Autoantibody profiles reveal ubiquilin 1 as a humoral immune response target in lung adenocarcinoma. Cancer Res. 2007;67:3461–3467. doi: 10.1158/0008-5472.CAN-06-4475. [DOI] [PubMed] [Google Scholar]
- 33.Lee ES, Son DS, Kim SH, et al. Prediction of recurrence-free survival in postoperative non-small cell lung cancer patients by using an integrated model of clinical information and gene expression. Clin Cancer Res. 2008;14:7397–7404. doi: 10.1158/1078-0432.CCR-07-4937. [DOI] [PubMed] [Google Scholar]
- 34.Lu Y, Lemon W, Liu PY, et al. A gene expression signature predicts survival of patients with stage I non-small cell lung cancer. PLoS Med. 2006;3:e467. doi: 10.1371/journal.pmed.0030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Skrzypski M, Jassem E, Taron M, et al. Three-gene expression signature predicts survival in early-stage squamous cell carcinoma of the lung. Clin Cancer Res. 2008;14:4794–4799. doi: 10.1158/1078-0432.CCR-08-0576. [DOI] [PubMed] [Google Scholar]
- 36.Wigle DA, Jurisica I, Radulovich N, et al. Molecular profiling of non-small cell lung cancer and correlation with disease-free survival. Cancer Res. 2002;62:3005–3008. [PubMed] [Google Scholar]
- 37.Group TTABPW. Expression profiling--best practices for data generation and interpretation in clinical trials. Nat Rev Genet. 2004;5:229–237. doi: 10.1038/nrg1297. [DOI] [PubMed] [Google Scholar]
- 38.Dupuy A, Simon RM. Critical review of published microarray studies for cancer outcome and guidelines on statistical analysis and reporting. J Natl Cancer Inst. 2007;99:147–157. doi: 10.1093/jnci/djk018. [DOI] [PubMed] [Google Scholar]
- 39.Ein-Dor L, Zuk O, Domany E. Thousands of samples are needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci U S A. 2006;103:5923–5928. doi: 10.1073/pnas.0601231103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Subramanian J, Simon R. Gene expression-based prognostic signatures in lung cancer: ready for clinical use? J Natl Cancer Inst. 2010;102:464–474. doi: 10.1093/jnci/djq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo L, Ma Y, Ward R, et al. Constructing molecular classifiers for the accurate prognosis of lung adenocarcinoma. Clin Cancer Res. 2006;12:3344–3354. doi: 10.1158/1078-0432.CCR-05-2336. [DOI] [PubMed] [Google Scholar]
- 42.Roepman P, Jassem J, Smit EF, et al. An immune response enriched 72-gene prognostic profile for early-stage non-small-cell lung cancer. Clin Cancer Res. 2009;15:284–290. doi: 10.1158/1078-0432.CCR-08-1258. [DOI] [PubMed] [Google Scholar]
- 43.Pang H, Datta D, Zhao H. Pathway analysis using random forests with bivariate node-split for survival outcomes. Bioinformatics. 2010;26:250–258. doi: 10.1093/bioinformatics/btp640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Z, Wigle DA, Yang P. Non-overlapping and non-cell-type-specific gene expression signatures predict lung cancer survival. J Clin Oncol. 2008;26:877–883. doi: 10.1200/JCO.2007.13.1516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.