Abstract
Members of the transforming growth factor-β superfamily play critical roles in controlling cell growth and differentiation. Effects of TGF-β family ligands are mediated by Smad proteins. To understand the mechanism of Smad function, we sought to identify novel interactors of Smads by use of a yeast two-hybrid system. A 396-amino acid nuclear protein termed SNIP1 was cloned and shown to harbor a nuclear localization signal (NLS) and a Forkhead-associated (FHA) domain. The carboxyl terminus of SNIP1 interacts with Smad1 and Smad2 in yeast two-hybrid as well as in mammalian overexpression systems. However, the amino terminus of SNIP1 harbors binding sites for both Smad4 and the coactivator CBP/p300. Interaction between endogenous levels of SNIP1 and Smad4 or CBP/p300 is detected in NMuMg cells as well as in vitro. Overexpression of full-length SNIP1 or its amino terminus is sufficient to inhibit multiple gene responses to TGF-β and CBP/p300, as well as the formation of a Smad4/p300 complex. Studies in Xenopus laevis further suggest that SNIP1 plays a role in regulating dorsomedial mesoderm formation by the TGF-β family member nodal. Thus, SNIP1 is a nuclear inhibitor of CBP/p300 and its level of expression in specific cell types has important physiological consequences by setting a threshold for TGF-β-induced transcriptional activation involving CBP/p300.
Keywords: TGF-β, Smad, CBP/p300, signal transduction, transcriptional suppression
TGF-β is the prototypic member of a large family of structurally related cytokines including the TGF-βs, activins, and bone morphogenetic proteins (BMPs), which regulate cell fate and extracellular matrix deposition through the transcriptional regulation of diverse gene targets. These ligands initiate intracellular signals by associating with two classes of interacting transmembrane receptor serine-threonine kinases (Derynck and Feng 1997; Massagué 1998). Smad proteins, which serve as direct targets of phosphorylation by the activated type I receptors, have been demonstrated to be downstream mediators of specific TGF-β/BMP-signaling pathways from the receptors to the nucleus.
Three classes of Smads have been identified and are referred to as the receptor-activated Smads, co-Smads, and inhibitory Smads. Receptor-activated Smads (R-Smads), Smad2, and Smad3 are specific mediators of TGF-β and activin signaling pathways, whereas Smad1, Smad5, and Smad8 are involved in BMP responses (Massagué 1998). Smad4 is a co-Smad and is not regulated by phosphorylation, but acts as a common and essential mediator of TGF-β, activin, and BMP-signaling responses (Candia et al. 1997; de Caestecker et al. 1997). A third class of Smads includes Smad6 and Smad7, and act as an inhibitor of this pathway by binding to the activated receptors or to R-Smads, thereby sequestering the transcriptionally active complex from the promoter.
Like many transcription factors, the R-Smad proteins interact with the paralogous transcriptional coactivators CBP and p300 through their MH2 domains (Feng et al. 1998; Janknecht et al. 1998; Pouponnot et al. 1998; Topper et al. 1998). Although these two proteins were characterized by different means, they are now considered to be orthologous and to act as a crucial scaffold to bring together transcription factors and basal factors in the transcriptional initiation complex. They are essential coactivators for a wide variety of transcriptional factors including nuclear hormone receptors and NF-κB (Mannervik et al. 1999). Besides possessing intrinsic histone acetyltransferase (HAT) activity, these proteins are also able to interact with other HATs such as p/CAF and steroid receptor coactivators (SRCs) (Torchia et al. 1998). Although recent publications have concentrated on the HAT activity of CBP/p300, these proteins also contain three highly conserved cysteine-histidine-rich domains (C/H1, C/H2, and C/H3). Among these, the C/H1 domain was shown to interact with the Smad activation domain (SAD) of Smad4, which is necessary for transcriptional activating activity of the Smads (de Caestecker et al. 2000), whereas the C/H3 domain is the locus of interaction with R-Smads (Feng et al. 1998; Janknecht et al. 1998; Nishihara et al. 1998; Shen et al. 1999).
Many recent investigations have focused on identification of proteins that interact with Smad proteins to modulate both their trafficking and stability in the cytoplasm and their transcriptional activating activity in the nucleus. Cytoplasmic interactors include Smurf1, which was found to modulate the BMP signal-transduction pathway by regulating the level of Smad1 and Smad5 in the cell (Zhu et al. 1999), and SARA, which interacts directly with Smad2 and Smad3 and functions to recruit Smad2 to the TGF-β receptor (Tsukazaki et al. 1998). An increasing number of nuclear activators including the AP-1 complex, vitamin D receptor, and TFE3 transcriptional factor (Hua et al. 1998; Zhang et al. 1998; Yanagi et al. 1999; Yanagisawa et al. 1999) have been shown to interact specifically with Smad proteins. In addition, a number of transcriptional inhibitors have been shown to interact with Smad proteins and to repress their transcriptional activating activity. These include the oncoproteins Evi-1 (Kurokawa et al. 1998), Ski and SnoN, (Luo et al. 1999), TGIF, which recruits the histone deacetylase HDAC (Wotton et al. 1999), and the δEF1/Zfh-1 family member SIP1 (Verschueren et al. 1999).
In this study, we characterize a novel nuclear protein, termed SNIP1 for Smad nuclear interacting protein 1, which also serves to suppress the TGF-β-signaling pathway. We demonstrate that, whereas this inhibitor can interact with the MH2 domain of Smad4 in a TGF-β dependent manner, its principal mechanism of action appears to be through inhibition of transcription by binding to CBP/p300 and interfering with the ability of these coactivators to interact with Smad4.
Results
Cloning and characterization of SNIP1
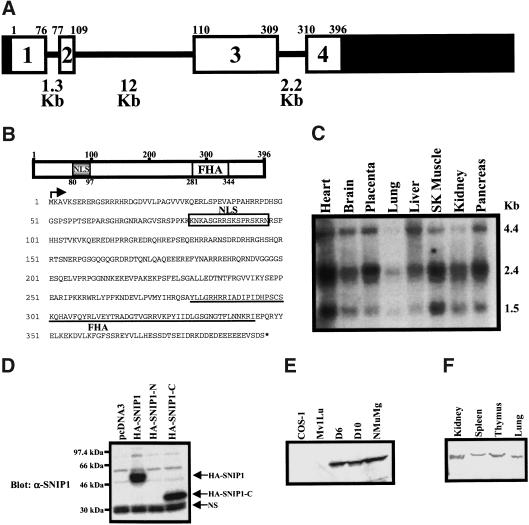
To identify proteins that could be involved in the downstream signaling of the Smad proteins, we screened a human fetal brain cDNA library using the full-length Smad1 as bait in the yeast two-hybrid system. Several positive clones were identified, one of which was SNIP1. The full-length cDNA was subsequently cloned from a human cardiac cDNA library using as a probe the 3′-fragment of SNIP1 cDNA that was cloned from the yeast two-hybrid assay. The full-length cDNA clone contained an insert of 2496 bp, in which the largest ORF encoded a protein of 396 amino acids with an estimated MW of ∼50kD. BLAST search of the Genbank database revealed that the SNIP1 gene (Accession no. AL034379) is located on chromosome 1 (1p32.2–1p32.3) (Altschul et al. 1997). The putative genomic structure of SNIP1 shows that the gene is composed of four exons interrupted by three introns (Fig. 1A). SNIP1 contains a sequence homologous to a bipartite nuclear localization signal (NLS) at the amino terminus and a Forkhead-associated (FHA) domain at its carboxyl terminus (Fig. 1B).
Figure 1.
Cloning and characterization of SNIP1. (A) Putative genomic structure of human SNIP1 gene is shown. Black boxes represent untranslated regions and the white boxes represent translated regions. The exon numbers are noted inside their respective white boxes with the sizes of introns written at the bottom. Numbers at the edges of the boxes denote the amino acid number from the human protein sequence. (B) Schematic diagram of human SNIP1 protein is shown with two conserved domains noted in shaded boxes, which are a bipartite NLS and a FHA. Numbers at the edges of the boxes denote the amino acid number from the deduced protein sequence. The sequence of the human SNIP1 protein is shown with the arrow indicating the starting methionine and an asterisk star representing the stop codon. The NLS is enclosed in a box and the FHA domain is underlined. (C) Commercially available Northern blot was purchased from Clontech and hybridized with the carboxy-terminal fragment of SNIP1. The sizes of three transcripts are indicated. (D) COS-1 cells were transfected with HA-tagged SNIP1 constructs as indicated. The specificity of an antibody against SNIP1 was monitored by Western blotting of the lysates. HA–SNIP1 and HA–SNIP1-C are indicated by arrows. (NS) Nonspecific band. (E) Western blot of cell lysates from various cell lysates. A single band of ∼50 kD was detected. (F) Western blot of lysates from various adult mouse tissues.
Northern hybridization was carried out to determine the expression pattern of SNIP1 mRNA in various human tissues (Fig. 1C). Three SNIP1 transcripts of 4.4, 2.4, and 1.5 kb are present at similar levels in multiple human tissues, with the highest transcript levels in heart and skeletal muscle (Fig. 1C). An antibody against SNIP1 was raised by injecting rabbits with a GST fusion protein of a region of SNIP1 from amino acids 215 to 324. Western blot analysis of COS-1 cell lysates transfected with various SNIP1 constructs showed that the antibody recognizes the full-length SNIP1 and SNIP1-C (amino acids 121–396), whereas SNIP1-N (amino acids 2–140), which does not contain the region of SNIP1 against which the antibody was raised, was not detected (Fig. 1D). The specificity of this antibody was also demonstrated in immunoprecipitation assays by use of this antibody and pre-immune serum from the same rabbit (data not shown). Use of this antibody for Western blot analysis of extracts from various cell lines identified a specific band of ∼50 kD (Fig. 1E), confirming that the predicted ORF from sequence analysis was correct. The monkey kidney cell line COS-1 and mink lung epithelial cell line Mv1Lu do not express detectable levels of SNIP1, whereas spontaneously immortalized primary murine hepatocyte cell lines D6 and D10 (Williams et al. 1996), and the mouse mammary cell line NMuMg, show good expression of endogenous SNIP1 (Fig. 1E). The fact that SNIP1 was undetected in COS-1 and Mv1Lu cell lysates could result from specificity of the antibody for mouse and human proteins or could indicate a restricted pattern of expression of the protein. Western blot analysis of extracts of multiple tissues from adult mice showed that SNIP1 is expressed in all tissues examined (Fig. 1F). Whereas these data demonstrate that SNIP1 mRNA and protein are widely expressed, immunohistochemistry of rat kidney sections at various stages of development shows that SNIP1 is localized specifically to epithelial elements, suggesting that its expression may be tightly controlled in different cell types (A. Perantoni, pers. comm.).
SNIP1 interacts with Smads
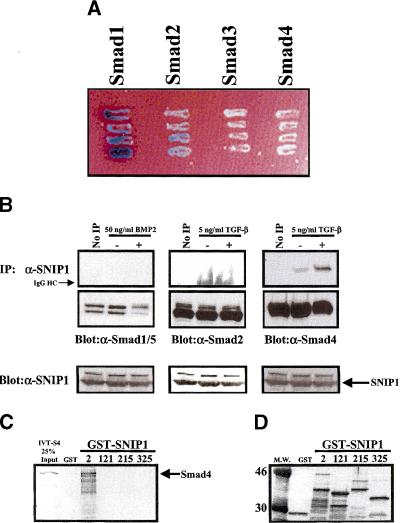
To test the specificity of the interaction of SNIP1 with Smad proteins in yeast, truncated SNIP1 (amino acids 142–396) cloned from the yeast two-hybrid assay, was cloned into the prey vector pJG4-5 and transformed into yeast. Transformants expressing this truncated SNIP1 were then transformed again with the indicated LexA–Smad fusion constructs in the bait construct pEG202. Four separate colonies from each group of transformants were streaked. Blue color, indicating a positive interaction was detected for the transformants of SNIP1/Smad1 and SNIP1/Smad2 (Fig. 2A). However, Smad 3 and Smad4 were unable to activate the assay indicating that the interaction of the carboxyl terminus of SNIP1 used in this assay was specific to Smad1 and Smad2.
Figure 2.
SNIP1 interacts with Smad proteins in vivo and in vitro. (A) Truncated SNIP1 cloned from the yeast two-hybrid assay was cloned into the prey vector pJG4-5 and was transformed into yeast. The truncated SNIP1 transformants were then transformed again with the indicated LexA–Smad fusion constructs in the bait construct pEG202. Four separate colonies from each group of transformants were streaked. (Blue) A positive interaction was detected for the transformants of SNIP1/Smad1 and SNIP1/Smad2. (B) NMuMg cell lysates treated with or without 5 ng/ml TGF-β or 50 ng/ml BMP2 for 1 hr was subjected to immunoprecipitation with α-SNIP1 and blotted with antibodies against indicated Smads. No IP lane was immunoprecipitated with rabbit IgG. The presence of SNIP1 and Smads in these cells were monitored by direct immunoblotting using antibodies against SNIP1 and Smads. (C) In vitro product of Smad4 made using reticulocyte lysate was incubated with various GST–SNIP1 deletion constructs. (Arrow) Smad4 product. (D) GST–SNIP1 (25%) constructs used in the reactions were subjected to SDS-PAGE and stained.
To ascertain the physiological relevance of these interactions, we next investigated whether complexes might be found between the endogenous Smad proteins and SNIP1 in mammalian cells. NMuMg cells were grown to confluence and serum starved overnight. These cells were treated with or without TGF-β or BMP2 for 1 hr, harvested, and lysates used for immunoprecipitation with the antibody against SNIP1. As shown in Figure 2B, endogenous Smad4 and SNIP1 interact strongly following TGF-β treatment. In contrast, no interaction between SNIP1 and endogenous Smad1 or Smad2 could be detected following treatment with the appropriate ligand, although these Smads were expressed at detectable levels in NMuMg cells (Fig. 2B). Next, we sought to determine a direct interaction between Smad4 and SNIP1. Incubation of the in vitro-transcribed and translated product of Smad4 with various deletion constructs of SNIP1 linked to GST and expressed in bacteria (Fig. 3A) showed that Smad4 was able to interact directly with the amino terminus of SNIP1 (Fig. 2C). On the basis of this strong and direct interaction of SNIP1 and Smad4, we decided to focus on the interaction of SNIP1 with Smad4.
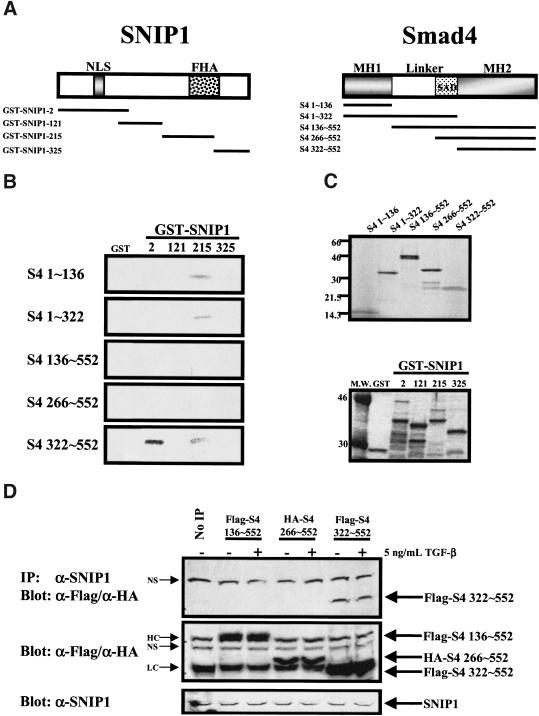
Figure 3.
Deletion mapping of the interacting regions of SNIP1 and Smad4. (A) Schematic diagram of the SNIP1 protein with the GST fusion protein of SNIP1 deletion constructs used in these experiments shown below (left). Numbers indicate the starting amino acid number of the deletion constructs. Smad4 and its deletion constructs used to produce in vitro transcribed/translated products used in the experiment (right). (B) Smad4 deletion constructs from the above diagram were used to make protein using reticulocyte lysate. The Smad4 deletion proteins were used in in vitro-binding assay using various bacterially expressed GST fusion of SNIP1 deletions constructs as indicated. After extensive washing, the beads were subjected to SDS-PAGE and autoradiography. (C) Twenty-five percent of the reticulocyte lysates used in the reaction was subjected to SDS-PAGE and autoradiography to monitor for expression of Smad4 constructs (top) GST fusion proteins (25%) were also subjected to standard SDS-PAGE and stained as per experimental protocol to control for equal loading of proteins in the experiments (bottom). (D) NMuMg cell lysates transfected with the indicated deletion constructs Smad4 were treated with or without 5 ng/ml TGF-β for 1 hr. These lysates were subjected to immunoprecipitation with α-SNIP1 and blotted with α-Flag and α-HA. No IP lane was immunoprecipitated with rabbit IgG. The presence of SNIP1 in these cells was monitored by direct immunoblotting using antibodies against SNIP1. These lysates were also used for direct immunoblotting with antibodies against Flag and HA to determine the expression of transfected Smad4 deletion constructs. HC and LC denote heavy and light chain of mouse IgG, respectively. (NS) Nonspecific band.
To begin to understand the interaction of SNIP1 and Smad4 in more detail, various Smad4 deletion constructs were produced by use of a coupled in vitro transcription/translation reaction (Fig. 3A,C) and tested for interaction with various GST–SNIP1 fusion proteins expressed in bacteria (Fig. 3A,C). As shown in Figure 3B, SNIP1 interacts with Smad4 through two different regions. The middle region of SNIP1 (GST–SNIP1–215) interacts with the MH1 domain (S4 1–136) of Smad4 and somewhat less strongly with the MH2 domain (S4 322–552). However, the amino terminus of SNIP1, which contains the NLS (GST–SNIP1–2), clearly interacts with the MH2 domain of Smad4 (S4 322–552), whereas inclusion of middle linker sequences precludes this interaction (Fig. 3B).
To delineate the region in Smad4 that interacts with SNIP1 in vivo, NMuMg cells were transfected with various deletion fragments of Smad4. Immunoprecipitation with the antibody against SNIP1 demonstrated that endogenous SNIP1 binds to the MH2 domain (Flag-S4 322–552) of Smad4 in a ligand-independent fashion (Fig. 3D). Similar to the results obtained in vitro, inclusion of the middle linker region (136–552) or simply the SAD domain (266–552) prevented the SNIP1 interaction. This suggests that these elements may be altering the conformation of the MH2 domain in such a way as to interfere with the ability of SNIP1 to interact both directly in vitro and in the cell.
SNIP1 is a nuclear protein involved in transcriptional suppression
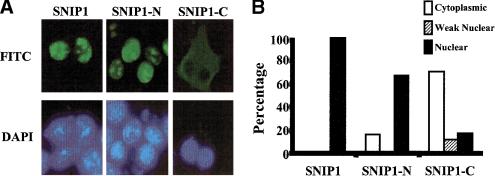
To begin to understand its physiological function, we investigated the pattern of subcellular localization of SNIP1. Indirect immunofluorescence demonstrates that full-length SNIP1 localizes to the nucleus constitutively (Fig. 4A). Indirect immunofluorescence was also used to test whether the NLS located at the amino terminus of SNIP1 was functional. As shown in Figure 4A, the construct HA–SNIP1-N, which encompasses the region of SNIP1 from amino acids 2 to 140 and contains the NLS, localizes predominantly to the nucleus (>70%, Fig. 4B), whereas the construct HA–SNIP1-C, that stretches from amino acids 121 to the carboxyl terminus and lacks this NLS, is preferentially localized to the cytoplasm (>70%, Fig. 4B). On close examination, the full-length SNIP1 has a punctuate pattern of localization in the nuclei that suggests a possible involvement of SNIP1 in transcriptional complexes. These data suggest that the TGF-β-dependent interaction of SNIP1 with Smad4 may result from the signal-dependent nuclear translocation of Smad4. In contrast, the MH2 domain of Smad4 (322–552) has been shown to be constitutively nuclear (R.H. Kim et al., unpubl.).
Figure 4.
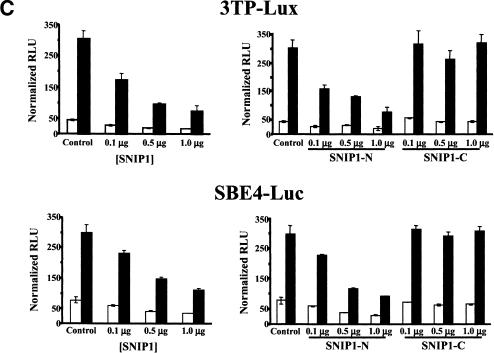
SNIP1 localizes to the nucleus constitutively and is involved in transcriptional inhibition. (A) HA–SNIP1, HA–SNIP1-N, and HA-SNIP1-C expression vectors were transiently cotransfected into NMuMg cells. After an overnight serum starvation, cells were analyzed by indirect immunofluorescence using α-HA monoclonal antibody. (Top) FITC staining of the cells to localize transfected SNIP1 and its deletion constructs. (Bottom) DAPI staining for the nuclei. (B) Cellular localization was determined by an independent observer, blinded as to the construct transfected, counting 200 cells per well to determine the percentage of each SNIP1 construct present in each of the sub-cellular compartments. Cytoplasmic (open bars) represents fluorescent signal present only in the cytoplasm of the cell, weak nuclear (hatched bars) represent signal in both the cytoplasm and nucleus, and nuclear (solid bars) represent signal present only in the nucleus. Results are presented from a single experiment, representative of three independent experiments. (C) NMuMg cells were cotransfected with 500 ng of 3TP–Lux, or SBE4–Luc with indicated amounts of HA–SNIP1 and its deletions. TGF-β was either not added (open bars) or added (solid bars) at a final concentration of 5 ng/ml, 24 hr before lysis, and analyzed for luciferase activity. Results are expressed as means (±s.d.) of triplicate assays, normalized for transfection efficiency using β-galactosidase activity.
To examine the role of SNIP1 in the regulation of transcriptional responses induced by TGF-β different TGF-β-responsive reporters were used in transient transfection assays. Both the basal and ligand-induced activity of 3TP-Lux, which contains TGF-β-responsive elements from the plasminogen activator inhibitor-1 and collagenase promoters (Wrana et al. 1992), and SBE4-Luc, which contains four copies of a Smad consensus-binding element CAGA (Zawel et al. 1998), were suppressed in a dose-dependent fashion by overexpression of SNIP1 (Fig. 4C). Moreover, SNIP1-N was able to inhibit the TGF-β-dependent activation of these reporters with activity similar to that of the full-length protein (Fig. 4C), whereas SNIP1-C was inactive. Interestingly, all of these reporters have been shown to be p300 dependent (Janknecht et al. 1998), suggesting that the inhibitory action of SNIP1 may result from interference with the functional interaction of CBP/p300 with the activated Smad complex.
These data indicate that binding of Smad4 to SNIP1 through the carboxy-terminal three-quarters of SNIP1 (GST–SNIP1–215) is not sufficient to inactivate TGF-β-dependent transcriptional responses. However, because this carboxy-terminal SNIP1 construct lacks a NLS, failure to inhibit TGF-β-dependent transcriptional responses could result instead from its exclusion from the nucleus (Fig. 4A,B). Taken together, these data indicate that the NLS is essential for the transcriptional suppressor function of SNIP1.
SNIP1 acts in vivo to inhibit TGF-β signal transduction pathway
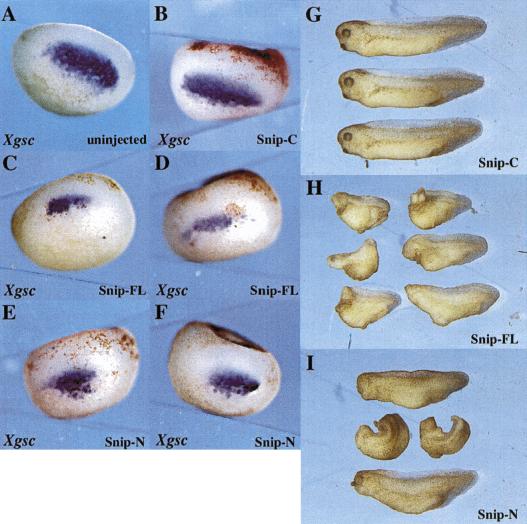
In vitro and coimmunoprecipitation data indicate that SNIP1 can interact with components of the TGF-β-signaling pathway, and transient transfection assays using reporter constructs responsive to TGF-β suggest a role for SNIP1 in modulating signals induced by TGF-β. To determine whether SNIP1 could play a role in regulating physiological responses to TGF-β family signaling pathways in vivo, we injected RNA for SNIP1 or various deletion constructs produced in vitro into early stage Xenopus embryos. In these assays, inhibition of TGF-β family signaling in the dorsal embryo inhibits notochord and head formation, presumably through inhibition of nodal signaling (Harland and Gerhart 1997). When RNA from SNIP1 or SNIP1-N was injected into the two dorsal blastomeres of 4-cell Xenopus embryos, dose-dependent truncation of anterior structures resulted, whereas SNIP1-C was without effect in this assay (Fig. 5, G–I). Marker gene analysis by in situ hybridization showed that goosecoid, a marker of dorsal mesoderm induced by nodal, was suppressed following injection of either SNIP1 or SNIP1-N, but that SNIP1-C had no effect (Fig. 5, A–F). These data demonstrate that ectopic expression of SNIP1 in vivo inhibits developmental processes attributed to the TGF-β family member nodal, correlating with in vitro findings of inhibition of reporter gene activity.
Figure 5.
Injection of SNIP1 into Xenopus embryos leads to a suppression of Gsc expression and anterior truncations. A total of 1 ng of SNIP1-FL (C,D,H), or SNIP1-N (E,F,I) or 2 ng of SNIP1-C (B,G) RNA were injected into the two dorsal blastomeres of a 4-cell embryo. Embryos were allowed to develop until stage 10.5 and fixed for whole mount in situ analysis for Xgsc expression, or embryos were cultured until stage 33, fixed, and photographed. SNIP1-FL or SNIP1-N-injected embryos exhibited a suppression of Xgsc expression as compared with the uninjected control (A) or to the SNIP1-C injected embryos. At stage 33, SNIP1-C injected embryos were phenotypically normal, whereas SNIP1-FL and SNIP1-N-injected embryos often showed loss of anterior structures.
SNIP1 interacts with endogenous CBP and p300
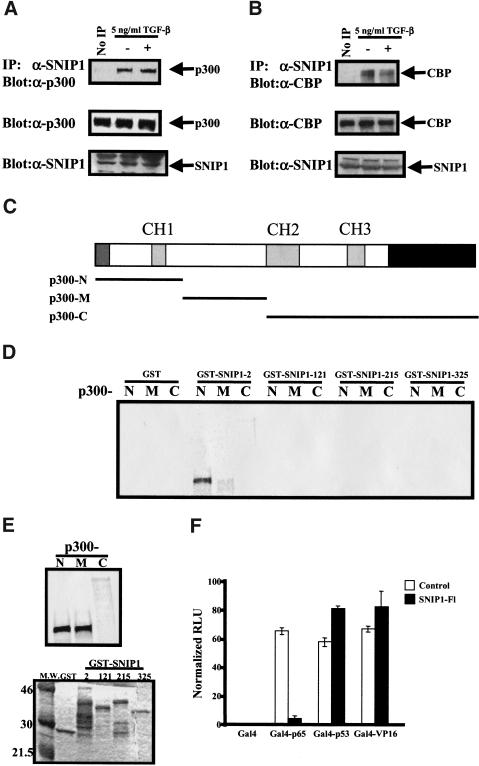
Because p300 has been shown to be an important coactivator that interacts with the Smads in the nucleus and is critical for their transcriptional activating activity (Feng et al. 1998), and because SNIP1 was shown to inhibit the activation by TGF-β signaling of reporter constructs that are p300 dependent (Fig. 4C; Janknecht et al. 1998; Pouponnot et al. 1998; Topper et al. 1998), we next sought to determine whether SNIP1 could also interact with CBP and/or p300. NMuMg cells were grown to confluence and treated with TGF-β. As shown in Figure 6 lysates of cells immunoprecipitated with α-SNIP1 and subjected to Western blotting using antibodies against either CBP or p300 showed that endogenous SNIP1 interacts with both endogenous CBP and p300 in a ligand-independent manner (Fig. 6A,B). An in vitro interaction experiment carried out by use of various bacterially expressed GST fusion constructs of SNIP1 and p300 constructs (Ogryzko et al. 1996) prepared with a coupled in vitro transcription/translation reaction showed that the amino terminus of SNIP1 (GST–SNIP1-2) is able to interact directly with the amino terminus of p300 (Fig. 6D).
Figure 6.
Endogenous SNIP1 interacts with endogenous CBP/p300 through its amino terminus. NMuMg cell lysates treated with or without TGF-β were subjected to immunoprecipitation using α-SNIP1 and blotted with α-p300 (A) or α-CBP (B). The expression of these proteins in the cells was monitored by direct immunoblotting as shown at bottom. (C) Schematic diagram of p300 and location of the deletion constructs used to make an in vitro-transcribed/translated product. (D) p300 deletion constructs from the above diagram were used to make protein using reticulocyte lysate. The p300 deletion proteins were used in in vitro-binding assay using various bacterially expressed GST fusion of SNIP1 deletions constructs as indicated. After extensive washing, the beads were subjected to SDS-PAGE and autoradiography. (E) A total of 25% of the reticulocyte lysates used in the reaction was subjected to standard SDS-PAGE and autoradiography to detect the amounts used in each reactions (top). A total of 25% of GST fusion proteins used in the experiments was subjected to standard SDS-PAGE and stained as per experimental protocol to control for equal loading of GST proteins in the experiments (bottom). (F) NMuMg cells were cotransfected with pG5E1B with (+) or without (−) 0.5 μg of HA–SNIP1-FL, as indicated along with Gal4 fusion proteins of p65, p53, and VP16. Results are expressed as means (±s.d) of triplicate assays, normalized for transfection efficiency using β-Galactosidase activity.
Functional assays were also used to determine the region of CBP/p300 through which SNIP1 exerts its inhibitory function. Cotransfection of various SNIP1 constructs with Gal4 constructs of full-length p300 or its deletions (Snowden et al. 2000) showed that full-length SNIP1 as well as SNIP1-N are able to specifically suppress the activity of the C/H1 domain of p300, which was also the locus of the SNIP1 interaction (data not shown). This domain of p300 interacts with a number of transcription factors such as Stat2 and Stat3 (Bhattacharya et al. 1996; Nakashima et al. 1999), ets-1 (Yang et al. 1998), p53, MDM2 (Grossman et al. 1998), and the p65 subunit of NF-κB (Perkins et al. 1997), suggesting that the inhibition of p300-dependent transcriptional activity by SNIP1 may not be restricted to Smad-dependent signaling downstream of TGF-β family receptors. In fact, cotransfection of SNIP1 inhibits the transcriptional activity to the carboxy-terminal transcriptional activation domain of the p65 subunit of NF-κB linked to the Gal4 DNA-binding domain, whereas the transcription of Gal4–VP16, which is independent of p300, is not affected by SNIP1 (Fig. 6F). Interestingly, the transcription of Gal4–p53, which, like Smad4 and p65, is dependent on the C/H1 domain of p300, is not inhibited by SNIP1 (Fig. 6F), suggesting that the region of p300 to which SNIP1 binds is different from that of p53.
Competition between SNIP1 and Smad4 for binding to p300
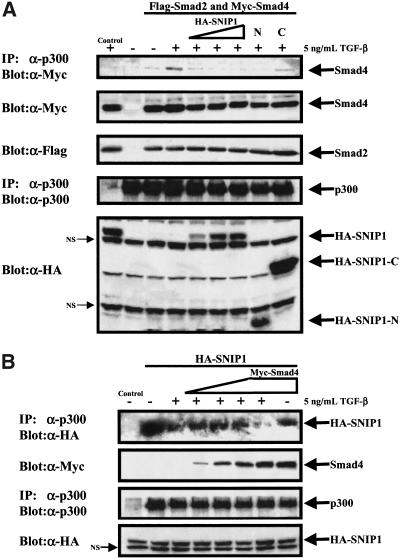
To address the mechanism whereby SNIP1 suppresses the transcriptional activation of the TGF-β signal-transduction pathway, cells were transfected with Smad2, Smad4, and SNIP1, and the interaction between Smad4 and the endogenous p300 examined. Immunoprecipitation of endogenous p300 followed by Western blotting with α-Myc antibody to detect transfected Smad4 showed that overexpression of SNIP1 interfered with the interaction between p300 and Smad4 in the presence of Smad2 (Fig. 7A). SNIP1-N was as effective as full-length SNIP in preventing the Smad4–p300 interaction, whereas SNIP1-C had little effect (Fig. 7A). These data suggest that SNIP1 inhibits Smad signaling activity by competing with Smad4 for binding to p300. Importantly, SNIP1 had no effect on the interaction of R-Smads with p300 (data not shown), consistent with the finding that R-Smads interact with the C/H3 domain of p300, which is not important for the SNIP1–p300 interaction. Moreover, the reverse experiment to determine whether Smad4 could interfere with the interaction of SNIP1 and p300 was also carried out. Cells were transfected with Smad4 and SNIP1 and the interaction between SNIP1 and endogenous p300 was examined. Immunoprecipitation of endogenous p300, followed by Western blotting with α-HA antibody to detect transfected SNIP1, showed that overexpression of Smad4 interfered with the interaction between SNIP1 and p300 in a dose-dependent manner and only in the presence of TGF-β (Fig. 7B).
Figure 7.
SNIP1 is able to inhibit the interaction of Smad4 with p300. (A) NMuMg cells were transfected with HA-tagged SNIP1 constructs and indicated Flag-tagged full-length Smad2 and Myc-tagged Smad4 expression constructs. The activation of the system was achieved by treating the cells with (+) or without (−) 5ng/ml TGF-β for 1 hr before lysis. Interaction between p300 and Smad4 was analyzed by immunoblotting the α-p300 immunoprecipitates with α-Myc antibody. The amount of immunoprecipitated p300 was monitored by immunoblotting 25% of p300 immunoprecipitate with α-p300 antibody. The expression of the transfected constructs was monitored by immunoblotting with antibodies against HA for SNIP1 or Myc and Flag for Smads. The control lane was immunoprecipitated with normal rabbit IgG. (B) COS-1 cells were transfected with HA-tagged SNIP1 constructs and increasing amounts of Myc-tagged Smad4 expression construct. The activation of the system was achieved by treating the cells with (+) or without (−) 5ng/ml TGF-β for 1 hr before lysis. Interaction between p300 and SNIP1 was analyzed by immunoblotting the α-p300 immunoprecipitates with α-HA antibody. The amount of immunoprecipitated p300 was monitored by immunoblotting 25% of p300 immunoprecipitate with α-p300 antibody. The expression of the transfected constructs was monitored by immunoblotting with antibodies against HA for SNIP1 or Myc Smad4. The control lane was immunoprecipitated with normal rabbit IgG.
Discussion
We now describe the isolation and characterization of a new transcriptional suppressor of TGF-β signal-transduction pathways. SNIP1 is a novel nuclear protein, located on chromosome 1 (1p32.2–1p32.3), that has no previously identified homologs. Recognizable motifs include a NLS, which we have shown to be critical to the overall function of SNIP1, and a FHA domain, which is dispensable for the transcriptional inhibitory activity of SNIP1. The FHA domain has recently been shown to be a modular phosphothreonine recognition motif expressed on a variety of nuclear proteins, and suggested to play a docking role analogous to that of the modular phosphotyrosine domain recognition site, SH2 (Durocher et al. 1999). It will be important to determine whether this motif might mediate protein–protein interactions that regulate the selectivity or modulate the activity of SNIP1.
An amino-terminally truncated version of SNIP1 was originally isolated from a yeast two-hybrid screen as a novel interactor of Smad1 and was also shown to bind Smad2 in a yeast two-hybrid system. Although similar interactions can be shown in mammalian cells when SNIP1-C and Smad1 and Smad2 are transiently overexpressed (data not shown), interaction of endogenous SNIP1 appears to be restricted to the common mediator, Smad4, suggesting that it represents the physiologically relevant interaction. The ligand-dependent nature of the SNIP1–Smad4 association is consistent with nuclear localization of SNIP1 and the ligand-dependent translocation of Smad4 to the nucleus in association with R-Smads. (Lagna et al. 1996; de Caestecker et al. 1997, 2000). Whereas weak interactions of SNIP1 215–324 with the MH1 domain of Smad4 are detectable, the principal interaction, and the interaction that is sufficient for transcriptional repression by SNIP1, is the association of the amino-terminal domain of SNIP1 with the carboxy-terminal MH2 domain of Smad4. The Smad4 MH2 domain, which is necessary for interaction with R-Smads and essential for mediating transcriptional responses to signals from members of the TGF-β superfamily (Candia et al. 1997; Hata et al. 1997), is also the locus for the interactions of Smad4 with two other recently described transcriptional inhibitors, the oncoproteins Ski and SnoN (Luo et al. 1999; Sun et al. 1999). This hypothesis suggests that interference with the structure or activity of the MH2 domain of Smad4 could be the basis of a broader mechanism of transcriptional repression of Smad pathways, not confined to SNIP1.
Our laboratory has previously identified a region of Smad4 called the SAD (de Caestecker et al. 1997). This region, which is located in the middle linker region just amino-terminal to the MH2 domain (amino acids 275–322), has been shown to be both necessary and sufficient for the transcriptional-activating activity of Smad4 through its interaction with CBP/p300 (de Caestecker et al., 2000). It is noteworthy that Smad4 deletion constructs of the MH2 domain inclusive of the SAD motif were unable to bind SNIP1 either in vitro or in vivo. A recent crystallographic structural analysis of the transcriptionally active domain of Smad4 including the SAD (amino acids 276–552) has shown that inclusion of the SAD alters the previously published structure of the inactive MH2 domain (Shi et al. 1997) by stabilizing a glutamine-rich helical extension from the core (Qin et al. 1999). Thus, we suggest that inclusion of this domain, in the absence of other constraints imposed by the MH1 domain, may restrict interactions of SNIP1 with the MH2 domain.
Importantly, SNIP1 also interacts constitutively with CBP/p300 through the same amino-terminal domain that mediates its principal interaction with Smad4. On this basis, we propose a model in which the relative levels of SNIP1 and nuclear Smad4 contribute to setting limits on both the basal activity as well as the maximum level of transcriptional activation of target genes that can be achieved in a cell following ligand activation. In this model, high levels of SNIP1 relative to nuclear Smad4 will restrict the interaction between the activated Smad complex and CBP/p300 with resultant inhibition of the ligand-dependent transcriptional response. However, high levels of Smad4 relative to that of SNIP1 will favor the formation of transcriptionally active Smad4/coactivator complexes and Smad4/SNIP1 complexes, which may initiate proteasomal degradation of SNIP1 as discussed below (Lin et al., in prep.). In the case of the homeodomain repressor TGIF, which, like SNIP1, suppresses both basal and TGF-β-activated transcription, a competitive mechanism has also been proposed whereby the relative levels of TGIF and Smad2 determine formation of mutually exclusive Smad2/inhibitor and Smad2/coactivator complexes (Wotton et al. 1999). Suppression of signaling responses from TGF-β family ligands by the adenoviral oncoprotein E1A, which interacts with the MH2 domain R-Smads, has also been shown recently to involve competition for binding of these Smads to the C/H3 domain of p300 (Nishihara et al. 1999). Our data suggest that SNIP1 acts in a similar fashion to inhibit interaction of Smad4 with the C/H1 domain of p300.
SNIP1 mRNA and protein are broadly expressed, and, in the limited number of cell lines examined, did not change with TGF-β treatment (data not shown). However, our preliminary observations of highly selective patterns of immunohistochemical staining for SNIP1 in tissues suggest that its expression is under stringent control (A. Perantoni, pers. comm.). In closely related findings (Lin et al., in prep.) show that activation of BMP or TGF-β-signaling pathways can lead to degradation of SNIP1 through a process involving antizyme and the proteasome β subunit HsN3. Proteolytic degradation has also been suggested to be important in regulation of the suppressor activity of Ski and SnoN, where it has been proposed that a TGF-β-signal leads to activation of Smad3, which then mediates degradation of SnoN and Ski (Luo et al. 1999). Mechanisms such as these suggest that the balance of activated Smad complexes and repressor proteins in the nucleus is critical to regulation of the signal-transduction pathways from TGF-β family ligands.
Although the cloning and characterization of SNIP1 was based on its ability to bind to Smad proteins and to inhibit Smad-dependent signaling, its ability to inhibit the transcriptional activating activity of the NF-κB transcriptional activator p65 shows that its action is not limited to Smad-dependent signaling pathways. Because NF-κB, like Smad4, interacts with p300 through the C/H1 domain, we speculated that SNIP1 might also suppress transcription dependent on other factors interacting with this same domain of p300 including Stat2 and Stat3 (Bhattacharya et al. 1996; Nakashima et al. 1999), ets-1 (Yang et al. 1998), p53 and MDM2 (Grossman et al. 1998). However, the inability of SNIP1 to inhibit the p300-dependent activity of Gal4–p53 now suggests that the specificity of SNIP1 may be more narrowly defined in terms of the region of p300 with which it interacts and may possibly also be restricted by other parameters such as direct binding to the transcription factor itself. As such, SNIP1 could both limit the magnitude of a particular cellular response and serve to mediate an additional level of crosstalk between various transcriptional regulators that interact with it to fine tune cellular proliferation, differentiation, and response to injury and stress.
Materials and methods
Cell lines, antibodies, and expression constructs
NMuMg and COS-1 cells were maintained in DMEM (Life Technologies) with 10% FBS (Life Technologies) and antibiotics.
Antibodies used in these experiments are as follows: mouse monoclonal α-HA (clone 12CA5), α-Myc (clone 9E10), α-Flag-M2 (Sigma), α-Smad4 (clone B8, Santa Cruz Biotechnoloy, CA), and α-p300,CT (Upstate Biotechnology); rabbit polyclonal α-Smad2 (clone 51–1400, Zymed), α-p300 (clone N15), α-p300 (clone C20), α-CBP (clone C20, Santa Cruz Biotechnology). Antibody against SNIP1 (rabbit polyclonal 1413-B) was a kind gift of Dr. Steve Tronick (Santa Cruz Biotechnology, CA).
HA-tagged SNIP1 constructs were generated by use of pfu polymerase (Stragene) with oligonucleotides corresponding to the specified regions of SNIP1 and subcloned into pcDNA3 mammalian expression vector (Invitrogen). GST fusion proteins of SNIP1 constructs were also generated by the same protocol and subcloned into pGEX 4T3 bacterial expression vector (Pharmacia). Smad4 deletion constructs used in GST pull-down assays were generated by use of pfu polymerase (Stratagene) with oligonucleotides corresponding to the specified regions of Smad4 and cloned into pCRII-TOPO (Invitrogen) vector. Deletion constructs of p300 were obtained from Dr. John Brady (Ogryzko et al. 1996). All PCR-generated products were sequenced by the dideoxynucleotide method. Gal4–p300 constructs used for Luciferase assays were described previously (Snowden et al. 2000)
Northern hybridization
Multiple tissue Northern blot (Clontech) was hybridized with the carboxyl terminus of SNIP1 (amino acids 121–396).
Indirect immunofluorescence
NMuMg cells were transiently transfected in 6-well plates with the HA–SNIP1, HA–SNIP1-N, or HA–SNIP1-C using Lipofectamine (Life Technologies) according to the manufacturer's protocol. After 24 hr, cells were serum starved overnight, fixed, and permeabilized as described previously (de Caestecker et al. 1998). The epitopes were detected by incubating with α-HA at room temperature for 1 hr, followed by incubation with goat α-mouse FITC secondary antibody (Kirkegaard & Perry Laboratories, MD) for 1 hr at room temperature. The cells were mounted with medium containing DAPI (Vectashield, Vector Labs). Cells were visualized by use of a fluorescence microscope.
Yeast two-hybrid assay
SNIP1 was isolated as a Smad1 interactor in the yeast two-hybrid system. The full-length Smad1 was used as the bait and a human fetal brain library as prey. To test the ability of SNIP1 to interact with other Smads, B42–SNIP1 fusion in the prey vector pJG4-5 was transformed into the yeast strain EGY48 (Leu-2, His-3, Trp-1, Ura-3), which contains both an integrated LexAop–Leu-2 reporter and a transfected LexAop–LacZ reporter with a Ura-3 selective marker. Yeast transformants were selected on U-W- plates. The B42–SNIP1 transformants were then transformed again with the indicated LexA–Smad fusion constructs in the bait construct pEG202. Yeast transformants were then selected on U-H-W- plates. Four separate colonies from each group of transformants were streaked onto two sets of X-Gal plates, U-H-W- glucose and U-H-W- galactose plate. The expression of B42–SNIP1 fusion protein is under the control of the GAL1 promoter. The glucose plate serves as the negative control.
Immunoprecipitation and Western blots
To detect in vivo interaction of transfected constructs, COS-1 cells were transfected with the indicated constructs by use of Lipofectamine as described above. After 24 hr, cells were switched to 0.2% serum overnight, and lysed in 0.5 ml of Triton X-100 lysis buffer A (25 mn HEPES at pH 7.5, 150 mm NaCl, 10% Glycerol, 5 mm EDTA, 1% Triton X-100) in the presence of phosphatase and protease inhibitors for Smad interaction with SNIP1, or 0.5 ml of Triton X-100 lysis buffer B (25 mn HEPES at pH 7.5, 75 mm NaCl, 10% Glycerol, 5mm EDTA, 1% Triton X-100) in the presence of phosphatase and protease inhibitors for CBP/p300 interaction with SNIP1. Lysates were either directly separated by SDS-PAGE and transferred onto Immobilon-P membranes (Millipore), and/or first immunoprecipitated with the indicated antibody. Western blots were detected using the appropriate HRP-conjugated secondary antibody, and visualized by chemiluminescence (Pierce).
GST pull-down assay
For GST pull-down assays, GST fusion proteins for the various SNIP1 deletion constructs were expressed in bacteria, quantitated by SDS-PAGE, and stained using GELCODE Blue Stain (Pierce) following affinity purification on glutathione beads (Pharmacia). Equal amounts of fusion protein were incubated in Triton X-100 lysis buffer B with in vitro transcribed/translated Smad4 and its deletions or p300 deletions made with TnT Master mix (Promega) according to manufacturer's protocol. Products were incubated with GST fusion proteins for 2 hr and washed with the same lysis buffer before subjecting them to autoradiography using X-OMAT MR films (Kodak).
Transcriptional assays
The 3TP-Lux, SBE4-Luc, and p800 reporter vectors were used to measure TGF-β-induced gene expression. Transfection, TGF-β treatment, and luciferase assays were performed as described previously, cotransfecting pSV-β-galactosidase to allow for normalization of transfection efficiency (de Caestecker et al. 1997). The total amount of transfected DNA was standardized by addition of pcDNA3 control vector as required, and all assays were performed in triplicate, and represented as mean (± s.e.m.) of three independent transfections.
Acknowledgments
We thank Dr. Steve Tronick from Santa Cruz Biotechnology for graciously providing the antibody against SNIP1. Flag-Smad2 and Myc-Smad4 were kindly provided by Drs. Jeff Wrana and Rik Derynck, respectively. The reporter vectors 3TP-Lux, SBE4-Luc, and p800-Luc were kind gifts from Drs. Joan Massagué, Scott Kern, and D.J. Loskutoff, respectively. Many thanks go to Dr. Neil Perkins for providing the Gal4–p65, Gal4–p53, and Gal4–VP16 constructs. We also thank Drs. Stephanie Reffey and Colin Duckett for the NF-κB results. The authors also thank Corina Bassity and David Frank for their expert technical assistance.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Robertsa@dce41.nci.nih.gov; FAX (301) 496-8395.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Eckner R, Grossman S, Oldread E, Arany Z, D'Andrea A, Livingston DM. Cooperation of Stat2 and p300/CBP in signalling induced by interferon-alpha. Nature. 1996;383:344–347. doi: 10.1038/383344a0. [DOI] [PubMed] [Google Scholar]
- Candia AF, Watabe T, Hawley SH, Onichtchouk D, Zhang Y, Derynck R, Niehrs C, Cho KW. Cellular interpretation of multiple TGF-β signals: Intracellular antagonism between activin/BVg1 and BMP-2/4 signaling mediated by Smads. Development. 1997;124:4467–4480. doi: 10.1242/dev.124.22.4467. [DOI] [PubMed] [Google Scholar]
- de Caestecker MP, Hemmati P, Larisch-Bloch S, Ajmera R, Roberts AB, Lechleider RJ. Characterization of functional domains within Smad4/DPC4. J Biol Chem. 1997;272:13690–13696. doi: 10.1074/jbc.272.21.13690. [DOI] [PubMed] [Google Scholar]
- de Caestecker MP, Parks WT, Frank CJ, Castagnino P, Bottaro DP, Roberts AB, Lechleider RJ. Smad2 transduces common signals from receptor serine-threonine and tyrosine kinases. Genes & Dev. 1998;12:1587–1592. doi: 10.1101/gad.12.11.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Caestecker MP, Yahata T, Wang D, Parks WT, Huang S, Hill CS, Shioda T, Roberts AB, Lechleider RJ. The Smad4 activation domain (SAD) is a proline-rich, p300-dependent transcriptional activation domain. J Biol Chem. 2000;275:2115–2122. doi: 10.1074/jbc.275.3.2115. [DOI] [PubMed] [Google Scholar]
- Derynck R, Feng XH. TGF-β receptor signaling. Biochim Biophys Acta. 1997;1333:F105–F150. doi: 10.1016/s0304-419x(97)00017-6. [DOI] [PubMed] [Google Scholar]
- Durocher D, Henckel J, Fersht AR, Jackson SP. The FHA domain is a modular phosphopeptide recognition motif. Mol Cell. 1999;4:387–394. doi: 10.1016/s1097-2765(00)80340-8. [DOI] [PubMed] [Google Scholar]
- Feng XH, Zhang Y, Wu RY, Derynck R. The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-β-induced transcriptional activation. Genes & Dev. 1998;12:2153–2163. doi: 10.1101/gad.12.14.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Perez M, Kung AL, Joseph M, Mansur C, Xiao ZX, Kumar S, Howley PM, Livingston DM. p300/MDM2 complexes participate in MDM2-mediated p53 degradation. Mol Cell. 1998;2:405–415. doi: 10.1016/s1097-2765(00)80140-9. [DOI] [PubMed] [Google Scholar]
- Harland R, Gerhart J. Formation and function of Spemann's organizer. Annu Rev Cell Dev Biol. 1997;13:611–667. doi: 10.1146/annurev.cellbio.13.1.611. [DOI] [PubMed] [Google Scholar]
- Hata A, Lo RS, Wotton D, Lagna G, Massagué J. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature. 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- Hua X, Liu X, Ansari DO, Lodish HF. Synergistic cooperation of TFE3 and smad proteins in TGF-β-induced transcription of the plasminogen activator inhibitor-1 gene. Genes & Dev. 1998;12:3084–3095. doi: 10.1101/gad.12.19.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janknecht R, Wells NJ, Hunter T. TGF-β-stimulated cooperation of smad proteins with the coactivators CBP/p300. Genes & Dev. 1998;12:2114–2119. doi: 10.1101/gad.12.14.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa M, Mitani K, Irie K, Matsuyama T, Takahashi T, Chiba S, Yazaki Y, Matsumoto K, Hirai H. The oncoprotein Evi-1 represses TGF-β signalling by inhibiting Smad3. Nature. 1998;394:92–96. doi: 10.1038/27945. [DOI] [PubMed] [Google Scholar]
- Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. Partnership between DPC4 and SMAD proteins in TGF-β signalling pathways. Nature. 1996;383:832–836. doi: 10.1038/383832a0. [DOI] [PubMed] [Google Scholar]
- Luo K, Stroschein SL, Wang W, Chen D, Martens E, Zhou S, Zhou Q. The ski oncoprotein interacts with the smad proteins to repress TGF-β signaling. Genes & Dev. 1999;13:2196–2206. doi: 10.1101/gad.13.17.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannervik M, Nibu Y, Zhang H, Levine M. Transcriptional coregulators in development. Science. 1999;284:606–609. doi: 10.1126/science.284.5414.606. [DOI] [PubMed] [Google Scholar]
- Massagué J. TGF-β signal transduction. Annu Rev Biochem. 1998;67:753–791. doi: 10.1146/annurev.biochem.67.1.753. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T. Synergistic signaling in fetal brain by STAT3-smad1 complex bridged by p300. Science. 1999;284:479–482. doi: 10.1126/science.284.5413.479. [DOI] [PubMed] [Google Scholar]
- Nishihara A, Hanai JI, Okamoto N, Yanagisawa J, Kato S, Miyazono K, Kawabata M. Role of p300, a transcriptional coactivator, in signalling of TGF-β. Genes Cells. 1998;3:613–623. doi: 10.1046/j.1365-2443.1998.00217.x. [DOI] [PubMed] [Google Scholar]
- Nishihara A, Hanai J, Imamura T, Miyazono K, Kawabata M. E1A inhibits transforming growth factor-κ signaling through binding to Smad proteins. J Biol Chem. 1999;274:28716–28723. doi: 10.1074/jbc.274.40.28716. [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–959. doi: 10.1016/s0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Perkins ND, Felzien LK, Betts JC, Leung K, Beach DH, Nabel GJ. Regulation of NF-κB by cyclin-dependent kinases associated with the p300 coactivator. Science. 1997;275:523–527. doi: 10.1126/science.275.5299.523. [DOI] [PubMed] [Google Scholar]
- Pouponnot C, Jayaraman L, Massagué J. Physical and functional interaction of SMADs and p300/CBP. J Biol Chem. 1998;273:22865–22868. doi: 10.1074/jbc.273.36.22865. [DOI] [PubMed] [Google Scholar]
- Qin B, Lam SS, Lin K. Crystal structure of a transcriptionally active Smad4 fragment. Structure Fold Des. 1999;7:1493–1503. doi: 10.1016/s0969-2126(00)88340-9. [DOI] [PubMed] [Google Scholar]
- Shen X, Hu PP, Liberati NT, Datto MB, Frederick JP, Wang XF. TGF-β-induced phosphorylation of Smad3 regulates its interaction with coactivator p300/CREB-binding protein. Mol Biol Cell. 1999;9:3309–3319. doi: 10.1091/mbc.9.12.3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Hata A, Lo RS, Massagué J, Pavletich NP. A structural basis for mutational inactivation of the tumour suppressor Smad4. Nature. 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- Snowden AW, Webster GA, Perkins ND. A novel transcriptional repression domain mediates p21WAF/CIP1 induction of p300 transactivation. Mol Cell Biol. 2000;20:2676–2686. doi: 10.1128/mcb.20.8.2676-2686.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liu X, Ng-Eaton E, Lodish HF, Weinberg RA. SnoN and Ski protooncoproteins are rapidly degraded in response to transforming growth factor-β signaling. Proc Natl Acad Sci. 1999;96:12442–12447. doi: 10.1073/pnas.96.22.12442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topper JN, DiChiara MR, Brown JD, Williams AJ, Falb D, Collins T, Gimbrone MAJ. CREB binding protein is a required coactivator for Smad-dependent, transforming growth factor-β transcriptional responses in endothelial cells. Proc Natl Acad Sci. 1998;95:9506–9511. doi: 10.1073/pnas.95.16.9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia J, Glass C, Rosenfeld MG. Co-activators and co-repressors in the integration of transcriptional responses. Curr Opin Cell Biol. 1998;10:373–383. doi: 10.1016/s0955-0674(98)80014-8. [DOI] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a FYVE domain protein that recruits Smad2 to the TGF-β receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- Verschueren K, Remacle JE, Collart C, Kraft H, Baker BS, Tylzanowski P, Nelles L, Wuytens G, Su MT, Bodmer R, et al. SIP1, a novel zinc finger/homeodomain repressor, interacts with Smad proteins and binds to 5′-CACCT sequences in candidate target genes. J Biol Chem. 1999;274:20489–20498. doi: 10.1074/jbc.274.29.20489. [DOI] [PubMed] [Google Scholar]
- Williams AO, Knapton AD, Geiser A, Letterio JJ, Roberts AB. The liver in transforming growth factor-β1 (TGF-β1) null mutant mice. Ultrastruct Pathol. 1996;20:477–490. doi: 10.3109/01913129609016352. [DOI] [PubMed] [Google Scholar]
- Wotton D, Lo RS, Lee S, Massagué J. A Smad transcriptional corepressor. Cell. 1999;97:29–39. doi: 10.1016/s0092-8674(00)80712-6. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Attisano L, C:arcamo J, Zentella A, Doody J, Laiho M, Wang XF, Massagué J. TGF-β signals through a heteromeric protein kinase receptor complex. Cell. 1992;71:1003–1014. doi: 10.1016/0092-8674(92)90395-s. [DOI] [PubMed] [Google Scholar]
- Yanagi Y, Suzawa M, Kawabata M, Miyazono K, Yanagisawa J, Kato S. Positive and negative modulation of vitamin D receptor function by transforming growth factor-β signaling through smad proteins. J Biol Chem. 1999;274:12971–12974. doi: 10.1074/jbc.274.19.12971. [DOI] [PubMed] [Google Scholar]
- Yanagisawa J, Yanagi Y, Masuhiro Y, Suzawa M, Watanabe M, Kashiwagi K, Toriyabe T, Kawabata M, Miyazono K, Kato S. Convergence of transforming growth factor-β and vitamin D signaling pathways on SMAD transcriptional coactivators. Science. 1999;283:1317–1321. doi: 10.1126/science.283.5406.1317. [DOI] [PubMed] [Google Scholar]
- Yang C, Shapiro LH, Rivera M, Kumar A, Brindle PK. A role for CREB binding protein and p300 transcriptional coactivators in Ets-1 transactivation functions. Mol Cell Biol. 1998;18:2218–2229. doi: 10.1128/mcb.18.4.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Mol Cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng XH, Derynck R. Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature. 1998;394:909–913. doi: 10.1038/29814. [DOI] [PubMed] [Google Scholar]
- Zhu H, Kavsak P, Abdollah S, Wrana JL, Thomsen GH. A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature. 1999;400:687–693. doi: 10.1038/23293. [DOI] [PubMed] [Google Scholar]