Abstract
Genetic analysis was applied to identify novel genes involved in G protein-linked pathways controlling development. Using restriction enzyme-mediated integration (REMI), we have identified a new gene, Pianissimo (PiaA), involved in cAMP signaling in Dictyostelium discoideum. PiaA encodes a 130-kD cytosolic protein required for chemoattractant receptor and G protein-mediated activation of the 12 transmembrane domain adenylyl cyclase. In piaA− null mutants, neither chemoattractant stimulation of intact cells nor GTPγS treatment of lysates activates the enzyme; constitutive expression of PiaA reverses these defects. Cytosols of wild-type cells that contain Pia protein reconstitute the GTPγS stimulation of adenylyl cyclase activity in piaA− lysates, indicating that Pia is directly involved in the activation. Pia and CRAC, a previously identified cytosolic regulator, are both essential for activation of the enzyme as lysates of crac− piaA− double mutants require both proteins for reconstitution. Homologs of PiaA are found in Saccharomyces cerevisiae and Schizosaccaromyces pombe; disruption of the S. cerevisiae homolog results in lethality. We propose that homologs of Pia and similar modes of regulation of these ubiquitous G protein-linked pathways are likely to exist in higher eukaryotes.
Keywords: Chemoattractant receptor, G protein, adenylyl cyclase, signal transduction, Dictyostelium
Cells are capable of sensing their environment and altering biological functions in response to external stimuli. In one well-known signal transduction pathway, external signals trigger the production of the second messenger cAMP. Stimulation or inhibition of adenylyl cyclase in response to extracellular signals is part of the re-pertoire of cellular regulation in diverse organisms. In mammals, for example, the enzyme is activated or inhibited in response to hormones, odorants, neurotransmitters, and chemoattractants (Gilman 1984, 1987; Levitzki 1988). The cell surface receptors that detect these stimuli possess seven transmembrane domains and are coupled to heterotrimeric G proteins (Dohlman et al. 1991). When excited, receptors activate G proteins, catalyzing the exchange of GTP for GDP on the α-subunit and the dissociation of the α-subunit from the βγ-subunit complex.
The regulation of adenylyl cyclase has been the subject of extensive studies in mammalian cells. There are 10 different types of mammalian adenylyl cyclases (ACI-ACX) known to date (Sunahara et al. 1996). They share a predicted structure of 12 transmembrane segments and two large cytoplasmic domains, but differ in tissue distribution and mode of regulation. Although all eight of the isozymes characterized thus far are stimulated by the GTP-bound α-subunit of Gs (Gsα), they respond differently to coregulators. For instance, the G protein βγ-subunit complex is a potent inhibitor of type I adenylyl cyclase but a striking stimulator of type II and type IV adenylyl cyclases (Tang and Gilman 1991). Similarly, there is type-specific regulation by Ca2+-calmodulin and protein kinases PKA and PKC (for review, see Sunahara et al. 1996).
In Dictyostelium, cAMP controls multiple stages of a developmental program triggered by depletion of nutrients, functioning as a chemoattractant, a morphogen, and an intracellular second messenger (Kay 1994; Firtel 1995; Parent and Devreotes 1996). Within a few hours after starvation, aggregation centers emerge spontaneously when the central cells within each territory begin to secrete cAMP at 6-min intervals. The periodic bursts of cAMP attract surrounding cells and also stimulate them to synthesize and secrete additional cAMP, which relays the signal distally as a propagated cAMP wave. The periodic stimulation also induces optimal expression of aggregation-specific genes. After the cells aggregate, cAMP continues to play a role within the multicellular structures as they undergo further morphogenesis to form slugs and differentiate into either stalk or spore cells in fruiting bodies.
In analogy to the hormone-activated mammalian systems, the cAMP signaling system in Dictyostelium involves surface receptor/G protein-linked signal transduction pathways (Devreotes et al. 1987, 1994). Excitation of the cAMP receptor cAR1 activates the heterotrimeric G protein G2, leading to an elevation of intracellular cGMP (Kesbeke et al. 1988), an activation of the cytoskeletal components involved in chemotaxis (Hall et al. 1989), and an increase in the activity of the adenylyl cyclase ACA (Pitt et al. 1992). Similar to type II and IV mammalian adenylyl cyclases, ACA is activated by the βγ-subunit complex, rather than the α-subunit, from G2. Structurally, ACA resembles the mammalian adenylyl cyclases; it has two sets of predicted six transmembrane spans and two homologous cytoplasmic domains. The crystal structure of the mammalian adenylyl cyclase catalytic core has been solved recently (Zhang et al. 1997); the active site is formed jointly by cytosolic domain monomers upon dimerization. Many mutations rendering ACA catalytically inactive or G protein insensitive (Parent and Devreotes 1995) map to a region in or adjoining the interface of the dimer.
There are differences between the regulation of ACA and its mammalian counterpart. The stimulatory effects of the βγ-subunit complex on type II and IV mammalian adenylyl cyclases depend on the presence of activated Gsα, whereas no Gsα has been identified in Dictyostelium. ACA does not contain the Gln-X-X-Glu-Arg sequence suggested to be the βγ-subunit contact site (Chen et al. 1995). Unlike other effectors/regulators that interact with Gβγ-subunits, adenylyl cyclases do not have pleckstrin homology (PH) domains (Musacchio et al. 1993) but, interestingly, a PH domain-containing cytosolic protein, CRAC, is required for both receptor and GTPγS stimulation of ACA (Insall et al. 1994). CRAC is translocated to membranes after chemoattractant stimulation of intact cells or during GTPγS activation of lysates; the translocation does not take place in the gβ− mutant (Lilly and Devreotes 1995). It has been proposed that CRAC serves as an adapter linking the G protein βγ-subunits to activation of ACA.
Cytosolic regulators, other than calmodulin, PKC, and PKA, of mammalian adenylyl cyclases have not been reported. However, there are indications of unidentified components in adenylyl cyclase pathways. In human polymorphonuclear leukocytes (PMNs), for example, chemoattractant receptors, such as that for fMet-Leu-Phe (fMLP), which are linked to Gαi stimulate increased intracellular cAMP levels by activating adenylyl cyclase (Spisani et al. 1996). But fMLP is incapable of stimulating the enzyme in membrane preparations (Verghese et al. 1985). Similarly, in A9 L cells transfected with the m1 muscarinic receptor carbachol activates synthesis of cAMP in intact cells but not in cell membranes (Felder et al. 1989). Membrane fractions contain functionally coupled receptors, G proteins, and responsive adenylyl cyclase, as guanine nucleotides can regulate the binding affinity of the receptors and prostaglandin E1 activates or α2-adrenergic treatment inhibits adenylyl cyclase activity.
Using insertional mutagenesis by restriction enzyme-mediated integration (REMI; Kuspa and Loomis 1992), we have isolated a mutant, designated Pianissimo, that is defective in the cAMP signaling pathway. Genetic and biochemical analyses revealed that the product of the mutated gene is a cytosolic protein, distinct from CRAC. Pianissimo is required for receptor and G protein-mediated activation of ACA, as is CRAC. However, our results demonstrate that Pianissimo and CRAC do not function redundantly; both proteins are integral components of the adenylyl cyclase activation pathway. Interestingly, homologs of Pianissimo are present in yeasts and we have demonstrated that the Saccharomyces cerevisiae homolog is an essential gene. It is likely that homologs of Pianissimo and similar modes of regulation of adenylyl cyclase also exist in higher eukaryotes.
Results
Identification and isolation of the PiaA gene
To discover new genes involved in signal transduction at the early developmental stages, we isolated REMI mutants unable to aggregate on bacterial lawns. These mutants were further characterized and screened for those that failed to aggregate on non-nutrient agar plates but were able to express the known components of the signal transduction pathways. These mutants are likely to have specific novel defects; the mutant designated Pianissimo was among them.
We cloned the Pianissimo gene (PiaA), as described in Materials and Methods, and found the REMI insertion to be 300 bp upstream of a 3447-bp open reading frame (ORF). Extensive evidence demonstrated that the insertion was responsible for the developmental phenotype of the mutant. First, we linearized the rescued plasmid carrying flanking genomic fragments (pMYC32, Fig. 1A), transformed it into the wild-type cells, and recreated the mutated genomic structure by homologous recombination. The resulting cell line, MYC15, displayed the same phenotype as the original REMI mutant. We also made a knockout construct (pYL23, Fig. 1A) within the ORF using cDNA fragments and transformed it into wild-type cells. The resulting cell line, MYC28, had the same phenotype as the original REMI mutant. Second, using cDNA fragments as the probes, we carried out Northern blot analyses on RNA samples prepared from both wild-type and mutant cells at different time points of development. As shown in Figure 1B, PiaA was expressed as a 4.5-kb mRNA that, in wild-type cells, peaks between 2.5 and 5 hr of development. There was no PiaA transcript detectable in the mutant, suggesting the cloned cDNA is the PiaA gene. Furthermore, we prepared a rabbit polyclonal antiserum using a peptide with a 15-amino-acid sequence corresponding to the deduced carboxyl terminus of the Pia protein. As shown in Figure 1C, in growing stage wild-type cells, there is a significant amount of Pia protein. The protein level decreases slightly at 2.5 hr of development, then reaches maximum at 5 hr. There was no detectable signal in the piaA− mutant using the antiserum. In another experiment we examined the protein levels up to 32 hr of development. We noted that the maximum level remained for 12 hr when several degradation products began to show on the gel; at later time points (16, 20, and 32 hr) the amount of intact protein gradually decreased and the degradation products increased. Finally, we constructed an expression vector carrying the full-length cDNA under a constitutive promoter (Act15) and transformed it into the piaA− cells. The resulting cell line, PiaA/piaA−, overexpressed the Pia protein about three- to fivefold (data not shown) and was able to aggregate and make wild-type-appearing fruiting bodies (see below).
Figure 1.
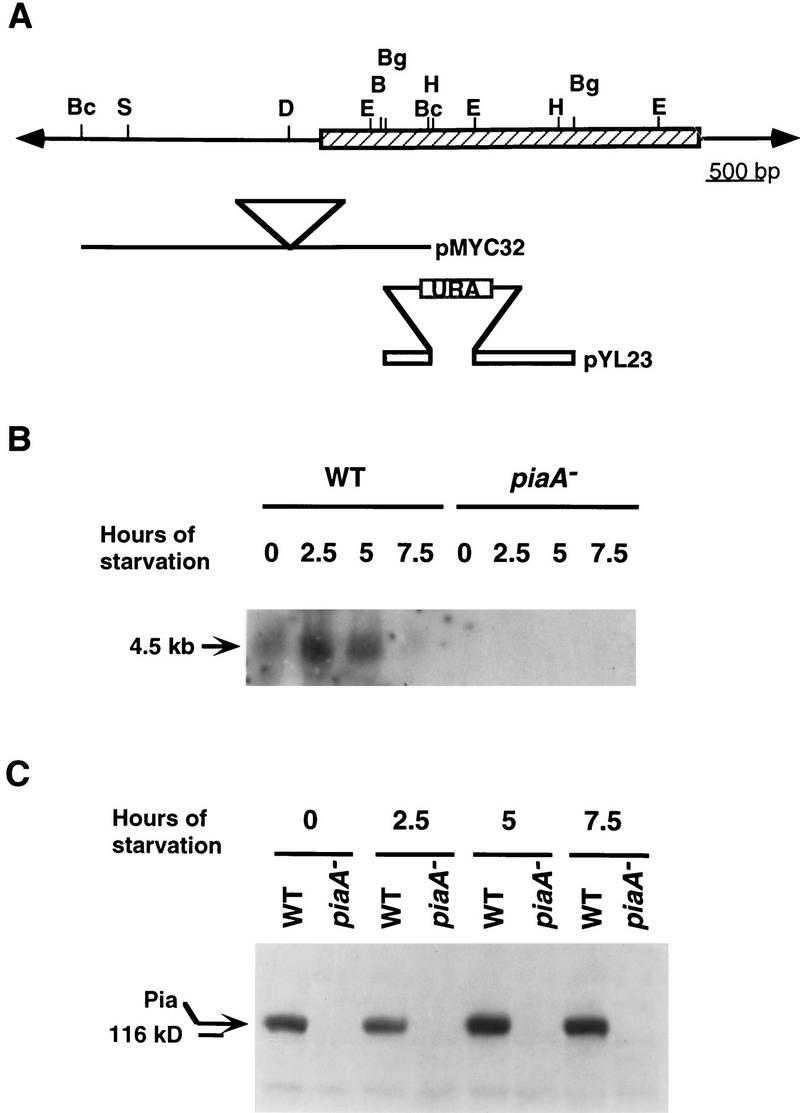
The structure of the PiaA gene and its expression during growth and early development. (A) Structure of the PiaA locus. The hatched box represents the PiaA coding region. (Bc) BclI; (S) SphI; (D) DpnII; (E) EcoRI; (Bg) BglII; (B) BamHI; (H) HindIII. Solid lines and open bars represent genomic DNA and cDNA fragments, respectively. The plasmid pMYC32 was rescued from the original REMI mutant using BclI digestion (see Materials and Methods). The triangle represents the insertion of a REMI vector, pRHI30, at a DpnII site. Plasmid pYL23 is a cDNA construct used for gene targeting; when linearized by BglII digestion and transformed into wild-type cells, it disrupted the PiaA gene by homologous recombination, replacing 0.4 kb of the coding region (a HindIII–EcoRI fragment) with a vector carrying the URA selectable marker. (B) Wild-type (WT) and piaA− cells were developed in suspension and samples were taken at times indicated. RNA was prepared and separated on a 1% agarose gel containing formaldehyde, blotted, and probed with a 2.4-kb 32P-labeled cDNA fragment. (C) Protein samples were prepared from the same cells as in B at times indicated, separated on a 7.5% SDS-PAGE gel, transferred onto a polyvinyldifluoride membrane, and probed with a rabbit polyclonal anti-Pia antibody.
Gene expression, chemotaxis, cGMP response, and actin polymerization in the piaA− cells
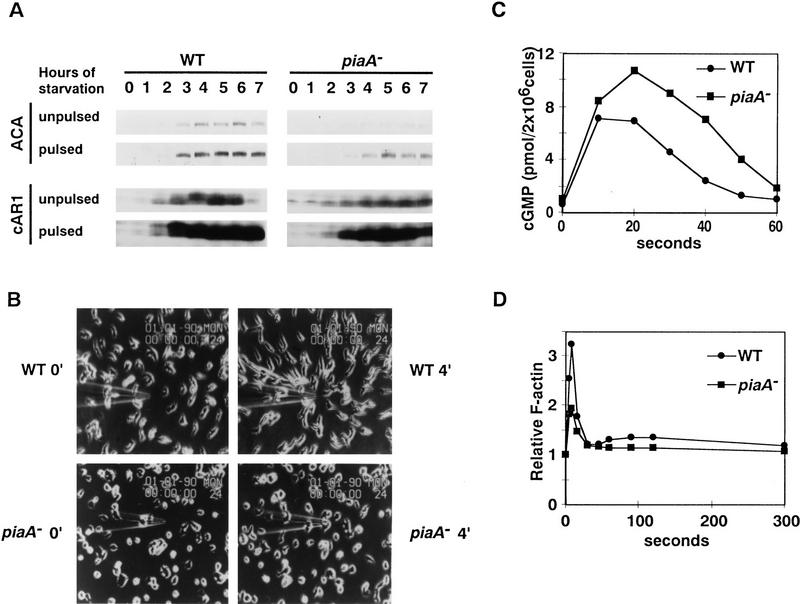
One possible explanation for the failure of piaA− cells to aggregate during development is an inability to express an essential component of the chemoattractant receptor signaling pathway. We allowed mutant and wild-type cells to develop in suspension, with or without the addition of 50–100 nm cAMP every 6 min, and examined several components of the pathway. As shown in Figure 2A, although the piaA− cells accumulated less ACA or cAR1 than wild-type levels in the absence of added cAMP pulses, they accumulated similar levels when stimulated repeatedly with cAMP. The expression of Gα2 and Gβ in piaA− cells was also comparable to that in wild-type cells (data not shown). Therefore, the phenotype of piaA− cells cannot be traced to a failure to express other known required genes.
Figure 2.
Gene expression, chemotaxis, cGMP production, and actin polymerization in the piaA− cells. (A) Wild-type and piaA− cells were developed in suspension with or without addition of 100 nm cAMP pulses. Samples were taken at times indicated, separated on 10% SDS-PAGE gels, and transferred onto a polyvinyldifluoride membrane. Blots were each cut horizontally, and respective halves were probed with polyclonal antisera against ACA and cAR1, respectively. (B) Wild-type and piaA− cells were developed for 5 hr with the addition of 100 nm cAMP at 6-min intervals. Cells were examined for chemotaxis to cAMP in a microneedle assay as described in Materials and Methods. At time 0 min, a microneedle filled with cAMP solution was positioned to stimulate the cells. The response of the cells at time 4 min is shown on the right. (C,D) Cells were developed as in B. cAMP-induced cGMP production (C) and actin polymerization (D) were assayed as described. Means of two to four experiments are shown.
To test whether the piaA− cells are able to complete the developmental program if appropriately stimulated, we performed a synergy experiment. Wild-type and piaA− cells were mixed at a 1:1 ratio and plated on non-nutrient agar. Spores from the fruiting bodies were collected into a buffer containing 10% glycerol and heated at 42°C for 1 hr to eliminate possible contamination by amebae. The treated spores were diluted and plated clonally on bacteria lawns. After several days, individual plaques were scored for developmental phenotype. From a total of approximately 1600 plaques scored, 12 were found to be derived from the mutant spores, showing an aggregationless phenotype. The result demonstrates that the piaA− cells can respond to exogenous signals by expression of developmental genes necessary for spore formation, although the efficiency of the process is reduced. Similar behavior has been observed in aca− and crac− cells. This suggests that the defect of the piaA− cells may be, as in aca− and crac− cells, in generating cAMP signals.
Is the failure of piaA− cells to aggregate a result of their inability to carry out chemotaxis toward cAMP? Using cells developed in suspension for 5 hr with repeated addition of cAMP, we performed a small-drop chemotaxis assay. In this assay, cAMP is spotted near a drop of cells on the surface of agar and after 20 min of incubation, cells are checked for movement toward the drop of chemoattractant. In 18 of 23 experiments done, piaA− cells showed a weaker chemotactic response, but in 5 experiments they responded as well as wild type (data not shown). Figure 2B shows the result of a different assay for chemotaxis. Cells developed for 5 hr were placed on a cover slide and a microneedle filled with 100 μm cAMP solution was brought to the vicinity of the cells. Mutant cells within 30 μm of the tip where the cAMP concentration is highest consistently responded. The wild-type cells typically responded from distances of >100 μm, indicating a lower sensitivity in the piaA− cells. Further experiments will be required to determine whether this behavior represents a primary defect in chemotaxis (see Discussion). The positive results, however, suggest the existence of other primary defects in the piaA− cells.
After 5 hr of development, piaA− cells were also able to produce cGMP and polymerize actin in response to cAMP stimulation (Fig. 2C,D). The observations described above indicate that piaA− cells possess the machinery to respond to cAMP signals. However, they are unable to aggregate in pure populations. This suggests that the defect may be in the production of the cAMP signals.
PiaA is required for receptor and G protein-mediated activation of ACA
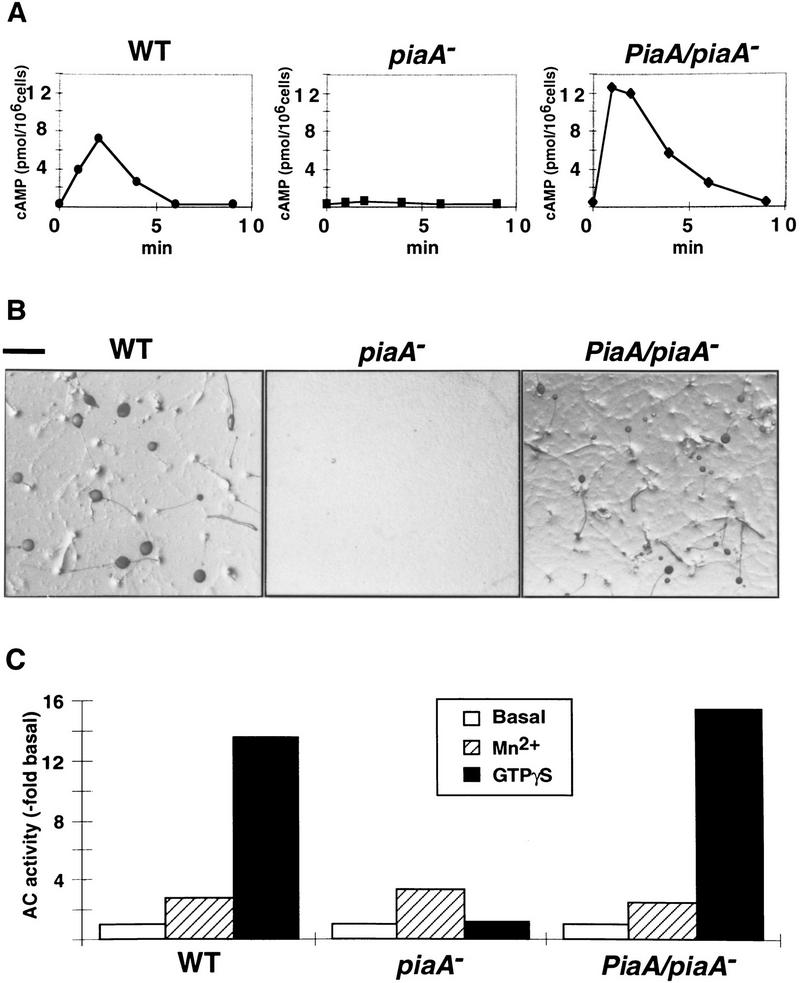
Therefore, we examined the cAMP production pathway in piaA− cells. Cells developed for 5 hr were stimulated with a cAMP analog, 2′-deoxy-cAMP. As shown in Figure 3A, in wild-type cells, the accumulation of cAMP peaked at about 2 min after addition of the chemoattractant and then subsided. In the piaA− cells, there was no detectable activation of the enzyme in response to the stimulus. The coupling between cAR1 and G2 is intact in the piaA− cells as chemotaxis, cGMP response, and actin polymerization still occurred. Therefore, the inability to synthesize cAMP could be attributable to inefficient activation of a pathway linking the activated G protein to ACA. We tested this possibility by assaying the activation of ACA in vitro in cell-free lysates. In this assay, GTPγS greatly stimulates ACA through a Gβ-dependent pathway (Theibert and Devreotes 1986; Wu et al. 1995). As shown in Figure 3C, GTPγS stimulated ACA activity about 13-fold in wild-type lysates, but failed to significantly stimulate the activity in piaA− lysates. However, in the presence of Mn2+ ions, which stimulate ACA directly, lysates of wild-type and piaA− cells were similar, indicating that the defect in piaA− cells does not affect the catalytic activity of the enzyme.
Figure 3.
Pia is required for receptor-mediated and GTPγS activation of ACA. (A) Chemoattractant receptor-mediated activation of adenylyl cyclase was assayed in 5-hr-developed wild-type, piaA− and PiaA/piaA− cells as described using 2′-deoxy-cAMP as the stimulus. (B) Wild-type, piaA−, and PiaA/piaA− cells were developed on non-nutrient agar plates and photographed at 48 hr. Bar, 1 mm. (C) Wild-type, piaA− and PiaA/piaA− cells were developed for 5 hr and assayed for adenylyl cyclase activity in the absence or presence of 5 mm MnSO4. GTPγS stimulation was determined in the presence of 40 μm GTPγS and 1 μm cAMP in the lysate. Means of 8–10 experiments are shown.
As stated above, expression of the full-length PiaA cDNA rescued the piaA− cells and the cells were able to complete the developmental program and make fruiting bodies (Fig. 3B). Consistently, all the biochemical defects were reversed. The rescued cell line PiaA/piaA− accumulated cAMP in response to 2′-deoxy-cAMP stimulation with kinetics similar to those of wild-type cells (Fig. 3A). GTPγS stimulation of ACA activity in the PiaA/piaA− lysates was also restored (Fig. 3C). These observations suggest that the failure of piaA− cells to aggregate is caused primarily by their inability to synthesize and secrete cAMP. The results also demonstrate that the cloned cDNA is sufficient for all of the functions of the PiaA gene.
PiaA has homologs in both S. cerevisiae and Schizosaccharomyces pombe
We sequenced the cDNA fragments, PCR fragments, and appropriate genomic fragments to assemble the full-length sequence of the PiaA gene. The ORF encodes a protein of 1148 amino acids with a molecular mass of 129.5 kD (Fig. 4). The predicted protein is generally hydrophilic, with scattered short hydrophobic segments. A motif search on the sequence did not yield possible functions of the protein. We used the TBLASTN program (Altschul et al. 1990) to search the National Center for Biotechnology Information (NCBI) nonredundant databank. Two homologous sequences were found; one is SPAC12C2.02C in S. pombe and the other is YER093C in S. cerevisiae (accession nos. for the sequences are emb/Z54140 and gb/U18839, respectively). Both were uncharacterized ORFs identified through genome sequencing. The D. discoideum PiaA gene is more homologous to the S. pombe gene than to the S. cerevisiae gene (BLAST P value of 10−110 for the S. pombe gene compared with 10−51 for the S. cerevisiae gene). The two yeast homologs are slightly larger (147.4 kD for the S. pombe protein and 164.4 kD for the S. cerevisiae protein). When sequences of the three proteins are aligned, the homology is distributed throughout nearly the entire length; the size difference lies in the very amino-terminal region (Fig. 4).
Figure 4.
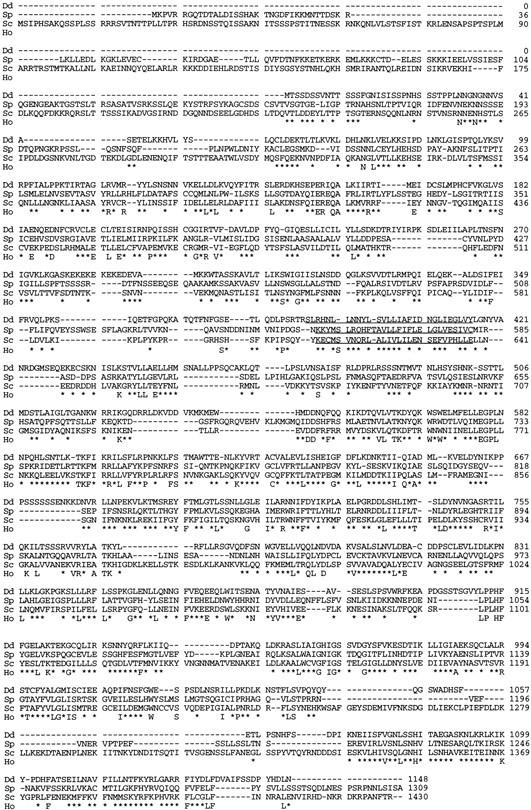
Amino acid sequences of Dictyostelium Pia and two yeast homologs. The alignment of three sequences is shown. Numbers on the right indicate amino acid positions. (Dd) Dictyostelium; (Sp) S. pombe; (Sc) S. cerevisiae; (Ho) homology between sequences. Residues identical in all three homologs are indicated by letters. Residues similar in all three homologs are indicated by an asterisk. Residues identical or similar in two sequences only are not marked. Segments identified by TMAP are underlined (see text for details).
We have performed a computer analysis on the multiple sequence alignments using the TMAP algorithm (Persson and Argos 1994; Milpetz et al. 1995) and the results predict that all three proteins have a transmembrane segment (residues 387–415 in the D. discoideum Pia; see Fig. 4). The membrane localization is proven wrong for the D. discoideum Pia (see below), whereas the localization for the two yeast proteins remains to be determined. If the yeast proteins are also cytosolic, the transmembrane segments predicted by TMAP may simply indicate a buried hydrophobic region common in all three proteins.
PIA1 is an essential gene in S. cerevisiae
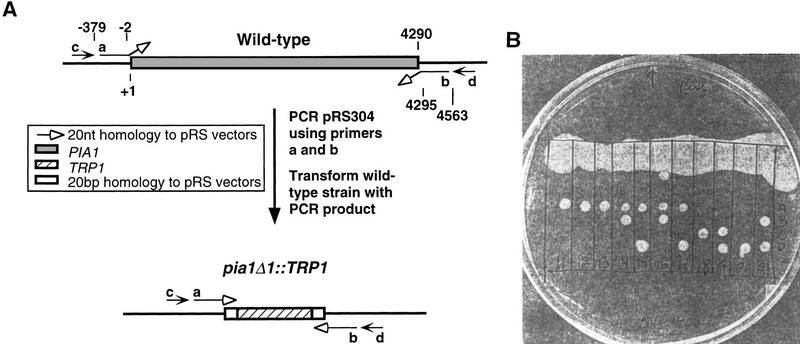
We disrupted the homologous gene PIA1 in S. cerevisiae by using the deletion technique of Lorenz et al. (1995) (Fig. 5A; see Materials and Methods). A wild-type diploid Trp auxotroph was transformed with a PCR fragment consisting of the TRP1 marker and 40 bp of sequence homology to the regions upstream of the 5′ end and downstream of the 3′ end of the PIA1 gene and Trp+ colonies were selected. The heterozygous pia1 deletion strain (YMC1; pia1Δ1::TRP1/PIA1), as verified by both PCR analysis and Southern blotting, was sporulated. The resulting asci were dissected and the spore viability was determined. Among the 21 sets of tetrads analyzed, 9 gave rise to one and 12 produced two viable colonies. An example is shown in Figure 5B. Subsequent plating found all the viable colonies to be trp−. This result indicates that the deletion of PIA1 is lethal and that PIA1 is an essential gene in S. cerevisiae. The spots on the tetrad dissection plate where no gross colonies formed contained microcolonies formed by clusters of cells. This observation argues against the possibility that the PIA1 gene is required for germination.
Figure 5.
Disruption of the S. cerevisiae PIA1 gene results in lethality. (A) A schematic diagram of the disruption of the PIA1 gene. Numbers indicate nucleotide positions. Primers a and b were both 60 nucleotides in length; each contained 40 nucleotides homologous to the locus, and 20 nucleotides homologous to pRS (indicated by open arrows). These two primers were used to amplify the TRP1 gene from pRS304 and the PCR product was transformed into a wild-type strain. The 40-bp region flanking TRP1 allowed homologous recombination at the PIA1 locus and deletion clones were first identified by PCR analysis (using primers a and d or primers b and c), then verified by Southern blot analysis (probed with oligonucleotide c). (B) The heterozygous pia1 deletion strain was sporulated and the tetrads were dissected by micromanipulation. The four spores from individual asci were aligned vertically and allowed to germinate on a YPD plate at 30°C. The picture was taken 5 days after dissection.
Reconstitution of GTPγS activation of adenylyl cyclase in piaA− lysates
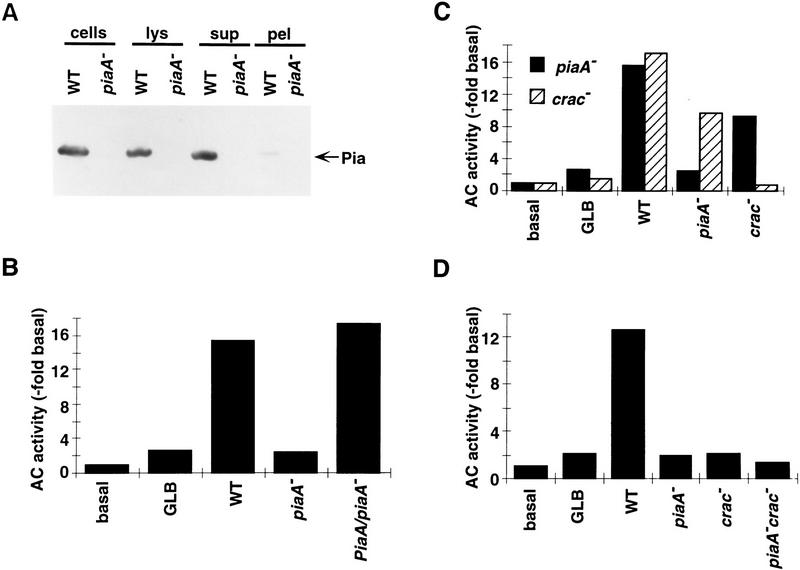
To further define the function of the D. discoideum Pia protein, we performed a crude subcellular fractionation to localize the protein. Cells were lysed by passage through a 5-μm nucleopore filter and the lysates were analyzed by differential centrifugation. The particulate and soluble fractions were analyzed by SDS-PAGE and Western blot analysis with a carboxy-terminal antiserum of Pia. As shown in Figure 6A, the protein was located quantitatively to the soluble fraction. This finding immediately raised the possibility that the defect in GTPγS stimulation of ACA in piaA− lysates might be reconstituted by the addition of supernatants containing Pia protein. To test this possibility, various supernatants or buffer were added into lysates prepared from piaA− cells in the presence of GTPγS and ACA activity was assayed after a short incubation. As shown in Figure 6B, neither the buffer nor the supernatants prepared from piaA− cells corrected the defect, whereas supernatants prepared from either wild-type cells or PiaA/piaA− cells did. This suggests that the Pia protein acts as a cytosolic activator of adenylyl cyclase.
Figure 6.
Reconstitution of GTPγS activation of ACA in mutant lysates. (A) Protein samples of whole cells (cells), filter lysates (lys), soluble (sup), and particulate (pel) fractions of lysates were separated on a 6% SDS-PAGE gel, blotted and probed with a rabbit antiserum directed against the carboxyl terminus of the Pia protein. Each lane was loaded with a sample equivalent to 4 × 106 cells. (B) Reconstitution of GTPγS activation of ACA in piaA− lysate was performed, as described in Materials and Methods, on cells developed for 5.5 hr, using buffer (GLB) or supernatants prepared from different cell lines as indicated. Basal activity was assayed in the absence of GTPγS. Means of three to four experiments are shown. (C) Reconstitution of GTPγS activation of ACA in lysates prepared from piaA− or crac− cells. Buffer or supernatants from different cell lines were added as indicated. Shown are means of three to four experiments. (D) Reconstitution of GTPγS activation of ACA in lysates prepared from piaA−crac− cells. Buffer or supernatants from different cell lines were added as indicated. Means of two to four experiments are shown.
The function of the Pia protein in conferring GTPγS stimulation of ACA is not redundant with that of the previously identified cytosolic regulator of adenylyl cyclase CRAC. As shown in Figure 6C, the supernatants from crac− cells reconstituted piaA− lysates significantly and supernatants from piaA− cells reconstituted crac− lysates significantly. The activity of either protein in cytosols does not depend on the presence of the other protein. This suggests that both of these proteins are integral components of the pathway leading to activation of ACA; the conclusion is further supported by the observation presented in Figure 6D. We prepared a cell line lacking both the Pia and CRAC proteins by knocking out the PiaA gene in crac− cells (see Materials and Methods). Lysates from this piaA−crac− double knockout cell line were prepared and various supernatants added to test for reconstitution activity. Although wild-type supernatant was able to reconstitute GTPγS stimulation of ACA, supernatants lacking either one of the cytosolic regulators were ineffective.
Discussion
The discovery of the Pia protein identifies a second cytosolic regulator of adenylyl cyclase ACA. Pia and the previously identified cytosolic regulator CRAC have several common features. They both seem to act downstream of receptor/G protein coupling. Responses requiring cAR1/G2 interaction, such as cAMP-induced cGMP production and actin polymerization, can be measured in crac− (Insall et al. 1994) and piaA− mutants. However, chemoattractant receptor activation of ACA in vivo and GTPγS activation of ACA in vitro are completely absent in both mutants. Furthermore, GTPγS activation of ACA in lysates prepared from either crac− or piaA− cells can be reconstituted by providing supernatants containing the appropriate missing protein. Nevertheless, Pia and CRAC do not function redundantly in activating adenylyl cyclase; both are needed for responses to cAMP or GTPγS. Data from reciprocal reconstitution experiments and reconstitution of piaA−crac− lysates also suggest that both Pia and CRAC are components in the activation pathway, not that one is controlling the expression of the other.
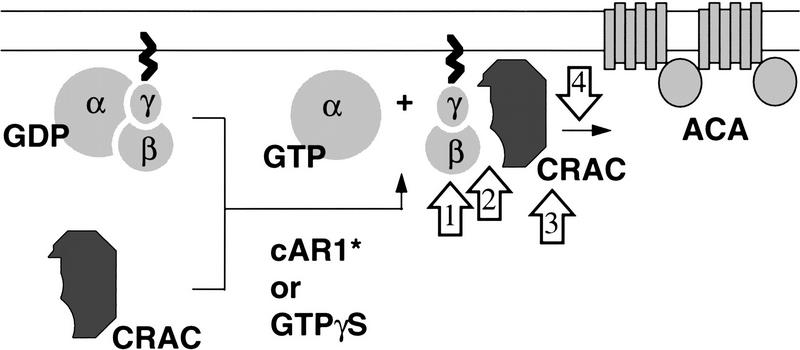
Figure 7 shows a schematic model of the activation of ACA. Because the other components of the adenylyl cyclase activation system are membrane proteins, it is expected that the cytosolic regulators somehow interact with the membrane for activation to occur. This is the case for CRAC. During receptor or GTPγS-mediated activation of ACA, there is an increase in the amount of CRAC that cosediments with membranes (Lilly and Devreotes 1995). The association of CRAC with membranes is time and GTPγS dependent and correlated with the activation of ACA. The binding of CRAC to membranes does not depend on cAR1, Gα2, or ACA, but in cells lacking the Gβ subunit it does not occur, suggesting either that βγ-subunits serve as CRAC-binding sites or are required for their generation. In preliminary experiments, negligible amounts of Pia have been observed to translocate to the membranes (M.-Y. Chen and P.N. Devreotes, unpubl.). Therefore, Pia is unlikely to be the CRAC-binding site. Pia may participate in the activation, beyond GTPγS binding and subunit dissociation, of the G protein (open arrow 1 in Fig. 7). Pia may be required in the generation of CRAC sites or act on Gβγ (open arrow 2 in Fig. 7) to facilitate the translocation of CRAC. Alternatively, optimal activation may require some interaction between Pia and CRAC (open arrow 3 in Fig. 7). Or Pia may act after CRAC has bound to the membranes (open arrow 4 in Fig. 7). We have developed an assay to separate the step involving GTPγS activation from that of CRAC binding and shown that CRAC can act after the removal of GTPγS (Lilly and Devreotes 1995). Using this assay and the piaA−crac− double knockout cells, we should be able to test the above possibilities.
Figure 7.
A schematic model of activation of ACA. The double lines represent the plasma membrane. cAR1* represents the activated surface receptor. Upon receptor or GTPγS activation, CRAC translocates onto the membrane. The binding of CRAC to the membrane is a Gβ-dependent process. Open arrows indicate possible points of action of Pia. See Discussion for details.
Genetic analysis suggests that the pathway leading from surface receptor to the activation of adenylyl cyclase involves further regulatory steps. Other than the two cytosolic regulators, two additional genes, ERK2 and AleA, are also involved in the activation of ACA (Segall et al. 1995; Insall et al. 1996). ERK2 is a mitogen-associated protein (MAP) kinase and AleA is a homolog of the yeast CDC25 gene, a Ras exchange factor (RasGEF). The erk2− and aleA− mutants are phenotypically similar to the piaA− and crac− mutants in that they are specifically defective in receptor/G protein-mediated activation of ACA. It is not yet known whether ERK2 and Ale act directly in the activation pathway; attempts to reconstitute the GTPγS stimulation of ACA in lysates from these two mutants have not been successful (B.J. Blacklock and P.N. Devreotes, unpubl.). Recently, ERK2 has been shown to be activated transiently by chemoattractants (Maeda et al. 1996). It remains to be determined whether ERK2 acts on, for instance, G protein βγ-subunits, ACA, CRAC, or Pia. We have noted that supernatants prepared from erk2− or aleA− cells can reconstitute lysates prepared from piaA− or crac− cells in GTPγS activation of adenylyl cyclase (M.-Y. Chen, B.J. Blacklock, and P.N. Devreotes, unpubl.), indicating that Pia and CRAC proteins are present in the erk2− and aleA− cells.
Whether piaA− cells have a primary chemotaxis defect requires further investigation. In the chemotaxis assay using microneedles, the response in the mutant was much weaker than that in wild-type cells. However, it was clearly a positive response when compared to gβ− cells, which are unable to carry out chemotaxis to any chemoattractant. The intermittent positive results from small-drop assays also demonstrate the ability of the cells to move toward the cAMP source. It is noted that another adenylyl cyclase pathway mutant, aleA−, also exhibits very weak chemotactic response (Insall et al. 1996). This may indicate that the pathways leading to activation of adenylyl cyclase and chemotaxis are intertwined and share common components or that intracellular cAMP somehow modulates the chemotaxis response.
Chemoattractants lead to many responses, besides activation of adenylyl cyclase, in Dictyostelium (Devreotes and Zigmond 1988; Caterina and Devreotes 1991; Chen et al. 1996). Evidence suggests that a single Gβ is required for most of the G protein-mediated responses in D. discoideum. The variety of Gα subunits may serve to specify the activation of the βγ-subunit complex by different chemoattractant receptors. The unique Gβ senses signals from different chemoattractant receptors and is a major transducer of signals to different effectors. For example, Gα2 and Gα4 subunits are responsible for the coupling of the release of βγ-subunits to cAR1 and the folic acid receptor, respectively (Kesbeke et al. 1988; Hadwiger et al. 1994). The gβ− cells respond to neither cAMP nor folic acid; receptor-mediated adenylyl cyclase, guanylyl cyclase, phospholipase C (PLC) activation, and actin polymerization are all absent (Wu et al. 1995). The signaling pathways leading to different effectors seem to branch at Gβ because mutants defective in activation of one specific effector exist. There are nonchemotactic mutants, such as KI8 and KI10 obtained from chemical mutagenesis, defective in cAMP-induced activation of guanylyl cyclase but not the activation of adenylyl cyclase and PLC (Kuwayama et al. 1993). Mutants crac−, aleA−, erk2−, and piaA− are all specifically defective in cAMP activation of adenylyl cyclase, whereas the cGMP response to cAMP stimulation is still present in these mutants. This indicates that the blockade in the signaling pathway in these mutants resides downstream of Gβ and specifically in the branch of the pathway leading to ACA. How are these multiple proteins involved in activating the same enzyme? Do they act sequentially in the pathway or do they form a complex and act simultaneously? Further biochemical analyses should help to answer these and other questions.
The target of Ale is likely to be a Ras-like protein. Several Ras genes have been identified in Dictyostelium, but little is known about the function of Ras proteins in D. discoideum. Whether there is a Ras pathway interacting with and modulating the adenylyl cyclase activation pathway in D. discoideum is currently under investigation. Interestingly, Ras proteins and CDC25 gene product are controlling elements of the adenylyl cyclase system in the yeast S. cerevisiae (Broek et al. 1985; Toda et al. 1985). RAS1 and RAS2 proteins regulate this adenylyl cyclase in a GTP-dependent manner (Toda et al. 1985). Ras activity is controlled by IRA1/IRA2 (GTPase activating proteins; Tanaka et al. 1990) and CDC25/SCD25 (nucleotide exchange factors; Crechet et al. 1990; Jones et al. 1991). The Ras/cyclase pathway regulates a range of cellular events, including cell growth, glycogen metabolism, cell cycle progression, and heat shock sensitivity (Thevelein 1992). Haploid spores of S. cerevisiae lacking adenylyl cyclase give rise to microcolonies and haploid spores lacking both RAS1 and RAS2 genes are not viable (Wigler et al. 1988). The phenotype of S. cerevisiae PIA1 deletion mutant is reminiscent of that of adenylyl cyclase pathway mutants; further experiments are required to determine whether the S. cerevisiae Pia protein is involved in this pathway.
The S. pombe adenylyl cyclase, not regulated by Ras (Nadin-Davis et al. 1986), is likely regulated by a heterotrimeric G protein-linked pathway as the Gα subunit encoded by the GPA2 gene is involved in the determination of the cAMP level according to nutritional conditions (Isshiki et al. 1992). In S. pombe, the FBP1 gene, encoding fructose-1,6-bisphosphatase, is repressed transcriptionally by glucose and this glucose repression involves a cAMP signaling pathway. Genetic and molecular analyses of FBP1 transcriptional regulation have led to the identification of 1 GIT (glucose-insensitive-transcription) genes (Hoffman and Winston 1990). Among these 10 genes, GIT2 encodes an adenylyl cyclase (Hoffman and Winston 1991) and GIT8 is the GPA2 gene (Nocero et al. 1994); the rest of the GIT genes are likely to encode components of the cAMP signal transduction pathway in S. pombe. It will be interesting to see whether the S. pombe PIA homolog gene is one of the other GIT genes.
It is intriguing that PiaA, a D. discoideum regulator of adenylyl cyclase, has homologs in yeasts, where the structure and regulation of the adenylyl cyclases appear to be very different (Kataoka et al. 1985; Yamawaki-Kataoka et al. 1989; Young et al. 1989). It is possible that certain subtypes of adenylyl cyclase in mammals are regulated by a similar pathway involving cytosolic regulators. But it is perhaps more likely that the Pia genes play some more fundamental role. Our studies position the site of action of PiaA between the G protein βγ-subunits and the enzyme, perhaps in regulation or modification of the βγ-subunits. In yeasts, the Pia pathway targets effectors involved in lethality; it will be interesting to determine whether heterotrimeric G proteins are involved in this pathway. Further studies, such as cloning of mammalian PiaA homologs and yeast proteins interacting with Pia, are required to address these questions.
Materials and methods
Dictyostelium growth, development, and transformation
D. discoideum strains were grown axenically in HL5 medium (Sussman and Sussman 1967) with appropriate selection at 22°C. Development of cells in the development buffer (DB) (5 mm Na2HPO4, 5 mm KH2PO4, 2 mm MgSO4, 0.2 mm CaCl2) was done as described (Devreotes et al. 1987). Transformation of cells with DNA was performed essentially as described (Howard et al. 1988).
Cloning of Dictyostelium PiaA
Molecular cloning procedures were performed essentially as described (Sambrook et al. 1989), unless otherwise noted.
The initial step of cloning the PiaA gene was to isolate the genomic DNA flanking the REMI vector. Genomic DNA was isolated as described (Sun et al. 1990) from the REMI mutant and digested with BclI, an enzyme that does not cut in the inserted REMI vector. Subsequent DNA ligation and transformation into Escherichia coli were performed essentially as described (Kuspa and Loomis 1992). Transformants, carrying the rescued plasmid pMYC32 (Fig. 1A), were selected on ampicillin plates.
Three rounds of cDNA walks were performed in a λgt11 Dictyostelium cDNA library, using a digoxiginin (DIG)-labeled genomic fragment obtained from pMYC32 as the probe for the first round of screening. DIG labeling was done by using the Genius nonradiolabeling system (Boehringer Mannheim) according to the protocol of the manufacturer. Inserts of lambda clones were subcloned and sequenced. A cDNA contig of 3.5 kb was assembled according to restriction maps and partial sequences of these fragments. Complete sequence analysis of this 3.5-kb contig revealed a partial ORF, missing its 5′ portion.
To obtain DNA fragments containing the start codon, we performed PCR amplifications on a pACTII Dictyostelium cDNA library (a kind gift of Dr. Adam Kuspa, Baylor College of Medicine, Houston, TX). Primary amplifications were done using the library as the template, an antisense oligonucleotide (antiE; 5′-TGAGATCTCTGTTAGACATTCAAGAC), and an oligonucleotide carrying vector sequence (L1736; 5′-CTATCTATTCGATGATG) as the primers. Secondly, amplifications on the products of primary amplifications were done using a more 5′ (when compared with antiE) antisense primer (antiF; 5′-GCTTGAATTCTTTCAGGTTCTGAATG) and the same vector primer. We subcloned the products of the secondary PCR amplifications and sequenced three clones. Sequences of the three clones differ slightly in the very 5′ region yet they share an in-frame start codon. This initiation codon was verified by sequences from relevant genomic fragments.
Full-length cDNA clones were constructed by splicing together cDNA fragments and the 5′ PCR fragments using convenient restriction sites.
Construction of cell lines
piaA− cells
The original REMI piaA− mutant, HM440 (a kind gift of Dr. R. Kay, Medical Research Council, Cambridge, UK) was generated by DpnII REMI of pRHI30 (Insall et al. 1996) into DH1, a uracil axotroph strain derived from AX3 by deleting the entire pyr5-6 sequence. Two knockout constructs, pMYC32 and pYL23 (Fig. 1A), were used to create the piaA− mutants MYC15 and MYC28, respectively, by homologous recombination. Briefly, pMYC32 was linearized by BclI or pYL23 by BglII digestion and transformed into DH1; uracil prototrophs were selected in FM medium with no uracil supplement. Genomic DNA was isolated from Ura+ clones and digested with BclI or BglII, respectively. Southern analysis was performed using appropriate DIG-labeled cDNA fragments as the probes to verify the disruption of PiaA locus. Both MYC15 and MYC28 cells were used in further characterizations.
PiaA/piaA− cells
The full-length cDNA of PiaA was inserted into the Dictyostelium integrating expression vector pB18 (Johnson et al. 1991) in a sense orientation. The resulting plasmid was transformed into the piaA− cells; transformants were selected in HL5 plus 20 μg/ml of G418. The expression of the Pia protein was verified by Western blot analysis with an antibody directed against the carboxyl terminus of Pia (see below).
piaA−crac− cells
The PiaA gene was disrupted in the previously existing crac− cell line BB1 (obtained from B. Blacklock, this laboratory) by gene targeting. The disruption construct, pYL44, was similar to pYL23 (Fig. 1A), except that the URA fragment was replaced with a blastocidin-S resistance gene expression cassette (1.4-kb EcoRI–XbaI fragment from pJH280, a kind gift of Dr. Jeffrey A. Hadwiger, Oklahoma State University, Stillwater). The plasmid pYL44 was linearized and electroporated into BB1; transformants were selected in HL5 plus 10 μg/ml of blastocidin S and the disruption of PiaA locus was verified by PCR and Southern blot analyses.
Northern and Western blot analyses
Total cellular RNA was prepared from either growing cells or cells developed in suspension using catrimox-14 (Dahle and Macfarlane 1993) (Iowa Biotech Corp., no. IBC 010) as described (Insall et al. 1996). Forty micrograms of total RNA for each time point was electrophoresed on a formaldehyde-containing 1% agarose gel, transferred to Hybond-N+ membrane (Amersham), and fixed by baking at 80°C for 2 hr. A 32P−labeled 2.4-kb PiaA cDNA fragment (BamHI–XbaI) was used as the probe. Prehybridization and hybridization were carried out at 42°C in 50% formamide, 10% dextran sulfate, 1 m NaCl, 1% SDS, and 250 μg/ml sonicated salmon sperm DNA for 1 hr and overnight, respectively. After hybridization, the filter was washed in 2× SSC, 0.1% SDS at room temperature for 15 min; in 2× SSC, 0.1% SDS at 60°C for 15 min; and twice in 0.2× SSC, 0.1% SDS at 65°C for 15 min.
Whole cell protein samples from either growing or developed cells were prepared by resuspending cell pellets in SDS sample buffer. Fractions of lysates were prepared by first forcing the cell suspension at a density of 8 × 107 cells/ml in glycerol lysis buffer [GLB; 10 mm Tris-HCl (pH 8), 1 mm MgSO4, 0.2 mm EGTA, and 10% glycerol] through a 5-μm nucleopore filter and then centrifuged the lysates at 12,000 rpm in a SS34 rotor at 4°C for 30 min or 36,000 rpm in a SW60 rotor at 4°C for 1 hr. Both supernatants and pellets were collected and GLB was used to resuspend the pellets. Protein samples were separated by SDS-PAGE and transferred onto the Immobilon-P membrane (Millipore). cAR1, Gα2, Gβ, and ACA were probed with polyclonal antisera, as previously described (Chen et al. 1994; Klein et al. 1988; Lilly et al. 1993; Parent and Devreotes 1995). Pia was probed with a polyclonal antiserum raised against a peptide (H2N-CFDVAIFSSDPYHDLN-COOH) corresponding to the carboxy-terminal sequence of Pia protein. The peptide was coupled to BSA using the Inject Activated Immunogen Conjugation kit (PIERCE) according to the protocol of the manufacturer and used to immunize a rabbit.
Disruption of PiaA homolog in S. cerevisiae
The pia1 mutant strain was generated using the PCR-mediated gene deletion technique previously described (see Fig. 5A; Lorenz et al. 1995). In brief, two PCR primers, each 60 nucleotides in length, were synthesized. Primer a (5′-CTTCGTGCTGTACCGCTTCTATTAAGTTTTTGAAATTCACAGATTGTACTGAGAGTGCAC) consists of 40 nucleotides of sequence homologous to the region upstream of the start codon of PIA1, followed by 20 nucleotides of sequence homologous to the pRS series of yeast shuttle vectors. Primer b (5′-ATTGTGACTATATACATTTATACATGCGGCCCTTTTTTGCCTGTGCGGTATTTCACACCG) consists of 40 nucleotides of sequence homologous to the region downstream of the stop codon of PIA1, followed by 20 nucleotides of sequence from the opposing side of the selectable marker within the pRS vectors. The two primers were used in PCRs to amplify the TRP1 marker from one of the pRS vectors, pRS304. PCRs were performed using the following cycling protocol: one cycle for 3 min at 94°C; 35 cycles of 1 min at 94°C, 2 min at 55°C, 3 min at 72°C; followed by one cycle of 8 min at 72°C. The PCR product consists of linear double-stranded DNA containing the selectable marker TRP1 and 40 bp of sequence homologous to the region flanking the PIA1 locus. After phenol/chloroform extraction and ethanol precipitation, this PCR product was transformed into diploid SM1060 (MATa/α can1/can1 his4/his4 leu2/leu2 trp1/trp1 ura3/ura3) yeast cells by the lithium acetate procedure (Ito et al. 1983). Homologous recombination replaced the PIA1 locus with the TRP1 marker. Trp+ clones were colony-purified and genomic DNA was isolated from them. PCR amplifications on the genomic DNA using diagnostic primer sets (primers a and d or primers b and c; see Fig. 5A) were performed. The sequences of the oligonucleotides c and d are 5′-CCGACACGAGCATGGACGAAG and 5′-CTGCTGAAACGGAACTCCCAC, respectively. The knockout genotype was verified by Southern blot analysis using DIG-labeled oligonucleotide c.
The heterozygous pia1 deletion strain, designated YMC1, was allowed to sporulate on a minimal sporulation plate at 30°C for 1 week. The asci formed were dissected under a microscope using the micromanipulator; the spores were placed on a YPD plate and incubated at 30°C. Colonies formed were replica-plated onto SC–Trp plates to test for Trp axotrophy.
Assays
The small-drop chemotaxis assay was performed essentially as described (Konijn and Van Haastert 1987; Insall et al. 1996). The microneedle chemotaxis experiment was performed as follows. Cells were developed in shaking suspension for 5–6 hr, washed and resuspended in PM buffer (5 mm Na2HPO4, 5 mm KH2PO4, and 2 mm MgSO4) at 106 cells/ml. A 20-μl drop of cell suspension was placed in a chamber made up of a glass cover slide and a rectangular metal frame of 8 mm in height. Cells were allowed to settle at room temperature for 5–10 min and attach onto the glass surface. A gentle wash was done by adding and removing 1 ml of DB. Two milliliters of DB was then added to the chamber and the chemotactic stimulation was provided by a microneedle, filled with 100 μm cAMP solution, positioned with the aid of an inverted microscope and a micromanipulating system. Movement of cells was monitored and recorded with a TV camera.
Cyclic GMP accumulation in response to cAMP stimulation was measured as described (Mato et al. 1977) using an isotope dilution assay kit (Amersham International plc, TRK 500). Each time point was assayed in duplicate and the assay was repeated at least twice for each cell line with similar results.
F-actin levels were measured by a modification of the method of Hall et al. (1988) as described (Insall et al. 1996).
To examine the effects of chemoattractant stimulation of adenylyl cyclase in vivo, cells starved for 5 hr were stimulated with 10 μm 2′-deoxy-cAMP and cAMP accumulation measured essentially as described (Segall et al. 1995) using an isotope dilution kit (Amersham International plc, TRK 432). In vitro adenylyl cyclase assays were performed essentially as described (Theibert and Devreotes 1986) on 5-hr developed cells except that the concentration of unlabeled ATP and cAMP in the reaction were increased to 0.3 and 0.5 mm, respectively. Mn2+ stimulated activity was assayed with the presence of 5 mm MnSO4 in the reaction. GTPγS stimulation was determined with the presence of 40 μm of GTPγS and 1 μm of cAMP in the lysate.
To reconstitute the GTPγS stimulation of ACA in lysates, supernatants from various cell lines were prepared in GLB at 8 × 107 cells/ml as described above in the Northern and Western blot analyses section. In typical reconstitution assays, either fresh supernatants (prepared at 8 × 107 cells/ml) or supernatants frozen at −70°C and thawed immediately before reconstitution were mixed with equal volume of lysates freshly prepared at 4 × 107 cells/ml by filter lysis in the presence of 40 μm of GTPγS. The reconstitution mixtures were incubated on ice for 8–12 min and 200-μl aliquots of the mixtures were assayed for adenylyl cyclase activity as described above. High speed supernatants gave the same results as the low speed supernatants; in most experiments low speed supernatants were used because they were more readily prepared. In the controls, GLB was used in place of supernatants from cells.
Acknowledgments
We thank Dr. Rob Kay for the original Pianissimo REMI mutant HM440. We thank Dr. Susan Michaelis and Dr. Konomi Fujimura-Kamada for help in disruption of the S. cerevisiae Pianissimo homolog. M.-Y. C. was supported by a Merck predoctoral fellowship. This work was supported by grants (GM28007 and GM34933) from the National Institutes of Health to P. N. D.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL pnd@welchlink.welch.jhu.edu; FAX (410) 955-5759.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Broek D, Samiy N, Fasano O, Fujiyama A, Tamanoi F, Northup J, Wigler M. Differential activation of yeast adenylate cyclase by wild-type and mutant RAS proteins. Cell. 1985;41:763–760. doi: 10.1016/s0092-8674(85)80057-x. [DOI] [PubMed] [Google Scholar]
- Caterina MJ, Devreotes PN. Molecular insights into eukaryotic chemotaxis. FASEB J. 1991;5:3078–3085. [PubMed] [Google Scholar]
- Chen J, DeVivo M, Dingus J, Harry A, Li J, Sui J, Carty DJ, Blank JL, Exton JH, Stoffel RH, Inglese J, Lefkowitz RJ, Logothetis DE, Hildebrandt JD, Iyengar R. A region of adenylyl cyclase 2 critical for regulation by G protein βγ subunits. Science. 1995;268:1166–1169. doi: 10.1126/science.7761832. [DOI] [PubMed] [Google Scholar]
- Chen M-Y, Devreotes PN, Gundersen RE. Serine 113 is the site of receptor-mediated phosphorylation of the Dictyostelium G protein α-subunit Gα2. J Biol Chem. 1994;269:20925–20930. [PubMed] [Google Scholar]
- Chen M-Y, Insall RH, Devreotes PN. Signaling through chemoattractant receptors in Dictyostelium. Trends Genet. 1996;12:52–57. doi: 10.1016/0168-9525(96)81400-4. [DOI] [PubMed] [Google Scholar]
- Crechet J-B, Poullet P, Mistou M-Y, Parmeggiani A, Camonis J, Boy-Marcotte E, Damak F, Jacquet M. Enhancement of the GDP-GTP exchange of RAS proteins by the carboxyl-terminal domain of SCD25. Science. 1990;248:866–868. doi: 10.1126/science.2188363. [DOI] [PubMed] [Google Scholar]
- Dahle C, Macfarlane D. Isolation of RNA from cells in culture using catrimox-14 cationic surfactant. BioTechniques. 1993;15:1102–1105. [PubMed] [Google Scholar]
- Devreotes PN. G protein-linked signaling pathways control the developmental program of Dictyostelium. Neuron. 1994;12:235–241. doi: 10.1016/0896-6273(94)90267-4. [DOI] [PubMed] [Google Scholar]
- Devreotes PN, Zigmond SH. Chemotaxis in eukaryotic cells: A focus on leukocytes and Dictyostelium. Annu Rev Cell Biol. 1988;4:649–686. doi: 10.1146/annurev.cb.04.110188.003245. [DOI] [PubMed] [Google Scholar]
- Devreotes P, Fontana D, Klein P, Sherring J, Theibert A. Transmembrane signaling in Dictyostelium. Methods Cell Biol. 1987;28:299–331. doi: 10.1016/s0091-679x(08)61653-2. [DOI] [PubMed] [Google Scholar]
- Dohlman HG, Thorner J, Caron MG, Lefkowitz RJ. Model system for the study of seven-transmembrane-segment receptors. Annu Rev Biochem. 1991;60:653–688. doi: 10.1146/annurev.bi.60.070191.003253. [DOI] [PubMed] [Google Scholar]
- Felder CC, Kanterman RY, Ma AL, Axelrod J. A transfected m1 muscarinic acetylcholine receptor stimulates adenylate cyclase via phosphatidylinositol hydrolysis. J Biol Chem. 1989;264:20356–20362. [PubMed] [Google Scholar]
- Firtel RA. Integration of signaling information in controlling cell-fate decisions in Dictyostelium. Genes & Dev. 1995;9:1427–1444. doi: 10.1101/gad.9.12.1427. [DOI] [PubMed] [Google Scholar]
- Gilman AG. G proteins and dual control of adenylate cyclase. Cell. 1984;36:577–579. doi: 10.1016/0092-8674(84)90336-2. [DOI] [PubMed] [Google Scholar]
- ————— G proteins: Transducers of receptor-generated signals. Annu Rev Biochem. 1987;56:615–649. doi: 10.1146/annurev.bi.56.070187.003151. [DOI] [PubMed] [Google Scholar]
- Hadwiger JA, Lee S, Firtel RA. The Gα subunit Gα4 couples to pterin receptors and identifies a signaling pathway that is essential for multicellular development in Dictyostelium. Proc Natl Acad Sci. 1994;91:10566–10570. doi: 10.1073/pnas.91.22.10566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AL, Schlein A, Condeelis J. Relationship of pseudopod extension to chemotactic hormone-induced actin polymerization in amoeboid cells. J Cell Biochem. 1988;37:285–299. doi: 10.1002/jcb.240370304. [DOI] [PubMed] [Google Scholar]
- Hall AL, Warren V, Condeelis J. Transduction of the chemotactic signal to the actin cytoskeleton of Dictyostelium discoideum. Dev Biol. 1989;136:517–525. doi: 10.1016/0012-1606(89)90277-7. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. Glucose repression of transcription of the Schizosaccharomyces pombe fbp1 gene occurs by a cAMP signaling pathway. Genes & Dev. 1991;5:561–571. doi: 10.1101/gad.5.4.561. [DOI] [PubMed] [Google Scholar]
- Hoffman CS, Winston F. Isolation and characterization of mutants constitutive for expression of the fbp1 gene of Schizosaccharomyces pombe. Genetics. 1990;124:807–816. doi: 10.1093/genetics/124.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard PK, Ahern KG, Firtel RA. Establishment of a transient expression system for Dictyostelium discoideum. Nucleic Acids Res. 1988;16:2613–2623. doi: 10.1093/nar/16.6.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall R, Kuspa A, Lilly PJ, Shaulsky G, Levin LR, Loomis WF, Devreotes P. CRAC, a cytosolic protein containing a pleckstrin homology domain, is required for receptor and G protein-mediated activation of adenylyl cyclase in Dictyostelium. J Cell Biol. 1994;126:1537–1545. doi: 10.1083/jcb.126.6.1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall RH, Borleis J, Devreotes PN. The aimless RasGEF is required for processing of chemotactic signals through G-protein-coupled receptors in Dictyostelium. Curr Biol. 1996;6:719–729. doi: 10.1016/s0960-9822(09)00453-9. [DOI] [PubMed] [Google Scholar]
- Isshiki T, Mochizuki N, Maeda T, Yamamoto M. Characterization of a fission yeast gene, gpa2, that encodes a Gα subunit involved in the monitoring of nutrition. Genes & Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- Ito H, Funkuda Y, Murata K, Kimura A. Transformation of intact cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RL, Vaughan RA, Caterina MJ, Van Haastert PJ, Devreotes PN. Overexpression of the cAMP receptor 1 in growing Dictyostelium cells. Biochemistry. 1991;30:6982–6986. doi: 10.1021/bi00242a025. [DOI] [PubMed] [Google Scholar]
- Jones S, Vignais M-L, Broach JR. The CDC25 protein of Saccharomyces cerevisiae promotes exchange of guanine nucleotides bound to Ras. Mol Cell Biol. 1991;11:2641–2646. doi: 10.1128/mcb.11.5.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka T, Broek D, Wigler M. DNA sequence and characterization of the S. cerevisiae gene encoding adenylate cyclase. Cell. 1985;43:493–505. doi: 10.1016/0092-8674(85)90179-5. [DOI] [PubMed] [Google Scholar]
- Kay RR. Differentiation and patterning in Dictyostelium. Curr Opin Genet Dev. 1994;4:637–641. doi: 10.1016/0959-437x(94)90128-p. [DOI] [PubMed] [Google Scholar]
- Kesbeke F, Snaar-Jagalska BE, Van Haastert PJM. Signal transduction in Dictyostelium fgd A mutants with a defective interaction between surface cAMP receptors and a GTP-binding regulatory protein. J Cell Biol. 1988;107:521–528. doi: 10.1083/jcb.107.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein PS, Sun TJ, Saxe III CL, Kimmel AR, Johnson RL, Devreotes PN. A chemoattractant receptor controls development in Dictyostelium discoideum. Science. 1988;241:1467–1472. doi: 10.1126/science.3047871. [DOI] [PubMed] [Google Scholar]
- Konijn TM, Van Haastert PJM. Measurement of chemotaxis in Dictyostelium. Meth Cell Biol. 1987;28:283–298. doi: 10.1016/s0091-679x(08)61652-0. [DOI] [PubMed] [Google Scholar]
- Kuspa A, Loomis WF. Tagging developmental genes in Dictyostelium by restriction enzyme-mediated integration of plasmid DNA. Proc Natl Acad Sci. 1992;89:8803–8807. doi: 10.1073/pnas.89.18.8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwayama H, Ishida S, Van Haastert PJM. Non-chemotactic Dictyostelium discoideum mutants with altered cGMP signal transduction. J Cell Biol. 1993;123:1453–1462. doi: 10.1083/jcb.123.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitzki A. Transmembrane signalling to adenylate cyclase in mammalian cells and in Saccharomyces cerevisiae. Trends Biochem Sci. 1988;13:298–301. doi: 10.1016/0968-0004(88)90122-3. [DOI] [PubMed] [Google Scholar]
- Lilly PJ, Devreotes PN. Chemoattractant and GTPγS-mediated stimulation of adenylyl cyclase in Dictyostelium requires translocation of CRAC to membranes. J Cell Biol. 1995;129:1659–1665. doi: 10.1083/jcb.129.6.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly P, Wu L, Welker DL, Devreotes PN. A G-protein β-subunit is essential for Dictyostelium development. Genes & Dev. 1993;7:986–995. doi: 10.1101/gad.7.6.986. [DOI] [PubMed] [Google Scholar]
- Lorenz MC, Muir RS, Lim E, McElver J, Weber SC, Heitman J. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene. 1995;158:113–117. doi: 10.1016/0378-1119(95)00144-u. [DOI] [PubMed] [Google Scholar]
- Maeda M, Aubry L, Insall R, Gaskins C, Devreotes PN, Firtel RA. Seven helix chemoattractant receptors transiently stimulate mitogen-activated protein kinase in Dictyostelium. J Biol Chem. 1996;271:3351–3354. doi: 10.1074/jbc.271.7.3351. [DOI] [PubMed] [Google Scholar]
- Mato JM, Krens FA, van Haastert PJ, Konijn TM. 3′:5′-cyclic AMP-dependent 3′:5′-cyclic GMP accumulation in Dictyostelium discoideum. Proc Natl Acad Sci. 1977;74:2348–2351. doi: 10.1073/pnas.74.6.2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milpetz F, Argos P, Persson B. TMAP: A new email and WWW service for membrane-protein structural predictions. Trends Biochem Sci. 1995;20:204–205. doi: 10.1016/s0968-0004(00)89009-x. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Gibson T, Rice P, Thompson J, Saraste M. The PH domain: A common piece in the structural patchwork of signalling proteins. Trends Biochem Sci. 1993;18:343–348. doi: 10.1016/0968-0004(93)90071-t. [DOI] [PubMed] [Google Scholar]
- Nadin-Davis SA, Nasim A, Beach D. Involvement of ras in sexual differentiation but not in growth control in fission yeast. EMBO J. 1986;5:2963–2971. doi: 10.1002/j.1460-2075.1986.tb04593.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocero M, Isshiki T, Yamamoto M, Hoffman CS. Glucose repression of fbp1 transcription of Schizosaccharomyces pombe is partially regulated by adenylate cyclase activation by a G protein α subunit encoded by gpa2 (git8) Genetics. 1994;138:39–45. doi: 10.1093/genetics/138.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent CA, Devreotes PN. Isolation of inactive and G protein-resistant adenylyl cyclase mutants using random mutagenesis. J Biol Chem. 1995;270:22693–22696. doi: 10.1074/jbc.270.39.22693. [DOI] [PubMed] [Google Scholar]
- ————— Molecular genetics of signal transduction in Dictyostelium. Annu Rev Biochem. 1996;65:411–440. doi: 10.1146/annurev.bi.65.070196.002211. [DOI] [PubMed] [Google Scholar]
- Persson B, Argos P. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J Mol Biol. 1994;237:182–192. doi: 10.1006/jmbi.1994.1220. [DOI] [PubMed] [Google Scholar]
- Pitt GS, Milona N, Borleis J, Lin KC, Reed RR, Devreotes PN. Structurally distinct and stage-specific adenylyl cyclase genes play different roles in Dictyostelium development. Cell. 1992;69:305–315. doi: 10.1016/0092-8674(92)90411-5. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Segall JE, Kuspa A, Shaulsky G, Ecke M, Maeda M, Gaskins C, Firtel RA, Loomis WF. A MAP kinase necessary for receptor-mediated activation of adenylyl cyclase in Dictyostelium. J Cell Biol. 1995;128:405–413. doi: 10.1083/jcb.128.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spisani S, Pareschi MC, Buzzi M, Colamussi M, Biondi C, Traniello S, Zecchini GP, Paradisi MP, Torrini I, Ferretti ME. Effect of cyclic AMP level reduction on human neutrophil responses to formylated peptides. Cell Signal. 1996;8:269–277. doi: 10.1016/0898-6568(96)00049-6. [DOI] [PubMed] [Google Scholar]
- Sun TJ, Van Haastert PJM, Devreotes PN. Surface cAMP receptors mediate multiple responses during development in Dictyostelium: Evidenced by antisense mutagenesis. J Cell Biol. 1990;110:1549–1554. doi: 10.1083/jcb.110.5.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunahara RK, Dessauer CW, Gilman AG. Complexity and diversity of mammalian adenylyl cyclases. Annu Rev Pharmacol Toxicol. 1996;36:461–480. doi: 10.1146/annurev.pa.36.040196.002333. [DOI] [PubMed] [Google Scholar]
- Sussman R, Sussman M. Cultivation of Dictyostelium discoideum in axenic medium. Biochem Biophys Res Commun. 1967;29:53–55. doi: 10.1016/0006-291x(67)90539-6. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nakafuku M, Satoh T, Marshall MS, Gibbs JB, Matsumoto K, Kaziro Y, Toh-e A. S. cerevisiae genes IRA1 and IRA2 encode proteins that may be functionally equivalent to mammalian ras GTPase activating protein. Cell. 1990;60:803–807. doi: 10.1016/0092-8674(90)90094-u. [DOI] [PubMed] [Google Scholar]
- Tang W-J, Gilman AG. Type-specific regulation of adenylyl cyclase by G protein βγ subunits. Science. 1991;254:1500–1503. doi: 10.1126/science.1962211. [DOI] [PubMed] [Google Scholar]
- Theibert A, Devreotes PN. Surface receptor-mediated activation of adenylate cyclase in Dictyostelium. Regulation by guanine nucleotides in wild-type cells and aggregation deficient mutants. J Biol Chem. 1986;261:15121–15125. [PubMed] [Google Scholar]
- Thevelein JM. The RAS-adenylate cyclase pathway and cell cycle control in Saccharomyces cerevisiae. Antonie Leeuwenhoek. 1992;62:109–130. doi: 10.1007/BF00584466. [DOI] [PubMed] [Google Scholar]
- Toda T, Uno I, Ishikawa T, Powers S, Kataoka T, Broek D, Cameron S, Broach J, Matsumoto K, Wigler M. In yeast, RAS proteins are controlling elements of adenylate cyclase. Cell. 1985;40:27–36. doi: 10.1016/0092-8674(85)90305-8. [DOI] [PubMed] [Google Scholar]
- Verghese MW, Fox K, McPhail LC, Snyderman R. Chemoattractant-elicited alterations of cAMP levels in human polymorphonuclear leukocytes require a Ca2+-dependent mechanism which is independent of transmembrane activation of adenylate cyclase. J Biol Chem. 1985;260:6769–6775. [PubMed] [Google Scholar]
- Wigler M, Field J, Powers S, Broek D, Toda T, Cameron S, Nikawa J, Michaeli T, Colicelli J, Ferguson K. Studies of RAS function in the yeast Saccharomyces cerevisiae. Cold Spring Harbor Symp Quant Biol. 1988;53:649–655. doi: 10.1101/sqb.1988.053.01.074. [DOI] [PubMed] [Google Scholar]
- Wu L, Valkema R, Van Haastert PJ, Devreotes PN. The G protein β subunit is essential for multiple responses to chemoattractants in Dictyostelium. J Cell Biol. 1995;129:1667–1675. doi: 10.1083/jcb.129.6.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamawaki-Kataoka Y, Tamaoki T, Choe H-R, Tanaka H, Kataoka T. Adenylate cyclases in yeast: A comparison of the genes from Schizosaccharomyces pombe and Saccharomyces cerevisiae. Proc Natl Acad Sci. 1989;86:5693–5697. doi: 10.1073/pnas.86.15.5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young D, Riggs M, Field J, Vojtek A, Broek D, Wigler M. The adenylyl cyclase gene from Schizosaccharomyces pombe. Proc Natl Acad Sci. 1989;86:7989–7993. doi: 10.1073/pnas.86.20.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Liu Y, Ruoho AE, Hurley JH. Structure of the adenylyl cyclase catalytic core. Nature. 1997;386:247–253. doi: 10.1038/386247a0. [DOI] [PubMed] [Google Scholar]