Abstract
We have cloned and characterized a human gene encoding TP2 (telomerase-associated protein 2), a protein with similarity to reverse transcriptases and the catalytic telomerase subunits from Saccharomyces cerevisiae and Euplotes aediculatus. Indirect immunofluorescence revealed that TP2 was localized to the nucleus. Using antibodies to endogenous and epitope-tagged TP2, we found that TP2 was associated specifically with human telomerase activity and the recently identified telomerase-associated protein TP1. Mutation of conserved residues within the reverse transcriptase domain of TP2 severely reduced associated telomerase activity. These results suggest that telomerase is an evolutionarily conserved multisubunit complex composed of both structural and catalytic subunits.
Keywords: Reverse transcriptase, telomere, cancer, senescence
Unicellular organisms such as yeast and ciliates require telomerase and telomere-binding proteins for telomere maintenance and cell viability (for review, see Greider 1996). Telomerase is a multisubunit ribonucleoprotein that uses an RNA template to catalyze the addition of telomeric DNA to chromosome ends (for review, see Greider 1996). The RNA component of telomerase has been cloned in several organisms, including ciliates, yeast, human, and mouse (Greider and Blackburn 1989; Shippen-Lentz and Blackburn 1990; Romero and Blackburn 1991; Lingner et al. 1994; Blasco et al. 1995; Feng et al. 1995; McEachern and Blackburn 1995; Singer and Gottschling 1995; McCormick-Graham and Romero 1996). In each organism, the telomerase RNA contains a template complementary to its cognate G-rich telomeric repeat. Telomerase uses the RNA template to prime the synthesis of telomeric repeats to the 3′ end of single-stranded, G-rich telomeric DNA (for review, see Greider 1996). The RNA components do not share extensive sequence homology outside the template domain; however, the ciliate telomerase RNAs do share a common secondary structure (Romero and Blackburn 1991; ten Dam et al. 1991; Bhattacharyya and Blackburn 1994; Lingner et al. 1994; McCormick-Graham and Romero 1995).
The protein components of telomerase have only recently been characterized (for review, see Lustig 1997; Shore 1997). In Tetrahymena, two telomerase proteins, p95 and p80, copurify with telomerase activity (Collins et al. 1995; Harrington et al. 1995). The p80 subunit binds to telomerase RNA, whereas the p95 subunit binds to single-stranded, telomeric DNA (Collins et al. 1995; Harrington et al. 1995). The first mammalian telomerase-associated protein, TP1/TLP1, was identified based on amino acid similarity to the Tetrahymena p80 protein (Harrington et al. 1997; Nakayama et al. 1997). Murine TP1 binds telomerase RNA and is associated with mouse telomerase activity in vivo (Harrington et al. 1997), and rat TLP1 copurifies with rat telomerase activity (Nakayama et al. 1997). The absence of an obvious RNA-dependent DNA polymerase domain in TP1 suggests this protein may be associated with a catalytic subunit in a multicomponent telomerase complex (Harrington et al. 1997; Nakayama et al. 1997).
Purification of telomerase from the ciliate Euplotes aediculatus revealed two proteins, of 43 and 123 kD, that are apparently unrelated to the Tetrahymena proteins (Lingner and Cech 1996; Counter et al. 1997; Hammond et al. 1997; Lingner et al. 1997). p123 is a homolog of Est2; in Saccharomyces cerevisiae, EST2 is essential for telomere maintenance and telomerase activity (Lendvay et al. 1996; Counter et al. 1997; Lingner et al. 1997). Est2 and p123 are 20% similar and contain six of seven conserved motifs within the reverse transcriptase (RT) domain. Mutation of conserved amino acids within the RT motifs in Est2 result in loss of telomerase function in vivo and in vitro (Lendvay et al. 1996; Counter et al. 1997; Lingner et al. 1997). These results suggest that p123 and its homologs are the proteins that catalyze the telomerase RNA-dependent synthesis of telomeric DNA.
Deletion of EST2 and TLC1 (the telomerase RNA component) in yeast leads to telomere shortening and cessation of cell division (Singer and Gottschling 1995; Lendvay et al. 1996; Counter et al. 1997; Lingner et al. 1997). However, in mammals, the requirement for telomerase activity does not extend to all cell types (for review, see Harley 1996). Several normal adult human tissues lack telomerase activity, and their telomeres shorten with each round of cell division. In contrast, germ cells, immortalized cells, and the majority of primary tumors do contain telomerase activity and maintain their average telomere length. These observations have led to the hypothesis that telomerase activity may be required for the continued division of “immortal” cells (for review, see Harley 1996). There are some exceptions to this generalization. Some immortalized cell lines do not contain telomerase activity and possess aberrantly long telomeres (for review, see Bryan and Reddel 1997). Also, telomerase activity is not always sufficient to maintain telomeres in some human cell types. Some hematopoietic cells exhibit gradual telomere sequence loss despite a low level of telomerase activity (Vaziri et al. 1993, 1994; Counter et al. 1995).
An understanding of telomerase regulation and its role in tumor cell growth requires characterization of the molecular components of mammalian telomerase. We have identified and characterized TP2 (telomerase-associated protein 2), a human homolog of the Est2 and p123 telomerase proteins. The TP2 protein localized to the nucleus and associated with telomerase activity in human cells. The telomerase activity associated with TP2 was virtually abolished upon introduction of point mutations within the putative RT domain. In addition, TP2 coimmunoprecipitated with TP1, suggesting that these two proteins form part of a single telomerase complex in humans.
Results
TP2, a human homolog of p123 and Est2p
We identified a human EST exhibiting 30% amino acid identity to the p123 protein. Oligonucleotides derived from this sequence were used as probes to clone and assemble a contiguous cDNA sequence that contained an open reading frame (ORF) of 1132 amino acids (Fig. 1A). A BLAST 1.48 search of the nonredundant protein database revealed similarity to several different RTs (data not shown). Based on the results below and to retain consistent nomenclature with the previously identified TP1, the gene was named TP2. BLAST, FASTA, and Clustal W alignment revealed some homology to both p123 and EST2 throughout the ORF. The region of highest homology includes the RT motifs (Fig. 1B). Although the RT family is highly divergent, several amino acid residues are highly conserved; these are present in TP2. One of these motifs was found in TP2 (hLGXXh, amino acids 914–919) but not in p123 or EST2. After completion of the characterization of TP2, two other groups also reported the cloning of the same gene, named TRT1 and hEST2, respectively (Meyerson et al. 1997; Nakamura et al. 1997). In these studies, the expression pattern of the TRT1/hEST2 mRNA was examined, but no functional characterization of the protein product was reported (Meyerson et al. 1997; Nakamura et al. 1997).
Figure 1.
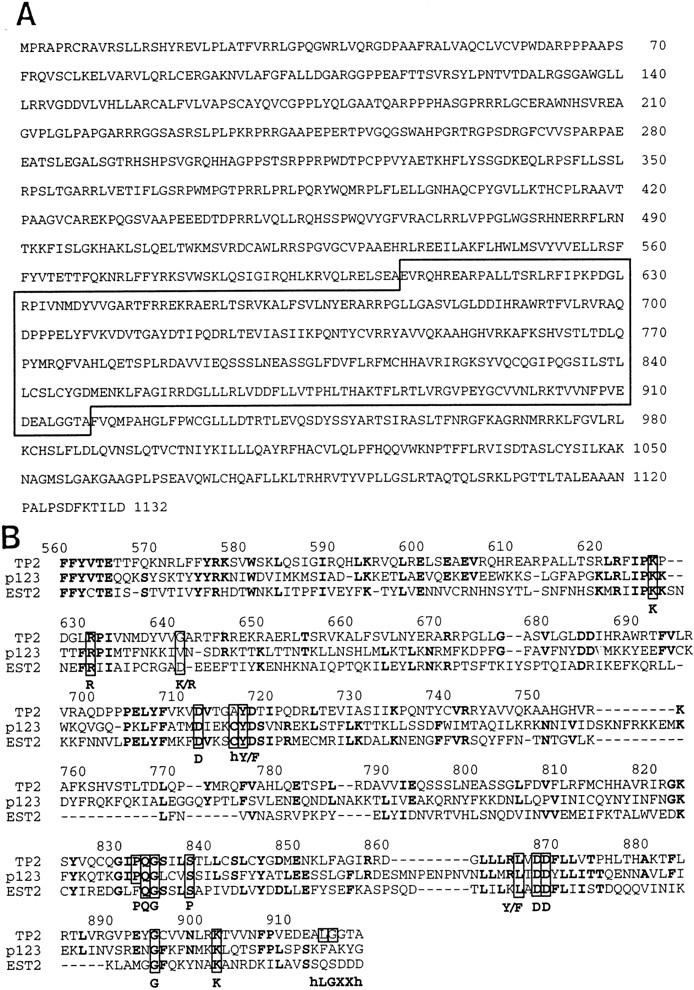
Amino acid sequence of human TP2. (A) Predicted amino acid sequence of human TP2. Homology to the RT domain is boxed with a solid line. (B) Domain of highest homology between TP2, p123, and Est2. Alignment was generated by Clustal W using a PAM350 matrix, then adjusted manually. Residues conserved in at least two of the aligned proteins are shown in boldface type. Residues conserved among RT are boxed, with consensus residues shown below.
TP2 is localized to the nucleus
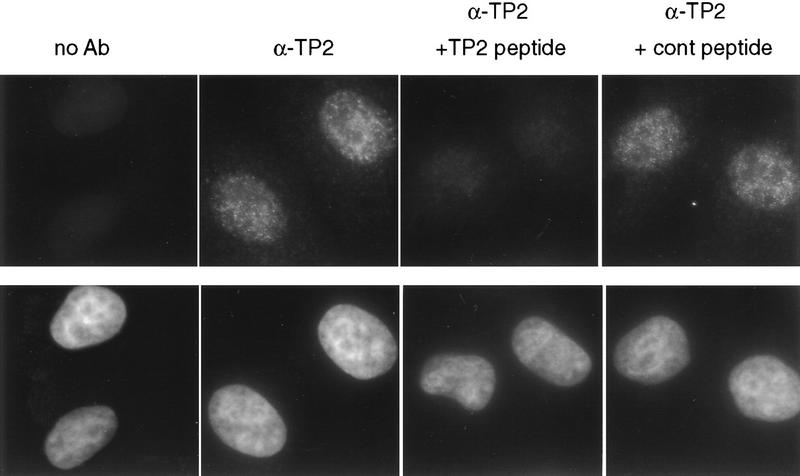
Because telomerase activity is present in HeLa nuclear fractions (Morin 1989), we examined the subcellular localization of TP2 protein. An antipeptide antiserum was generated against a region of the human TP2 protein (amino acids 602–626) and used in immunofluorescence analysis of HeLa cells (Fig. 2). Punctate nuclear staining was observed, which was specifically blocked by the presence of the TP2 peptide, but not by a nonspecific peptide (Fig. 2). Interestingly, a similar punctate nuclear staining pattern was also observed with antiserum to TP1 (R. Oulton, unpubl.). However, we have not yet determined whether the two proteins precisely colocalize within the nucleus.
Figure 2.
Immunofluorescence of TP2 protein. HeLa cells were stained with DAPI and anti-TP2 or control sera as indicated: (no Ab) No primary antibody was added; (α-TP2) the TP2 antipeptide antibody; (α-TP2 + TP2 peptide) antibody incubated with TP2 peptide; (α-TP2 + cont peptide) antibody incubated with nonspecific peptide (see Materials and Methods).
TP2 is associated with human telomerase activity
To determine whether human TP2 was associated with the telomerase complex, the telomere repeat amplification protocol (TRAP) assay was used to detect telomerase activity in TP2 immunoprecipitates from HeLa cell lysates (Fig. 3). TRAP activity has been characterized previously in many human cell lines and is dependent on the human telomerase RNA component (Kim et al. 1994). We found that telomerase activity associated specifically with TP2 immunoprecipitates and was sensitive to preincubation with ribonuclease A (Fig. 3B). Little or no telomerase activity was detected in immunoprecipitations using non-TP2 antibodies (anti-Myc and anti-GST). Prior incubation of the peptide antiserum with TP2 peptide reduced the associated telomerase activity to background levels, whereas a nonspecific peptide (amino acids 536–553 of murine TP1) did not compete for this association (Fig. 3B). Incubation of HeLa lysates with control or TP2 antisera and peptides did not nonspecifically inhibit telomerase activity (Fig. 3A). Western blot analysis of the TP2 immunoprecipitates using anti-TP2 antiserum revealed immunoreactivity against three major species, ranging in molecular mass from 130 to 180 kD. The 130-kD species corresponds to the predicted mass of the TP2 ORF and comigrated with epitope-tagged, exogenously expressed full-length TP2 (Figs. 4 and 5; data not shown). Although the pattern of TP2 species appeared complex, all three species were blocked by preincubation with TP2 peptide but not by the presence of nonspecific peptide (Fig. 3C). The abundance of TP2 may be low, because these species were not detectable by Western analysis of crude cell lysates (Figs. 4 and 5; data not shown). TP2 therefore specifically associates with telomerase activity in human cell extracts.
Figure 3.
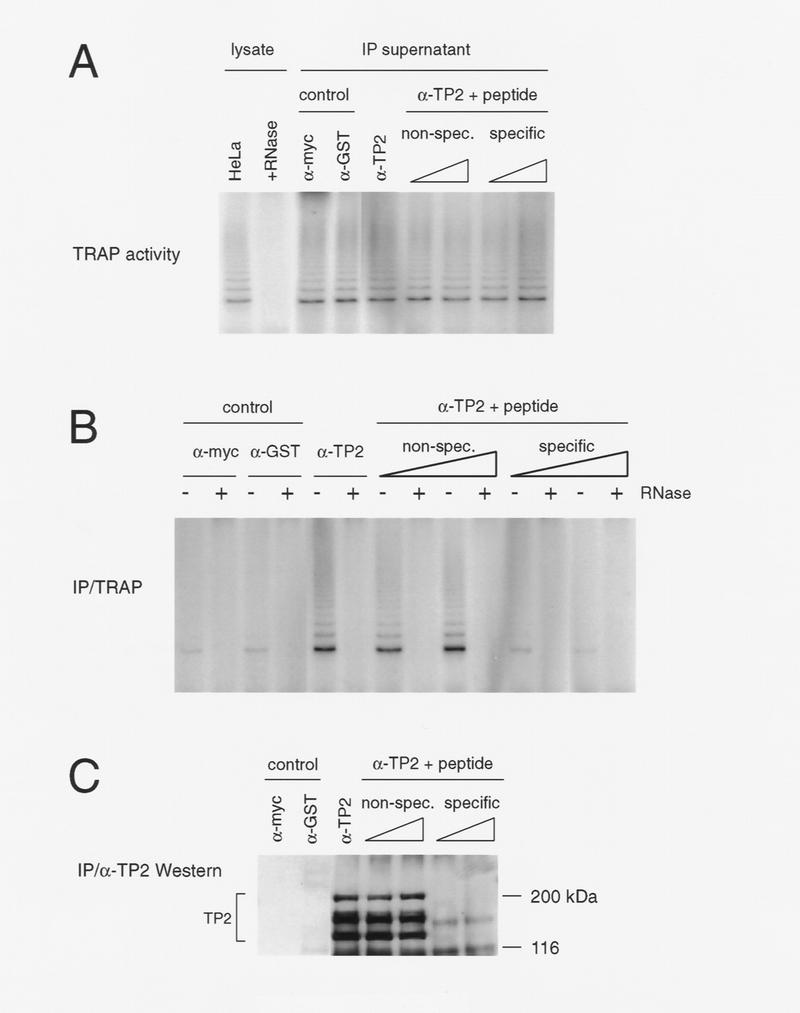
TP2 associates with telomerase activity in HeLa cells. (A) HeLa cell lysate (5 μg) (lysate) was assayed for telomerase activity by the TRAP protocol in the absence (HeLa) and presence (+RNase) of ribonuclease. Telomerase activity of supernatants after incubation (IP supernatant) with the control antibodies (α-Myc and α-GST), anti-TP2 peptide antiserum (α-TP2), and anti-TP2 peptide antiserum incubated with 30 and 60 μg each of nonspecific (non-spec.) or TP2-specific peptide (specific). Under these conditions, the TP2 antisera does not significantly immunodeplete telomerase acitivity from the supernatant. (B) The immunoprecipitates as shown in A were assayed for telomerase activity using TRAP in the absence (−) and presence (+) of RNase A. (C) The remainder of the immunoprecipitates shown in B were analyzed by Westen blotting with the anti-TP2 peptide antiserum. The position of the protein markers are indicated at right in kD.
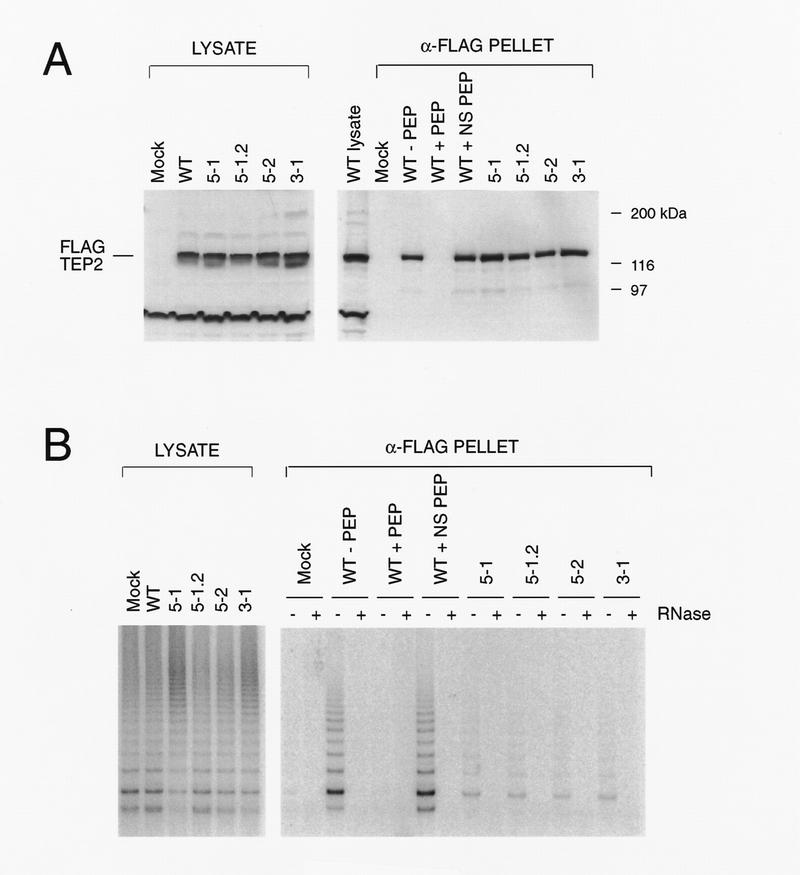
Figure 4.
Analysis of mutations in the putative catalytic residues of TP2. (A) 293 cells were transiently transfected with no plasmid (Mock), wild-type TP2 (WT), and the following TP2 point mutations in the third and fifth conserved RT motifs: 5-1, 5-1.2, 5-2, 3-1 (see Materials and Methods). Cell lysates were immunoprecipitated with anti-FLAG antibody, and the lysates (LYSATE) and immunoprecipitates (α-FLAG PELLET) were resolved by SDS-PAGE and probed with TP2 antiserum. The specificity of the FLAG immunoprecipitate for tagged TP2 was demonstrated by competiton of FLAG-tagged, wild-type TP2 as follows: WT − PEP, no peptide added; WT + PEP, preincubation with FLAG peptide; and WT + NS PEP, preincubation with a nonspecific (TP1) peptide. (B) The samples as in A were analyzed for telomerase activity. The anti-FLAG immunoprecipitates were assayed in the absence (−) and presence (+) of RNase A.
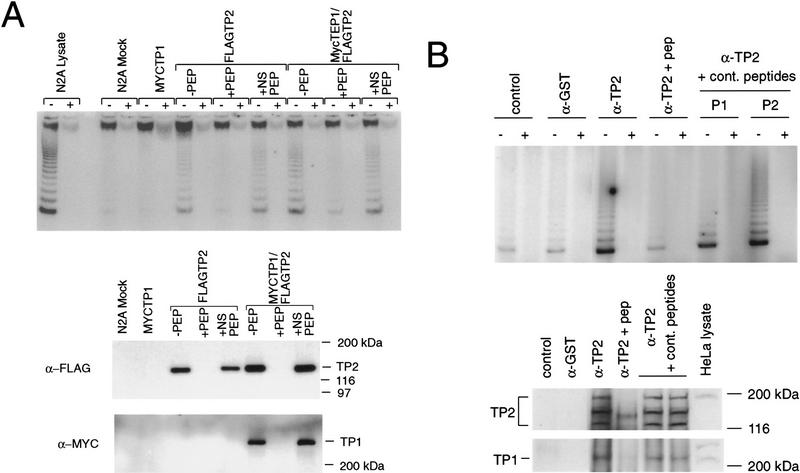
Figure 5.
TP2 interacts with TP1 in HeLa cells and transfected neuroblastoma cells. (A) Neuroblastoma cells were mock-transfected (N2A Mock) or transfected with MYC–TP1 alone, FLAG–TP2 alone, or MYC–TP1 and FLAG–TP2. Lysates plus (+) or minus (−) RNase were immunoprecipitated with anti-FLAG antibody, the precipitates were resolved by SDS-PAGE, and the Western blot was probed with anti-Myc and anti-FLAG antiserum. The peptide competitions on the FLAG immunoprecipitates were −PEP, no peptide added; +PEP, plus FLAG peptide; and +NS PEP, plus nonspecific peptide (Myc peptide). The mobility of the protein markers is shown at right. (B) Immunoprecipitations with TP2 (α-TP2) or control antiserum (rabbit α-mouse; control) and anti-GST (α-GST) were assayed for telomerase activity (top panel), Western blotting using anti-TP2 antiserum (middle), and anti-TP1 antiserum (bottom). Immunoprecipitation with anti-TP2 antiserum was also performed in the presence of TP2-specific peptide (α-TP2 + pep) and two different nonspecific peptides (to TP1; see Materials and Methods). In the bottom two panels, the samples are in the same order as above; the position of the protein markers are indicated at right, and the endogenous TP1 and TP2 proteins are indicated at left of each panel.
TP2 is important for catalytic telomerase activity
To determine the role of TP2 in the catalytic activity of telomerase, we introduced point mutations in the conserved aspartate residues also shared within EST2 and p123. Wild-type TP2 and four different TP2 point mutants (in the analogous residues introduced into EST2; see Materials and Methods) were tagged with the FLAG epitope and transiently expressed in human 293 cells. Western analysis using TP2 antisera revealed efficient and equal expression of both wild-type and mutant TP2 proteins (Fig. 4A). To differentiate the activity associated with exogenous TP2 from endogenous telomerase activity, we performed immunoprecipitations using anti-FLAG antibodies and examined them for telomerase activity. Both wild-type and mutant TP2 proteins were efficiently immunoprecipitated with the anti-FLAG antibody (Fig. 4A). The specificity of the immunoprecipitation was confirmed by preincubation of the anti-FLAG antibody with FLAG peptide, which completely blocked the immunoprecipitation of FLAG–TP2 (Fig. 4A). Nonspecific peptide did not block the association of FLAG–TP2 with the anti-FLAG antibody. Robust telomerase activity was associated with the immunoprecipitated wild-type TP2, but all four of the TP2 point mutations contained very little associated telomerase activity (Fig. 4B). These results show that as little as a single amino acid substitution within the RT domain of TP2 can impair significantly telomerase catalytic activity. Although we cannot rule out nonspecific effects of these mutations on protein conformation, the observation that four separate substitutions all significantly hinder telomerase activity suggests that TP2 contributes to the enzymatic activity of telomerase and is likely the catalytic subunit.
TP1 and TP2 associate with active telomerase
We showed previously that endogenous and epitope-tagged TP1 is associated with telomerase activity in mouse cell lysates (Harrington et al. 1997). To determine whether human TP1 and TP2 are associated, MYC–TP1 and FLAG–TP2 were transiently transfected into mouse N2A neuroblastoma cells. Lysates of single transfectants (TP1 or TP2) or double transfectants (TP1 and TP2) were immunoprecipitated using an anti-FLAG monoclonal antibody, and the immunoblots were probed with anti-MYC and anti-FLAG antisera (Fig. 5A). Immunoprecipitates from cells transfected with FLAG–TP2 contained a single FLAG–TP2 protein species at ∼130 kD, which was specifically competed by preincubation with FLAG peptide (Fig. 5A). MYC–TP1 migrated at ∼240 kD, as shown previously (Fig. 5A) (Harrington et al. 1997). Both MYC–TP1 and telomerase activity were associated with immunoprecipitates containing FLAG–TP2. The interaction of MYC–TP1 with FLAG–TP2 was specific, because FLAG peptide, but not nonspecific peptide, competed for MYC–TP1 association.
We then examined the association of endogenous TP1, TP2, and telomerase activity in HeLa cell lysates. Immunoprecipitations with TP2 antipeptide antiserum were analyzed by Western blotting and TRAP assays. TP2 protein and telomerase activity were specifically present in TP2 immunoprecipitates (Fig. 5B). TP1 was also present in the TP2 immunoprecipitates, as detected by TP1 polyclonal antiserum, but absent from the immunoprecipitates in which TP2 protein and telomerase activity were competed with TP2 peptide. RNase A treatment of TP2 immunoprecipitates did not disrupt the association between TP2 and TP1, suggesting that this interaction is telomerase RNA independent (L. Harrington, unpubl.). Thus, TP1 and TP2 interact physically in a complex that contains telomerase activity.
Discussion
We have cloned and characterized a human telomerase-associated protein that contains conserved sequence motifs characteristic of a reverse transcriptase. We have demonstrated that TP2 protein is associated with telomerase activity in human cells and that mutation of conserved residues within the RT domain severely impair telomerase activity. TP2 is present in a complex with TP1. These results represent the first functional evidence that TP2 contributes to the catalytic activity of human telomerase.
Evidence for a single telomerase complex
Unicellular ciliates have been invaluable in the identification and characterization of telomerase, owing in part to the abundance of telomerase activity in these organisms (Greider 1996). Two separate biochemical approaches in different ciliates resulted in the cloning of apparently unrelated telomerase proteins: p95 and p80 from Tetrahymena (Collins et al. 1995) and p123 and p43 from Euplotes (Lingner and Cech 1996; Hammond et al. 1997; Lingner et al. 1997). Lingner et al. (1997) postulated the existence of two different telomerases or, alternatively, that a catalytic subunit analogous to p123 was under-represented in the purified Tetrahymena telomerase. Two separate cross-linking methods were used to reveal proteins of ∼100 kD in purified Tetrahymena telomerase fractions (Collins et al. 1995; Harrington et al. 1995); because these cross-linked species were not sequenced directly, one or both techniques may have identified a low-abundance protein that was not cloned. In Euplotes, the mobility of telomerase in nondenaturing gels changed at one purification step, suggesting that other subunits may have been lost during the purification (Lingner and Cech 1996).
The amino acid sequence similiarity between the Est2, p123, and TP2/TRT1/hEST2 and the telomerase RNA-binding proteins (p80 and TP1) from different organisms suggest that the components of the telomerase complex are evolutionarily conserved. Although there are no obvious sequence homologs to p80 or p95 present in the S. cerevisiae genome, further analysis of yeast telomerase may yield functional homologs. We note that the two double-stranded telomere-binding proteins, S. cerevisiae Rap1 and human TRF, are not related in primary sequence yet contain a structurally and functionally related Myb-like DNA-binding domain (Chong et al. 1995; Konig et al. 1996; Bianchi et al. 1997). Alternatively, the function of certain telomerase-associated proteins may be dispensable in some organisms.
Potential roles for the TP proteins
On the basis of the association of TP2 with telomerase activity and the dramatic impairment in activity upon mutation of the RT domain, we suggest that TP2 is the functional equivalent of the catalytic telomerase subunits from S. cerevisiae and Euplotes. Proof that EST2 was essential for catalytic activity was demonstrated (Lendvay et al. 1996; Counter et al. 1997; Lingner et al. 1997). In mammals, further proof of the role of TP2 in telomerase function will require additional genetic and biochemical analysis. As with ciliate telomerase, extensive purification of human telomerase will yield important information regarding the stoichiometry of TP1, TP2, and telomerase RNA within the complex (Collins et al. 1995; Lingner and Cech 1996). The function of the TP1 protein in the telomerase enzyme complex is not yet clear. As the interaction of TP1 and TP2 does not appear to require the telomerase RNA component, TP1 and TP2 may interact either directly or indirectly through other telomerase-associated components. For example, the human telomerase complex may contain additional evolutionarily conserved components, such as homologs of the Euplotes 43-kD telomerase subunit or the Tetrahymena telomerase protein p95.
The identification of a human telomerase complex containing TP1, TP2, and the telomerase RNA will facilitate elucidation of the role of telomerase in telomere maintenance, cancer, and senescence. A functional reconstitution assay for human telomerase activity has been developed using partially purified telomerase extracts (Autexier et al. 1996). Thus, it may soon be possible to reconstitute telomerase, in vitro and in vivo, with cloned components. These experiments will allow a direct test of the hypothesis that telomerase reactivation plays a causal role in human cancer and senescence.
Materials and methods
Cloning and overexpression of TP2
A Smith Waterman search of the GenBank EST database using the Euplotes p123 sequence identified a human EST (AA281296) from a B-lymphocyte library that exhibited 30% identity to the p123 sequence. 32P-labeled oligonucleotides (5′-CCAAGTTCCTGCACTGGCTGAT-3′ and 5′-GCTCGTAGTTGAGCACGCTGAA-3′) were generated from the EST sequence and used as probes to identify cDNA clones from a human colon carcinoma LIM1863 cell line. Two overlapping clones were identified and sequenced to produce a contiguous DNA sequence that contained an ORF of 1132 amino acids. To produce recombinant TP2 protein, the ORF was cloned into the pCR3 expression vector (Invitrogen). The 3′ end of the TP2 ORF was tagged by the PCR-based addition of an oligonucleotide coding for the 8-amino-acid FLAG epitope (DYKDDDDK) followed by a termination codon. Point mutations in the FLAG–TP2 pCR3 plasmids were introduced by PCR and confirmed by sequencing. In each mutation, aspartate was changed to alanine, and the mutations were labeled according to their location in each respective RT motif (i.e., third and fifth motifs, also termed motifs A and C) (Counter et al. 1997; Lingner et al. 1997): 5-1, D868/A868; 5-2, D869/A869; 5-1,2, D868,D869/A868,A869; 3-1, D712/A712.
Immunofluorescence using TP2 antipeptide antiserum
HeLa cells were grown overnight on Labtek chamber slides (Nunc), washed twice with PBS, and fixed with 2% formaldehyde/PBS for 10 min at 4°C. The cells were washed three times and incubated for 10 min at 4°C. TP2 polyclonal antibody, diluted to 0.2 μg/ml in 0.3% Triton X-100/PBS, was incubated with or without 50 μg of specific (TP2) or nonspecific (P3 from TP1; see below) peptide on ice for 1.5 hr. The antibody was applied to the cells and incubated overnight at 4°C. The cells were washed three times in PBS and incubated for 10 min at 4°C. Biotinylated anti-rabbit antibody (Amersham), diluted 1:100 in 0.3% Triton-X 100/PBS, was applied for 45 min at 37°C. The cells were washed as before and then incubated with Cy2–Streptavidin (Amersham) diluted 1:100 in PBS for 45 min at 37°C. The cells were washed once with PBS, incubated with DAPI (0.2 μg/ml in PBS) for 5 min at 4°C and then washed in PBS for 10 min at 4°C. The cells were mounted in Permafluor (Fisher) and visualized with a Leica DMLB; images were captured with Northern Exposure software (Empix, Mississauga, Ontario, Canada).
Immunoprecipitation analysis of TP1 and TP2
HeLa cell extracts (S100) were prepared as described (Prowse et al. 1993). Approximately 1 mg of total lysate was incubated with TP2 antipeptide antiserum bound to protein G–Sepharose (5 μg of affinity purified antiserum bound per 10 μl of protein G beads) for 1 hr at 4°C, and the beads were washed three times with 2.3× hypobuffer containing 20% glycerol and 0.1 m NaCl (Harrington et al. 1997). For peptide competition, the peptide was incubated with TP2 antibody-bound protein G beads for 30 min at room temperature before the addition of lysate. Peptides used were the TP2-specific peptide (DEAEVRQHREARPALLTSRLRFIPKC) and nonspecific peptides P1 and P3, derived from TP1 (Harrington et al. 1997). Transfection and protein lysate preparation of mouse neuroblastoma (N2A) and 293 cells was carried out as in Harrington et al. (1997), except that SUPERFECT was used instead of Pfx-6 in the lipofection, and 29 μg of tagged TP2 plasmid DNA (FLAG–TP2, wild type, or mutants) was used to tranfect each 100-mm culture dish. Approximately 5 μl of beads bound with FLAG antibody (Kodak) was incubated with 1 mg of cell lysate transfected with MYC-tagged, full-length TP1 (MYC–TP1), FLAG-tagged TP2 (FLAG–TP2), or no plasmid (Mock) for 1 hr, and washed as described above. The lysates (5 μg of total protein) and immunoprecipitates (2 μl of protein G beads) were assayed by TRAP as described (Kim et al. 1994), and the remainder was subjected to SDS-PAGE and transferred to a PVDF membrane. The membrane was incubated with 0.1 μg/ml of anti-FLAG or anti-Myc antibody, followed by a HRP-conjugated secondary antibody and visualized by ECL. The TP1 antiserum was generated by immunization of rabbits with a denatured, fusion protein containing glutathione S-transferase (GST) and the amino-terminal 870 amino acids of TP1. The antiserum was affinity-purified against the purified GST–TP1 fusion protein bound to glutathione–Sepharose and was used at 0.2 μg/ml on Western blots. The specificity of the TP1 antiserum was confirmed by Western blot analysis using purified recombinant TP1. For the TP2 antipeptide antiserum, rabbits were immunized with the TP2 peptide corresponding to amino acids 602–626. TP2 peptide coupled to Affigel was used to affinity purify the antiserum and was used at 0.2 μg/ml on Western blots. The specificity of the TP2 antiserum was confirmed by Western blot analysis using recombinant TP2 and epitope-tagged, immunoprecipitated TP2 from cell lysates.
Acknowledgments
We thank I. Arruda, A. Hessel, and L. Ulias for technical and computation assistance; C. Cole, A. Colombero, M. Rihanek, H. Yamane, and T. Zamborelli for reagents; and B. Bosselman, C. Craig, N. Oleski, T. Mak, L. Souza, and M. Tyers for helpful advice. This work was supported in part by a grant from the Medical Research Council of Canada to L.H.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Leah@amgen.com; mrobinso@amgen.com; FAX (416) 204-2278.
References
- Autexier A, Pruzan R, Funk WD, Greider CW. Reconstitution of human telomerase activity and identification of a minimal functional region of the human telomerase RNA. EMBO J. 1996;15:5928–5935. [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya A, Blackburn EH. Architecture of telomerase RNA. EMBO J. 1994;13:5721–5731. doi: 10.1002/j.1460-2075.1994.tb06910.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco MA, Funk W, Villeponteau B, Greider CW. Functional characterization and developmental regulation of mouse telomerase RNA. Science. 1995;269:1267–1270. doi: 10.1126/science.7544492. [DOI] [PubMed] [Google Scholar]
- Bryan TM, Reddel RR. Telomere dynamics and telomerase activity in in vitro immortalized cell lines. Eur J Canc. 1997;33:767–773. doi: 10.1016/S0959-8049(97)00065-8. [DOI] [PubMed] [Google Scholar]
- Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- Collins K, Kobayashi R, Greider CW. Purification of Tetrahymena telomerase and cloning of genes encoding two protein components of the enzyme. Cell. 1995;81:677–686. doi: 10.1016/0092-8674(95)90529-4. [DOI] [PubMed] [Google Scholar]
- Counter CM, Gupta J, Harley CB, Leber B, Bacchetti S. Telomerase activity in normal leukocytes and in hematologic malignancies. Blood. 1995;85:2315–2320. [PubMed] [Google Scholar]
- Counter CM, Meyerson M, Eaton EN, Weinberg RA. The catalytic subunit of yeast telomerase. Proc Natl Acad Sci. 1997;94:9202–9207. doi: 10.1073/pnas.94.17.9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Funk WD, Wang SS, Weinrich SL, Avilion AA, Chiu C-P, Adams RR, Chang E, Allsopp RC, Yu J, et al. The RNA component of human telomerase. Science. 1995;269:1236–1241. doi: 10.1126/science.7544491. [DOI] [PubMed] [Google Scholar]
- Greider CW. Telomere length regulation. Annu Rev Biochem. 1996;65:337–365. doi: 10.1146/annurev.bi.65.070196.002005. [DOI] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. A telomeric sequence in the RNA of Tetrahymena telomerase required for telomere repeat synthesis. Nature. 1989;337:331–337. doi: 10.1038/337331a0. [DOI] [PubMed] [Google Scholar]
- Hammond P, Lively T, Cech TR. The anchor site of telomerase from Euplotes aediculatus revealed by photo-cross-linking to single- and double-stranded DNA primers. Mol Cell Biol. 1997;17:296–308. doi: 10.1128/mcb.17.1.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CB. Telomeres and aging. In: Blackburn EH, Greider CW, editors. Telomeres. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 247–263. [Google Scholar]
- Harrington L, Hull C, Crittenden J, Greider C. Gel shift and UV cross-linking analysis of Tetrahymena telomerase. J Biol Chem. 1995;270:8893–8901. doi: 10.1074/jbc.270.15.8893. [DOI] [PubMed] [Google Scholar]
- Harrington L, McPhail T, Mar V, Zhou W, Oulton R, Bass MB, Arruda I, Robinson MO. A mammalian telomerase-associated protein. Science. 1997;275:973–977. doi: 10.1126/science.275.5302.973. [DOI] [PubMed] [Google Scholar]
- Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific association of human telomerase activity with immortal cells and cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- Konig P, Giraldo R, Chapman L, Rhodes D. The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA. Cell. 1996;85:125–136. doi: 10.1016/s0092-8674(00)81088-0. [DOI] [PubMed] [Google Scholar]
- Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Cech TR. Purification of telomerase from Euplotes aediculatus: Requirement of a primer 3′ overhang. Proc Natl Acad Sci. 1996;93:10712–10717. doi: 10.1073/pnas.93.20.10712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner J, Hendrick LL, Cech TR. Telomerase RNAs of different ciliates have a common secondary structure and a permuted template. Genes & Dev. 1994;8:1984–1998. doi: 10.1101/gad.8.16.1984. [DOI] [PubMed] [Google Scholar]
- Lingner J, Hughes TR, Shevchenko A, Mann M, Lundblad V, Cech TR. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science. 1997;276:561–567. doi: 10.1126/science.276.5312.561. [DOI] [PubMed] [Google Scholar]
- Lustig A. The identification of telomerase subunits: Catalysing telomere research. Trends Cell Biol. 1997;7:299–302. doi: 10.1016/S0962-8924(97)01110-0. [DOI] [PubMed] [Google Scholar]
- McCormick-Graham M, Romero DP. Ciliate telomerase RNA structural features. Nucleic Acids Res. 1995;23:1091–1097. doi: 10.1093/nar/23.7.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A single telomerase RNA is sufficient for the synthesis of variable telomeric DNA repeats in ciliates of the genus Paramecium. Mol Cell Biol. 1996;16:1871–1879. doi: 10.1128/mcb.16.4.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEachern MJ, Blackburn EH. Runaway telomere elongation caused by telomerase RNA gene mutations. Nature. 1995;376:403–409. doi: 10.1038/376403a0. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Counter CM, Eaton EN, Ellisen LW, Steiner P, Caddle SD, Ziaugra L, Beijersbergen RL, Davidoff MJ, Liu Q, et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell. 1997;90:785–795. doi: 10.1016/s0092-8674(00)80538-3. [DOI] [PubMed] [Google Scholar]
- Morin GB. The human telomere terminal transferase enzyme is a ribonucleoprotein that synthesizes TTAGGG repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- Nakamura TM, Morin GB, Chapman KB, Weinrich SL, Andrews WH, Lingner J, Harley CB, Cech TR. Telomerase catalytic subunit homologs from fission yeast and human. Science. 1997;277:955–959. doi: 10.1126/science.277.5328.955. [DOI] [PubMed] [Google Scholar]
- Nakayama J, Saito M, Nakamura H, Matsuura A, Ishikawa F. TLP1: A gene encoding a protein component of mammalian telomerase is a novel member of WD repeats family. Cell. 1997;88:875–884. doi: 10.1016/s0092-8674(00)81933-9. [DOI] [PubMed] [Google Scholar]
- Prowse KR, Avilion AA, Greider CW. Identification of a nonprocessive telomerase activity from mouse cells. Proc Natl Acad Sci. 1993;90:1493–1497. doi: 10.1073/pnas.90.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero DP, Blackburn EH. A conserved secondary structure for telomerase RNA. Cell. 1991;67:343–353. doi: 10.1016/0092-8674(91)90186-3. [DOI] [PubMed] [Google Scholar]
- Shippen-Lentz D, Blackburn EH. Functional evidence for an RNA template in telomerase. Science. 1990;247:546–552. doi: 10.1126/science.1689074. [DOI] [PubMed] [Google Scholar]
- Shore D. Telomerase and telomere-binding proteins: Controlling the endgame. Trends Biochem Sci. 1997;22:233–235. doi: 10.1016/s0968-0004(97)01082-7. [DOI] [PubMed] [Google Scholar]
- Singer M, Gottschling DE. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science. 1995;266:404–408. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- ten Dam E, van Belkum A, Pleij K. A conserved pseudo-knot in telomerase RNA. Nucleic Acids Res. 1991;19:6951. doi: 10.1093/nar/19.24.6951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Schachter F, Uchida I, Wei L, Zhu X, Effros R, Cohen D, Harley CB. Loss of telomeric DNA during aging of normal and trisomy 21 human lymphocytes. Am J Hum Genet. 1993;52:661–667. [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dragowski W, Allsopp RC, Thomas TD, Harley CB, Lansdorp PM. Evidence for a mitotic clock in human hematopoietic stem cells: Loss of telomeric DNA with age. Proc Natl Acad Sci. 1994;91:9857–9860. doi: 10.1073/pnas.91.21.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]