Abstract
Promoter–proximal pausing during transcriptional elongation is an important way of regulating many diverse loci, including the human hsp70 gene. Pausing of RNA polymerase can be enhanced by chromatin structure. We demonstrate that activation of hsp70 leads to disruption of transcribed chromatin in front of RNA polymerase. In vivo, disruption of chromatin in the first 400 bp of the transcribed region of hsp70 following heat shock is resistant to the transcriptional inhibitor α-amanitin. Disruption of chromatin farther downstream also occurs following activation but is sensitive to α-amanitin, suggesting that polymerase movement is needed to disrupt distal portions of the hsp70 gene. In vitro, disruption of transcribed chromatin is dependent on the presence of the human heat shock factor 1 (HSF1) activation domains. These experiments demonstrate that HSF1 can direct disruption of chromatin in transcribed regions. We suggest that this is one of the mechanisms used by HSF1 to facilitate transcriptional elongation.
Keywords: Chromatin, elongation, transcription, pausing, heat shock
Many eukaryotic genes are regulated at the level of transcriptional elongation (for review, see Spencer and Groudine 1990). One of the best understood examples of eukaryotic elongational regulation is the hsp70 heat shock response gene. Both human and Drosophila hsp70 genes exhibit a regulatory block to transcription near their 5′ ends. On the uninduced gene, a paused, transcriptionally engaged RNA polymerase ternary complex is present over a narrow region centered at +21–35 in Drosophila (Rougvie and Lis 1988; Giardina et al. 1992; Rasmussen and Lis 1993) and at +45 in humans (Brown et al. 1996). In response to heat shock, not only does the rate of initiation increase but the transit time of polymerase through the pause is drastically reduced.
Several recent studies have begun to address the mechanism by which transcriptional pausing might be regulated, both at the hsp70 gene and at other loci. These studies show that certain transcriptional activators can release pausing, that general transcription factors are likely to be involved in forming and regulating the pause, that pausing can be recreated in vitro in the apparent absence of chromatin formation, that nucleosomes enhance pausing in vitro, and that regulation of pausing can be observed in vitro on nucleosomal templates (Izban and Luse 1991; Yankulov et al. 1994; Krumm et al. 1995; Blau et al. 1996; Brown et al. 1996; Li et al. 1996). Although these studies have been done with diverse promoters and in diverse experimental systems, they are all consistent with a single model for regulation of promoter–proximal transcriptional pausing: General transcription factors and certain activators promote initiation of transcription and formation of a pause, and chromatin structure lengthens the duration of the pause. Release of the pause in this model requires specific activators to alter general factor function and also to enhance the ability of RNA polymerase to transcribe through chromatin.
In this context activators might enhance elongation through chromatin in several ways. In addition to altering interactions between RNA polymerase and general transcription factors, they might direct a modification that allows RNA polymerase to traverse chromatin. Activators might also direct the remodeling of downstream chromatin to eliminate its inhibition of transcriptional elongation. In this study we show that the transcriptional activator heat shock factor 1 (HSF1) can alter chromatin in a manner that supports the latter hypothesis.
Results
Proximal downstream chromatin on the hsp70 gene is disrupted in the absence of new transcription in vivo
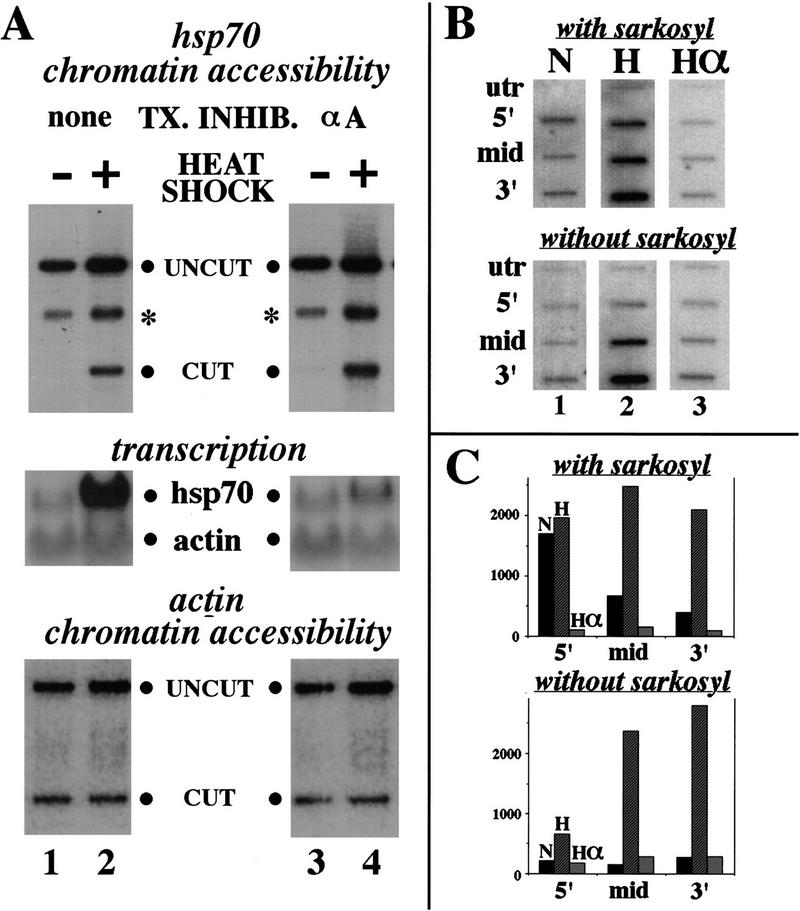
To look at in vivo changes in the chromatin accessibility of transcribed human hsp70 regions upon gene induction, we digested nuclei from normal or heat-shocked HeLa cells with the restriction enzyme BamHI, which cuts at +154 relative to the start of transcription. Restriction enzyme accessibility has been used previously in numerous systems to probe chromatin structure (e.g., Morse 1989). Cleavage products were visualized relative to a second, complete restriction cut in vitro by using ligation-mediated PCR (Mueller and Wold 1989). Although little or no accessibility to restriction enzyme was seen in uninduced cells, cutting was greatly increased in heat-shocked cells (Fig. 1A, top panel, lanes 1,2). In contrast, cutting in the promoter–proximal downstream region of the β-actin gene was identical in both cases (Fig. 1A, bottom panel).
Figure 1.
Restriction enzyme accessibility and transcription of the hsp70 gene in vivo. (A) Duplicate plates of HeLa cells were treated for 1 hr with 50 μg/ml of α-Amanitin (lanes 3,4) or left untreated (lanes 1,2). One plate of each set was then heat-shocked for 1 hr at 43°C (lanes 2,4), or left at 37°C (lanes 1,3). Northern blots were probed with fragments of the hsp70 and β-actin-transcribed regions (middle). Nuclei were incubated with either BamHI or PstI; DNA was then deproteinized and cleaved with SacII for normalization purposes, and cleavages were visualized by ligation-mediated PCR with primers specific for hsp70 (top) or actin (bottom). (B) Nuclear run-on analysis. HeLa cells were treated as above with amanitin and then heat-shocked (lane 3), heat-shocked for 1 hr without prior amanitin treatment (lane 2), or left untreated at 37°C (lane 1). Nuclei were harvested and used in nuclear run-on analysis in the presence (top) or absence (bottom) of 0.6% Sarkosyl. Probes contain untranscribed hsp70 sequences from −150 to −6 (utr); from −150 to +154 (5′); from +228 to +570 (mid), and from +570 to +1263 (3′). (C) Signals from B were quantitated and normalized for the number of thymidines in the coding strand of each probe. Probe utr has 19, probe 5′ has 33, probe mid has 47, and probe 3′ has 103.
Remodeling at +154 might be caused by RNA polymerase movement through this site or might precede RNA polymerase movement. To address this issue, the same experiment was repeated after pretreating the cells with α-amanitin, a transcriptional inhibitor that blocks or inordinately slows RNA polymerase II (Pol II) elongation. Although Northern blots showed that new full-length transcripts were virtually eliminated under these conditions (Fig. 1A, middle panel), the increase in restriction enzyme accessibility at +154 was unchanged (Fig. 1A, lanes 3,4). (Actin transcript signal, shown for normalization purposes in the Northern analysis, does not decrease significantly because of the stability of the actin message.)
To show more thoroughly that amanitin inhibited transcription successfully, nuclear run-on assays were done on the human hsp70 gene (Fig. 1B, quantitated in C). Run-on transcripts from the uninduced gene were significantly greater in the +1 to +150 region than in the +229 to +575 or later regions, demonstrating the presence of paused polymerases near the promoter. As expected from studies on the Drosophila hsp70 promoter (Rougvie and Lis 1988), when Sarkosyl was eliminated from the run-on transcription buffer, much less promoter–proximal transcription was observed, suggesting that Sarkosyl removes an inhibitor or block to RNA polymerase movement in this region. Heat shock resulted in large increases in run-on transcripts across the gene, but when cells were incubated with amanitin prior to heat shock, decreased rather than increased run-on transcripts were observed in the presence of Sarkosyl in all regions probed. This observation suggests either that amanitin can partially disrupt paused polymerases on hsp70, an observation consistent with our previous work (Brown et al. 1996), or that residual amanitin still inhibited elongation after cells were washed and nuclei were isolated. Regardless, amanitin inhibited transcription effectively in the promoter–proximal region in this system.
A further indication that amanitin treatment is effective is provided by the kinetics of downstream chromatin disruption: In both the presence and the absence of amanitin, this disruption is complete by 4 min, the shortest accurate time point possible given the time required to harvest nuclei (data not shown). Amanitin is thought to block or slow polymerase movement at least 100-fold (Rudd and Luse 1996), and polymerase movement on the Drosophila hsp70 gene has been estimated at 1200 bp/min (O’Brien and Lis 1993). Hence, in the 4 min required for disruption, polymerase might move 48 bp at most. This consideration butresses the demonstration above that polymerase movement in the promoter–proximal portion of the gene is blocked by amanitin and indicates further that downstream chromatin rearrangement is not caused by a passing RNA polymerase molecule.
Disruption of distal downstream regions depends on polymerase movement
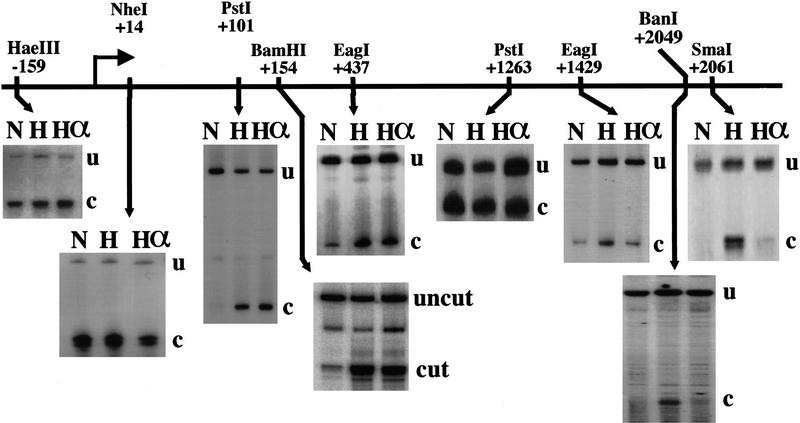
To determine whether the entire hsp70 transcribed region was similarly disrupted, the restriction enzyme accessibility assay described above was employed using different enzymes cutting across the coding region of the hsp70 gene (Fig. 2). Promoter–proximal sites from +101 to +437 all showed amanitin-insensitive accessibility upon heat shock. As expected, sites upstream of the paused polymerase at +14 and at −159 showed constitutive accessibility. Interestingly, a site at +1263 showed increased accessiblity on the uninduced gene that was only mildly augmented by heat shock. Sites still farther downstream (+1429, +2049, +2061) again showed heat shock–induced accessibility, but this accessibility was eliminated by amanitin. Altogether, these data demonstrate that although disruption of chromatin in far downstream regions of hsp70 is transcription dependent, disruption across a broad promoter–proximal region is not.
Figure 2.
Chromatin accessibility across the hsp70 gene. HeLa cells were treated at 37°C for 1 hr with 50 μg/ml of amanitin, and then heat-shocked for 1 hr (lanes Hα), heat-shocked for 1 hr without prior amanitin treatment (lanes H), or left untreated at 37°C (lanes N). Nuclei were harvested and digested for 1 hr with the restriction enzyme indicated. DNA was isolated and then digested a second time in vitro with another enzyme cutting nearby for normalization purposes. Cleavages were visualized by ligation-mediated PCR.
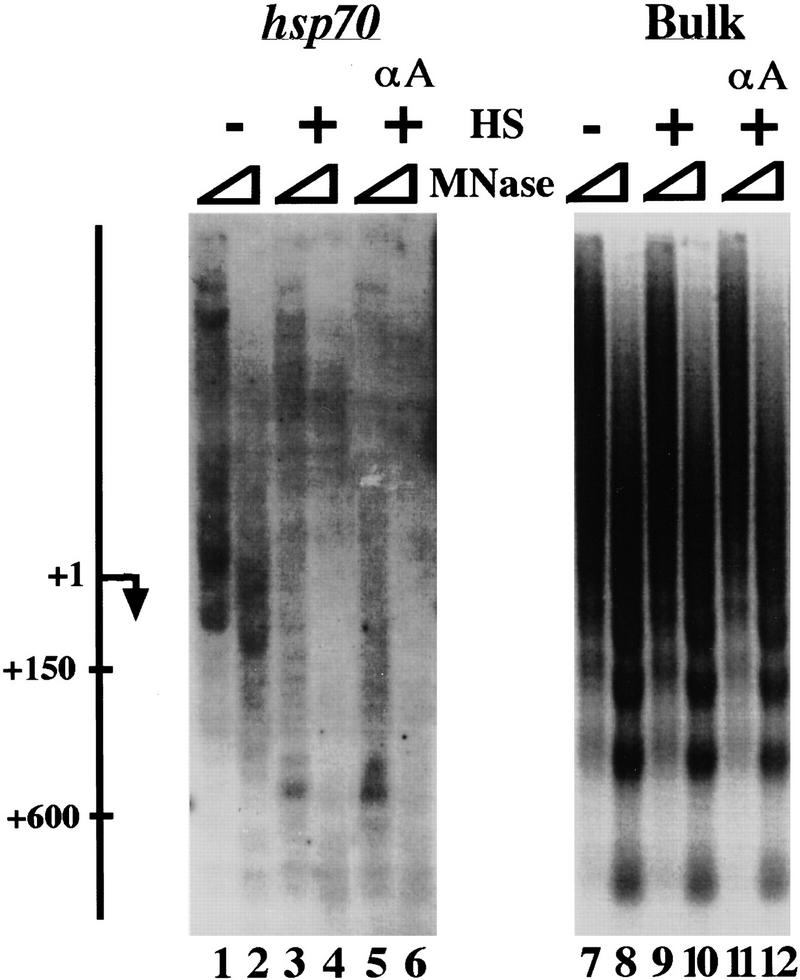
This region can also be seen by micrococcal nuclease (MNase) accessibility (as visualized by indirect end labeling). Nucleosomes are present in this region of the uninduced hsp70 gene (Brown et al. 1996); however, indirect end-labeling shows that these nucleosomes are not precisely positioned, as judged by a general decrease in nuclease sensitivity relative to the promoter region rather than specific banding in the transcribed region (Fig. 3, lanes 1,2). An increase in accessibility of transcribed and promoter regions to MNase was observed upon heat shock (lanes 3,4). This increase extended as far as the probe at +575–930, and was independent of prior amanitin treatment (lanes 5,6). Bulk DNA was equivalently digested by MNase in each of these samples (lanes 7–12), demonstrating the specificity of the changes in nuclease sensitivity of the hsp70 gene. We conclude that significant RNA polymerase movement is not required for rearrangement of chromatin in promoter–proximal areas of the hsp70-transcribed region.
Figure 3.
MNase analysis of hsp70 chromatin structure. HeLa cells were treated at 37°C for 1 hr with 50 μg/ml of amanitin and then heat-shocked for 1 hr (lanes 5,6,11,12), heat-shocked for 1 hr without prior amanitin treatment (lanes 3,4,9,10), or left untreated at 37°C (lanes 1,2,7,8). Nuclei were harvested and digested with 12 units (lanes 1,3,5,7,9,11) or 36 units (lanes 2,4,6,8,1,12) of MNase. DNA was isolated and then digested in vitro with XmnI. Bulk ethidium bromide-stained DNA is shown in lanes 7–12; hsp70 DNA, as visualized by indirect end-labeling, is shown in lanes 1–6. Approximate location markers were generated by following the above procedure with restriction-cut rather than with MNase-cut DNA.
In vitro remodeling is dependent on the HSF1 transcriptional activation domains
To more thoroughly examine the transcription-independent remodeling observed on the hsp70 gene in vivo, we employed an in vitro system that mimics polymerase pausing and release on hsp70 to investigate the same phenomenon (Brown et al. 1996). RNA polymerase was artifically stalled at +15 on a modified human hsp70 template—mutated in 4 bases to make the first 15 bases of the transcript lack guanosine—by initiating transcription in the absence of guanosine triphosphate. This template was assembled into nucleosomes, and then RNA polymerase was permitted to continue elongation by adding all 4 nucleotides. The template is bound to polystyrene beads, which allowed us to wash the template as needed in the following experiments. We used this system to determine whether activator increased the accessibility of BamHI to a site at +154 prior to addition of nucleotides and thus prior to any movement of RNA polymerase.
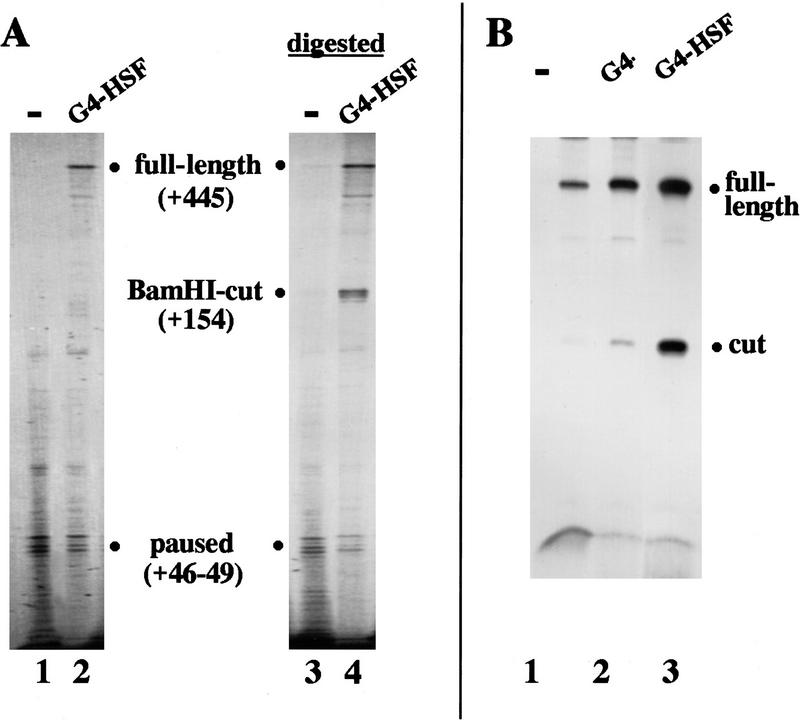
As seen previously in this system (Brown et al. 1996), in the absence of the HSF1 activation domains, polymerase paused stably at +46–49 in nucleosome-dependent fashion, but in the presence of activator full-length transcript was also made (Fig. 4A, lanes 1,2). If BamHI restriction enzyme (cutting at +154) was added to reactions prior to the addition of all 4 nucleotides, but after activator addition, and then washed away, subsequent transcription resulted in both full-length and shorter readthrough transcripts (lane 4), demonstrating that downstream chromatin was remodeled in the absence of transcription, as seen in vivo. Although reactions in which no activator was present demonstrated no transcription past +46–49, and hence did not indicate whether cutting at +154 had occurred (lane 3), nucleosomes could be stripped from the templates with 1% Sarkosyl after cutting with restriction enzyme. Following Sarkosyl treatment, addition of all 4 nucleotides leads to permissive elongation, and the fraction of transcribed templates that are cleaved in the absence and presence of activator can be directly examined by comparing the amount of full-length and truncated transcript. Although very few truncated transcripts were seen in the absence of activator (Fig. 4B, lane 1) or in the presence of the GAL4 DNA-binding domain alone (lane 2), one-third of the transcripts were truncated in the presence of GAL4–HSF1 (lane 3), showing that downstream remodeling is activator dependent.
Figure 4.
Restriction enzyme accessibility and transcription of the hsp70 gene in vitro. (A) In the absence (lane 1,3) or presence (lane 2,4) of GAL4–HSF1, transcription complexes were stalled at +15 on human hsp70 template pSAB8 (containing a short G-less region from +1 to +15). The template was assembled into nucleosomes and treated with restriction enzyme BamHI (lanes 3,4) or left untreated (lanes 1,2). Elongation was then continued, and transcripts were examined by gel electrophoresis. (B) The same protocol of artificially stalling RNA polymerase complexes, assembling nucleosomes, and digesting template with restriction enzyme was carried out. Nucleosomes were then removed by washing templates with 1% Sarkosyl, and elongation was permitted to continue. These reactions were carried out either in the absence of activator (lane 1), in the presence of the GAL4 DNA-binding domain (lane 2), or in the presence of GAL4–HSF1 (lane 3). In four experiments, cutting of transcribed templates in the presence of the GAL4 DNA-binding domain varied from 10%–14%, and cutting in the presence of GAL4–HSF varied from 30%–42%.
Discussion
Previous experiments have shown that remodeling of chromatin near promoter and enhancer regions accompanies transcriptional activation in both yeast and mammalian systems (e.g., Hirschhorn et al. 1992; Axelrod et al. 1993; Mymryk and Archer 1995; Gerber et al. 1997). Remodeling of transcribed regions could easily be a consequence of elongation of RNA polymerase through chromatin following activation, rather than a causal phenomenon that precedes elongation. We show here that the HSF1 activator directs remodeling of chromatin in front of RNA polymerase on the human hsp70 promoter. This conclusion is based on both in vivo studies, where remodeling occurs over a broad transcribed region even in the presence of the drug amanitin, and in vitro studies, where we are able to observe activation domain-directed remodeling before nucleotides are added to allow elongation.
We propose that the ability of HSF1 to direct remodeling of the transcribed region of hsp70 is an important component of the mechanisms used by HSF1 to stimulate transcriptional elongation. Others have suggested that RNA polymerase may initially pause on hsp70 because of interactions with general transcription factors (Lis and Wu 1993; Purnell et al. 1994), and our own previous work suggests that a nucleosome can greatly enhance this pause. HSF1 was required for transcriptional elongation past this chromatin-enhanced pause site in vitro (Brown et al. 1996). Hence, relief of the pause might require both an appropriate signal from the activator to the general machinery and a remodeling of the blocking nucleosomal structure.
To remodel nucleosomal DNA, HSF1 might direct acetylases to post-translationally modify histones. Alternatively, HSF1 might recruit ATP-dependent nucleosome-disrupting complexes like SWI/SNF, NURF, RSC, CHRAC, or ACF. Indeed, SWI/SNF-containing fractions have already been shown to augment readthrough of pausing on hsp70 in an activator-dependent fashion (Brown et al. 1996) and NURF has been shown to be capable of remodeling the Drosophila hsp70 promoter in vitro (Tsukiyama and Wu 1995). Both of these complexes are present in the crude transcription factor fractions used to initiate transcription in the experiments of Figure 4 (data not shown), and fractions enriched in SWI/SNF were present during elongation as well. Definitive analysis of whether these or other factors are recruited by HSF1 to perform downstream remodeling on the hsp70 gene, however, will require an in vitro system that is sufficiently defined that it can be depleted of these activities, allowing analysis of the effects of readdition of individual complexes.
Although the results above imply that extensive RNA polymerase movement is not required to remodel promoter–proximal transcribed regions, it is still possible that an activator-directed remodeling event might require the presence of RNA polymerase on the gene. It is possible, for example, that activators might signal RNA polymerase to enter an elongation-competent configuration that might activate an associated remodeling activity. Alternatively, activators could signal directly to the remodeling activity.
Our results also show a fundamental difference between promoter–proximal and promoter–distal remodeling of the transcribed region of the hsp70 gene. Although the former requires extensive movement by RNA polymerase, the latter does not. Multiple in vitro studies have shown differences in elongation capability between just-started RNA polymerase complexes and more distal ones (see Dvir et al. 1996 and references therein; Jiang et al. 1996). It is possible that once clear of promoter contacts, RNA Pol II is able to move independently through nucleosomes in a fashion similar to that demonstrated by SP6 polymerase (Clark and Felsenfeld 1992; Studitsky et al. 1994), thereby directly altering chromatin structure. It is also possible that nucleosome-remodeling complexes might move with the polymerase, altering nucleosomes to help it traverse them, as SWI/SNF can be a component of the yeast Pol II holoenzyme (Wilson et al. 1996).
Overall, these studies begin to characterize broader roles for both activators and chromatin than defined previously. Not only can activators remodel promoter architecture and/or directly recruit components of the basal transcription machinery, but they can also direct downstream remodeling to perhaps affect later steps in the transcription process.
Materials and methods
Restriction enzyme accessibility assays
HeLa cells were treated as described in the legends, then trypsinized and lysed in buffer L (5 mm PIPES at pH 8, 85 mm KCl, 1 mm CaCl2, 5% sucrose, 0.5% NP-40). Nuclei were resuspended in buffer M (15 mm Tris at pH 7.5, 15 mm NaCl, 60 mm KCl, 5 mm 2-mercaptoethanol, 1 mm CaCl2, 3 mm MgCl2, 0.34 m sucrose) at a concentration of 1 mg/ml as measured by A260. Restriction digestions were carried out in reactions containing 50 μl nuclei, 150 units of the indicated restriction enzyme (New England Biolabs), and 1× recommended NEB buffer. Reactions were incubated 1.5 hr at 37°C, and then RNase-treated and deproteinized. DNA was digested again with a second restriction enzyme as an internal reference, and all overhanging ends were filled in with Klenow. Ligation-mediated PCR was then performed as described previously (Brown et al. 1996), except that the first Sequenase extension step was skipped. To visualize cleavages on hsp70, the following primer sets were used: For sites at +68, +101, +154, and +228, the first primer was −73 to −46 and the second was −65 to −38; for sites at −158 and +14, primer 1 was +154 to +129 and primer 2 was +147 to +121; for the site +437, primer 1 was +550 to +525 and primer 2 was +540 to +513; for sites at +1263 and +1429, primer 1 was +1089 to +1114 and primer 2 was +1097 to +1024; for sites at +2049 and +2061, primer 1 was +2140 to +2113 and primer 2 was +2131 to +2103. For actin, primer 1 was +886 to 862 and primer 2 was +875 to +852.
Northern and Southern blots
RNA was made from HeLa cells according to the protocol of Xie and Rothblum (1991). Twenty micrograms per lane was loaded onto a 1% agarose/formaldehyde gel (Ausubel et al. 1995) and blotted to GeneScreen (New England Nuclear). Blots were cross-linked and probed according to the directions of the membrane manufacturer. Hybridization was carried out in the presence of two probes: 4 ng of a fragment containing the β-actin coding region (a gift of B. Seed, Massachusetts General Hospital), and 4 ng of a BamHI–EcoRI fragment containing the hsp70 coding region. For Southern blots, blotting was performed according to the protocol described in Brown et al. (1996), except that 300 μg of total nucleic acid at 1 mg/ml was used per MNase reaction (as measured by A260), and the genomic DNA used was treated with XmnI restriction enzyme (New England Biolabs) prior to electrophoresis. The resultant filter was probed with a XmnI–BglII restriction fragment from the human hsp70-transcribed region (+575–930). Probes were labeled using a Boehringer Mannheim Random Priming Kit.
Nuclear run-on assays
Run-on assays were performed exactly as described in Ausubel et al. (1995) except that each slot of the hybridization filter contained 12.5 μg of linearized whole-plasmid DNA. Probes were the hsp70 fragments BamHI–RsrII (−150 to −6), BamHI (−150 to +154), SacII–BglII (+228 to +570), and BglII–PstI (+570 to +1263).
Proteins and extracts used
The GAL4 DNA-binding domain (amino acids 1–94) was 17 μm in concentration of dimer active for DNA binding and was 80% pure and 80% active relative to total protein. The GAL4–HSF protein contains amino acids 1–147 of the GAL4 DNA-binding domain fused to the activation domains of human HSF1. It was 1.5 μm in concentration of active dimer as measured by DNA binding and was 90% pure and 80% active. Both proteins were dialyzed into buffer D (100 mm KCl, 20% glycerol, 20 mm HEPES at pH 7.9, 0.2 mm EDTA, 0.5 mm DTT, 0.2 mm PMSF). Activators, transcription factors, Xenopus heat-treated assembly extract (dHTE), histones, and hSWI/SNF protein were purified and used as described previously (Brown et al. 1996).
In vitro transcription/accessibility assay
Reactions were begun by prebinding activators to their cognate DNA sites: We incubated 0.5 μg of bead-bound pSAB12 template (Brown et al. 1996) for 15 min at room temperature in a reaction including a 1.2-fold molar excess of activator relative to DNA-binding sites, or 1.4 pmole. As described previously in Brown et al. (1996), transcription was initiated on these templates, initiation factors and nucleotides were washed away, and nucleosomes were assembled using desalted, heat-treated Xenopus assembly extract. Reactions were resuspended in a 23-μl reaction including activator, 7 mm MgCl2, 4 mm ATP, 20 units of RNAsin (Promega), 15 μl of total of buffer D (modified to contain enough KCl that the concentration in the final reaction is 150 mm) plus 3-μl hSWI/SNF fraction. Reactions were incubated for 20 mins at 30°C and either elongated directly (see below) or supplemented with 20 units of the appropriate restriction enzyme (from New England Biochemical) in 2 μl, digested for 1 hr at 30°C, and stripped and washed, or simply washed. Stripped and washed reactions were washed once with 0.6× buffer D plus 1% Sarkosyl and 1 mm MgCl2 (a treatment that removes nucleosomes) and once with 0.6× buffer D plus 2 mm MgCl2. Washed reactions were subjected to only the latter step. Elongation was then continued, and reactions were analyzed electrophoretically as described previously (Brown et al. 1996).
Acknowledgments
This work was supported by grants to R.E.K. from the National Institutes of Health and from Hoechst A.G. We thank F. Winston, M. Gilman, L. Corey, and G. Schnitzler for their comments on this manuscript, A. Imbalzano for a gift of GAL4(1–94) protein, and G. Schnitzler and S. Sif for gifts of SWI/SNF fractions used in these studies.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kingston@frodo.mgh.harvard.edu; FAX (617) 726-5949.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current protocols in molecular biology. New York, NY: Wiley and Sons; 1995. [Google Scholar]
- Axelrod JD, Reagan MS, Majors J. GAL4 disrupts a repressing nucleosome during activation of GAL1 transcription in vivo. Genes & Dev. 1993;7:857–869. doi: 10.1101/gad.7.5.857. [DOI] [PubMed] [Google Scholar]
- Blau J, Xiao H, McCracken S, O’Hare P, Greenblatt J, Bentley D. Three functional classes of transcriptional activation domains. Mol Cell Biol. 1996;16:2044–2055. doi: 10.1128/mcb.16.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown SA, Imbalzano AN, Kingston RE. Activator-dependent regulation of transcriptional pausing on nucleosomal templates. Genes & Dev. 1996;10:1479–1490. doi: 10.1101/gad.10.12.1479. [DOI] [PubMed] [Google Scholar]
- Clark DJ, Felsenfeld G. A nucleosome core is transferred out of the path of a transcribing polymerase. Cell. 1992;71:11–22. doi: 10.1016/0092-8674(92)90262-b. [DOI] [PubMed] [Google Scholar]
- Dvir A, Conaway RC, Conaway JW. Promoter escape by RNA polymerase II: A role for an ATP cofactor in suppression of arrest by polymerase at promoter-proximal sites. J Biol Chem. 1996;271:23352–23356. doi: 10.1074/jbc.271.38.23352. [DOI] [PubMed] [Google Scholar]
- Gerber AN, Klesert TR, Bergstrom DA, Tapscott SJ. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: A mechanism for lineage determination in myogenesis. Genes & Dev. 1997;11:436–450. doi: 10.1101/gad.11.4.436. [DOI] [PubMed] [Google Scholar]
- Giardina C, Perez-Riba M, Lis JT. Promoter melting and TFIID complexes on Drosophila genes in vivo. Genes & Dev. 1992;6:2190–2200. doi: 10.1101/gad.6.11.2190. [DOI] [PubMed] [Google Scholar]
- Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes & Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: Inhibition of elongation with enhancement of sequence-specific pausing. Genes & Dev. 1991;5:683–696. doi: 10.1101/gad.5.4.683. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Yan M, Gralla JD. A three-step pathway of transcription initiation leading to promoter clearance at an activated RNA polymerase II promoter. Mol Cell Biol. 1996;16:1614–1621. doi: 10.1128/mcb.16.4.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm A, Hickey LB, Groudine M. Promoter-proximal pausing of RNA polymerase II defines a general rate-limiting step after transcription initiation. Genes & Dev. 1995;9:559–572. doi: 10.1101/gad.9.5.559. [DOI] [PubMed] [Google Scholar]
- Li B, Weber JA, Chen Y, Greenleaf AL, Gilmour DS. Analyses of promoter-proximal pausing by RNA polymerase II on the hsp70 heat shock gene promoter in a Drosophila nuclear extract. Mol Cell Biol. 1996;16:5433–5443. doi: 10.1128/mcb.16.10.5433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lis J, Wu C. Protein traffic on the heat shock promoter: parking, stalling, and trucking along. Cell. 1993;74:1–4. doi: 10.1016/0092-8674(93)90286-y. [DOI] [PubMed] [Google Scholar]
- Morse RH. Nucleosomes inhibit both transcriptional initiation and elongation by RNA polymerase III in vitro. EMBO J. 1989;8:2343–2351. doi: 10.1002/j.1460-2075.1989.tb08362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller PR, Wold B. In vivo footprinting of a muscle-specific enhancer by ligation-mediated PCR. Science. 1989;246:780–786. doi: 10.1126/science.2814500. [DOI] [PubMed] [Google Scholar]
- Mymryk JS, Archer TK. Dissection of progesterone receptor-mediated chromatin modeling and transcriptional activation in vivo. Genes & Dev. 1995;9:1366–1376. doi: 10.1101/gad.9.11.1366. [DOI] [PubMed] [Google Scholar]
- O’Brien T, Lis JT. Rapid changes in Drosophila transcription after instantaneous heat shock. Mol Cell Biol. 1993;13:3456–3463. doi: 10.1128/mcb.13.6.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purnell BA, Emanuel PA, Gilmour DS. TFIID recognition of the initiator and sequences farther downstream in Drosophila class II genes. Genes & Dev. 1994;8:830–842. doi: 10.1101/gad.8.7.830. [DOI] [PubMed] [Google Scholar]
- Rasmussen EB, Lis JT. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc Natl Acad Sci. 1993;90:7923–7927. doi: 10.1073/pnas.90.17.7923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell. 1988;54:795–804. doi: 10.1016/s0092-8674(88)91087-2. [DOI] [PubMed] [Google Scholar]
- Rudd MD, Luse DS. Amanitin greatly reduces the rate of transcription by RNA polymerase II ternary complexes but fails to inhibit some transcript cleavage modes. J Biol Chem. 1996;271:21549–21558. doi: 10.1074/jbc.271.35.21549. [DOI] [PubMed] [Google Scholar]
- Spencer CA, Groudine M. Transcription elongation and eukaryotic gene regulation. Oncogene. 1990;5:777–785. [PubMed] [Google Scholar]
- Studitsky VM, Clark DJ, Felsenfeld G. A histone octamer can step around a transcribing polymerase without leaving the template. Cell. 1994;76:371–382. doi: 10.1016/0092-8674(94)90343-3. [DOI] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Wilson CJ, Chao DM, Imbalzano AN, Schnitzler GR, Kingston RE, Young RA. RNA polymerase holoenzyme contains SWI/SNF regulators involved in chromatin remodeling. Cell. 1996;84:235–244. doi: 10.1016/s0092-8674(00)80978-2. [DOI] [PubMed] [Google Scholar]
- Xie W, Rothblum L I. Rapid, small-scale RNA isolation from tissue culture cells. BioTechniques. 1991;11:324–325. [PubMed] [Google Scholar]
- Yankulov K, Blau J, Purton T, Roberts S, Bentley DL. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell. 1994;77:749–759. doi: 10.1016/0092-8674(94)90058-2. [DOI] [PubMed] [Google Scholar]