Abstract
Electrostatic forces, acting between helices and modulated by the presence of the ion atmosphere, are key determinants in the energetic balance that governs RNA folding. Previous studies have employed Poisson-Boltzmann (PB) theory to compute the energetic contribution of these forces in RNA folding. However, the complex interaction of these electrostatic forces with RNA features such as tertiary contact formation, specific ion-binding, and complex interhelical junctions present in prior studies precluded a rigorous evaluation of PB theory, especially in physiologically important Mg2+ solutions. To critically assess PB theory, we developed a model system that isolates these electrostatic forces. The model system, composed of two DNA duplexes tethered by a polyethylene glycol junction, is an analog for the unfolded state of canonical helix-junction-helix motifs found in virtually all structured RNAs. This model system lacks the complicating features that have precluded a critical assessment of PB in prior studies, ensuring that interhelical electrostatic forces dominate the behavior of the system. The system’s simplicity allows PB predictions to be directly compared with small angle x-ray scattering experiments over a range of monovalent and divalent ion concentrations. These comparisons indicate that PB is a reasonable description of the underlying electrostatic energies for monovalent ions, but large deviations are observed for divalent ions. The validation of PB for monovalent solutions allows analysis of the change in the conformational ensemble of this simple motif as salt concentration is changed. Addition of ions allows the motif to sample more compact microstates, increasing its conformational entropy. The increase of conformational entropy presents an additional barrier to folding by stabilizing the unfolded state. Neglecting this effect will adversely impact the accuracy of folding analyses and models.
Introduction
Noncoding RNAs (ncRNA) play a number of roles in cellular function and control [1]. For many of these ncRNAs, folding from a disordered unfolded state is a critical first step in the acquisition of biological activity. The diverse and essential biological functions played by these ncRNAs underscore the need to understand their folding at a fundamental level. Superficially, RNA folding may resemble protein folding. However, RNA folding differs fundamentally from protein folding in ways that complicate the application of knowledge and intuition gained from protein folding research. In particular, the absence of a hydrophobic core and the many degrees of freedom and high charge density in the nucleic acid backbone present serious challenges in the analysis of RNA folding.
Thankfully, several simplifying features exist for RNA folding. The stability of RNA helices ensures that secondary structure is present even under conditions that preclude tertiary folding [2]. Therefore, it is natural to define the unfolded state of structured RNA as the fluctuating conformational ensemble containing helical elements of secondary structure and the junctions that join them. In this view, folding is the balance between the disparate stabilizing and resisting forces that act on the heterogeneous unfolded state, transforming it into a stable, folded structure [2, 3, 4]. Strong electrostatic repulsion between helices, modulated by solution ion concentration, resists folding. Conformational biases present in the junctions that join helical elements can assist in bringing distant regions of the molecule together, allowing tertiary contacts and specific ion-binding sites to form that further stabilize the folded structure. Understanding these forces individually will greatly advance our knowledge of the fundamental processes that govern folding.
Folding experiments have demonstrated that RNA structure is strongly dependent on interhelical electrostatic forces, modulated by solution ions; these forces exist independent of tertiary contact formation or specific ion-binding. These electrostatic forces are largely responsible for the conformational “relaxation” of various structured RNAs in response to increasing concentration of solution ions [5, 6, 7, 8, 9]. This strong ion dependence has long motivated researchers to model the roles that ions play in screening the electrostatic forces between helices.
A major advance in our understanding came with the realization that the ion atmosphere — a fluctuating and loosely structured sheath of ions that surrounds polyelectrolytes — is responsible for the bulk of the electrostatic screening supplied by solution ions. Draper and co-workers have written extensively on this subject and have introduced a framework for modeling the ion atmosphere and its energetic contribution to the stabilization of folded RNAs. This framework employs Poisson-Boltzmann (PB) theory to model the electrostatic interactions present in the ion atmosphere and typically treats the unfolded and folded conformational ensembles of the RNA as either two distinct structures or a small collection of structures [10, 11, 12, 13, 14].
The simplicity of this framework — PB theory is computationally tractable and a two-structure formalism obviates the need to construct physically-realistic conformational ensembles — has led to its application in modeling diverse RNAs such as transfer RNA, ribosomal RNA, RNA pseudoknots, the Tetrahymena ribozyme, and the hairpin ribozyme [10, 11, 12, 15, 16, 17]. Despite the reasonable agreement between framework predictions and experimental results found in these studies, there are reasons to question the underlying assumptions of the framework. Theoretical arguments and some experiments have suggested that simplifications in PB theory render it inaccurate for solutions containing physiological concentrations of Mg2+ and other multivalent salts [18, 19, 20, 21, 22, 23, 24]. Furthermore, it is unclear how well a single structure or small ensemble of structures represents the multiple conformations of the unfolded ensemble.
The validity of these assumptions has been difficult to test. Previous studies have focused on realistic RNAs containing tertiary contacts, specific ion-binding sites, and complex interhelical junctions. The difficulty in modeling these RNA elements computationally has made it impossible to compare with confidence the framework predictions obtained from PB theory and experimental results. Furthermore, the a priori assumption of a single unfolded structure in the framework has implicitly forced comparisons between experiments conducted on what are likely to be conformational ensembles and simulations computed on single structures.
Critical assessment of PB theory demands a molecular platform that permits both theoretical and experimental examination of the electrostatic forces that govern the conformational ensemble of structured nucleic acids. To this end, we have created a model system consisting of two DNA duplexes joined by a short polyethylene glycol (PEG) junction. Despite its simplicity, the system captures essential aspects of a fundamental and ubiquitous nucleic acid motif: the unfolded state of two regions of Watson-Crick base pairing joined by a junction. Such motifs are present in all structured RNAs with great variability in junction structure and sequence. However, in distinct contrast to the complex RNAs investigated in previous studies, our tethered duplex system lacks tertiary contacts, specific ion-binding sites, and complex junction regions. The absence of these complicated features ensured that we probed the interhelical electrostatic forces in isolation and not some convolution of these forces with tertiary contact formation, and specific ion-binding.
We utilized small angle x-ray scattering (SAXS) to probe the conformational ensemble of the tethered duplex system under a range of monovalent and divalent ion concentrations. SAXS is ideally suited for monitoring the tethered duplex conformational ensemble as it is a direct measurement of the solution state of the ensemble in thermodynamic equilibrium under different ionic conditions. In particular, the experimentally-measured SAXS profile is a direct sum over the scattering contributions from the individual ensemble conformers [9]. We employed Monte Carlo (MC) sampling, driven by energies computed from PB theory, to construct conformational ensembles at each experimental condition. SAXS observables were computed from the atomic models of the ensemble and compared to experimental profiles [25]. The use of MC sampling generates ensembles whose conformers are properly weighted with respect to the PB energy function and avoids the framework’s a priori assumption that a small collection of selected conformers can adequately represent the underlying conformational ensemble.
Our results show that PB is reasonably consistent with experiments over a wide range of concentrations for monovalent ions. However, PB introduces significant errors for divalent ions and thus its quantitative applicability in RNA-Mg2+ systems should be reconsidered. The validation of PB for monovalent ions allows us to determine what is lost in replacing the conformational ensemble with a single structure. We find that the conformational entropy of the tethered duplex — and by analogy, other structured RNAs — increases with ionic concentration, stabilizing the unfolded state. Neglecting this stabilization will adversely affect the accuracy of folding analyses that approximate the unfolded ensemble as a single static structure.
Experimental Section
Sample preparation and SAXS data acquisition
The tethered duplex model consisted of two 12 bp DNA duplexes joined by a flexible PEG tether of 6 ethylene glycol monomers (Fig. 1). The component oligonucleotides were synthesized, purified and annealed as described previously [9]. The DNA stock solution was dialyzed into 40 mM Na-MOPS buffer, pH 7.0, with additional monovalent or divalent salt at the indicated concentration. The final DNA concentration ranges from 0.04 to 0.08 mM. SAXS data were acquired at the BESSRC-CAT beamline 12-IDC at the Advanced Photon Source as described previously [9, 26].
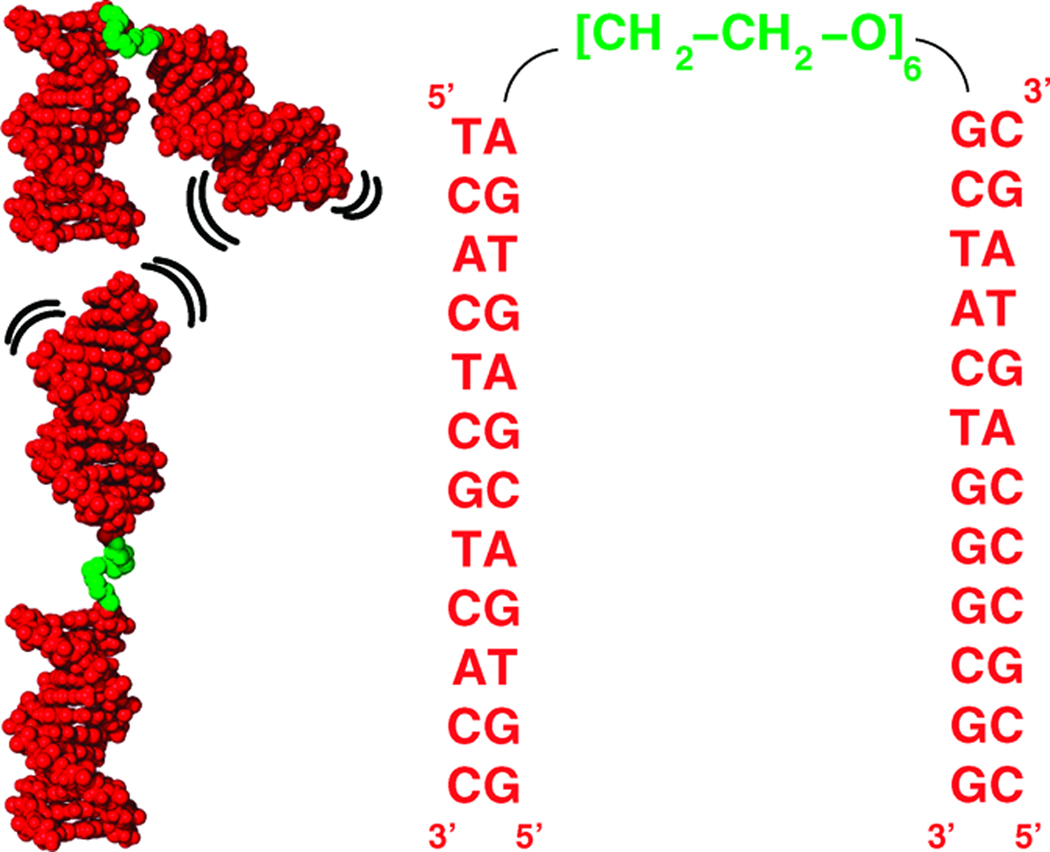
Figure 1.
Schematic and sequence of the tethered duplex system. The system consists of two 12 bp DNA duplexes (red) joined by a flexible PEG tether (green). Under low ionic conditions, extended conformers dominate the unfolded ensemble; as ions are added, the ensemble relaxes to more relaxed states.
Conformational sampling and calculation of SAXS profiles
We employed a MC algorithm to sample the energy landscape and construct unfolded ensembles at each ionic concentration. The energy of a candidate conformer was decomposed into two terms: HE, a term that describes the interduplex electrostatic repulsion and HPEG, a term that describes the contribution from the chain entropy of the PEG tether. HE was computed by solving the PB equation for all-atom representations of the DNA duplexes using the Adaptive Poisson-Boltzmann Solver v0.4.0 under ionic conditions identical to the experiments [27]. HPEG was computed as the logarithm of the probability distribution of end-to-end distances observed in a separate MC simulation of the PEG tether in isolation. Further details may be found in the Supplementary Materials.
Theoretical SAXS intensities for each structure in the ensemble were computed with CRYSOL [25]. The predicted ensemble scattering profile at each ion condition was computed as the average of the individual scattering profiles. Simulation convergence was tested using 10-fold cross validation (Supplementary Table 2). The magnitude of the scattering contribution from the ion atmosphere was estimated and removed from the scattering data using the procedure in Ref. [9]. The resulting scattering profiles were directly compared to theoretical predictions. Experimental errors were estimated using at least triplicate repeats for selected measurements.
Results
SAXS reveals an ion-dependent conformational change
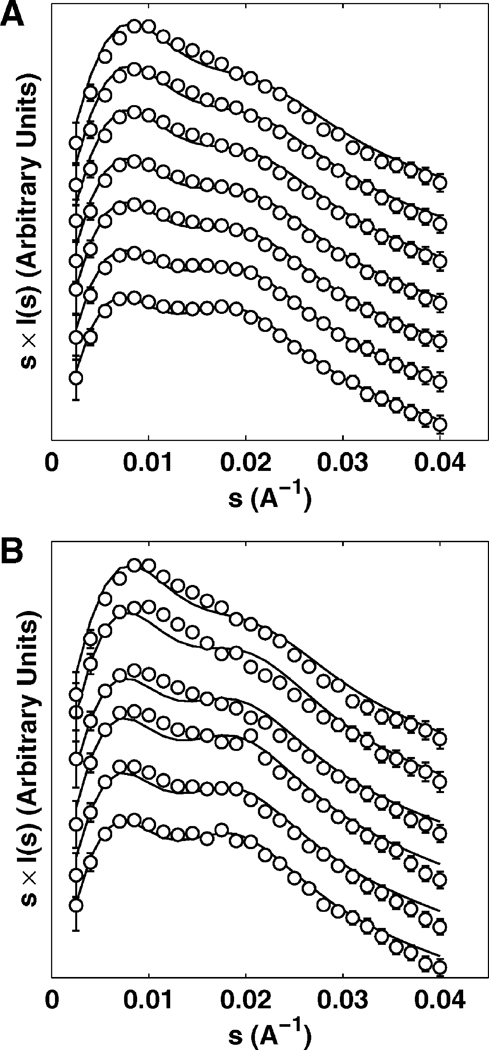
We applied SAXS to obtain a quantitative measure of the conformational ensemble of the tethered duplex over a wide range of ionic conditions. The tethered duplex SAXS profiles are plotted in the Holtzer representation (i.e., scattering intensity multiplied by momentum transfer s = 2 sin θ/λ, where 2θ is the total scattering angle and λ is the X-ray wavelength) over a range of Na+ (0 – 2 M, Fig. 2A, circles) and Mg2+ (0.05 – 500 mM, Fig. 2B, circles) concentrations.1 The Holtzer representation has been shown in previous studies to be sensitive to structural changes in the tethered duplex [9].2 At low ionic conditions (0 mM Na+ or 0.05 mM Mg2+, Fig. 2, bottom profiles), the “saddle” shape of the profile reflects an approximately collinear arrangement of two duplexes. As the ion concentration increases, the eventual disappearance of the second peak in the scattering profiles (Fig. 2 top profiles) reflects the gradual adoption of more globular conformations.
Figure 2.
Scattering intensities I(s), weighted by momentum transfer s at different ionic concentrations as a function of s for Na+ (A) and Mg2+ (B). Open circles represent experimental data and solid black lines represent predictions from PB theory. Na+ concentrations (top to bottom): 2, 0.6, 0.3, 0.15, 0.08, 0.02, 0 M (in 16 mM Na+ background from Na-MOPS buffer). Mg2+ concentrations (top to bottom): 100, 10, 2, 0.6, 0.2, 0.05 mM (in 16 mM Na+ background from Na-MOPS buffer). For clarity, profiles are vertically offset and the number of data points are reduced. Error bars are smaller than symbols for some points.
Similar conformational changes were observed for other monovalent (Li+, K+, Rb+, and TMA+) and divalent (Ca2+, Sr2+, Ba2+ and putrescine2+) ions (see “Critical assessment of PB theory”). The general dependence of structure on ionic concentration is consistent with nonspecific electrostatic interactions that are primarily responsible for the observed structural changes, as would be expected for the tethered duplex system. However, the specific molecular details of the ion-mediated structural, energetic, and entropic changes at the microstate level are missing from the experimental description, as SAXS only probes the mean structure of the unfolded ensemble. For insight into these critical aspects of the unfolded ensemble, we must turn to theoretical descriptions.
Monte Carlo sampling reproduces SAXS-observed conformational changes in monovalent ion solutions
To understand the changes in the SAXS profiles as a function of ionic conditions, we employed a MC approach (Experimental Section) to create detailed models of the unfolded ensemble over a wide range of ionic conditions. Our MC protocol sampled the conformations of the two duplexes using a simple polymer representation of the PEG tether and PB to compute the electrostatic energy. The ensemble models were used to predict the observed SAXS profile at each ionic condition. The SAXS profiles derived from MC exhibit structural changes similar to those observed experimentally, reflected by the gradual disappearance of the “saddle” in the SAXS profiles with increasing monovalent and divalent ion concentration (Fig. 2, lines). In particular, the predicted SAXS profiles for Na+ are in good agreement with the experimental data over the entire concentration range of 0 – 2 M (Fig. 2A), with a reduced-χ2 of 1.10.
The agreement between the predicted and observed Mg2+-induced conformational change is notably weaker than that observed in Na+, with a reduced-χ2 of 3.87. Systematic deviations between the prediction and experiment are pronounced at [Mg2+] ≥ 0.2 mM (Fig. 2B). In particular, the disappearance of the second peak in the predicted profiles is retarded relative to the experimental profiles and occurs at higher ionic concentration, suggesting that the ensemble derived using PB is more extended than the measured profiles indicate. The magnitude of the deviation is further analyzed in the discussion section (see “Critical assessment of PB theory”).
Despite the retarded structural changes in the PB-derived ensemble for Mg2+, the trend of the conformational change with increasing ion concentration is well captured by the MC predictions, illustrating the link between electrostatic repulsion and the spatial arrangement of the duplexes. Under low ionic strength, the large electrostatic repulsion between the duplexes forces the unfolded ensemble to adopt more extended conformations; at high ion concentration, the screening of the ion atmosphere allows more compact states to be explored. This conformational change of the tethered duplex ensemble with increasing ion concentration was previously termed “electrostatic relaxation, based on observations of folding initiation in large RNA molecules by Mg2+ addition [5, 7, 8].
Discussion
Critical assessment of PB theory
PB theory, widely applied in protein and nucleic acid systems, describes the electrostatic potential around biological molecules [13, 30, 31, 32]. In PB theory, ions are treated as point charges that do not interact with one another; in this framework, excluded volume and ion-ion correlation effects are assumed to be negligible. Extensive theoretical work has examined the conditions under which these assumptions hold [19, 20, 21, 33, 34, 35, 36]. These studies suggested that PB works reasonably well for monovalent ions, consistent with a series of successful PB applications in predicting the DNA melting behavior as a function of monovalent salt [37, 38]. In contrast, PB was predicted to be inaccurate for divalent or higher charged ions in simulations and theoretical treatments [18, 19, 20, 21, 22]; these observations are consistent with the inability of PB to account for duplex melting behavior as a function of divalent cation concentration [39, 40].
Nevertheless, direct and quantitative experimental tests of PB in divalent ions are rare. The aforementioned theoretical studies highlighting departures from PB theory typically compare PB against predictions from other simulations (e.g., Monte Carlo); such simulations, while incorporating some of the effects neglected by PB, still include simplifications and approximations whose importance in nucleic acid systems is unknown [19, 20, 21, 22, 36, 41]. Further, a series of recent analyses have led to the contradictory conclusion that PB is sufficient to explain many experimental observations in Mg2+-induced folding and stabilization of RNA [10, 11, 12, 13, 14, 17]. However, the interpretation of these experiments is complicated by the presence of tertiary contacts, multiple junctions, and specific ion-binding sites whose effects cannot currently be modeled accurately. Even the interpretation of the DNA duplex melting studies is complicated by the presence of multiple single-stranded states at different salt concentrations [42, 43, 44].
These contradictory conclusions highlight the need for a simple and quantitative assessment of PB theory in a model system accessible to both theory and experiment. The tethered duplex satisfies this requirement. Its component duplexes eliminate the ambiguities associated with the modeling of specific ion-binding sites or tertiary contacts. Moreover, its PEG junction is well characterized and readily simulated with a simple polymer model [45]. In contrast, modeling a realistic RNA junction would have introduced many more ambiguities as models of RNA junctions, using molecular dynamics or other approaches, are of unknown or limited accuracy.
To provide a quantitative basis for comparison of the experiment and simulation, we expressed the experimental and predicted scattering profiles at different ionic concentrations I(s; [M]) as the linear combination of low-ion (LI) and high-ion (HI) experimental scattering profiles, defined respectively as 0 M Na+ and 500 mM Mg2+ (added to the 16 mM Na+ background) (Eq. 1):
| (1) |
The resulting fit coefficients a, b measure the degree of relaxation of the ensemble over the range of ionic conditions. We fitted a simple empirical Hill model to the observed structural relaxation in both the experimental SAXS data and theoretical ensemble (Eq. 2) as a function of the ionic concentration [M]:
| (2) |
Although the relaxation of the tethered duplex system is patently not two-state, such a fit is useful in describing the relaxation and guiding intuition. We emphasize that the Hill coefficient n does not describe discrete ion-binding sites. The fitted midpoint [M]1/2 measures the ability of the ion to induce electrostatic relaxation; the stronger the screening, the lower the [M]1/2.
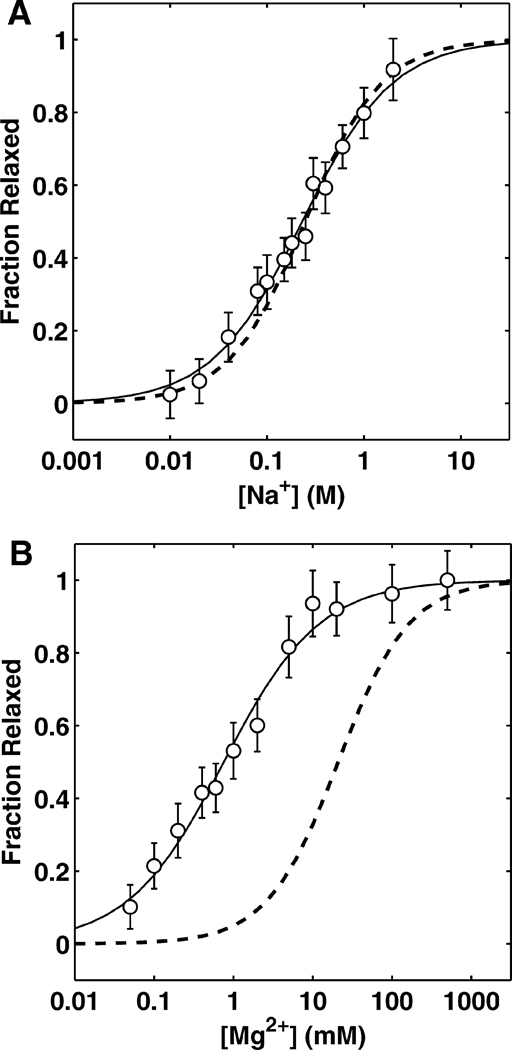
Fig. 3 shows the experimentally-measured conformational change of the tethered duplex as a function of Na+ and Mg2+ concentration (see Supplementary Fig. 5 for other monovalent and divalent ions). The change in conformational ensemble is similar for the various monovalent ions (Li+, Na+, K+, Rb+, TMA+), with the midpoints and Hill coefficients differing by <3 fold (Table 1). We note, however, the presence of a modest variation in the screening of the different monovalent cations. For instance, comparison of midpoints reveals that Li+ is able to induce structural relaxation at roughly half the concentration of Rb+ (Table 1). The trend in screening efficiency, Li+> Na+≳ K+> Rb+> TMA+, correlates inversely with ion size as approximated by the first order hydration shell [46] (see Supplementary Fig. 6). A similar correlation between ion size and the relative affinity of ions has been observed from a direct measurement of the number of associated competing ions around DNA duplexes [24]. In addition, the distribution of large ions is less dense in the vicinity of DNA [47]. Thus, the size-mediated modulation in screening is presumably due to the fact that larger cations associate more loosely around the DNA and cannot approach the DNA as closely as smaller ions, due to their larger excluded volume. Consistently, large polyamines have lower efficiencies in stabilizing DNA duplexes or folded RNA molecules than metal ions of equivalent charge [7, 48].
Figure 3.
Ion-mediated structural relaxation fit to the empirical Hill model for Na+ (A) and Mg2+ (B). Fraction relaxed reflects the overall relaxation of the ensemble as determined by projection onto the experimental scattering profiles taken at low- and high-ionic concentration (see Eq. 1). For both plots, solid lines show the Hill fit to the experimental data and the dashed lines show the same relaxation derived from PB.
Table 1.
Observed and PB-derived midpoints and Hill coefficients of the structural relaxation mediated by various monovalent and divalent ions
| [M]1/2 (mM) | n | |||
|---|---|---|---|---|
| Ion | Measured | PB | Measured | PB |
| Li+ | 150 ± 35 | 1.03 ± 0.10 | ||
| Na+ | 230 ± 58 | 0.93 ± 0.11 | ||
| K+ | 250 ± 59 | 250 | 0.90 ± 0.10 | 1.10 |
| Rb+ | 330 ± 92 | 0.83 ± 0.11 | ||
| TMA+ | 550 ± 170 | 0.78 ± 0.10 | ||
| Mg2+ | 0.76 ± 0.50 | 0.72 ± 0.10 | ||
| Ca2+ | 0.90 ± 0.58 | 0.72 ± 0.11 | ||
| Sr2+ | 1.47 ± 1.18 | 21.0 | 0.60 ± 0.08 | 0.97 |
| Ba2+ | 1.65 ± 0.83 | 0.54 ± 0.06 | ||
| Putr.2+ | 5.57 ± 3.47 | 0.45 ± 0.05 | ||
Similarly, the experimental divalent-modulated (Mg2+, Ca2+, Ba2+ and putrescine2+) relaxations are clustered and exhibit modest size-mediated differences (see Supplementary Fig. 6). Their midpoints are approximately 100 fold smaller than those of the monovalent ions (~ 0.8 mM for Mg2+, ~ 200 mM for Na+, Table 1), indicating that divalent ions are much more efficient in reducing electrostatic repulsion than monovalents. The high efficiency of divalent ions in neutralizing the repulsion has been evident. Mg2+ ions preferentially occupy the ion atmosphere around nucleic acids, even in the presence of large amounts of monovalent ions [24], and induce folding transitions at much lower concentrations relative to monovalent ions [3, 7, 8, 9, 11, 12, 13, 49, 50].
The Mg2+-induced relaxation is predicted quite poorly by PB theory with >10 fold difference between the experimentally (0.8 mM) and theoretically (21 mM) derived midpoints (Fig. 3B and Table 1). The lower value of the observed midpoint shows that the experimental ensemble is much more compact and relaxed at lower concentrations than the theoretically derived ensemble. This discrepancy indicates that Mg2+ and other divalent ions are much better at screening the electrostatic repulsion between duplexes than PB predicts (Table 1).
Although the electrostatic screening by Mg2+ can dominantly determine the midpoint of a folding transition, it is also the case that Mg2+ site-binding, tertiary contact formation, and alternative secondary structures in large RNAs can alter the apparent efficiency of Mg2+ in reducing electrostatic repulsion. In contrast, the tethered duplex model studied herein is free of such complicating effects; thus, comparing the midpoints of divalent and monovalent ions provides a reference for the screening ability of different ions to induce electrostatic relaxation. The apparent agreement of PB electrostatic calculations with prior RNA folding data in Mg2+ presumably arose from a coincidental cancellation of competing effects such as coupled tertiary folding, Mg2+ site-binding, and, in some cases, consideration of a single unfolded conformer rather than an ensemble of unfolded conformers.
The observed deviations for divalent cations likely stem from the neglect of ion-ion correlations in the mean-field PB theory. For divalent ions, correlations stabilize high Mg2+ concentrations near the duplex surface, allowing surface concentrations to exceed the PB prediction [18, 34]. This hypothesis is consistent with previous studies comparing ion distributions obtained from PB and explicit-ion MC around all-atom models of DNA [33, 51, 52, 53]. In these studies, PB was found to underestimate the surface concentration of divalent ions. The higher Mg2+ surface concentration ensures that the phosphate charges are more effectively screened, explaining the downward shift in midpoints.
Electrostatic relaxation at the microstate level
The ionic environment controls the relaxation of an unfolded RNA and regulates the transition of the molecule to the folded state in which the helices are closely assembled. The agreement of our simulated ensembles with experimental data for monovalent ion solutions (Fig. 2A, B) allows us to visualize and characterize the electrostatic relaxation of a helix-junction-helix motif in terms of its constituent microstates. With this model, we can specifically analyze the change in the distribution of microstates as a function of monovalent ion concentration over the course of the relaxation, gaining insight into the electrostatic relaxation of unfolded or loosely structured nucleic acid systems. Although quantitative analyses cannot be performed for divalent ion solutions as these simulated ensembles do not appear to be accurate upon comparison with experimental data, analogous trends are expected.
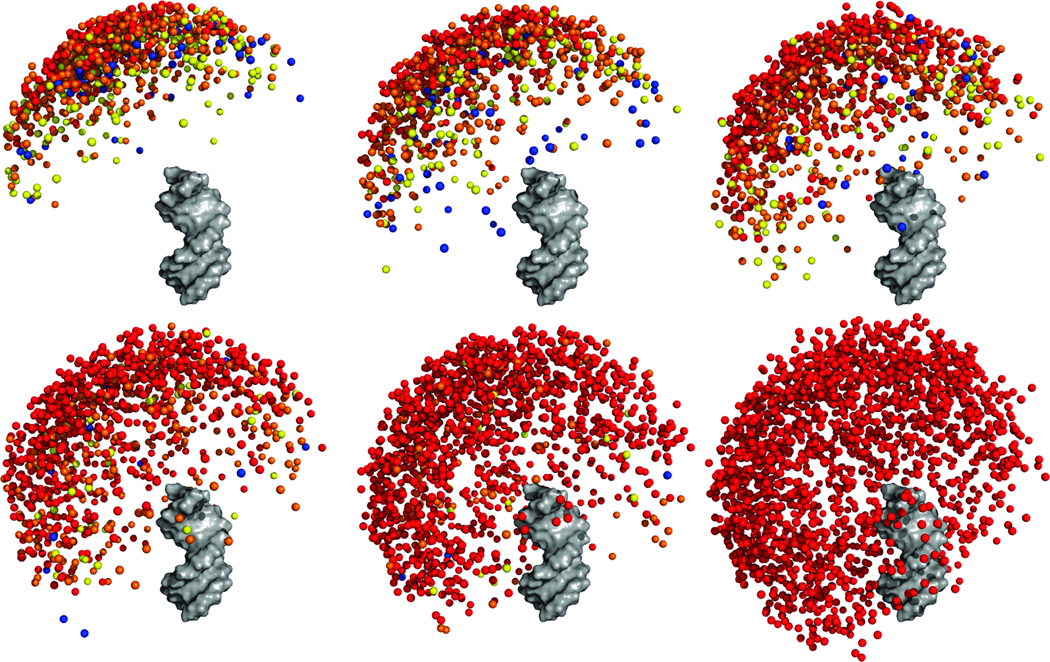
Fig. 4 and Supplementary Fig. 7 depict the energetic and structural modulation of the unfolded tethered duplex ensemble that results from increasing ion concentration. In Fig. 4, one duplex is designated as a fixed reference duplex and the colored spheres represent the distal end of the “mobile” duplex. The final frame of the plot (lower-right) depicts the ensemble in the sterics-only case (i.e., infinite screening) computed by omitting the electrostatics term (HE) in the MC modeling (Experimental Section).
Figure 4.
Visualization of the computationally-derived unfolded ensemble at various ionic conditions. From left to right, top to bottom: 0, 0.04, 0.15, 0.3, 2 M monovalent ions (in 16 mM monovalent background). The last figure shows the ensemble in the absence of electrostatics (i.e., steric effects only). One duplex is rendered in gray while the colored balls represent the distal end of the other duplex. Colors represent the energetic difference between the conformer and the minimum-energy conformer observed in the ensemble. Red, 0 – 1 kBT; Orange, 1 – 2kBT; Yellow, 2 – 3 kBT; Blue, > 3 kBT. The representation of each duplex by a single point occasionally results in the juxtaposition of a high-energy conformer close to a low-energy one; these cases correspond to conformers whose distal ends are in close proximity to low-energy conformers, but whose orientations (hidden in this representation) are energetically unfavorable (e.g., stretch the PEG tether unfavorably).
A salient feature of the ensembles is the marked conformational bias in the distribution of one duplex relative to another. The distribution of the distal end of the mobile duplex is not isotropic with respect to the azimuthal angle (defined by the helical axis of the reference duplex). Rather, the ensemble adopts a “tilted umbrella” shape with respect to the helical axis of the fixed reference duplex; the minimum energy conformer is one where the two duplexes are positioned with an interhelical angle of approximately 150°.
The asymmetry in the ensemble results from the junction attachment, which is not situated on the helical axis of the two duplexes. Rather, the PEG is attached off-axis to the 5’- and 3’-oxygens of the two duplexes, as is normal for nucleic acid attachment (Fig. 1). This attachment introduces the observed tilting of the ensemble by making certain conformers sterically unfavorable. This bias can be seen most clearly in the sterics-only ensemble (Fig. 4 and Supplementary Fig. 7, lower-right panel) in which the influence of electrostatics has been removed. The persistent asymmetry indicates that biases in the relative orientation of two helical regions can arise solely from the tether connectivity. Future work should explore these biases for more realistic RNA tethers.
The electrostatic environment can alter the relative orientation between helical regions. At low ionic concentration, electrostatic repulsion restricts the ensemble to a very small subset of the sterically-allowed conformations. Excursions away from the minimum are energetically costly; for example, placing the two helices at right angles requires approximately ≥ 2.0 kcal/mol. At high concentrations of monovalent ions, the ensemble explores a much larger volume of the sterically-allowed space due to the enhanced screening afforded by the higher ion concentrations. Ionic screening flattens the energy landscape, reducing the energy differences between states. Under such conditions, large excursions away from the minimum-energy conformer do not cost much energy (Fig. 4).
Changes in conformational entropy necessarily accompany the flattening of the energy landscape. To see this, consider the conformational ensemble in the absence of electrostatics. In this case, the spatial distribution of the duplexes is dictated by the tether alone; heuristically, the duplexes move inside an effective potential well whose shape is determined by the junction that joins them. Electrostatics acts upon this potential by changing its shape under different salt conditions. At low salt, the shape of the potential well is altered to make nonextended conformers extremely unfavorable; as salt concentration is increased the well broadens to include these more relaxed conformers, leading to a stabilizing increase in conformational entropy. From these basic results, we infer that the conformational entropy of the unfolded state of realistic RNAs with preformed secondary structure also increases as they relax electrostatically during in vitro folding experiments.
The substantial ion-dependent conformational changes in the unfolded state suggest that the ensemble is fundamental to the nature of the unfolded state and cannot be ignored a priori. For protein folding, it has been widely recognized that the unfolded state is critical for unraveling the structural and thermodynamic pathways for folding as it is the starting point for any folding event; moreover, the stability of the folded structure is determined by its stability relative to the unfolded state and other alternatively folded states. [54].
Despite this, RNA folding studies often assume, explicitly or implicitly, that the unfolded state is well-approximated by a single “average” structure [11, 17, 15]. In such a framework, the unfolded “ensemble” is composed of a single structure and the change in free energy of the unfolded state in response to changing ionic conditions is determined entirely by the change in electrostatic energy of a single conformer. Because this unfolded state model consists of a single structure, the contribution of conformational entropy to the free energy of the unfolded state is neglected.
The single-structure approximation may underestimate the electrostatic contribution to the free energy of the unfolded state as salt conditions are modulated. The change in electrostatic energy of the ensemble, which in reality is a change in an ensemble average, may be poorly approximated by the change in electrostatic energy of just a single conformer. Even the use of several randomly-selected conformers is unlikely to correct this deviation as the additional structures are not likely to give the correct weighting dictated by the underlying PB energy function. For the tethered duplex system, these errors may be estimated by comparing the results obtained from ensemble and single-structure representations of the unfolded state. The change in electrostatic energy afforded by adding 2 M monovalent ions to the 16 mM background was computed to be −1.7 kcal/mol for the single-structure representation; for the ensemble representation, the change in electrostatic energy was computed to be −2.7 kcal/mol, a deviation of ~ 1.0 kcal/mol (Supplementary Fig. 8). Correct consideration of the change conformational entropy will increase this deviation, further impacting estimates of folding free energies.
The PEG tether represents the limit of a flexible junction; realistic RNA junctions are expected to be less flexible (e.g., compact conformations can be eliminated by the presence of bases and the electrostatic repulsion between phosphate moieties [55]). Thus, we expect that this discrepancy will be reduced for realistic junctions. However, biological RNAs typically contain multiple junctions and the energetic discrepancy associated with each junction, introduced by ignoring the ensemble nature of the unfolded state, is expected to be roughly additive. Given the unknown accuracy of computational models for RNA, there is no straightforward estimation of the free energy discrepancy introduced by the single-structure approximation for realistic RNAs. Nevertheless, the tethered duplex represents an idealized model where the effects induced by altering junction topology (e.g., single- vs. double-stranded topologies) and electrostatics may be investigated in a straightforward and simple fashion. The alterations of junction topology or electrostatics in the tethered duplex model mirror those found in realistic RNA systems; for example, a double-stranded PEG junction is simply an idealized form of a double-stranded RNA junction. Insights gained from study of the tethered duplex model may help guide intuition into the molecular behavior of actual structured RNAs.
Conclusions and Implications
A critical assessment of PB theory requires a synthesis of experimental and theoretical approaches. The tethered duplex system studied herein allows precisely such a synthesis. Despite its simplicity, the tethered duplex approximates a fundamental and ubiquitous structural component of unfolded RNA, provides a conceptual foundation that can be applied to future studies of isolated helix-junction-helix systems extracted from natural RNAs, and has allowed direct and quantitative comparison of SAXS measurements and simulations derived from PB theory to address its accuracy. For monovalent ions, the results verify that PB is a reasonable description of the electrostatic interaction between the duplexes. However, PB significantly underestimates the screening efficiency of Mg2+ and other divalent cations by ~10 fold, presumably due to the neglect of ion correlation effects. Ion correlation effects are expected to increase with higher charge density [18, 34]. As most functional RNAs consist of multiple helix-junction-helix motifs, these effects and thus the discrepancy of PB may be exacerbated as folding progresses to increasingly compact states. Improved electrostatic models will be required for accurate modeling of RNA conformations and dynamics.
Electrostatic forces are a key component in determining the spatial organization of duplexes in structured RNAs. Herein, the electrostatic relaxation induced by increasing monovalent ion concentration was quantitatively dissected at the microstate level through a combination of theory and experiment. The description of electrostatic relaxation at the microstate level reveals that at low ionic concentration, interhelical repulsion results in an energy landscape that populates a small subset of nearly collinear conformational states; with increasing ionic concentration, these repulsive forces are reduced by counterion screening, resulting in the electrostatic relaxation of the system and a gain of conformational entropy.
RNA folding is typically controlled in vitro by varying the concentration of ions. Superficially, this bears some resemblance to control of protein folding in vitro by changing denaturant concentration. However, for RNA, increasing ion concentration allows RNAs with preformed secondary structures to gain conformational entropy through electrostatic relaxation. This gain presents an additional barrier to folding as it stabilizes the unfolded state and must be compensated by favorable energetic contributions in the folded structure from tertiary contacts. Notably, this feature does not hold in typical protein folding where formation of secondary structure or hydrophobic collapse in response to reduced denaturant concentration is associated with a loss of conformational entropy.
These results suggest minimally that the common simplification that a single “average” conformer can encapsulate the properties of the unfolded ensemble must be reexamined, especially as this assumption has been commonly used in previous studies. Single-structure representations of the unfolded state introduce errors by neglecting the ensemble nature of the unfolded state. The exact magnitude of these errors for realistic RNAs can be analyzed analogously via the tethered duplex system presented herein, provided that accurate simulations can be conducted for realistic RNAs. The development of such models will undoubtedly be a critical challenge.
Supplementary Material
Acknowledgments
The authors thank Sönke Seifert for assistance at the Advanced Photon Source and members of the Doniach and Herschlag groups for helpful discussion. VBC and YB were supported respectively in part by a Pfizer Bio-X and Stanford Graduate Fellowship. This research was supported by NIH grant PO1 GM066275 and NSF grant PHY-0140140 and used resources of the National Energy Research Scientific Computing Center, which was supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC03-76SF00098. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-06CH11357.
Footnotes
In all cases, the stated ion concentrations are in addition to the background 16 mM Na+ from the Na-MOPS buffer.
Conventional analyses typically derive the radius of gyration (Rg) or the atomic pairwise distribution function from SAXS data through fitting and extrapolation of all or part of the data [28, 29]. However, these protocols can introduce errors or distortions to the original data through the choice of fitting parameters. Furthermore, the derivation of Rg only uses data at extremely small scattering angles, discarding the bulk of the structural data at higher angles. We have used the full SAXS profiles to analyze the molecular structure, thereby avoiding any artificial disturbance of the structural information. The Holtzer representation is used solely to help graphically illustrate the changes of the profile.
References
- 1.Simons RW, Grunberg-Manago M, editors. RNA Structure and Function. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- 2.Brion P, Westhof E. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 3.Takamoto K, Das R, He Q, Doniach S, Brenowitz M, Herschlag D, Chance MR. J. Mol. Biol. 2004;343:1195–1206. doi: 10.1016/j.jmb.2004.08.080. [DOI] [PubMed] [Google Scholar]
- 4.Chu VB, Herschlag D. Curr. Opin. Struct. Biol. 2008;18:305–314. doi: 10.1016/j.sbi.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. Proc. Natl. Acad. Sci. USA. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russell R, Herschlag D. J. Mol. Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 7.Koculi E, Lee NK, Thirumalai D, Woodson SA. J. Mol. Biol. 2004;341:27–36. doi: 10.1016/j.jmb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Das R, Kwok LW, Millett IS, Bai Y, Mills TT, Jacob J, Maskel GS, Seifert S, Mochrie SGJ, Thiyagarajan P, Doniach S, Pollack L, Herschlag D. J. Mol. Biol. 2003;332:311–319. doi: 10.1016/s0022-2836(03)00854-4. [DOI] [PubMed] [Google Scholar]
- 9.Bai Y, Das R, Millett IS, Herschlag D, Doniach S. Proc. Natl. Acad. Sci. USA. 2005;102:1035–1040. doi: 10.1073/pnas.0404448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Misra VK, Draper DE. J. Mol. Biol. 2000;299:813–825. doi: 10.1006/jmbi.2000.3769. [DOI] [PubMed] [Google Scholar]
- 11.Misra VK, Draper DE. J. Mol. Biol. 2002;317:507–521. doi: 10.1006/jmbi.2002.5422. [DOI] [PubMed] [Google Scholar]
- 12.Misra VK, Shiman R, Draper DE. Biopolymers. 2003;69:118–136. doi: 10.1002/bip.10353. [DOI] [PubMed] [Google Scholar]
- 13.Draper DE. RNA. 2004;10:335–343. doi: 10.1261/rna.5205404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grilley D, Misra V, Caliskan G, Draper DE. Biochemistry. 2007;46:10266–10278. doi: 10.1021/bi062284r. [DOI] [PubMed] [Google Scholar]
- 15.Bokinsky G, Rueda D, Misra VK, Rhodes MM, Gordus A, Babcock HP, Walter NG, Zhuang X. Proc. Natl. Acad. Sci. USA. 2003;100:9302–9307. doi: 10.1073/pnas.1133280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grilley D, Soto AM, Draper DE. Proc. Natl. Acad. Sci. USA. 2006;103:14003–14008. doi: 10.1073/pnas.0606409103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soto AM, Misra VK, Draper DE. Biochemistry. 2007 [Google Scholar]
- 18.Grosberg AY, Nguyen TT, Shklovskii BI. Reviews of Modern Physics. 2002;74:329–345. [Google Scholar]
- 19.Solis FJ, Olvera de la Cruz M. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 1999;60:4496–4499. doi: 10.1103/physreve.60.4496. [DOI] [PubMed] [Google Scholar]
- 20.Diehl A, Carmona HA, Levin Y. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 2001;64:011804. doi: 10.1103/PhysRevE.64.011804. [DOI] [PubMed] [Google Scholar]
- 21.Stilck JF, Levin Y, Arenzon JJ. J. Stat. Phys. 2002;106:287–299. [Google Scholar]
- 22.Tan ZJ, Chen SJ. J. Chem. Phys. 2005;122:44903. doi: 10.1063/1.1842059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan ZJ, Chen SJ. Biophys. 2006;91:518–536. doi: 10.1529/biophysj.106.084285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai Y, Greenfeld M, Travers KJ, Chu VB, Lipfert J, Doniach S, Herschlag D. J. Am. Chem. Soc. 2007;129:14981–14988. doi: 10.1021/ja075020g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Svergun DI, Barberato C, Koch MHJ. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 26.Lipfert J, Millett IS, Seifert S, Doniach S. Rev. Sci. Instrum. 2006;77:046108. [Google Scholar]
- 27.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Proc. Natl. Acad. Sci. USA. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feigin LA, Svergun DI. Structure Analysis by Small-Angle X-Ray and Neutron Scattering. NY: NY Plenum Press; 1987. [Google Scholar]
- 29.Lipfert J, Doniach S. Ann. Rev. Biophys. Biomol. Struct. 2007;36:307–327. doi: 10.1146/annurev.biophys.36.040306.132655. [DOI] [PubMed] [Google Scholar]
- 30.Honig B, Nicholls A. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 31.Sharp KA, Honig B. Curr. Opin. Struct. Biol. 1995;5:323–328. doi: 10.1016/0959-440x(95)80093-x. [DOI] [PubMed] [Google Scholar]
- 32.Chen SW, Honig B. J. Phys. Chem. B. 1997;101:9113–9118. [Google Scholar]
- 33.Ni H, Anderson CF, Record MT. J. Phys. Chem. B. 1999;103:3489–3504. [Google Scholar]
- 34.Shklovskii BI. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 1999;60:5802–5811. doi: 10.1103/physreve.60.5802. [DOI] [PubMed] [Google Scholar]
- 35.Lyubartsev AP. Dekker Encyclopedia of Nanoscience and Nanotechnology. Marcel Dekker, Inc.; 2004. pp. 2131–2143. [Google Scholar]
- 36.Antypov D, Barbosa MC, Holm C. Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys. 2005;71:061106. doi: 10.1103/PhysRevE.71.061106. [DOI] [PubMed] [Google Scholar]
- 37.Bond JP, Anderson CF, Record MT. Biophys. J. 1994;67:825–836. doi: 10.1016/S0006-3495(94)80542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shkel IA, Record MT. Biochemistry. 2004;43:7090–7101. doi: 10.1021/bi036225e. [DOI] [PubMed] [Google Scholar]
- 39.Tan ZJ, Chen SJ. Biophys. J. 2006;90:1175–1190. doi: 10.1529/biophysj.105.070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan ZJ, Chen SJ. Biophys. J. 2007;92:3615–3632. doi: 10.1529/biophysj.106.100388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montoro JC, Abascal JL. J. Chem. Phys. 1998;109:6200–6210. [Google Scholar]
- 42.Jung J, Orden AV. J. Am. Chem. Soc. 2006;128:1240–1249. doi: 10.1021/ja0560736. [DOI] [PubMed] [Google Scholar]
- 43.Ma H, Proctor DJ, Kierzek E, Kierzek R, Bevilacqua PC, Gruebele M. J. Am. Chem. Soc. 2006;128:1523–1530. doi: 10.1021/ja0553856. [DOI] [PubMed] [Google Scholar]
- 44.Ma H, Wan C, Wu A, Zewail AH. Proc. Natl. Acad. Sci. USA. 2007;104:712–716. doi: 10.1073/pnas.0610028104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kienberger F, Pastushenko VP, Kada G, Gruber HJ, Riener C, Schindler H, Hinterdorfer P. Single Molecules. 2000;1:123–128. [Google Scholar]
- 46.Ohtaki H, Radńal T. Chem. Rev. 1993;93:1157–1204. [Google Scholar]
- 47.Andresen K, Das R, Park HY, Smith H, Kwok LW, Lamb JS, Kirkland EJ, Herschlag D, Finkelstein KD, Pollack L. Phys. Rev. Lett. 2004;93:248103. doi: 10.1103/PhysRevLett.93.248103. [DOI] [PubMed] [Google Scholar]
- 48.Anderson CF, Record MT. Annu. Rev. Phys. Chem. 1995;46:657–700. doi: 10.1146/annurev.pc.46.100195.003301. [DOI] [PubMed] [Google Scholar]
- 49.Pan J, Deras ML, Woodson SA. J. Mol. Biol. 2000;296:133–144. doi: 10.1006/jmbi.1999.3439. [DOI] [PubMed] [Google Scholar]
- 50.Record MT, Mazur SJ, Melancon P, Roe JH, Shaner SL, Unger L. Annu. Rev. Biochem. 1981;50:997–1024. doi: 10.1146/annurev.bi.50.070181.005025. [DOI] [PubMed] [Google Scholar]
- 51.Paulsen MD, Anderson CF, Record MT. Biopolymers. 1988;27:1249–1265. doi: 10.1002/bip.360270806. [DOI] [PubMed] [Google Scholar]
- 52.Pack GR, Wong L, Lamm G. Biopolymers. 1999;49:575–590. doi: 10.1002/(SICI)1097-0282(199906)49:7<575::AID-BIP4>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 53.Wang K, Yu YX, Gao GH, Luo GS. J Chem Phys. 2007;126:135102. doi: 10.1063/1.2713105. [DOI] [PubMed] [Google Scholar]
- 54.Dill KA, Shortle D. Annu. Rev. Biochem. 1991;60:795–825. doi: 10.1146/annurev.bi.60.070191.004051. [DOI] [PubMed] [Google Scholar]
- 55.Tan ZJ, Chen SJ. Nucleic Acids Res. 2006;34:6629–6639. doi: 10.1093/nar/gkl810. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.