Abstract
African trypanosomes such as Trypanosoma brucei undergo antigenic variation in the bloodstream of their mammalian hosts by regularly changing the variant surface glycoprotein (VSG) gene expressed. The transcribed VSG gene is invariably located in a telomeric expression site. There are multiple expression sites and one way to change the VSG gene expressed is by activating a new site and inactivating the previously active one. The mechanisms that control expression site switching are unknown, but have been suggested to involve epigenetic regulation. We have found previously that VSG genes in silent (but not active) expression sites contain modified restriction endonuclease cleavage sites, and we have presented circumstantial evidence indicating that this is attributable to the presence of a novel modified base β-d-glucosyl-hydroxymethyluracil, or J. To directly test this, we have generated antisera that specifically recognize J-containing DNA and have used these to determine the precise location of this modified thymine in the telomeric VSG expression sites. By anti J-DNA immunoprecipitations, we found that J is present in telomeric VSG genes in silenced expression sites and not in actively transcribed telomeric VSG genes. J was absent from inactive chromosome-internal VSG genes. DNA modification was also found at the boundaries of expression sites. In the long 50-bp repeat arrays upstream of the promoter and in the telomeric repeat arrays downstream of the VSG gene, J was found both in silent and active expression sites. This suggests that silencing results in a gradient of modification spreading from repetitive DNA flanks into the neighboring expression site sequences. In this paper, we discuss the possible role of J in silencing of expression sites.
Keywords: DNA modification, silencing, antigenic variation, VSG, sequence repeats
Trypanosoma brucei is a protozoan parasite that lives in the blood of mammals and causes sleeping sickness in man. By regularly changing the variant surface glycoprotein (VSG) coat, African trypanosomes can evade immunodestruction by the host, as reviewed in Cross (1996). Each trypanosome has hundreds of VSG genes but usually expresses only one at a time. The active VSG gene is exclusively located in one of the up to 20 telomeric VSG expression sites (for review, see Pays et al. 1994; Borst et al. 1997). These large transcription units are highly homologous and include several expression site-associated genes (ESAGs), besides a VSG gene (Revelard et al. 1990). The VSG coat can be changed by replacing the VSG gene in the active expression site, or by activating a new expression site and silencing the old one. Expression site switching can occur without any detectable DNA rearrangements (Zomerdijk et al. 1990; Horn and Cross 1997). How bloodstream trypanosomes silence all VSG expression sites but one and how the transcriptional states are stably inherited is not known (for review, see Borst et al. 1997). The promoter sequence independence of expression site control, however, suggests that an epigenetic mechanism such as telomere position effect might be involved (Horn and Cross 1995; Rudenko et al. 1995).
Silencing of an expression site is accompanied by DNA modifications in and around inactivated telomeric VSG genes (see Fig. 1A). These DNA modifications were deduced from partial cleavage of PstI, PvuII (Bernards et al. 1984b), and sometimes HindIII and SphI restriction sites (Pays et al. 1984). The restriction site polymorphisms were not found in transcribed VSG genes near telomeres or in silent chromosome-internal VSG genes, and were only present in bloodstream form (BF) trypanosomes. In insect form (or procyclic, PC) trypanosomes, which have a different coat protein and do not transcribe VSG genes, no modification was found (Pays et al. 1984). Modification at a given site was partial, that is, it was present in only a fraction of the cells in a clonal trypanosome population, and this fraction increased with the length of the associated telomeric repeat tract (Bernards et al. 1984b).
Figure 1.
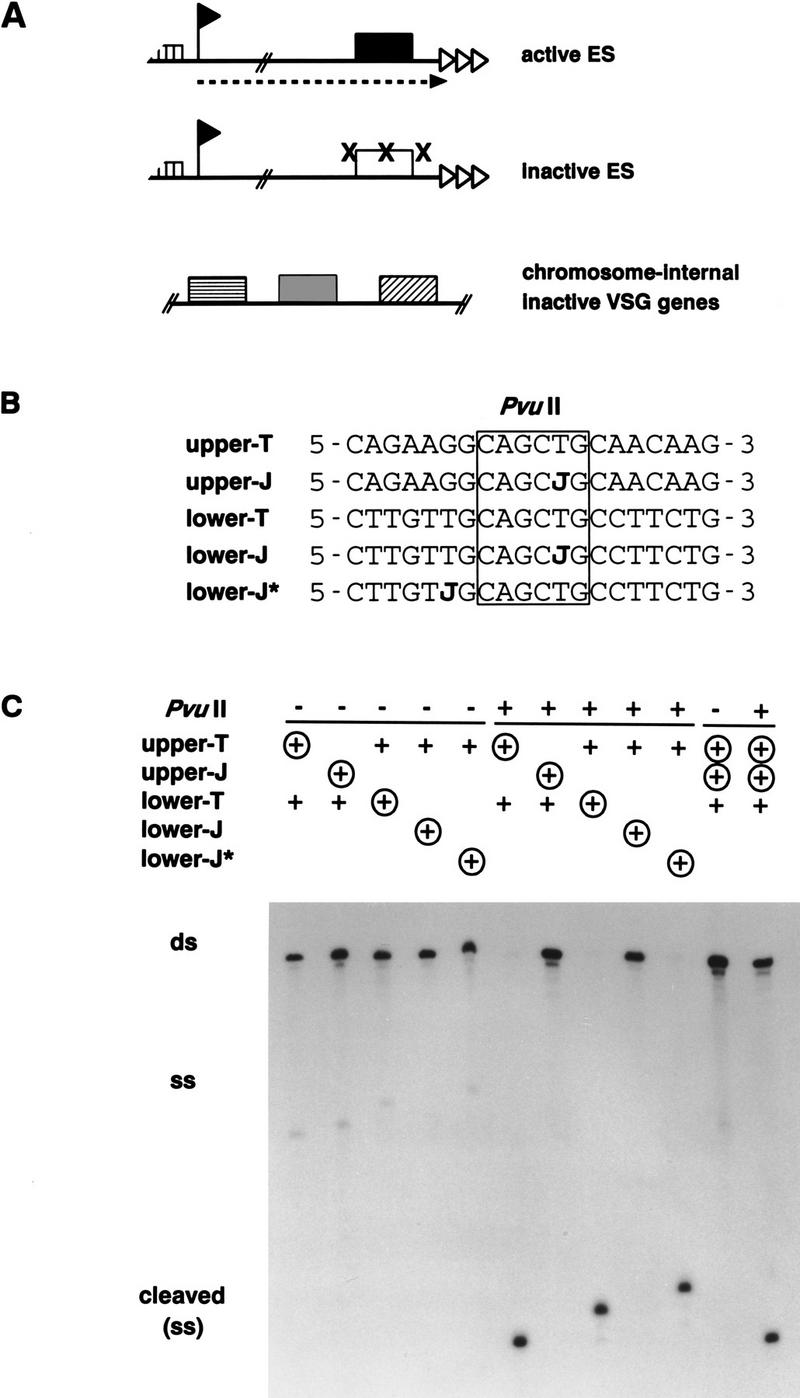
The modified base J prevents cleavage by PvuII. (A) Partial cleavage by restriction enzymes PstI, PvuII, and other enzymes (see text) suggested that silenced expression sites acquire a DNA modification (X) in and around the inactive telomeric VSG gene (box). These blocked restriction sites are not found in transcribed expression sites (broken line with arrowhead) or in silent chromosome-internal VSG genes. Telomeric repeats are indicated by triangles, the expression site promoter by a flag, and the imperfect tandem 50-bp repeats by a hatched box. (B) The effect of substitution of T by J on digestion of duplex DNA by PvuII restriction endonuclease; 20-mer oligonucleotides with a central PvuII restriction site (box) were used as substrates. If present, J replaced T in the PvuII site in either the upper or the lower strand, or replaced T two positions downstream of the PvuII site in the lower strand (lower-J*). (C) End-labeled oligomers (circled plus signs) were annealed to their nonlabeled complementary strands (+) to form duplex oligomers (ds), which were incubated without (−) or with (+) PvuII enzyme. Substrate and products were separated by native 20% polyacrylamide gel electrophoresis. Cleavage at 37°C resulted in short fragments that melted to single-stranded molecules at this temperature (cleaved ss). The last two lanes show a mixing control of duplexes with and without J. Incubation with PvuII (+) resulted in a noncleaved and a cleaved product.
The recently identified unusual base β-d-glucosyl-hydroxymethyluracil (β-gluc-HOMeU), called J (Gommers-Ampt et al. 1991, 1993), is a good candidate for this telomere-linked DNA modification. This modified base, detected by 32P-nucleotide postlabeling combined with separation on two-dimensional thin-layer chromatography, has only been found in DNA of African trypanosomes. It is present at low levels (0.2 mole%) in BF trypanosomes and is absent from PC trypanosomes (Gommers-Ampt et al. 1991). By nucleotide postlabeling analysis of purified telomeric tracts, we have shown recently that about half of all J is concentrated in both strands of the telomeric (GGGTTA)n repeats (van Leeuwen et al. 1996). Besides J and its putative precursor HOMeU, no other DNA modifications, such as DNA methylation, have been found in T. brucei (Crozatier et al. 1988; Gommers-Ampt et al. 1991; van Leeuwen et al. 1996). We have now verified that J prevents cleavage by restriction endonuclease PvuII. With antisera specific for J-containing DNA, we have located J in and around the telomeric VSG expression sites.
Results
J prevents cleavage by restriction endonuclease PvuII
Partial cleavage of restriction sites suggested the presence of a DNA modification in silenced telomeric VSG genes in bloodstream T. brucei (see Fig. 1A). The modified base J has the properties expected for the postulated modification, as a bulky base such as J may be expected to block cleavage by restriction enzymes (Huang et al. 1982). We have tested whether J blocks cleavage by PvuII, using DNA duplexes of short oligonucleotides encoding the PvuII site and its flanking sequences of VSG gene 221 (Fig. 1B). Figure 1C shows that duplexes with a hemimodified PvuII site were not cleaved, whereas duplexes without J were digested completely. J replacing T two positions downstream of the PvuII site did not block cleavage, showing that J at a short distance does not affect the endonuclease–DNA interaction. PstI has been shown already to be sensitive to the presence of HOMeU in the target sequence (McClelland et al. 1994) and is therefore expected to be also sensitive to J. These results confirm that J can block cleavage by restriction endonucleases and support further the correlation between J and the postulated modification in telomeric VSG genes. They do not, however, prove that J is present in VSG expression sites. Because restriction site polymorphisms and genomic sequencing (see Discussion) do not exclude the presence of other modifications, we set out to generate J-specific antisera to make it possible to detect low amounts of J in unique sequences in the genome.
Generation of antisera specific for J-containing DNA
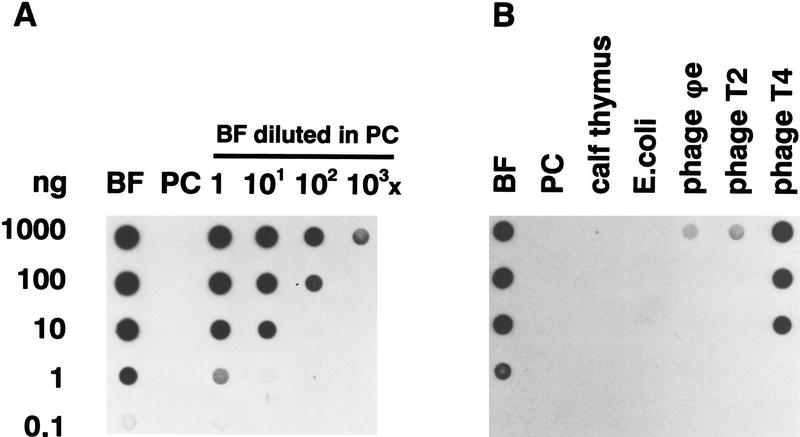
To obtain antinucleic acid antisera with a high specificity for DNA containing J in various sequence contexts, we induced antibodies with nucleotide–protein immunizing conjugates (see Materials and Methods). Immunization with J-5′-monophosphate (JMP) conjugated to keyhole limpet haemocyanin (KLH) or to bovine serum albumin (BSA) resulted in polyclonal rabbit antisera 538αJ and 539αJ, respectively. The specificity and sensitivity of the antisera were tested on DNA dot-blots with dilution series of various DNA samples, using immunodetection combined with enhanced chemiluminescence. With both antisera we could detect less than one J in 106 bases on dot-blots (Fig. 2A). This is at least 100-fold more sensitive than nucleotide postlabeling. 538αJ and 539αJ did not recognize DNA from Escherichia coli, calf thymus, or PC T. brucei, showing that they do not cross-react with nonmodified or methylated DNA, and confirming that PC trypanosomes are devoid of J (Fig. 2B). The antisera weakly recognized nonglucosylated HOMeU because some cross-reaction was found with phage φe DNA, in which all thymines are replaced by HOMeU. A stronger cross-reaction was found with β-glucosyl-hydroxymethylcytosine (β-gluc-HOMeC), but not with α-gluc-HOMeC, bases found in T-even phages. In bacteriophages T2 and T4, HOMeC replaces C (Kornberg et al. 1961). In both phages, 70% of HOMeC is α-glucosylated, in phage T4 another 30% is β-glucosylated. The glucosylated cytosine variants have only been found in T-even phage DNA and can be distinguished from J by 32P-nucleotide-postlabeling combined with two-dimensional thin-layer chromatography (Gommers-Ampt et al. 1991). Partially deaminated T2 DNA, in which a fraction of α-gluc-HOMeC was converted into α-gluc-HOMeU (0.1%–11% of T) did not cross-react, showing that the antibodies react specifically with β-glucose linked to HOMeU or HOMeC (data not shown).
Figure 2.
Detection of J-containing DNA on filters with polyclonal anti-J antisera. Dot-blots with dilution series of denatured DNA were incubated with polyclonal antiserum 539αJ and bound antibodies were detected with sheep α-rabbit-HRP in combination with enhanced chemiluminescence. (A) The sensitivity of detection was determined with a dilution series of BF trypanosome DNA (0.2 mole% J) and PC DNA (no J), and BF DNA diluted in PC DNA. Less than 0.0002 mole% J could be detected. Hybridization with a telomere repeat probe showed that equal amounts of DNA were present in each dilution series (data not shown). (B) Specificity of the antisera was tested with DNA samples with various DNA modifications: calf thymus (5-methylC), E. coli (6-methylA, 4-methylC, 5-methylC), phage φe (HOMeU), phage T2 (HOMeC, α-gluc- and β-gluc-α-gluc-HOMeC), phage T4 (α- and β-gluc-HOMeC). Pre-immune sera gave no signal at all (data not shown).
Immunoprecipitation of modified DNA
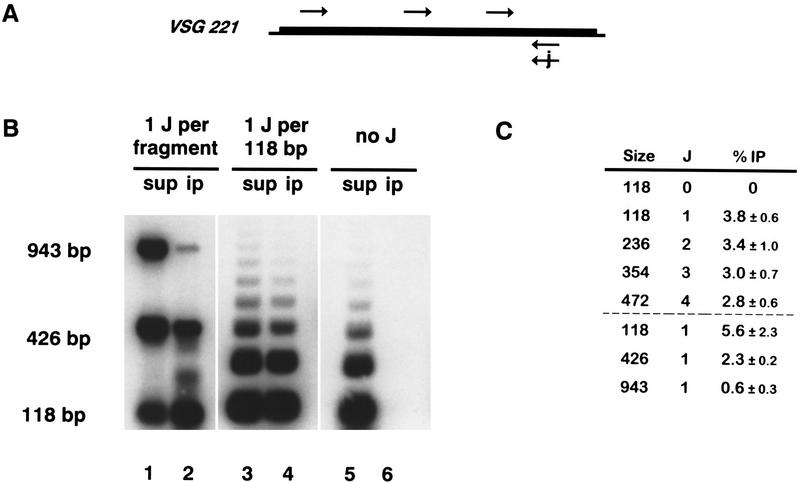
Immunoblots with denatured DNA provide a sensitive tool for the detection of J in DNA, but are less useful to study J-modification of specific sequences in a genome. We therefore tested whether the antibodies would immunoprecipitate J-containing double-stranded DNA fragments. Duplex DNA fragments of 118, 426, and 943 bp, each with one J residue, were generated by PCR amplification of part of the 221 VSG gene using one antisense primer with J and three different sense primers without J (Fig. 3A). As a negative control, the shortest fragment was amplified with two J-less primers. The 118-bp PCR products were ligated to each other to obtain a ladder of fragments of different sizes but with the same J density. Fragments were end-labeled, incubated with αJ antisera, and antibody–DNA complexes were captured by protein-A beads. Bound DNA was released by protease treatment and phenol extraction, separated by agarose gel-electrophoresis, and then blotted (Fig. 3B). One J residue was sufficient to immunoprecipitate a fraction of the duplex DNA molecules (Fig. 3B, lanes 1,2) and this fraction decreased with length. The effect of length was less if the density of modification was kept constant (Fig. 3B, lanes 3,4). DNA without J was not immunoprecipitated (Fig. 3B, lane 5,6). Quantitation of the relative efficiency of immunoprecipitation (IP) of the various fragments (Fig. 3C) showed that anti-J IP is dependent on the size of the target fragment and the degree of modification.
Figure 3.
Specific immunoprecipitation of J-containing double-stranded DNA depends on the length of the fragments and the density of modification. (A) DNA fragments of 118, 426, and 943 bp were generated by PCR-amplification with one antisense primer containing one J residue and three different sense primers without J. (B) The 118-bp fragment was used to generate a partial ligation ladder, resulting in fragments of different size with a constant J-density (one J per 118 bp, or no J). End-labeled fragments bound by the J-specific antisera were captured by protein-A beads, separated by 1.5% agarose gel electrophoresis, and blotted onto Hybond-N. 100% of the immunoprecipitated DNA (ip) and 10% of the supernatant (sup) was loaded. (C) Quantitation of the immunoprecipitated fraction as a percentage of the total input (average of three experiments with standard deviation).
Having the tools to select for J-containing DNA, we set out to analyze modified genomic restriction fragments from BF T. brucei DNA. Because we had found previously that the telomeric repeats contain about 4% J compared with 0.2% J in the total genome (van Leeuwen et al. 1996), we first tested the long telomeric repeat arrays and found that despite their length (2–26 kb) these were immunoprecipitated readily (see below). Furthermore, immunoprecipitation of sonicated T. brucei DNA resulted in up to 20-fold enrichment for J (data not shown).
J is present in silenced telomeric VSG genes
To test whether J is present in the silenced telomeric VSG genes, we used three related BF trypanosome clones, each expressing a different VSG gene. One PC clone, not expressing any VSG gene and devoid of any modification was used as a negative control. Maps of the VSG genes, which are present in all four clones, are depicted in Figure 4A. 121a BF cells express VSG gene 121 (expression linked copy or ELC) in the dominant expression site (Liu et al. 1985). All four clones have three additional silent chromosome-internal VSG 121 genes (basic copies or BCs). 221a BF cells, which express the single-copy VSG gene 221 in the 221 expression site, arose from clone 121a by an in situ switch. r5-1.1 BF cells express the single-copy VSG gene 1.1 and arose from clone 221a by a complex event (see Materials and Methods). The inactive 221 gene at its new location is not modified on its PvuII site and is not sensitive to Bal31 exonuclease treatment, showing that the 221 gene had been transposed to a chromosome-internal position in clone r5-1.1 (data not shown).
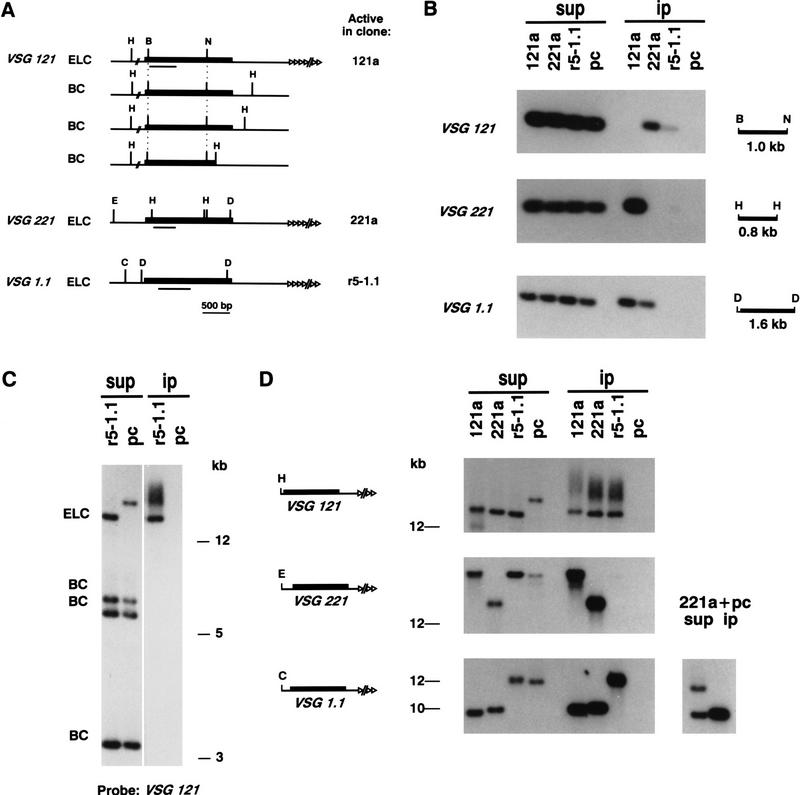
Figure 4.
Localization of J at the telomeric end of VSG expression sites by anti-J immunoprecipitation. (A) Restriction maps of VSG genes 121, 221, and 1.1, expressed in BF clones 121a, 221a, and r5-1.1 respectively. These genes are present in all clones analyzed here, including the PC clone, which was used as a negative control. In addition to the ELC of VSG 121, three chromosome-internal basic-copy (BC) 121 genes are present in all clones. In clone r5-1.1, the 221 gene has moved from its telomeric position in the expression site to a chromosome-internal position where the PvuII site is no longer modified (see text). (Solid boxes) Coding sequence; (open triangles) telomeric repeat arrays, of which the length varies per clone and per telomere. The VSG gene-specific probes are indicated underneath the coding sequences. (B) BglI; (C) ClaI; (D) DraI; (E) EcoRI; (H) HindIII; (N) NcoI. (B) Immunoprecipitation of VSG genes alone. DNA of the clones indicated at the top was digested with restriction enzymes shown on the right of each panel. Modified DNA fragments bound by the anti-J antibodies (ip) and 10% of the supernatant (sup) were analyzed by Southern blot hybridization. The VSG gene on the left of each panel indicates the probe used (probe fragments are shown in A). Note that the one telomeric copy and three basic copies of VSG gene 121 cannot be discriminated in the BglI–NcoI digest used for this VSG gene. (C) Analysis of modification of the three chromosome-internal BCs and the telomeric ELC of VSG gene 121 by αJ-IP of HindIII restriction fragments. A size marker is indicated on the right. (D) Analysis of the telomeric repeat arrays associated with expression sites. The fragments were detected by hybridization with probes specific for the VSG genes still linked to the telomeric tracts. The restriction digests, the VSG genes probed for, and a size marker are shown on the left of each panel. As a control, PC DNA was mixed with BF 221a DNA and probed for 1.1 to show that nonmodified PC telomeres do not specifically coimmunoprecipitate with modified BF DNA. Note that immunoprecipitation of VSG genes linked to telomeric repeats was consistently more efficient than that of VSG genes alone. Therefore, the smear upstream of the 121 telomere (sup), which is enriched on immunoprecipitation, is most likely attributable to cross-hybridization of the 121 probe to other VSG genes associated with telomeric tracts.
Figure 4B shows the results of immunoprecipitations of the three VSG genes studied. Genomic DNA of all clones was digested with various restriction enzymes to obtain small VSG gene fragments. These restriction fragments were analyzed by anti-J IP combined with Southern blot hybridization. In all BF clones, the actively transcribed VSG gene was not bound by antibodies, whereas silenced telomeric VSG genes were invariably immunoprecipitated. These results show a clear correlation between base J and telomeric gene repression.
Whereas the inactive telomeric 221 gene in 121a cells (Fig. 4B), and in 221aR12, 118a, and 118a′ cells (data not shown) was efficiently bound by antibodies, the silenced chromosome-internal 221 gene in r5-1.1 cells was not detectably bound by the antisera. This indicated that J is absent from chromosome-internal VSG genes. We confirmed this by analysis of the silent chromosome-internal 121 gene copies (BC), which can be separated from each other and from the ELC by a HindIII digest. None of the 121 BC genes was immunoprecipitated, whereas the ELC, here linked to the telomeric repeats, was pulled down (Fig. 4C; see below). The same lack of immunoprecipitation was found for the chromosome-internal basic copy of VSG gene 1.8 (data not shown). Together, the results from Figure 4, B and C, show that the inverse correlation between the presence of J and VSG gene activity holds true only for telomeric VSG genes, in agreement with the distribution of blocked restriction sites.
Telomere repeat arrays associated with inactive as well as active expression sites are modified
We have found previously that about half of J is present in telomeric repeats. Approximately 80% of the telomeres are part of minichromosomes (Van der Ploeg et al. 1984) and the analysis of telomeric repeats is therefore dominated by these minichromosomes, which do not contain functional VSG gene expression sites (Zomerdijk et al. 1990). To test whether the telomeric repeat arrays associated with expression sites were also modified, we analyzed restriction digests of genomic DNA in which the expression-linked VSG genes were still associated with the telomeric repeat arrays. This allowed the unique VSG sequences to be used as specific probes for individual telomere tracts. The length of these fragments varies between different clones because telomeres in trypanosomes grow and contract on cell division, resulting in clonal variation and heterogeneity of the size of the telomeric repeat arrays (Bernards et al. 1983). The results in Figure 4D show that VSG genes linked to telomeric repeats were immunoprecipitated efficiently. Unexpectedly, however, this occurred irrespective of the transcriptional state of the upstream expression site. Because transcribed VSG genes alone were not modified (Fig. 4B), these results show that the hexameric repeat tracts flanking active expression sites are modified. The efficient antibody binding of the long telomeric tracts of active and inactive sites shows that both must contain a similarly high degree of modification. This is in agreement with the 2%–4% J found in purified telomeric repeat arrays (van Leeuwen et al. 1996). The lack of IP of the PC 1.1 telomere in a mix of PC DNA and BF 221a DNA excludes the possibility that nonmodified telomeres nonspecifically coimmunoprecipitated with modified DNA (Fig. 4D). It has been suggested that transcription of the expression site reads through into the downstream telomeric repeats (Rudenko and Van der Ploeg 1989), and it is therefore possible that the first part of the repeat array is not modified. Transcribed repeats are thought to be sensitive to nucleases, resulting in formation of shorter telomeres during clonal propagation (Pays et al. 1983). The 12-kb smear associated with the 121 telomeric band in clone 121a (Fig. 4D, lane 1) could be an example of such an event and the absence of detectable immunoprecipitation of this smear might be caused by a higher proportion of transcribed repeats compared with the longer band.
Immunoprecipitation of VSG genes linked to telomeric repeats was always more efficient than that of (inactive) VSG genes alone. A silent VSG gene that is still linked to telomeric repeats because of partial cleavage caused by DNA modification will therefore be enriched by immunoprecipitation. This has allowed us to identify two additional restriction enzymes that yield partial cleavage products with silenced telomeric VSG genes—NcoI and DraI showed partial cleavage of VSG gene 121 and 1.1, and VSG gene 1.1, respectively (data not shown; see Discussion).
J in and around expression site promoters
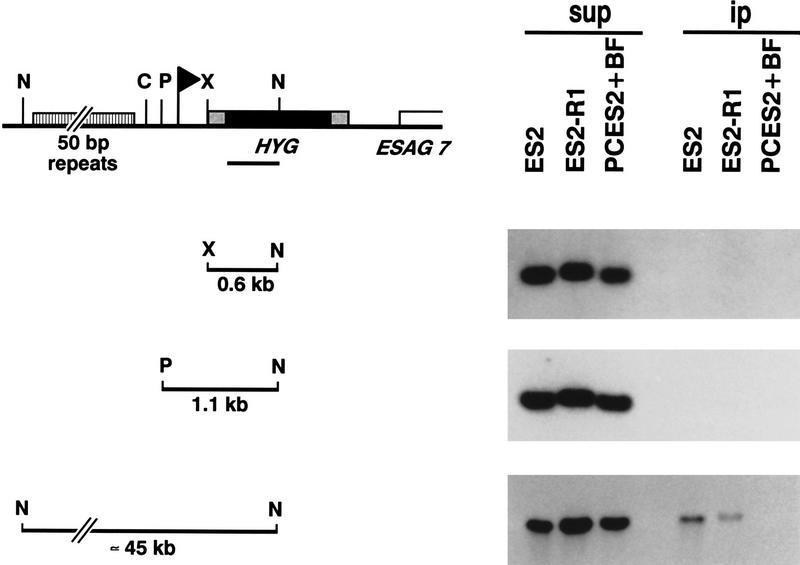
The experiments described above show that J is present in silenced telomeric VSG genes and in expression site-associated telomeric tracts, the most distal sequences in the expression site. The analysis of expression site sequences other than VSG genes is complicated by the high degree of homology between expression sites (Pays et al. 1989a; Kooter et al. 1987). To study the promoter region of expression sites, we used cell lines in which the 221 expression site was tagged with a unique sequence, the hygromycin resistance (HYG) gene (Rudenko et al. 1995; Blundell et al. 1996). In anti-J IP experiments, a small 0.6-kb segment of the HYG gene alone, either active (ES2) or silent (ES2-R1), did not bind to the antibodies (Fig. 5). A longer 1.1-kb fragment spanning the promoter element was also negative. The upstream part of the HYG gene linked to the long 50-bp repeat array, however, was immunoprecipitated, both from an inactive and an active expression site (Fig. 5). The absence of IP of the nonmodified HYG-marked 50-bp repeat fragment from PC cells (PCES2) mixed with BF 221a DNA (which does not contain a HYG gene) excludes nonspecific co-IP of these very long restriction fragments (∼45 kb). Cell lines in which a HYG gene was integrated in a 221 expression site, in which the expression site promoter was replaced by a ribosomal RNA promoter, gave the same results (data not shown). J is therefore present in the long repetitive DNA stretches closely upstream of the promoter, regardless of expression site activity. The function of the 50-bp repeat arrays is not known, but hybridization studies have shown that 50-bp repeats are invariably associated with expression site promoter sequences (Zomerdijk et al. 1990, 1991; G. Rudenko and P. Borst, unpubl.).
Figure 5.
Detection of J in the 50-bp repeats upstream of inactive and active expression sites. Cell lines with a HYG gene in an active (ES2) or inactive (ES2-R1) 221 expression site were used for anti-J immunoprecipitation of sequences in and around the expression site promoter (flag). (N) NcoI; (C) ClaI; (P) HpaI; (X) XbaI. The 5′ part of the HYG gene (line underneath the map) was used as a probe to specifically detect the tagged expression site sequences. The fragments analyzed are indicated on the left of each panel and include from top to bottom HYG gene alone, HYG gene linked to expression site promoter sequences, and HYG gene linked to the 50-bp repeat array. DNA of PC cells with a HYG gene downstream of the expression site promoter (PCES2) was mixed with wild-type BF 221a DNA (BF) as a negative control for nonspecific coimmunoprecipitation of nonmodified DNA. The solid box indicates the HYG-coding sequence, stippled boxes RNA-processing signals, and the striped box 50-bp repeats.
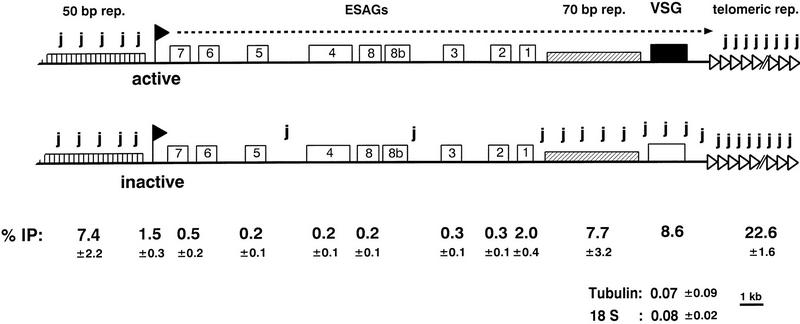
The presence of J at the borders of expression sites prompted us to test whether sequences in between the VSG gene and the promoter are modified in inactive expression sites. The lack of probes specific for individual expression sites in this region only allowed global analysis of the total pool of expression sites. Therefore, genomic DNA was sonicated and analyzed by αJ-immunoprecipitation combined with dot-blot hybridization. Because all sequences are sonicated to the same size range (0.5–3 kb), the relative efficiency of immunoprecipitation could be used as a measure for the density of modification (see also Fig. 3). Telomeric repeats, 50-bp repeats, inactive VSG genes, and also 70-bp repeats, which are just upstream of VSG genes, were immunoprecipitated efficiently (Fig. 6). Expression site promoter and ESAG 1 fragments bound inefficiently, and other ESAGs bound even more inefficiently to the antibodies, albeit still three to four times more than chromosome-internal DNA, such as tubulin genes or ribosomal 18S DNA (Fig. 6). These results show that expression site sequences are only sparsely modified outside the VSG gene and the repeats. Whether expression site promoter and ESAG 1 genes are really modified more densely than the other ESAGs, or whether the greater immunoselection is caused by linkage to modified 50- or 70-bp repeats is uncertain. We could not use sonicated DNA fragments shorter than 500 bp because this resulted in higher background IP. It should also be noted that 70-bp repeats and copies of some ESAGs (but not all) are also present outside of expression sites. Whether these outsiders are also modified and contribute to the immunoprecipitated fraction is not known.
Figure 6.
The modified base J in and around telomeric VSG expression sites. Schematic representation of the distribution of J in active and inactive VSG expression sites determined by αJ immunoprecipitation of sonicated DNA combined with dot-blot hybridizations (expression site adapted from Revelard et al. 1990). ES probes are described in Materials and Methods. ESAGs were studied as the cumulative signal of all copies in the genome using sonicated DNA. Telomeric repeats, 50-bp repeats, 70-bp repeats, VSG genes, and promoter regions were also studied in individual expression sites using restriction digests. % IP shows a quantitation of the IP efficiency (immunoprecipitated fraction of the input) of expression site sequences using sonicated DNA (average with standard deviation of two independent clones 221a and 221aR12). The numbers correspond to the expression site sequences shown above. IP of silent VSG genes in expression sites varied from 1 (±0.1) to 16.2 (±3.4) depending on the VSG gene studied. The absence of J in actively transcribed VSG genes and 70-bp repeats strongly suggests that J is also absent from active ESAGs, but this has not been tested directly.
We also analyzed the 70-bp repeats in a specific expression site using their linkage to the unique VSG pseudogene (Ψ), which is embedded in the 70-bp repeats in the 221 expression site (Bernards et al. 1985; Cornelissen et al. 1985). With restriction fragments of ∼9.5 kb (HindIII) and 6.5 kb (BglI, NcoI) containing the 70-bp repeat array and (part of) the pseudo gene, we found efficient immunoprecipitation (∼5%) with the inactive 221 expression site from clone 121a and no antibody binding of the transcribed fragment from clone 221a (data not shown). These results show that J is absent from VSG genes and 70-bp repeats in active expression sites and suggest that transcribed ESAGs also lack J.
Discussion
Partial cleavage by PstI, PvuII, and other restriction enzymes suggested previously the presence of DNA modifications in silenced telomeric VSG genes in BF T. brucei (Bernards et al. 1984b; Pays et al. 1984). To test whether this is caused by J, we first verified that the presence of J in a PvuII restriction site blocks cleavage by PvuII enzyme (Fig. 1). Interestingly, a J replacing T only two positions away from the PvuII site did not block cleavage, showing that J does not affect cleavage at a distance. By anti-J DNA immunoprecipitations, we subsequently found that J correlates with silencing of telomeric VSG genes. An inverse correlation between DNA modification and transcription of specific genes, as has been found for 5MeC in complex eukaryotes, has not been found before in simple eukaryotes (Rae and Steele 1978; Blackburn et al. 1983; Capowski et al. 1989; Bird 1995; Jablonka and Regev 1995; Tweedie et al. 1997).
By anti-J immunoprecipitations, J was also found in expression site sequences in which DNA modification previously remained undetected. The boundaries of expression sites, marked by long upstream 50-bp repeat arrays and long downstream telomeric repeat arrays, were modified substantially, regardless of expression site activity. By studying the total pool of expression sites with immunoprecipitations of sonicated DNA, we found low levels of J around the expression site promoter and in the ESAGs. No J was detected in a promoter fragment derived from a silent expression site and tagged with a HYG gene, but specific modification of a single thymine would not have been detected in these experiments.
We conclude that J has all the properties of the modification in PvuII and PstI restriction sites detected by Bernards et al. (1984b) and Pays et al. (1984)—both are developmentally regulated, that is, are present in BF trypanosomes and are absent from PC trypanosomes, both are found in silent telomeric VSG genes and not in active VSG genes or chromosome-internal VSG genes. The gradient of modification from telomere to chromosome-internal found for PstI and PvuII sites in VSG genes (Bernards et al. 1984b) correlates with the gradient of J found from telomeric repeats (high, to VSG genes, 70-bp repeats, ESAG 1, and other ESAGs (low). With the expression site telomeres studied here, no correlation was found between the length of the telomeric repeat array and the levels of J in VSG genes. Possibly the size difference of the individual telomeres studied was not great enough to cause a difference in immunoprecipitation.
The abundance of J in different repetitive DNA sequences, such as the telomeric, 50- and 70-bp repeats (Fig. 6), and the 177-bp repeats (data not shown), shows that the modifying enzyme (complex) that introduces J into DNA has a preference for repetitive DNA that is associated to telomeres. In addition to tandem repeats, however, VSG genes were also modified. The restriction of J to silent telomeric copies of VSG genes and the gradient of modification from telomere to chromosome-internal can be explained by an enzyme that recognizes telomeric repeats and that slides down the neighboring DNA, as proposed by Bernards et al. (1984b). Alternatively, the repetitive DNA surrounding VSG genes in expression sites might impose a specific chromatin structure or sub-nuclear localization on these VSG genes such that they also become a target for the modifying enzyme. The absence of J in transcribed telomeric VSG genes and 70-bp repeats is compatible with a competition between transcription and DNA modification in expression sites.
The J-synthesizing enzyme seems to have a preference for certain sites, but these sites do not show a clear consensus. In the telomeric repeats, both the (TAACCC)n and the (GGGTTA)n strand are modified, and in the G-rich strand only the second T is replaced by J (van Leeuwen et al. 1996). In and around telomeric VSG genes, mainly PvuII (CAGCTG) and PstI (CTGCAG) (Bernards et al. 1984b), but also HindIII (AAGCTT) and SphI (GCATGC) (Pays et al. 1984), and DraI (TTTAAA) and NcoI (CCATGG) (data not shown), showed partial cleavage. Unfortunately, a genomic sequencing method (Clark et al. 1994) developed to discriminate between T and J, was not specific enough to detect J at a certain site in a small fraction of a trypanosome population (J. Gommers-Ampt and P. Borst, unpubl.).
What could be the function of J in trypanosomes? If J is involved in expression site control, two functions could be envisaged. First, introduction of J causes expression site inactivation. Changes in modification, for example, after DNA replication, could allow activation of silent expression sites. Second, J is a consequence of expression site silencing and could help to stably maintain the repressed state of an inactive expression site. The latter model is supported by the observed abundance of J in repetitive DNA in T. brucei (described above), and by the recent finding of J in protozoans without antigenic variation (F. van Leeuwen and P. Borst, unpubl.). Those results suggest that J is not specifically involved in control of expression sites but is likely to have a more general function in the genome. It is not clear, however, why T. brucei would require such a function of J only in the mammalian stage of the life cycle. A critical test of the function of J awaits the identification and knock-out of the enzymes that make J or the identification of inhibitors that interfere with synthesis of J.
Materials and methods
Trypanosome clones and DNA
BF trypanosome clones 221a or MiTat 1.2 (Bernards et al. 1984a), 121a or MiTat 1.6a, 118a or MiTat 1.5a (Cross 1975), 221aR12 (Zomerdijk et al. 1990), or T. brucei strain 427 (Cross and Manning 1973) were grown and isolated as described (Gommers-Ampt et al. 1991). PC trypanosomes were grown in a semidefined medium (Brun and Schönenberger 1979). Clone r5-1.1 arose in a single relapse experiment from clone 221a, described as 221ar2 (Bernards et al. 1984a). From this polyclonal relapse population we cloned a 1.1 expresser (determined by Northern blot analysis), which was called r5-1.1. Pulsed-field gel analysis of this clone showed that the 1.1 gene and the 221 gene had exchanged their chromosomal position suggesting reciprocal telomeric exchange. VSG gene 1.1 had moved to chromosome band 15, while the 221 gene had moved to chromosome band 14. Further analysis showed that the silent 221 gene at its new position was not modified on the PvuII restriction site present in the coding sequence and showed that it was insensitive to Bal31 exonuclease treatment (data not shown). These results indicate that the 221 gene in clone r5-1.1 had moved to a chromosome-internal position. Total genomic DNA was isolated as described (Bernards et al. 1981) and resuspended in 10 mm Tris-HCl/1 mm EDTA (pH 7.4). Digested or sonicated DNA was transferred to nitrocellulose or Hybond-N (Amersham) by standard procedures (Sambrook et al. 1989). Probes were labeled with [α-32P]dATP by random priming. A 5′ 32P-labeled oligomer consisting of 5 telomeric GGGTTA repeats was used to probe for telomeric repeats. Probe fragments for subtelomeric sequence, 177-bp repeats, 70-bp repeats, 50-bp repeats, β-tubulin gene, ribosomal DNA, and kinetoplast DNA were all described in van Leeuwen et al. (1996). Other probes used were a 360-bp BamHI–NcoI 5′ HYG fragment (Blundell et al. 1996), a HindIII–BamHI ESAG 7 fragment (Zomerdijk et al. 1990), an 870-bp PstI pseudo VSG gene (Ψ) fragment (Cornelissen et al. 1985), and an 850-bp ESAG 1 fragment (McCulloch et al. 1997). Other ESAG-specific probe fragments were generated by PCR amplification (Taq polymerase with 1.5 mm MgCl2, 30 cycles of 1 min 94°C, 2 min 55°C, 2 min 72°C) of cloned ESAG sequences from the AnTat 1.3A expression site (Pays et al. 1989b; Alexandre et al. 1988) using sense primers (sp) with a 5′ BamHI site (underlined) flanked by a terminal CG dinucleotide and antisense primers (asp) with a 5′ XbaI site (underlined) flanked by CG. 5′ part of ESAG 2 (clone pBES 2000.1, sp CGGGATCCGATGAGTGTACGAGAGAGATGC, asp CGTCTAGATGATCAGCGTCTTTCCAACC); 5′ part of ESAG 3 (clone pES 200.5, sp CGGGATCCAACACAAGGATGGTGTAGGC, asp CGTCTAGACTAAATGCCCAGACTCTGGC); middle part of ESAG 8 (clone pES 200.8, sp CGGGATCCAGGGAGTTGGATATCTCCGG, asp CGTCTAGACCAGTCAAACACTGAAGTCC); 5′ part of ESAG 4 (clone lES 200.10, sp CGGGATCCACTTGAGCGACCGCAATGCC, asp CGTCTAGAAACTGGCATAGCGAATACCG); middle part of ESAG 5 (clone lES 200.10, sp CGGGATCCCATACATGTAGGGAGTTCGG, asp CGTCTAGATTAGGGACTTCAACCACGGG). VSG gene-specific probes were generated from cDNAs cloned into pBluescript—a 560-bp BglII–PstI fragment of VSG gene 121 (Liu et al. 1985), an 820-bp HindIII fragment of VSG 221, a 512-bp PstI–NcoI fragment of VSG 1.1, a 520-bp PstI fragment of VSG 1.8, and a 520-bp EcoRI–PstI fragment of VSG 118 (Michels et al. 1984), and a 600-bp EcoRI–HindIII fragment of VSG VO2 (Rudenko et al. 1995). Dot blots were scanned and quantitated on a PhosphorImager (Fujix BAS 2000, TINA 2.08b).
Endonuclease digestion of duplex oligonucleotides
Oligonucleotides encompassing the upper and the lower strand of the PvuII site (underlined) of the VSG 221 gene were used to generate non- or hemimodified duplex molecules. Oligos were end-labeled with [γ-32P]ATP, purified by exclusion chromatography, and annealed to their nonlabeled J-lacking complementary strand by gradually cooling down from 90°C to room temperature in 10 mm Tris-HCl (pH 7.5), 1 mm EDTA, 100 mm NaCl; upper-T (CAGAAGGCAGCTGCAACAAG) or upper-J (CAGAAGGCAGCJGCAACAAG) was annealed to lower-T (CTTGTTGCAGCTGCCTTCTG), lower-J (CTTGTTGCAGCJGCCTTCTG), or lower-J* (CTTGTJGCAGCTGCCTTCTG). The duplex oligos were incubated for 2 hr at 37°C in the appropriate restriction buffer with or without 10 units of PvuII. The products were separated by 20% native polyacrylamide gel electrophoresis (19:1, 1× TBE).
Generation of J-specific polyclonal antisera
Chemically synthesized JMP (Wijsman et al. 1994) was coupled to carrier proteins with a water-soluble carbodiimide [1-ethyl-3-(3-dimethylaminopropyl)carbodiimide HCl, or EDC, Sigma] according to a protocol modified from (Halloran and Parker 1966; Stollar 1980). Three micrograms of JMP and 400 mg of EDC were mixed with 4 mg of BSA (imject BSA, Pierce) or 4 mg of KLH (Calbiochem) in 1 ml of H2O, and incubated for 20 hr in the dark at room temperature or 37°C, respectively. This resulted in formation of phosphoramadite conjugates through the 5′-phosphate of JMP and the amino groups of the carrier proteins (Halloran and Parker 1966; Stollar 1980). The samples were dialyzed three times against 1000 volumes of PBS to remove the free JMP and EDC, monitored by UV absorbance (263/280 nm) to confirm crosslinking, and subsequently stored in 10% glycerol at −70°C. Twelve percent of the protein–nucleotide complex was injected into rabbits. Antisera were obtained against BSA–JMP (539αJ) and KLH–JMP (538αJ).
Anti J-DNA immunoblot
DNA was denatured for 20 min on ice in 0.4 n NaOH, neutralized by adding one volume of ice-cold 2.5 m ammonium acetate, and blotted onto nitrocellulose using a manifold dot-blot apparatus. The filters were baked for 2 hr at 80°C and blocked for 2 hr in TBST (10 mm Tris-HCl at pH 8.0, 150 mm NaCl, 0.02% Tween-20) with 5% milk powder. After three washes with TBST, the blots were incubated for 2 hr with antiserum 539αJ, diluted 1:10,000-fold in TBST with 2% milk powder, and then washed three times with TBST. Immunodetection was performed using a horseradish peroxidase (HRP) conjugated second antibody (CLB, The Netherlands) in 2% milk powder in TBST, in combination with enhanced chemiluminescence (ECL, Amersham).
Generation and ligation of J-containing PCR fragments
One antisense primer with or without one J residue and three sense primers without J were used to generate J-containing and J-less 221 VSG gene PCR fragments of different sizes with Pwo polymerase. The antisense primers used were CTTGTTGCAGCJGCCTTCTG and CTTGTTGCAGCTGCCTTCTG (221as1247), the sense primers used were 221s1129 (CGACTATATACTTGCCTATTACCG), 221s821 (ACCGTGGATCGACGACGCCTG), and 221s304 (CCAACCACTATGCCATGA). PCR fragments were purified by QIAEX gel extraction (Qiagen) and the presence or absence of J was confirmed by 32P-nucleotide postlabeling combined with two-dimensional TLC. For detection of anti-J immunoprecipitation the fragments were end-labeled and purified by exclusion chromatography. Part of the phosphorylated fragments were ligated for 16 hr at 16°C to generate ladders of fragments with a constant ratio of J/bp.
J–DNA immunoprecipitation
Digested or sonicated DNA (2–5 μg) was added to 5 μl antiserum 538αJ in a final volume of 500 μl IP buffer [TBST with 2 mm EDTA (TBSTE), 0.1 mg/ml of tRNA, and 1 mg/ml of BSA], and incubated for 2 hr at room temperature. ProtA beads (20–30 μl, Repligen) were washed twice with TBSTE, preblocked for 30 min in 100 μl IP buffer, and incubated for 1 hr with the IP reaction. Ten to 20% of the supernatant was taken and used as a control for the DNA input. The bead–antibody–DNA complexes were washed four times with TBSTE and finally proteinase K-treated at 58°C to release the bound DNA, which was phenol-extracted, and ethanol-precipitated with 20 μg glycogen.
Acknowledgments
We thank Magali Berberof, Pat Blundell, Inês Chaves, Mike Cross, Anita Dirks, Herlinde Gerrits, Gloria Rudenko, Ronald Plasterk, and Anton Berns for helpful discussions and critical reading of the manuscript, and P. Blundell and G. Rudenko for providing HYG-marked cell lines. We thank Ben Floot for helpful suggestions for the generation of the anti-J antisera, and E. Pays (Université Libre de Bruxelles, Brussels) for kindly providing AnTat1.3 genomic clones. This work was supported by grants from the Netherlands Foundation for Chemical Research (SON), with financial support of the Netherlands Organization for Scientific Research (NWO).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
FAX 31-20-669-1383.
References
- Alexandre S, Guyaux M, Murphy NB, Coquelet H, Pays A, Steinert M, Pays E. Putative genes of a variant-specific antigen gene transcription unit in Trypanosoma brucei. Mol Cell Biol. 1988;8:2367–2378. doi: 10.1128/mcb.8.6.2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A, Van der Ploeg LH, Frasch AC, Borst P, Boothroyd JC, Coleman S, Cross GA. Activation of trypanosome surface glycoprotein genes involves a duplication-transposition leading to an altered 3′ end. Cell. 1981;27:497–505. doi: 10.1016/0092-8674(81)90391-3. [DOI] [PubMed] [Google Scholar]
- Bernards A, Michels PAM, Lincke CR, Borst P. Growth of chromosome ends in multiplying trypanosomes. Nature. 1983;303:592–597. doi: 10.1038/303592a0. [DOI] [PubMed] [Google Scholar]
- Bernards A, de Lange T, Michels PA, Liu AY, Huisman MJ, Borst P. Two modes of activation of a single surface antigen gene of Trypanosoma brucei. Cell. 1984a;36:163–170. doi: 10.1016/0092-8674(84)90085-0. [DOI] [PubMed] [Google Scholar]
- Bernards A, van Harten-Loosbroek N, Borst P. Modification of telomeric DNA in Trypanosoma brucei; a role in antigenic variation? Nucleic Acids Res. 1984b;12:4153–4170. doi: 10.1093/nar/12.10.4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A, Kooter JM, Borst P. Structure and transcription of a telomeric surface antigen gene of Trypanosoma brucei. Mol Cell Biol. 1985;5:545–553. doi: 10.1128/mcb.5.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- Blackburn EH, Pan WC, Johnson CC. Methylation of ribosomal RNA genes in the macronucleus of Tetrahymena thermophila. Nucleic Acids Res. 1983;11:5131–5145. doi: 10.1093/nar/11.15.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell PA, Rudenko G, Borst P. Targeting of exogenous DNA into Trypanosoma brucei requires a high degree of homology between donor and target DNA. Mol Biochem Parasitol. 1996;76:215–229. doi: 10.1016/0166-6851(95)02560-x. [DOI] [PubMed] [Google Scholar]
- Borst P, Bitter W, Blundell PA, Cross MA, McCulloch R, Rudenko G, Taylor MC, van Leeuwen F. The expression sites for variant surface glycoproteins of Trypanosoma brucei. In: Hide G, Mottram JC, Coombs GH, Holmes PH, editors. Trypanosomiasis and Leishmaniasis. Oxford, UK: BSP/CAB International; 1997. pp. 109–131. [Google Scholar]
- Brun R, Schönenberger M. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- Capowski EE, Wells JM, Harrison GS, Karrer KM. Molecular analysis of N6-methyladenine patterns in Tetrahymena thermophila nuclear DNA. Mol Cell Biol. 1989;9:2598–2605. doi: 10.1128/mcb.9.6.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen AW, Johnson PJ, Kooter JM, Van der Ploeg LH, Borst P. Two simultaneously active VSG gene transcription units in a single Trypanosoma brucei variant. Cell. 1985;41:825–832. doi: 10.1016/s0092-8674(85)80063-5. [DOI] [PubMed] [Google Scholar]
- Cross GA. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology. 1975;71:393–417. doi: 10.1017/s003118200004717x. [DOI] [PubMed] [Google Scholar]
- ————— Antigenic variation in trypanosomes: Secrets surface slowly. BioEssays. 1996;18:283–291. doi: 10.1002/bies.950180406. [DOI] [PubMed] [Google Scholar]
- Cross GA, Manning JC. Cultivation of Trypanosoma brucei sspp. in semi-defined and defined media. Parasitology. 1973;67:315–331. doi: 10.1017/s0031182000046540. [DOI] [PubMed] [Google Scholar]
- Crozatier M, De Brij RJ, Den Engelse L, Johnson PJ, Borst P. Nucleoside analysis of DNA from Trypanosoma brucei and Trypanosoma equiperdum. Mol Biochem Parasitol. 1988;31:127–131. doi: 10.1016/0166-6851(88)90163-6. [DOI] [PubMed] [Google Scholar]
- Gommers-Ampt J, Lutgerink J, Borst P. A novel DNA nucleotide in Trypanosoma brucei only present in the mammalian phase of the life-cycle. Nucleic Acids Res. 1991;19:1745–1751. doi: 10.1093/nar/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers-Ampt JH, van Leeuwen F, de Beer AL, Vliegenthart JF, Dizdaroglu M, Kowalak JA, Crain PF, Borst P. β-D-glucosyl-hydroxymethyluracil: A novel modified base present in the DNA of the parasitic protozoan T. brucei. Cell. 1993;75:1129–1136. doi: 10.1016/0092-8674(93)90322-h. [DOI] [PubMed] [Google Scholar]
- Halloran MJ, Parker CW. The preparation of nucleotide-protein conjugates: Carbodiimides as coupling agents. J Immunol. 1966;96:373–378. [PubMed] [Google Scholar]
- Horn D, Cross GAM. A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell. 1995;83:555–561. doi: 10.1016/0092-8674(95)90095-0. [DOI] [PubMed] [Google Scholar]
- ————— Analysis of Trypanosoma brucei vsg expression site switching in vitro. Mol Biochem Parasitol. 1997;84:189–201. doi: 10.1016/s0166-6851(96)02794-6. [DOI] [PubMed] [Google Scholar]
- Huang LH, Farnet CM, Ehrlich KC, Ehrlich M. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 1982;10:1579–1591. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E, Regev A. Gene number, methylation and biological complexity. Trends Genet. 1995;11:383–384. doi: 10.1016/s0168-9525(00)89117-9. [DOI] [PubMed] [Google Scholar]
- Kooter JM, van der Spek HJ, Wagter R, d’Oliveira CE, van der Hoeven F, Johnson PJ, Borst P. The anatomy and transcription of a telomeric expression site for variant-specific surface antigens in T. brucei. Cell. 1987;51:261–272. doi: 10.1016/0092-8674(87)90153-x. [DOI] [PubMed] [Google Scholar]
- Kornberg SR, Zimmerman SB, Kornberg A. Glucosylation of deoxyribonucleic acid by enzymes from bacteriophage-infected Escherichia coli. J Biol Chem. 1961;236:1487–1493. [PubMed] [Google Scholar]
- Liu AY, Michels PA, Bernards A, Borst P. Trypanosome variant surface glycoprotein genes expressed early in infection. J Mol Biol. 1985;182:383–396. doi: 10.1016/0022-2836(85)90198-6. [DOI] [PubMed] [Google Scholar]
- McClelland M, Nelson M, Raschke E. Effect of site-specific modification on restriction endonucleases and DNA modification methyltransferases. Nucleic Acids Res. 1994;22:3640–3659. doi: 10.1093/nar/22.17.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch R, Rudenko G, Borst P. Gene conversions mediating antigenic variation in Trypanosoma brucei can occur in VSG expression sites lacking 70 bp repeat sequences. Mol Cell Biol. 1997;17:833–843. doi: 10.1128/mcb.17.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels PA, Van der Ploeg LH, Liu AY, Borst P. The inactivation and reactivation of an expression-linked gene copy for a variant surface glycoprotein in Trypanosoma brucei. EMBO J. 1984;3:1345–1351. doi: 10.1002/j.1460-2075.1984.tb01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E, Laurent M, Delinte K, Van Meirvenne N, Steinert M. Differential size variations between transcriptionally active and inactive telomeres of Trypanosoma brucei. Nucleic Acids Res. 1983;11:8137–8147. doi: 10.1093/nar/11.23.8137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E, Delauw MF, Laurent M, Steinert M. Possible DNA modification in GC dinucleotides of Trypanosoma brucei telomeric sequences; relationship with antigen gene transcription. Nucleic Acids Res. 1984;12:5235–5247. doi: 10.1093/nar/12.13.5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E, Coquelet H, Pays A, Tebabi P, Steinert M. Trypanosoma brucei: Posttranscriptional control of the variable surface glycoprotein gene expression site. Mol Cell Biol. 1989a;9:4018–4021. doi: 10.1128/mcb.9.9.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pays E, Tebabi P, Pays A, Coquelet H, Revelard P, Salmon D, Steinert M. The genes and transcripts of an antigen gene expression site from T. brucei. Cell. 1989b;57:835–845. doi: 10.1016/0092-8674(89)90798-8. [DOI] [PubMed] [Google Scholar]
- Pays E, Vanhamme L, Berberof M. Genetic controls for the expression of surface antigens in African trypanosomes. [Review] Annu Rev Microbiol. 1994;48:25–52. doi: 10.1146/annurev.mi.48.100194.000325. [DOI] [PubMed] [Google Scholar]
- Rae PM, Steele RE. Modified bases in the DNAs of unicellular eukaryotes: An examination of distributions and possible roles, with emphasis on hydroxymethyluracil in dinoflagellates. Biosystems. 1978;10:37–53. doi: 10.1016/0303-2647(78)90027-8. [DOI] [PubMed] [Google Scholar]
- Revelard P, Lips S, Pays E. A gene from the VSG expression site of Trypanosoma brucei encodes a protein with both leucine-rich repeats and a putative zinc finger. Nucleic Acids Res. 1990;18:7299–7303. doi: 10.1093/nar/18.24.7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko G, Blundell PA, Dirks-Mulder A, Kieft R, Borst P. A ribosomal DNA promoter replacing the promoter of a telomeric VSG gene expression site can be efficiently switched on and off in T. brucei. Cell. 1995;83:547–553. doi: 10.1016/0092-8674(95)90094-2. [DOI] [PubMed] [Google Scholar]
- Rudenko G, Van der Ploeg LH. Transcription of telomere repeats in protozoa. EMBO J. 1989;8:2633–2638. doi: 10.1002/j.1460-2075.1989.tb08403.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Stollar BD. The experimental induction of antibodies to nucleic acids. Methods Enzymol. 1980;70:70–85. doi: 10.1016/s0076-6879(80)70042-3. [DOI] [PubMed] [Google Scholar]
- Tweedie S, Charlton J, Clark V, Bird A. Methylation of genomes and genes at the invertebrate-vertebrate boundary. Mol Cell Biol. 1997;17:1469–1475. doi: 10.1128/mcb.17.3.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Ploeg LH, Schwartz DC, Cantor CR, Borst P. Antigenic variation in Trypanosoma brucei analyzed by electrophoretic separation of chromosome-sized DNA molecules. Cell. 1984;37:77–84. doi: 10.1016/0092-8674(84)90302-7. [DOI] [PubMed] [Google Scholar]
- van Leeuwen F, Wijsman ER, Kuyl-Yeheskiely E, van der Marel G, van Boom JH, Borst P. The telomeric GGGTTA repeats of Trypanosoma brucei contain the modified base J in both strands. Nucleic Acids Res. 1996;24:2476–2482. doi: 10.1093/nar/24.13.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijsman ER, van den Berg O, Kuyl-Yeheskiely E, van der Marel GA, van Boom JH. Synthesis of 5-(β-D-glucopyranosyloxymethyl)-2′-deoxyuridine and derivatives thereof. A modified d-nucleoside from the DNA of Trypanosoma brucei. Rec Trav Chim Pays-Bas. 1994;113:337–338. [Google Scholar]
- Zomerdijk JC, Ouellette M, ten Asbroek AL, Kieft R, Bommer AM, Clayton CE, Borst P. The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. EMBO J. 1990;9:2791–2801. doi: 10.1002/j.1460-2075.1990.tb07467.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomerdijk JC, Kieft R, Duyndam M, Shiels PG, Borst P. Antigenic variation in Trypanosoma brucei: A telomeric expression site for variant-specific surface glycoprotein genes with novel features. Nucleic Acids Res. 1991;19:1359–1368. doi: 10.1093/nar/19.7.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]