Abstract
The Arabidopsis gai mutant allele confers a reduction in gibberellin (GA) responsiveness. Here we report the molecular cloning of GAI and a closely related gene GRS. The predicted GAI (wild-type) and gai (mutant) proteins differ only by the deletion of a 17-amino-acid segment from within the amino-terminal region. GAI and GRS contain nuclear localization signals, a region of homology to a putative transcription factor, and motifs characteristic of transcriptional coactivators. Genetic analysis indicates that GAI is a repressor of GA responses, that GA can release this repression, and that gai is a mutant repressor that is relatively resistant to the effects of GA. Mutations at SPY and GAR2 suppress the gai phenotype, indicating the involvement of GAI, SPY, and GAR2 in a signaling pathway that regulates GA responses negatively. The existence of this pathway suggests that GA modulates plant growth through derepression rather than through simple stimulation.
Keywords: Arabidopsis thaliana, gai, gibberellin, mutant, signal transduction
Gibberellins (GAs) are tetracyclic diterpenoid growth factors that are essential regulators of stem elongation and other plant developmental processes (Hooley 1994; Swain and Olszewski 1996). GA-related mutants have been identified in several plant species, including Arabidopsis (Ross 1994). GA-deficient Arabidopsis mutants display characteristic phenotypes, including dark green leaves and a dwarf growth habit attributable to reduced stem elongation (Koornneef and van der Veen 1980; Talon et al. 1990a; Sun and Kamiya 1994; Peng and Harberd 1997). gai is a semidominant mutation of Arabidopsis, which also confers a dark green, dwarf phenotype (Koornneef et al. 1985; Peng and Harberd 1993, 1997; Wilson and Somerville 1995). The gai mutation affects GA reception or subsequent signal transduction, and does not result in GA deficiency (Koornneef et al. 1985; Talon et al. 1990b; Wilson et al. 1992; Peng and Harberd 1993; Wilson and Somerville 1995).
Dominant mutations conferring visible phenotypes resembling those attributable to GA deficiency are also known in other plants, including maize (D8 allelic series; Harberd and Freeling 1989; Winkler and Freeling 1994) and wheat (Rht homeoallelic series; Gale et al. 1975). Previous genetic and physiological analyses of gai, D8, and Rht indicate that all are gain-of-function mutations (Gale et al. 1975; Harberd and Freeling 1989; Peng and Harberd 1993; Winkler and Freeling 1994; Wilson and Somerville 1995) conferring reduced GA responses and increased endogenous GA levels (Lenton et al. 1987; Fujioka et al. 1988; Talon et al. 1990b). The increased endogenous GA levels found in gai, D8, and Rht mutants are likely to arise through perturbation of the feedback control mechanisms by which GAs regulate in planta GA levels negatively (Croker et al. 1990; Chiang et al. 1995; Phillips et al. 1995; Xu et al. 1995). These dominant GA-response mutations are of considerable agricultural significance. The Rht mutations are especially important because they are the genetic basis of the high-yielding, semi-dwarf wheat varieties of the “green revolution” (Gale and Youssefian 1985). We cloned GAI to enhance our understanding of the mechanisms of GA signal transduction, and because of the potential use for gai in crop improvement.
Previous experiments had identified a T-DNA insertion, genetically linked to GAI, which contained a Ds transposable element (Peng and Harberd 1993). This Ds was used to clone GAI through targeted insertional mutagenesis. Comparison of GAI and gai DNA sequences shows that the predicted mutant protein (gai) lacks a short (17-amino-acid) segment of the GAI protein sequence. We propose that this structural alteration is responsible for the dominant, gain-of-function properties of gai. In addition, presumed null alleles of GAI confer increased resistance to the growth-retarding effects of paclobutrazol (PAC), an inhibitor of GA biosynthesis. These observations suggest the following hypotheses to explain the role of GAI in GA signaling. First, GAI is proposed to be a negative regulator that represses GA responses but whose activity is opposed by GA. Second, gai is proposed to be a mutant repressor that is relatively resistant to the effects of GA and, therefore, maintains repression irrespective of the presence of GA.
Several recent publications have described extragenic mutations that suppress the phenotype conferred by gai (Carol et al. 1995; Wilson and Somerville 1995; Jacobsen et al. 1996) or by GA deficiency mutations (Jacobsen and Olszewski 1993; Silverstone et al. 1997). Here we extend the analysis of the phenotypes conferred by two of these suppressors (spy-7 and gar2-1). First, we compare the effects of spy-7 and gar2-1 (alone and in combination) on the growth of and PAC resistance of plants containing gai. We have also investigated the effects of spy-7 and gar2-1 on the regulation of GA biosynthesis, by comparing the steady-state levels of gene transcripts encoding GA C-20 oxidase, the enzyme that catalyzes the penultimate step in the synthesis of biologically active GAs (Phillips et al. 1995; Xu et al. 1995). Finally, we have investigated the effects of spy-7 and gar2-1 on steady-state levels of gai transcripts.
The results of the above experiments indicate that GAI, SPY, and GAR2 operate within, or modulate, a signal-transduction pathway that represses growth and whose activity is opposed by GA. Because of the existence of mutations having comparable effects to gai and spy in other plant species (Swain and Olszewski 1996), and because GA is an essential growth regulator in a wide variety of plant species (Hooley 1994), it seems likely that the Arabidopsis GAI, SPY, and GAR2 genes define a system for GA-mediated growth regulation that is common to all higher plants.
Results
Cloning of gai through insertional mutagenesis
gai maps to chromosome 1 of Arabidopsis (Koornneef et al. 1985; Peng and Harberd 1993), ∼11 cM from a T-DNA insertion (A264) carrying a Ds transposon (Peng and Harberd 1993; Balcells et al. 1991). Genetic analyses suggested that loss-of-function alleles of gai confer a tall plant phenotype, similar to that conferred by the wild-type allele (GAI; Peng and Harberd 1993; Wilson and Somerville 1995). Therefore, we isolated Ds insertion loss-of-function alleles of gai, exploiting the tendency of Ds (in the presence of the Ac transposase) to transpose preferentially to linked sites (Bancroft and Dean 1993; Jones et al. 1994). We constructed plant lines homozygous for both Ds-bearing T-DNA insertion A264 (Peng and Harberd 1993) and gai, and also containing a transgene expressing Ac transposase (ΔNaeI–sAc(GUS)-1; Bancroft and Dean 1993). Potential Ds insertion gai alleles were isolated from F1 to F4 generations of this material. The plants were screened for reduction or loss of the dark green, dwarf phenotype conferred by gai, by searching for rare individuals that were paler green and taller than expected for a gai homozygote (Peng and Harberd 1993). NA735B-1 was one such plant, being taller and paler green than a gai homozygote (gai/gai), but not as tall or pale as a wild-type (GAI/GAI) homozygote. In accord with previous observations (Peng and Harberd 1993), NA735B-1 was identified provisionally as a gai heterozygote of genotype gai/gai-t6, where gai-t6 was a new allele possibly carrying a Ds insertion. Self-pollination of NA735B-1 resulted in a progeny population that segregated for gai homozygotes, gai heterozygotes (gai/gai-t6), and a new class of plants (gai-t6/gai-t6) displaying a tall phenotype similar to that of wild type. Plants homozygous for GAI, gai, and gai-t6 are shown in Figure 1A.
Figure 1.
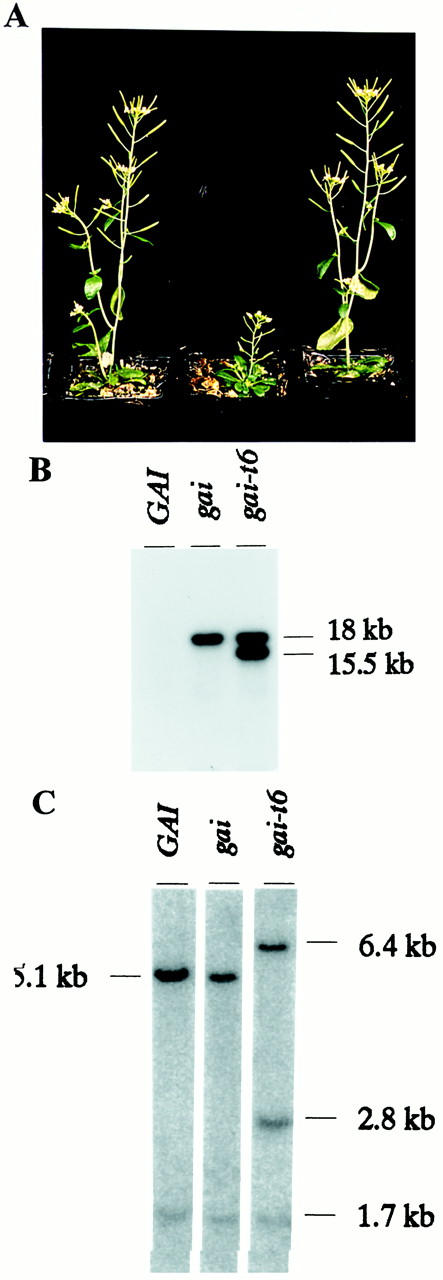
The gai-t6 line contains a transposed Ds that interrupts a transcribed gene. (A) Plants shown are (left to right) homozygous for GAI, gai, and gai-t6. GAI and gai-t6 plants display a tall phenotype. (B) DNA gel-blot hybridization using a radiolabeled Ds probe. DNA in the GAI lane lacks Ds. The gai lane contains DNA from plants homozygous for gai and for T-DNA A264, which contains Ds (18.0-kb EcoRI fragment). The gai-t6 lane contains DNA from plants homozygous for A264 and for a transposed Ds (15.5-kb fragment). gai-t6 has lost ΔNaeI–sAc(GUS)-1 through genetic segregation. (C) DNA gel-blot hybridization using a radiolabeled GAI cDNA probe. The cDNA (insert of pPC1) hybridizes with a 5.1-kb BclI fragment in DNA from GAI and gai, replaced in gai-t6 by 6.4- and 2.8-kb fragments. Because BclI cuts once within Ds, the Ds insertion is flanked on either side by the gene (GAI) encoding the cDNA. The fainter hybridization at 1.7 kb identifies GRS.
DNA gel-blot experiments (using a Ds hybridization probe; see Materials and Methods) revealed that gai-t6 contains two Ds elements, one in the original position (as in A264), the other (transposed Ds) in a new position (Fig. 1B). Using map-based cloning methods we had previously isolated Arabidopsis genomic DNA fragments spanning an ∼200-kb segment of chromosome 1 known to contain GAI (see Materials and Methods; P. Carol, D.E. Richards, R. Cowling, J. Peng, and N.P. Harberd, unpubl.). An IPCR fragment (JP95) containing genomic DNA adjacent to the 3′ end of the transposed Ds in gai-t6 (see Materials and Methods) hybridized specifically to DNA from within this segment, suggesting that this Ds is inserted into, or in the vicinity of, gai (P. Carol, J. Peng, D.E. Richards, and N.P. Harberd, unpubl.). A cDNA (insert of pPC1) was identified through hybridization to cosmid JP2 (see Materials and Methods) and JP95. DNA gel-blot experiments showed that the transposed Ds in gai-t6 interrupts the gene (GAI) encoding the mRNA represented in pPC1 (Fig. 1C).
In addition to GAI, the pPC1 cDNA also hybridizes with a genomic fragment containing a second gene GRS (for GAI-related sequence) (Fig. 1C). A cDNA clone containing GRS sequence was identified through hybridization with probes containing the GAI sequence. Although GAI maps to chromosome 1 (see above), GRS is located close to the top of chromosome 2 (K. King, P. Carol, and N.P. Harberd, unpubl.).
The gai mutant allele encodes an altered product
The DNA sequences of two overlapping GAI cDNAs revealed an open reading frame (ORF) encoding a protein (GAI) of 532 amino-acid residues. DNA fragments containing this ORF were amplified from GAI and gai genomic DNA; their sequences showed that the GAI ORF is not interrupted by introns. The predicted primary sequence of the GAI and gai proteins is shown in Figure 2A. The gai allele contains a deletion of 51-bp from within the GAI ORF. This in-frame deletion results in the absence of a 17-amino-acid residue segment situated close to the amino terminus of the predicted protein sequence (Fig. 2A). There are no other differences between the proteins encoded by GAI and gai.
Figure 2.
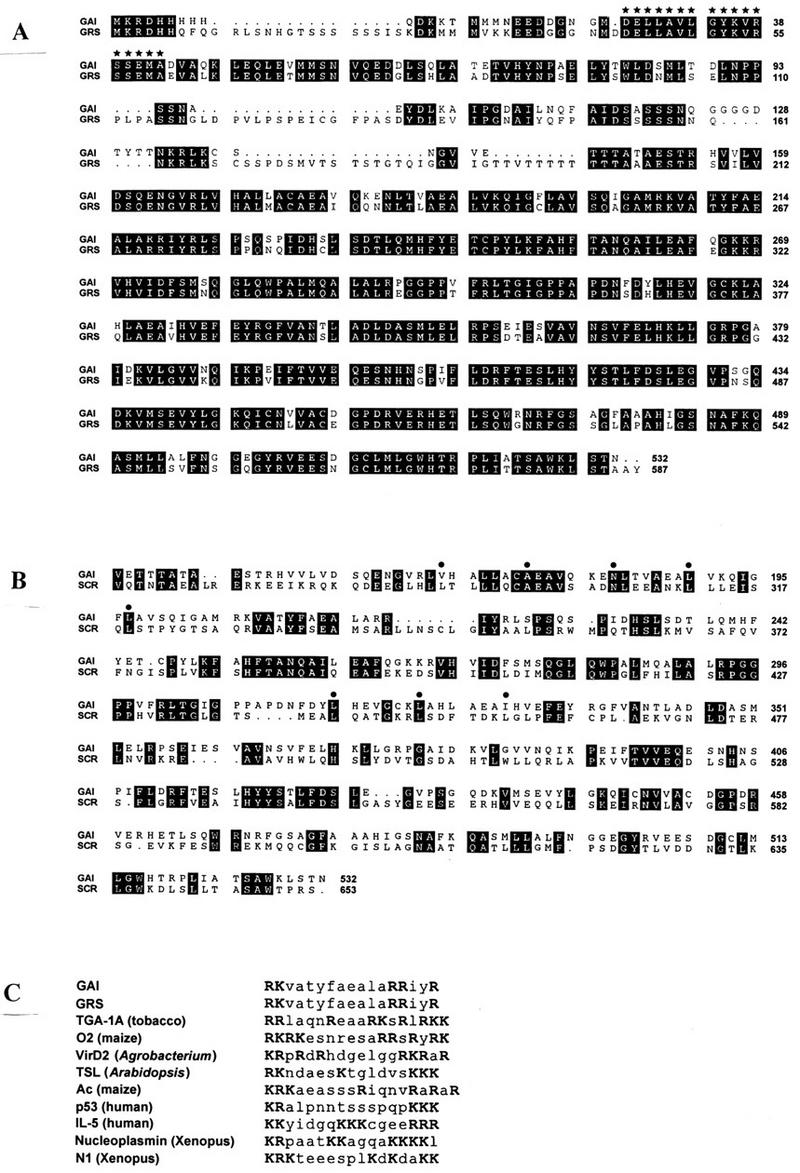
Analysis of GAI, gai and GRS amino acid sequences. (A) Alignment of the amino acid sequences (single-letter code) of GAI (predicted from the genomic DNA sequence of GAI) and GRS (from cDNA sequence) is shown. Gaps are introduced to maximize alignment, and identical amino acid residues are highlighted in black. The 17 amino acids missing in the gai mutant protein (D27–A43) have black stars above them. The Ds in gai-t6 is inserted between the E182 and N183 codons. (B) Alignment of the carboxyl termini of GAI (from V143) and SCR (from V263) is shown. The third row of the comparison shows the VHIID domain described in Di Laurenzio et al. (1996). Residues defining leucine heptad repeats are identified by closed circles. (C) Comparison of bipartite nuclear localization signals in GAI, GRS, and other proteins. Basic amino acids are shown in bold uppercase letters. Sequence information is from the following: for TGA-1A, Opaque-2 (NLS-B; O2) and VirD2, see Raikhel (1992); for TSL, see Roe et al. (1993, 1997); for Ac see Boehm et al. (1995); for p53, see Dingwall and Laskey (1991); for IL-5, see Jans et al. (1997); for Nucleoplasmin and N1, see Robbins et al. (1991).
Figure 2A also shows the predicted primary sequence of the GRS gene product. GRS encodes a protein (GRS) of 587 amino acids, somewhat larger than GAI. GRS shares a high degree of sequence similarity with GAI (83% amino acid identity) and contains a region of identical amino acid sequence to the segment that is deleted in the gai mutant protein.
The gai-derivative alleles contain mutations that disrupt the GAI ORF
A series of presumed gai-derivative alleles conferring tall phenotypes similar to that conferred by GAI were isolated after γ-irradiation mutagenesis of gai (Peng and Harberd 1993). These alleles (gai-d1, gai-d2, gai-d5, and gai-d7) contain the 51-bp deletion characteristic of gai (thus confirming that they are derived from gai), together with additional mutations that disrupt the GAI ORF (Table 1). Thus, loss of gai mutant phenotype is associated with the occurrence of mutations that may result in a nonfunctional gene product. Furthermore, in reversion experiments, excision of Ds from gai-t6 was associated with restoration of a genetically dominant, dwarf phenotype (J. Peng, P. Carol, D.E. Richards, and N.P. Harberd, unpubl.). These observations confirm that the transposed Ds in gai-t6 is inserted within GAI, and that GAI has been cloned. They are also consistent with predictions that the gai-d alleles would be null alleles (Peng and Harberd 1993; Wilson and Somerville 1995).
Table 1.
Mutations in GAI alleles
| Allele
|
Nature of mutationa
|
Position in coding sequence
|
Consequence of mutation
|
|---|---|---|---|
| gai-d1 | CAG to TAG | Gln239 | stop codon, truncated polypeptide |
| gai-d2 | GAT to GA, 1-base deletion | Asp274 | frameshift, addition of two novel amino acids, truncated polypeptide |
| gai-d5 | 7-base deletion, also C to G | follows Leu281 | frameshift, addition of 18 novel amino acids, truncated polypeptide |
| gai-d7 | GTT to GT, 1-base deletion | Val156 | frameshift, addition of 27 novel amino acids, truncated polypeptide |
Underlining denotes nucleotide substitution in each allele.
GAI contains a consensus nuclear localization signal, a LXXLL motif, and is a new member of the VHIID domain family
Searches of the DNA and protein sequence databases using the BLAST program (Altschul et al. 1990) revealed that GAI is closely related in sequence to the predicted product (SCR) of a recently cloned Arabidopsis gene SCARECROW (SCR; Di Laurenzio et al. 1996), a member of a novel family of candidate transcription factors (Fig. 2B). GAI has homology to the carboxyl terminus of SCR, especially to the VHIID domain that characterizes the new family. GAI contains two heptad repeat regions similar to leucine zippers (GAI amino acid residues 169–203 and 316–336), as described previously for SCR (Fig. 2B; Di Laurenzio et al. 1996). GAI contains the leucine zipper region of the proposed basic leucine zipper (bZIP) domain in SCR, but lacks the basic domain (Di Laurenzio et al. 1996). There is no significant homology between GAI and SCR in regions amino terminal to the area shown in Figure 2B. A short segment of GAI (amino acid residues 403–427), and also of SCR, shows homology with cdr 29 (BLAST Poisson probability score for GAI: 1.2 × e−5), a barley homolog of peroxisomal acyl CoA oxidase genes (Grossi et al. 1995). The significance of this finding is unknown.
Using PSORT, a program for the prediction of protein localization in cells (Nakai and Kanehisa 1992), the highest score assigned to GAI was for nuclear localization (certainty value = 0.760). The GAI protein contains two basic regions that are characteristic of nuclear localization signals (NLSs). The first region (206RKVATYFAEALARRIYR222) exactly fits the consensus for bipartite NLSs, which has been defined as two basic amino acid residues, a spacer region of ∼10 residues, and at least three basic residues out of the next five (Fig. 2C; Dingwall and Laskey 1991; Robbins et al. 1991; Raikhel 1992). In addition, GAI contains a second basic region (134KRLK137) that conforms to the consensus (K-R/K-X-R/K) proposed for nontypical SV40-like NLSs (Boulikas 1994; LaCasse and Lefebvre 1995). The presence of these sequences suggests that GAI may be targeted to the nucleus. Interestingly, GAI also contains two motifs, 169VHALL173 and 370LHKLL374, which are, respectively, closely related and identical to a consensus motif (LXXLL) that has been shown recently to mediate binding of transcriptional coactivators to nuclear receptors (Heery et al. 1997).
All of the features described above for GAI (SCR homology, cdr 29 homology, NLSs, LXXLL motifs), are also found in GRS, suggesting that GAI and GRS have similar functions.
GAI null alleles confer increased resistance to PAC
PAC is a triazole derivative that inhibits GA biosynthesis at the kaurene oxidase reaction (Hedden and Graebe 1985; Davis and Curry 1991). Wild-type Arabidopsis plants require GA for seed germination and stem elongation (Koornneef and van der Veen 1980), and depletion of endogenous GA levels by PAC inhibits these processes (Jacobsen and Olszewski 1993). There are several plant mutants that display increased resistance to the effects of PAC. Among these are the la crys and slender mutants of pea and barley (Brian 1957; Potts et al. 1985; Chandler 1988; Lanahan and Ho 1988; Croker et al. 1990), and the spy mutants of Arabidopsis (Jacobsen and Olszewski 1993; Jacobsen et al. 1996). These mutants exhibit growth that, to varying degrees, is less dependent on GA than is the growth of wild-type plants. Thus, in the la crys, slender, and spy mutants, stem elongation is at least partially uncoupled from the GA-mediated control characteristic of normal plants.
Experiments to determine whether the gai-t6 allele confers greater PAC resistance than the GAI allele were performed. The purpose of these experiments was to determine whether a loss-of-function allele of GAI confers a reduction in the GA dependency of growth. We chose gai-t6 for these experiments because it is a Ds insertion mutation, and a likely null allele. Initial experiments showed that gai-t6 does not germinate on 10−4 m PAC, a PAC concentration that permits germination of spy mutants (data not shown; Jacobsen and Olszewski 1993). However, as shown in Figure 3, gai-t6 does confer increased PAC resistance; when grown on medium containing 10−6 m PAC, gai-t6 mutants display longer floral bolt stems than GAI control plants. This result suggests that loss of GAI function causes a reduction in the GA dependency of stem elongation, that plants lacking GAI require less GA than normal plants for equivalent growth, and that GAI is a negative regulator of GA responses. However, the degree of PAC resistance conferred by gai-t6 is less than that conferred by the currently available spy alleles. One explanation for the fact that gai-t6 does not confer strong PAC resistance is that GRS can compensate substantially for loss of GAI.
Figure 3.

gai-t6 confers increased resistance to PAC. Bolt stem elongation of gai-t6 (rear two plants) and GAI (front two plants) plants grown on 10−6 m PAC is shown. The plants were photographed 57 days after sowing. Because PAC inhibits GA biosynthesis both classes of plant are dwarfed and darker green than when grown on medium lacking PAC. However, gai-t6 plants grow taller than the GAI plants and, therefore, are more resistant to the effects of 10−6 m PAC. As shown, the gai-t6 plants have open flowers, with the petals clearly visible, whereas the flowers on the GAI plants are not open (retarded petal and stamen elongation is characteristic of severe GA deficiency; Koornneef and van der Veen 1980). gai-d1 and gai-d5 mutants behave like gai-t6 under these conditions, and not like GAI. When grown on medium lacking PAC, gai-t6, gai-d1, and gai-d5 are indistinguishable from GAI.
gai-suppressor mutations have additive effects on stem elongation and PAC resistance
Screens for extragenic suppressors of the dwarf phenotype conferred by gai identified mutations at the GAS1 and GAR2 loci (Carol et al. 1995; Wilson and Somerville 1995). Complementation analysis showed that gas1-1 is a spy allele (now renamed spy-7; data not shown). This result is consistent with the observation that the spy-4 and spy-5 alleles suppress the dwarf phenotype conferred by gai (Wilson and Somerville 1995; Jacobsen et al. 1996). gar2-1, however, identifies a distinct genetic locus that segregates independently of SPY. A recent report describes the isolation of multiple alleles at the Arabidopsis RGA locus, which partially suppress the phenotype conferred by the GA deficiency mutation ga1-3 (Silverstone et al. 1997). Although complementation tests have not been performed, it is unlikely that RGA and GAR2 are allelic, because all known rga alleles are recessive (Silverstone et al. 1997), whereas gar2-1 is dominant (Wilson and Somerville 1995).
gai mutants exhibit reduced GA responses (Koornneef et al. 1985; Wilson and Somerville 1995). One possible explanation for the suppression of the gai phenotype by spy-7 and gar2-1 is that they restore partially the GA responses of gai. However, previous experiments showed that spy-7 does not increase the response of gai mutants to exogenous GA (Carol et al. 1995). Similarly, we have shown that the gai gar2-1 double mutant also does not exhibit a significant growth response to exogenous GA (data not shown). Thus, gar2-1, like spy-7, does not restore GA responses to gai mutant plants.
On their own, spy-7 and gar2-1 cause partial suppression of the phenotype conferred by gai. As shown in Figure 4A, gai spy-7 homozygotes are taller and paler than gai homozygotes, but not as tall as wild-type plants (Carol et al. 1995). gai gar2-1 homozygotes are taller, although not paler, than gai homozygotes, and not as tall as wild-type plants (Fig. 4A; Wilson and Somerville 1995). When combined, in a gai spy-7 gar2-1 homozygote, the two gai-suppressor mutations confer complete suppression of the gai phenotype (Fig. 4A). gai spy-7 gar2-1 plants exhibit increased internode length and apical dominance, are paler than, and at least as tall as, wild-type plants.
Figure 4.

Additive suppression of gai phenotype by spy-7 and gar2-1 alleles. (A) Final adult growth of GAI, gai, gai spy-7, gai gar2-1, and gai spy-7 gar2-1 plants in standard glasshouse conditions is shown. As previously described, gai spy-7 and gai gar2-1 plants are taller than gai, but less tall than GAI (Carol et al. 1995; Wilson and Somerville 1995). In gai spy-7 gar2-1 plants the effects of gai are suppressed completely, resulting in plants at least as tall as GAI. (B) Growth of plants grown on 10−6 m PAC. Plants were grown as in Fig. 3, and photographed 30 days after sowing. gai spy-7 gar2-1 displays the greatest resistance to PAC, followed by gai gar2-1 and gai spy-7, which are themselves more resistant than GAI or gai.
The SPY locus was identified originally by mutations that confer PAC-resistant seed germination (Jacobsen and Olszewski 1993). gar2-1 (on a GAI/GAI rather than a gai/gai background) also confers PAC-resistant seed germination (data not shown). Accordingly, we investigated the effects of PAC on the growth of plants carrying the gai-suppressor mutations. As shown in Figure 4B, growth of gai spy-7 gar2-1 plants is more resistant to PAC than is that of gai gar2-1 plants or gai spy-7 plants (which are themselves more resistant than gai or GAI). gai spy-7 gar2-1, gai gar2-1, and gai spy-7 all grow taller than gai-t6 on 10−6 m PAC (plants shown in Fig. 3 are more mature than those shown in Fig. 4B).
GA C-20 oxidase transcript abundance is affected by gai and by gai-suppressor mutations
The GA C-20 oxidase gene family encodes enzymes that catalyze the penultimate step in the biosynthesis of biologically active GAs. Steady-state levels of C-20 oxidase transcripts are regulated negatively by GA (or the GA signal), and are elevated in gai (Phillips et al. 1995; Xu et al. 1995). RNA gel-blot analysis was used to visualize C-20 oxidase transcripts in gai and in the gai-suppressor mutants (Fig. 5A,C). As previously reported, C-20 oxidase transcript levels are greater in gai than in the GAI control. Interestingly, the abundance of the C-20 oxidase transcripts is restored to approximately wild-type levels in gai spy-7, gai gar2-1, and gai spy-7 gar2-1. Thus, in addition to suppressing the dwarf phenotype conferred by gai, spy-7 and gar2-1 also suppress the elevated C-20 oxidase transcript levels conferred by gai.
Figure 5.
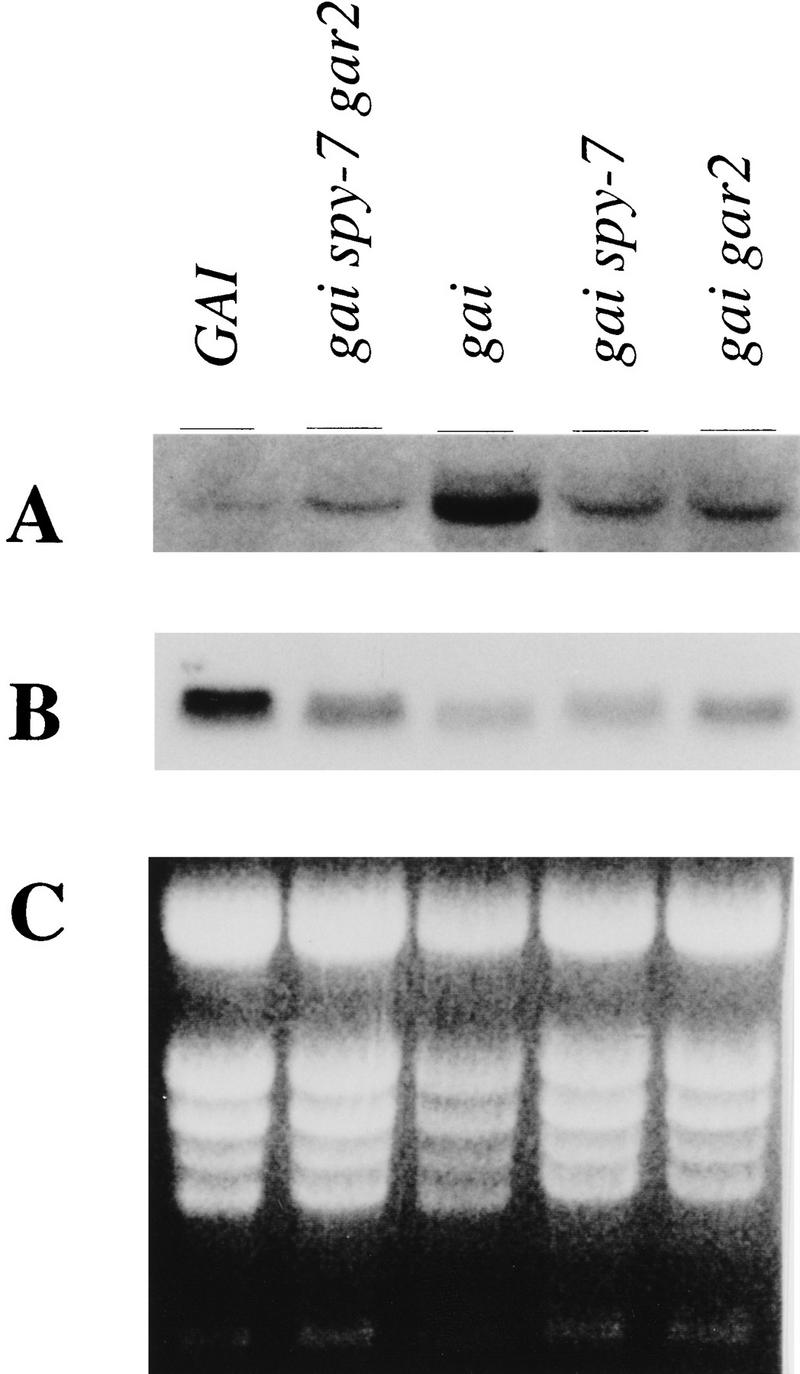
Effect of gai, gai spy-7, gai gar2-1, and gai spy-7 gar2-1 on GA C-20 oxidase and GAI/gai transcript levels. (A) A radiolabeled GA C-20 oxidase probe detects the C-20 oxidase message in total RNA isolated from seedlings of the indicated genotypes. As previously described, gai accumulates higher levels of C-20 oxidase transcript than does GAI (Xu et al. 1995). In gai spy-7, gai gar2-1, and gai spy-7 gar2-1 mutants, C-20 oxidase levels are similar to those found in GAI. (B) A radiolabeled GAI probe detects GAI or gai messages in total RNA from seedlings of the indicated genotypes. Levels of gai message in gai are similar to, and may be slightly lower than, levels of GAI message in GAI. There are no large differences between the gai message levels found in the gai, gai spy-7, gai gar2-1, and gai spy-7 gar2-1 samples. (C) UV fluorescence of ethidium bromide-stained gel blotted for use in the hybridizations shown in A and B. Note that there was less RNA in the gai lane.
GAI transcript abundance is not significantly affected by gai or gai-suppressor mutations
RNA gel-blot analysis was also used to visualize GAI (or gai) transcripts in wild-type, gai, and gai-suppressor mutants (Fig. 5B,C). Comparison of wild-type and gai samples shows that gai transcript levels in the gai mutant are similar to GAI transcript levels in the wild type, indicating that overexpression is an unlikely explanation for the genetic dominance of gai. If anything, gai transcript levels are actually slightly lower in gai than are GAI transcript levels in the wild type.
As shown above (see Fig. 3), apparent null alleles of gai confer a tall plant phenotype. It could be thought that spy-7 and gar2-1 might cause suppression of the gai phenotype through a reduction in gai transcript levels. However, this is unlikely, because gai transcript levels are not detectably different in gai, gai spy-7, gai gar2-1, or gai spy-7 gar2-1, although the latter genotype confers a phenotype that is at least as tall as wild type. Thus, spy-7 and gar2-1 are unlikely to suppress gai phenotype by an effect on gai transcript levels.
Discussion
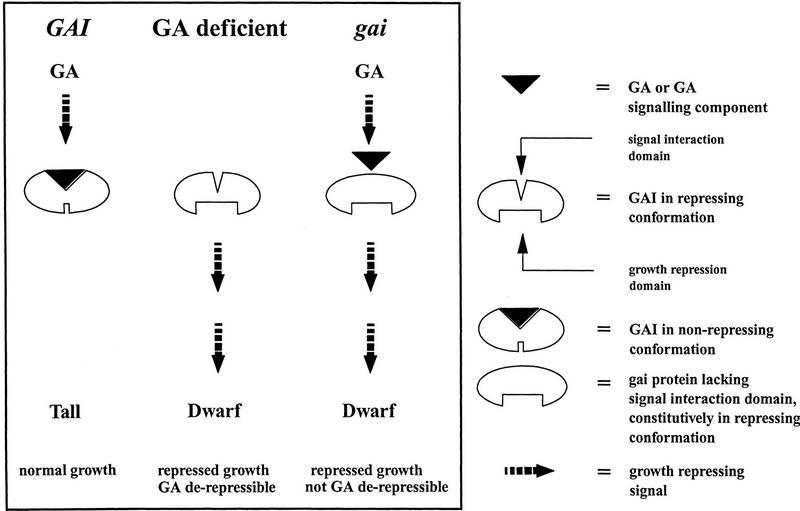
The cloning of GAI through insertional mutagenesis of the gai allele demonstrates that gai is a gain-of-function, rather than a dominant loss-of-function mutation. Gain-of-function mutations can have dominant effects for a variety of reasons, including ectopic or increased expression of a normal gene product, or altered function of a mutant gene product. Here we show that gai does not confer a detectable increase in gai transcript abundance, suggesting that increased expression is not the explanation for the dominance of gai. We also show that gai encodes an altered product, suggesting that gai is dominant because this alteration in structure results in an altered function. Thus, deletion of a 17-amino-acid segment from GAI results in a mutant protein (gai) that, in a genetically dominant fashion, causes a reduction in GA responses. We also show that loss of GAI function results in increased resistance to the growth-retarding effects of the GA biosynthesis inhibitor PAC. This observation is significant, because it demonstrates that the wild-type gene product GAI is a GA signal-transduction component. To explain these observations we propose that GAI is a repressor of stem elongation, and that GA derepresses stem elongation by opposing GAI action (Fig. 6). The segment missing in the mutant gai protein could be responsible for interacting with the GA signal (or with GA itself); gai would then constitutively repress stem elongation because it can no longer interact with GA or with the GA signal. Alternatively, the segment deleted in gai may have some other function, and the gai mutant protein may be locked into a repressive conformation for reasons other than the loss of a segment that interacts with the GA signal.
Figure 6.
Derepression model for regulation of plant stem elongation by GA. GA derepresses stem elongation because it (or a GA signaling component) opposes the activity of GAI, a protein that represses stem elongation. GAI contains signal interaction and growth-repressing domains, and exists in one of two distinct conformations. Interaction between GA (or the GA signal) and GAI transforms GAI into the nonrepressing conformation. Normal plants (GAI) grow tall because the level of endogenous GA is sufficiently high to oppose the activity of the GAI repressor. GA-deficient plants contain insufficient GA to oppose GAI repression to the same degree and, therefore, are thus dwarfed. gai mutant plants are dwarfed because the mutant gai protein is relatively resistant to the effects of GA, and represses growth in a dominant fashion. Null alleles at GAI (eg., gai-t6) confer a tall, PAC-resistant phenotype, because absence of GAI results in loss of its growth repression function. gai-t6 mutant plants are not totally PAC resistant because of the probable activity of GRS. This model provides a general explanation for the regulation of plant stem elongation by GA.
An alternative explanation for the dominance of gai is that gai interferes with the activity of a signal-transduction pathway that activates stem elongation in response to GA. We prefer the GAI repression explanation because it predicts that loss-of-function alleles of GAI should confer increased resistance to GA biosynthesis inhibitors, whereas the gai interference explanation does not. As shown above, the gai-t6 allele does indeed confer increased resistance to PAC. Previous studies have suggested that a repressor function is involved in GA signal transduction (Brian 1957; Potts et al. 1985; Chandler 1988; Lanahan and Ho 1988; Harberd and Freeling 1989; Croker et al. 1990). Our work provides direct evidence that such a repressor exists, and that it is GAI. A further consequence of these findings is that GA regulates stem elongation not through activation but by derepression.
GAI contains leucine heptad repeats, NLSs, and the LXXLL motif characteristic of transcriptional coactivators. All of these features are found in proteins that modify transcription (Montminy 1997; Torchia et al. 1997). Perhaps GAI acts as a transcriptional regulator, repressing transcription of genes that promote stem elongation. GAI lacks any obvious membrane-spanning domain and, therefore, is unlikely to be the plasma membrane-associated GA receptor implicated in the cereal aleurone GA response (Hooley et al. 1991; Smith et al. 1993; Gilroy and Jones 1994; Jacobsen et al. 1995).
Here we show that the spy-7 and gar2-1 mutations cause partial, and, when combined, total suppression of the dwarf phenotype conferred by gai, and also suppress the effect of gai on the accumulation of C-20 oxidase transcripts. Suppressed gai mutants (gai spy-7, gai gar2-1, gai spy-7 gar2-1) accumulate gai transcripts to levels similar to that found in plants carrying the gai mutation alone. These observations suggest that the SPY and GAR2 gene products do not modify GAI expression, but rather act as GA signal transduction components upstream or downstream of the GAI gene product. This idea may be an oversimplification with respect to GAR2 because the gar2-1 allele is a dominant, potential gain-of-function mutation (Wilson and Somerville 1995). Perhaps a mutant gar2-1 gene product interferes with the function of an unidentified GA signal transduction component.
The additive effects of spy-7 and gar2-1, together with their differing effects on paleness, might suggest that they identify different branches of the GA signaling pathway. Alternatively, because the spy-7 allele is weaker in its effects than spy-5 (spy-5 itself is not a strong spy allele; J. Peng and N.P. Harberd, unpubl.; Wilson and Somerville 1995; Jacobsen et al. 1996), spy-7 is unlikely to be a null allele. The combination of two partial blocks in a single (unbranched) pathway may be the equivalent of a complete block in that pathway.
The observation that spy mutations suppress the gai phenotype has led to suggestions that SPY acts downstream of GAI (Jacobsen et al. 1996; Swain and Olszewski 1996). However, interpretation of epistasis relationships in terms of the ordering of gene functions in pathways is not always a simple matter (Avery and Wasserman 1992). Recent evidence suggests that SPY encodes an O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT; Kreppel et al. 1997), an activity that is involved in the dynamic modification of regulatory proteins in a manner analogous to that of protein phosphorylation (Kreppel et al. 1997; Lubas et al. 1997). SPY may modify GAI, rather than being a component of the GA signaling pathway downstream of GAI.
The resistance to PAC conferred by gai-t6 is not as strong as that observed in the gai spy-7 gar2-1 mutant. This observation is consistent with the hypothesis that at least one other Arabidopsis gene product has a function that substantially duplicates that of GAI. As described above, the primary sequence of GRS is very similar to that of GAI, and is identical to GAI in the region deleted in gai. Perhaps GRS shares functional properties in common with GAI. If so, mutants lacking GRS, like those lacking GAI, might be predicted to exhibit PAC resistance. Also, because gai spy-7 gar2-1 is more PAC resistant than gai-t6, it seems that GA signaling from both GAI and GRS (assuming that GAI and GRS do have overlapping functions) is mediated or modulated by GAR2 and SPY.
The GA signaling system outlined in this paper has several intriguing properties. First, although GA is an essential regulator of various stages of the life cycle of normal plants (e.g., seed germination in Arabidopsis; Koorneef and van der Veen 1980), it is no longer essential if the GA signaling system is compromised. Second, the different degrees of PAC resistance exhibited by the various mutants described in this paper show that the GA signaling system is capable of eliciting a graduated, rather than an all-or-nothing response, such that a partially compromised system yields a partial reduction in GA dependency. Ethylene is another factor that plays an important part in the control of plant growth and development. Genetic dissection of ethylene signal transduction in Arabidopsis suggests that the ETR and ERS genes encode redundant ethylene receptors, that CTR1 acts downstream of ETR and ERS, and that the ETR, ERS, and CTR gene products define a pathway that operates as a negative regulator of ethylene responses (Bleeker et al. 1988; Chang et al. 1993; Kieber et al. 1993; Hua et al. 1995; Schaller and Bleeker 1995). Genetic analysis of GA signal transduction in Arabidopsis has identified the involvement of GAI, SPY, and GAR2. It appears that GAI encodes largely redundant functions, and that GAI, SPY, and GAR2 are components (or modulators) of a pathway that acts as a negative regulator of GA responses. Negative, derepressible regulatory systems may be common features of the mechanisms by which plant growth factors regulate plant development.
Materials and methods
Genetic nomenclature
In this paper genotypes are written in italics; the wild-type genotype is in capitals (e.g., GAI), and the mutant genotype is in lowercase letters (e.g., gai). The polypeptide product of the GAI gene is written as GAI, and of the gai gene as gai.
Plant materials
Mutant plant lines were obtained as previously described (Peng and Harberd 1993; Carol et al. 1995). The gar2-1 mutant was obtained from Ruth Wilson. Seeds were chilled on moistened filter paper at 4°C for 4 days (to break dormancy) and then planted on two parts Levington’s M3 potting compost to one part grit/sand. Plants were then grown in standard greenhouse conditions or in controlled environment chambers. Transgenic plants were grown according to United Kingdom Ministry of Agriculture, Fisheries and Food (MAFF) regulations (License no. PHF 1418/8/22).
Growth of plants on medium containing PAC
Seeds were surface-sterilized and germinated on GM medium, with or without supplementary PAC (obtained from Zeneca Agrochemicals), and seedlings were maintained on this medium, under the same conditions as previously described (Peng and Harberd 1993).
DNA and RNA gel-blot hybridizations, DNA sequencing
Genomic DNA preparation and gel-blot hybridizations were performed as described (Peng and Harberd 1993). The Ds probe was a radiolabeled 3.4-kb XhoI–BamHI subfragment of Ac. RNA was extracted from 27-day-old seedlings (grown under natural photoperiod), and RNA gel-blot transfers were performed as described (Whitelam et al. 1993). DNA–RNA hybridizations were in 0.3 m Na phosphate (pH 7.2), 7% SDS, 1% BSA, and 1 mm EDTA, at 65°C, with two subsequent washes for 30 min each in 0.5 m Na phosphate (pH 7.2), 5% SDS, 50 mm EDTA at 65°C. For DNA–RNA hybridizations, the GAI (gene specific) probe was a 369-bp PCR amplified fragment containing a 150-bp 5′ noncoding sequence and a portion encoding the amino terminus of GAI. The C-20 oxidase probe was an ∼800-bp PCR amplified fragment from GA5 (Xu et al. 1995). DNA sequences were determined using the dideoxynucleotide chain termination method.
Isolation of genomic DNA flanking the transposon insertion in gai-t6
JP95 is an ∼2.5-kb IPCR fragment containing DNA flanking the transposed Ds in gai-t6. This fragment extends from the 3′ end of Ds into the adjacent Arabidopsis chromosomal DNA and terminates at the next BclI site. To make JP95, genomic DNA from gai-t6 was digested with BclI, recircularized, and then amplified using primers DL5 and B39 for the first round and DL6 and D71 for the second, nested, round of amplification (Long et al. 1993).
Identification of cosmid clones containing GAI
Using methods described elsewhere (Putterill et al. 1993, 1995; Macknight et al. 1997) we established a contig of yeast artificial chromosomes (YACs; supplied by J. Ecker, University of Pennsylvania, Philadelphia) that contained GAI. This contig was based in part on unpublished hybridization data from the laboratories of J. Ecker and G. Jürgens (University of Tübingen, Germany). Cosmids containing DNA from a subregion of this contig [shown by restriction fragment length polymorphism (RFLP)-marked recombinant analysis to contain GAI; P. Carol, J. Peng, D.E. Richards, R. Cowling, and N.P. Harberd, unpubl.] were isolated from a Landsberg erecta DNA cosmid library (gift of C. Lister and C. Dean, John Innes Centre, Norwich, UK).
Identification and characterization of GAI and GRS cDNAs and genomic DNAs from GAI, gai, and gai-derivative alleles
A 10-kb subfragment of a cosmid (JP2) containing GAI, previously shown to hybridize with the IPCR fragment JP95, was used to screen a cDNA library made from young seedling aerial parts (Columbia ecotype). We identified cDNA clones pPC1 (GAI) and pPC2 (GRS). Part of the DNA sequence of pPC1 was identical with that of an ∼150-bp region of genomic DNA flanking the Ds insertion in gai-t6 (from JP95; J. Peng, P. Carol, D.E. Richards, and N.P. Harberd, unpubl.). In addition, searches of the dbEST database (Boguski et al. 1993) with the BLAST programme (Altschul et al. 1990) revealed an Arabidopsis-expressed sequence tag (EST; GenBank Identifier ATTS3217) containing sequence identical to that of JP95. cDNA ATTS3217 was obtained from the Arabidopsis Biological Resources Centre, and the complete DNA sequences of the pPC1 insert and of ATTS3217 were determined. These overlapping sequences revealed an ORF, together with 5′ and 3′ noncoding regions, for GAI. Oligonucleotide primers derived from 5′ and 3′ noncoding sequence of GAI were used to amplify, with PCR, 1.7-kb fragments from GAI, gai, gai-d1, gai-d2, gai-d5, and gai-d7 genomic DNA. The DNA sequences of these fragments were determined from duplicate amplifications, thus avoiding potential errors introduced by PCR. The GAI genomic sequence was almost identical with that of the overlapping cDNAs. Three nucleotide substitutions were detected, which could be attributable to differences between ecotypes (the GAI genomic sequence is from Landsberg erecta, the cDNAs from Columbia) and do not alter the predicted amino acid sequence of GAI. Amino-acid sequence alignments in Figure 2 were performed using the PILEUP and PRETTYBOX programs (Wisconsin Package, Genetics Computer Group, Madison, WI), using default parameters.
Acknowledgments
cDNAs were obtained from the Arabidopsis Stock Centre, Max Planck Institute, Köln, and from the Arabidopsis Biological Resource Center, Ohio State University. We thank Joe Ecker for the YUP YAC library, the laboratories of Joe Ecker and Gerd Jürgens for sharing unpublished contig information, Ruth Wilson for gar2-1, and Neil Olszewski for helpful discussions about SPY function. We thank Clare Lister and Caroline Dean for the cosmid library containing Arabidopsis thaliana genomic DNA (Landsberg erecta strain). We thank George Coupland and Caroline Dean for transposon-tagging lines and for useful discussions, and Ian Bancroft, Thierry Desnos, Nigel Hartley, Clare Lister, Debbie Long, Jo Putterill, Fran Robson, and Renate Schmidt for advice on techniques. Paul Sinacola assisted with plant care. Helpful discussions with Hiroshi Ezura, Pilar Puente, and Thierry Desnos are gratefully acknowledged. J.P. was supported by the Sino-British Friendship Scholarship Scheme, P.C. by a European Community Human Capital and Mobility Fellowship, and R.J.C. and K.K. by John Innes Foundation Studentships. This work was made possible by financial support from the Biotechnology and Biological Sciences Research Council (Core Strategic Grant to the John Innes Centre and Agricultural and Food Research Council/Biotechnology and Biological Science Research Council/Plant Molecular Biology grants PG208/520 and PG208/0600), and the Gatsby Charitable Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Note added in proof
Nucleotide sequences corresponding to the amino acid sequences described in this paper have been submitted to the EMBL data library under accession nos. Y15193 and Y15194.
Footnotes
E-MAIL harberd@bbsrc.ac.uk; FAX +44 1603 505725.
References
- Altschul SF, Warren G, Gish W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Avery L, Wasserman S. Ordering gene function: The interpretation of epistasis in regulatory hierarchies. Trends Genet. 1992;8:312–316. doi: 10.1016/0168-9525(92)90263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balcells L, Swinburne J, Coupland G. Transposons as tools for the isolation of plant genes. Trends Biotechnol. 1991;9:31–37. [Google Scholar]
- Bancroft I, Dean C. Transposition pattern of the maize element Ds in Arabidopsis thaliana. Genetics. 1993;134:1221–1229. doi: 10.1093/genetics/134.4.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleeker AB, Estelle MA, Somerville C, Kende H. Insensitivity to ethylene conferred by a dominant mutation in Arabidopsis thaliana. Science. 1988;241:1086–1089. doi: 10.1126/science.241.4869.1086. [DOI] [PubMed] [Google Scholar]
- Boehm U, Heinlein M, Behrens U, Kunze R. One of the three nuclear localization signals of maize Activator (Ac) transposase overlaps the DNA-binding domain. Plant J. 1995;7:441–451. doi: 10.1046/j.1365-313x.1995.7030441.x. [DOI] [PubMed] [Google Scholar]
- Boguski MS, Lowe TM, Tolstoshev CM. dbEST: Database for “expressed sequence tags.”. Nature Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- Boulikas T. Putative nuclear localization signals (NLS) in protein transcription factors. J Cell Biochem. 1994;55:32–58. doi: 10.1002/jcb.240550106. [DOI] [PubMed] [Google Scholar]
- Brian PW. The effects of some microbial metabolic products on plant growth. Symp Soc Exp Biol. 1957;11:166–182. [PubMed] [Google Scholar]
- Carol P, Peng J, Harberd NP. Isolation and preliminary characterization of gas1-1, a mutation causing partial suppression of the phenotype conferred by the gibberellin-insensitive (gai) mutation in Arabidopsis thaliana (L.) Heyhn. Planta. 1995;197:414–417. doi: 10.1007/BF00202665. [DOI] [PubMed] [Google Scholar]
- Chandler PM. Hormonal regulation of gene expression in the “slender” mutant of barley (Hordeum vulgare L.) Planta. 1988;175:115–120. doi: 10.1007/BF00402888. [DOI] [PubMed] [Google Scholar]
- Chang C, Kwok SF, Bleecker AB, Meyerowitz EM. Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science. 1993;262:539–544. doi: 10.1126/science.8211181. [DOI] [PubMed] [Google Scholar]
- Chiang H-H, Hwang I, Goodman HM. Isolation of the Arabidopsis GA4 locus. Plant Cell. 1995;7:195–201. doi: 10.1105/tpc.7.2.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croker SJ, Hedden P, Lenton JR, Stoddart JL. Comparison of gibberellins in normal and slender barley seedlings. Plant Physiol. 1990;94:194–200. doi: 10.1104/pp.94.1.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis TD, Curry EA. Chemical regulation of vegetative growth. Crit Rev Plant Sci. 1991;10:151–188. [Google Scholar]
- Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell. 1996;86:423–433. doi: 10.1016/s0092-8674(00)80115-4. [DOI] [PubMed] [Google Scholar]
- Dingwall C, Laskey RA. Nuclear targeting sequences—A consensus? Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- Fujioka S, Yamane H, Spray CR, Katsumi M, Phinney BO, Gaskin P, MacMillan J, Takahashi N. The dominant non-gibberellin-responding dwarf mutant (D8) of maize accumulates native gibberellins. Proc Natl Acad Sci. 1988;85:9031–9035. doi: 10.1073/pnas.85.23.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gale MD, Law CN, Marshall GA, Worland AJ. The genetic control of gibberellic acid insensitivity and coleoptile length in a “dwarf” wheat. Heredity. 1975;34:393–399. [Google Scholar]
- Gale MD, Youssefian S. Dwarfing genes in wheat. In: Russell GE, editor. Progress in plant breeding. London, UK: Butterworths; 1985. pp. 1–35. [Google Scholar]
- Gilroy S, Jones RL. Perception of gibberellin and abscisic acid at the external face of the plasma membrane of barley (Hordeum vulgare L.) aleurone protoplasts. Plant Physiol. 1994;104:1185–1192. doi: 10.1104/pp.104.4.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi M, Gulli M, Stanca AM, Cattivelli L. Characterization of two barley genes that respond rapidly to dehydration stress. Plant Sci. 1995;105:71–80. [Google Scholar]
- Harberd NP, Freeling M. Genetics of dominant gibberellin-insensitive dwarfism in maize. Genetics. 1989;121:827–838. doi: 10.1093/genetics/121.4.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P, Graebe JE. Inhibition of gibberellin biosynthesis by paclobutrazol in cell-free homogenates of Cucurbita maxima endosperm and Malus pumila embryos. J Plant Growth Regul. 1985;4:111–122. [Google Scholar]
- Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- Hooley R. Gibberellins: Perception, transduction and responses. Plant Mol Biol. 1994;26:1529–1555. doi: 10.1007/BF00016489. [DOI] [PubMed] [Google Scholar]
- Hooley R, Beale MH, Smith SJ. Gibberellin perception at the plasma membrane of Avena fatua aleurone protoplasts. Planta. 1991;183:271–280. doi: 10.1007/BF00197799. [DOI] [PubMed] [Google Scholar]
- Hua J, Chang C, Sun Q, Meyerowitz EM. Ethylene insensitivity conferred by Arabidopsis ERS gene. Science. 1995;269:1712–1714. doi: 10.1126/science.7569898. [DOI] [PubMed] [Google Scholar]
- Jacobsen JV, Gubler F, Chandler PM. Gibberellin action in germinated cereal grains. In: Davies PJ, editor. Plant hormones: Physiology, biochemistry and molecular biology. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. pp. 246–271. [Google Scholar]
- Jacobsen SE, Olszewski NE. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. Plant Cell. 1993;5:887–896. doi: 10.1105/tpc.5.8.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proc Natl Acad Sci. 1996;93:9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans DA, Briggs LJ, Gustin SE, Jans P, Ford S, Young IG. A functional bipartite nuclear localization signal in the cytokine interleukine-5. FEBS Lett. 1997;406:315–320. doi: 10.1016/s0014-5793(97)00293-7. [DOI] [PubMed] [Google Scholar]
- Jones DA, Thomas CM, Hammond-Kosack KE, Balint-Kurti PJ, Jones JDG. Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science. 1994;266:789–793. doi: 10.1126/science.7973631. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell. 1993;72:1–20. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Koornneef M, van der Veen JH. Induction and analysis of gibberellin sensitive mutants in Arabidopsis thaliana (L.) Hehyn. Theor Appl Genet. 1980;58:257–263. doi: 10.1007/BF00265176. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Elgersma A, Hanhart CJ, van Loenen-Martinet EP, van Rign L, Zeevaart JAD. A gibberellin insensitive mutant of Arabidopsis thaliana. Physiol Plant. 1985;65:33–39. [Google Scholar]
- Kreppel LK, Blomberg MA, Hart GW. Dynamic glycosylation of nuclear and cytosolic proteins: Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J Biol Chem. 1997;272:9308–9315. doi: 10.1074/jbc.272.14.9308. [DOI] [PubMed] [Google Scholar]
- LaCasse EC, Lefebvre YA. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanahan MB, Ho T-HD. Slender barley: A constitutive gibberellin response mutant. Planta. 1988;175:107–114. doi: 10.1007/BF00402887. [DOI] [PubMed] [Google Scholar]
- Lenton JR, Hedden P, Gale MD. Gibberellin insensitivity and depletion in wheat—consequences for development. In: Hoad GV, Lenton JR, Jackson MB, Atkin RK, editors. Hormone action in plant development: A critical appraisal. London, UK: Butterworths; 1987. pp. 145–160. [Google Scholar]
- Long D, Martin M, Sundberg E, Swinburne J, Puangsomlee P, Coupland G. The maize transposable element system Ac/Ds as a mutagen in Arabidopsis: Identification of an albino mutation induced by Ds insertion. Proc Natl Acad Sci. 1993;90:10370–10374. doi: 10.1073/pnas.90.21.10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas WA, Frank DW, Krause M, Hanover JA. O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J Biol Chem. 1997;272:9316–9324. doi: 10.1074/jbc.272.14.9316. [DOI] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love C, Westphal L, Murphy G, Sherson S, Cobbett C, Dean C. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- Montminy M. Something new to hang your HAT on. Nature. 1997;387:654–655. doi: 10.1038/42594. [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Harberd NP. Derivative alleles of the Arabidopsis gibberellin-insensitive (gai) mutation confer a wild-type phenotype. Plant Cell. 1993;5:351–360. doi: 10.1105/tpc.5.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol. 1997;113:1051–1058. doi: 10.1104/pp.113.4.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AL, Ward DA, Uknes S, Appleford NEJ, Lange T, Huttly AK, Gaskin P, Graebe JE, Hedden P. Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 1995;108:1049–1057. doi: 10.1104/pp.108.3.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potts WC, Reid JB, Murfet IC. Internode length in Pisum. Gibberellins and the slender phenotype. Physiol Plant. 1985;63:357–364. [Google Scholar]
- Putterill J, Robson F, Lee K, Coupland G. Chromosome walking with YAC clones in Arabidopsis: Isolation of 1700 kb of contiguous DNA on chromosome 5, including a 300-kb region containing the flowering-time gene CO. Mol Gen Genet. 1993;239:145–157. doi: 10.1007/BF00281613. [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G. The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell. 1995;80:847–857. doi: 10.1016/0092-8674(95)90288-0. [DOI] [PubMed] [Google Scholar]
- Raikhel N. Nuclear targeting in plants. Plant Physiol. 1992;100:1627–1632. doi: 10.1104/pp.100.4.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- Roe JL, Rivin CJ, Sessions RA, Feldmann KA, Zambryski PC. The Tousled gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell. 1993;75:939–950. doi: 10.1016/0092-8674(93)90537-z. [DOI] [PubMed] [Google Scholar]
- Roe JL, Durfee T, Zupan JR, Repetti PP, McLean BG, Zambryski PC. TOUSLED is a nuclear serine/threonine protein kinase that requires a coiled-coil region for oligomerization and catalytic activity. J Biol Chem. 1997;272:5838–5845. doi: 10.1074/jbc.272.9.5838. [DOI] [PubMed] [Google Scholar]
- Ross JJ. Recent advances in the study of gibberellin mutants. Plant Growth Regul. 1994;15:193–206. [Google Scholar]
- Schaller GE, Bleecker AB. High-affinity binding sites for ethylene are generated in yeast expressing the Arabidopsis ETR1 gene. Science. 1995;270:1809–1811. doi: 10.1126/science.270.5243.1809. [DOI] [PubMed] [Google Scholar]
- Silverstone AL, Mak PYA, Martínez EC, Sun T-p. The new RGA locus encodes a negative regulator of gibberellin response in Arabidopsis thaliana. Genetics. 1997;146:1087–1099. doi: 10.1093/genetics/146.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SJ, Walker RP, Beale MH, Hooley R. Biological activity of some gibberellins and gibberellin derivatives in aleurone cells and protoplasts of Avena fatua. Phytochemistry. 1993;33:17–20. [Google Scholar]
- Sun T-p, Kamiya Y. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. Plant Cell. 1994;6:1509–1518. doi: 10.1105/tpc.6.10.1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Olszewski NE. Genetic analysis of gibberellin signal transduction. Plant Physiol. 1996;112:11–17. doi: 10.1104/pp.112.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon M, Koornneef M, Zeevaart JAD. Endogenous gibberellins in Arabidopsis thaliana and possible steps blocked in the biosynthetic pathways of the semidwarf ga4 and ga5 mutants. Proc Natl Acad Sci. 1990a;87:7983–7987. doi: 10.1073/pnas.87.20.7983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talon M, Koornneef M, Zeevaart JAD. Accumulation of C19-gibberellins in the gibberellin-insensitive dwarf mutant gai of Arabidopsis thaliana (L.) Hehyn. Planta. 1990b;182:501–505. doi: 10.1007/BF02341024. [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Somerville C. Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiol. 1995;108:495–502. doi: 10.1104/pp.108.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Heckman JW, Somerville C. Gibberellin is required for flowering in Arabidopsis thaliana under short days. Plant Physiol. 1992;100:403–408. doi: 10.1104/pp.100.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler RG, Freeling M. Physiological genetics of the dominant gibberellin-nonresponsive maize dwarfs, Dwarf8 and Dwarf9. Planta. 1994;193:341–348. [Google Scholar]
- Xu Y-L, Li L, Wu K, Peeters AJM, Gage DA, Zeevaart JAD. The GA5 locus of Arabidopsis thaliana encodes a multifunctional gibberellin 20-oxidase: Molecular cloning and functional expression. Proc Natl Acad Sci. 1995;92:6640–6644. doi: 10.1073/pnas.92.14.6640. [DOI] [PMC free article] [PubMed] [Google Scholar]