Abstract
We investigated the potential (d)NDP/(d)NTP discrimination mechanisms in nucleotide pyrophosphatases. Here, we report that dUTPase, an essential nucleotide pyrophosphatase, uses a C-terminal P-loop-like sequence in a unique mechanism for substrate discrimination and efficient hydrolysis. Our spectroscopy and transient kinetics results on human dUTPase mutants combined with previous structural studies indicate that (i) H-bond interactions between the γ-phosphate and the P-loop-like motif V promote the catalytically competent conformation of the reaction center at the α-phosphate group; (ii) these interactions accelerate the chemical step of the kinetic cycle and that (iii) hydrolysis occurs very slowly or not at all in the absence of the γ-phosphate—motif V interactions, i.e., in dUDP, dUDP.BeFx, or in the motif V-deleted mutant. The physiological role of dUTPase is to set cellular dUTP∶dTTP ratios and prevent injurious uracil incorporation into DNA. Based upon comparison with related pyrophosphate generating (d)NTPases, we propose that the unusual use of a P-loop-like motif enables dUTPases to achieve efficient catalysis of dUTP hydrolysis and efficient discrimination against dUDP at the same time. These specifics might have been advantageous on the appearance of uracil-DNA repair. The similarities and differences between dUTPase motif V and the P-loop (or Walker A sequence) commonly featured by ATP- and GTPases offer insight into functional adaptation to various nucleotide hydrolysis tasks.
Keywords: NTP hydrolysis, nucleotide discrimination, dUTP pyrophosphatase, Walker A motif, evolutionary adaptation
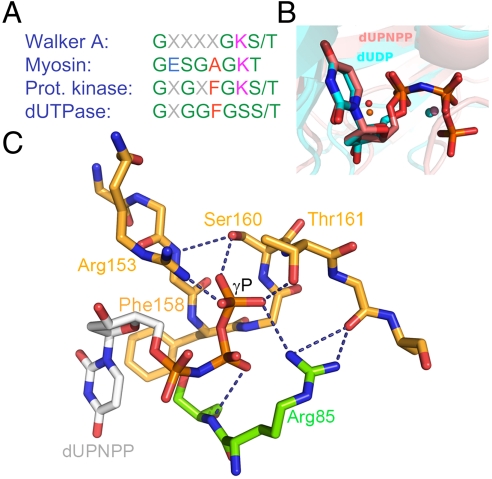
It is an intriguing question how pyrophosphate generating nucleotide hydrolases distinguish between (d)NDP and (d)NTP both containing the α-β phosphoanhydride bond to be hydrolyzed. In the present paper, we investigate two fundamental questions related to the nucleotide pyrophosphatase enzymatic activity exhibited by the enzyme dUTPase and by other pyrophosphatases: (i) the mechanism of discrimination between nucleoside di- and triphosphate ligands; (ii) the potential contribution of a P-loop-like motif to such discrimination. The enzyme dUTPase naturally evoked these questions as it specifically performs the hydrolysis of dUTP between the α-β phosphates with no further coupled reactions and it contains a P-loop-like motif (Fig. 1A). In addition, the structural comparison between Mg:dUDP- and Mg:dUTP analog-bound enzymes does not offer a straightforward explanation to why dUDP is not hydrolyzed because the scissile bond and the nucleophile adopt the same conformation in both (Fig. 1B and in stereo in Fig. S1A, the sole Mg:dUDP-complexed structure is superposed with a human Mg:dUPNPP complex).
Fig. 1.
The P-loop-like C-terminal motif V and its interactions with the substrate, (A), Consensus P-loop motifs from well known P-loop NTPases compared with the P-loop-like sequence of dUTPases. Coloring is as follows, polar residues, green; apolar r., red; negatively charged r., blue; and positively charged r., magenta (B), Alignment of two dUTPase active sites containing uridine di- and triphosphate ligands [atomic coloring with cyan and pink carbons, PDB IDs 1SLH (9) and 2HQU (8) from M. tuberculosis and Homo sapiens, respectively]. The positions of the α-β phosphates are identical and the catalytic water (orange for the tri- and magenta for the diphosphate structure) and Mg2+ ions (spheres in colors corresponding to the related nucleosides) are in place for putative hydrolysis of the α-β phosphate bond in both structures. (C), Coordination of the γ-P of the substrate by amino acids subjected to mutation in this study. The homotrimeric protein (2, PDB ID: 2HQU) is colored by subunits (subunit C, atomic coloring with orange carbons; subunit B atomic coloring with green carbons). dUPNPP is depicted as atomic colored sticks with gray carbons. Dashed lines depict hydrogen bonds partially abolished by the mutations thus hampering the coordination of the γ-P. Arg85 of subunit B is shown to complete the H-bonding network around the γ-P. Phe158, which engages in an aromatic stacking with uracil, is exchanged to a Trp to yield the specific fluorescence signal throughout this study. For better viewing, these structural images are also shown in stereo mode in Fig. S1.
dUTPase hydrolyzes dUTP to yield dUMP (a precursor for dTTP biosynthesis) and pyrophosphate (PPi). The action of dUTPase is the only known direct mechanism to minimize uracil incorporation into DNA (1). Most dUTPases are homotrimers and confer three active sites. The substrate in each active site is bound by conserved sequence motifs from all three subunits. Therefore, although each subunit contains all necessary residues for substrate binding, trimer formation is indispensable to bring these residues in proximity for the cognate binding site (1, 2). dUTPase, similar to many other nucleotide binding proteins, contains a conserved loop motif (motif V) to coordinate the phosphate chain of the protein. The C terminus conferring this loop is not part of the globular enzyme core, it reaches far from its protomer to isolate a remote active site from the solvent (Fig. S2). The sequence of the C-terminal motif V shares a limited number of features with those of the P-loop motifs (Fig. 1A) (3, 4) present in a large number of ATPase and GTPase enzyme families known as P-loop NTPases including kinases, cytoskeleton and DNA motors, membrane pumps, and transporters.
It has been shown that the C terminus of dUTPase is necessary for dUTP hydrolysis but not for nucleotide binding or structural integrity (3, 5–7). Surprisingly, the active site architecture with bound substrate in the crystal structure of the C-terminally truncated inactive enzyme is identical to that of the wild-type (WT) (7). On the basis of activity measurements on full length and enzymatically truncated Escherichia coli enzymes, it was concluded that ordering of the flexible C terminus upon the active site, which occurs only in the presence of the gamma phosphate (γ-P) containing substrate analog dUPNPP is responsible for dUDP/dUTP discrimination (3, 5). There seems to be a clear difference, however, between the conformational freedom of the C terminus of E. coli and several other investigated dUTPases as evidenced by structural data in the crystal (8–11) and in solution (3, 5, 6, 12). Our previous results in human dUTPase suggest that the C-terminal motif V remains close to the active site even in apo state (13) whereas the E. coli motif V becomes loose without the substrate (12).
It was earlier proposed that closing of the C-terminal motif V upon the active site forces an unfavorable eclipsed conformation on the nucleotide substrate which may be responsible for catalysis (4). However, the dUPNPP substrate analog maintains its eclipsed [either gauche or trans, (14)] conformation in several crystal structures in which the C-terminal arm is disordered (i.e., flexible) (8–10, 15, 16). The peculiar eclipsed conformation thus seems to be independent of the flexibility of the C-terminal motif V. It remained therefore an open question as to how this P-loop-like sequence contributes to the hydrolytic event which occurs via nucleophilic attack on the α-phosphate (15).
To gain insight into the mechanism of discrimination potentially related to the P-loop-like sequence (as earlier proposed in ref. 5), we designed mutants which fully or partially lost contact between the γ-P of the substrate nucleotide and the P-loop-like sequence. The coordination of the γ-P of the nucleotide and the amino acid sites which were mutated are depicted in Fig. 1C (and in stereo in Fig. S1B). The mutations were intended to partially (Ser160Ala/Thr161Ala termed hDUTST/AA, and Arg153Lys termed hDUTR/K) or completely (Thr151Stop termed hDUTarmless) disrupt the secondary interaction network between the γ-P and the protein as also shown in an earlier work by Freeman et al. (7). These mutant enzymes were then subjected to kinetic and ligand binding analysis. Fortunately, the P-loop-like sequence of dUTPase is situated at the very C terminus of the polypeptide chain (Fig. S2) and therefore, its mutation or deletion does not endanger the overall fold and conformation of the full protein (Fig. S3) (3, 5, 10, 12). We also analyzed the available structural and kinetics data on the structurally and/or functionally related bifunctional dUDP/dUTPase, dCTP deaminase (DCD), and bifunctional dCTP deaminase/dUTPase (DCD-DUT) enzymes and compared them to our results.
Here we present a mechanism by which the P-loop-like motif of dUTPase promotes catalysis and discriminates against dUDP at the same time. We demonstrate that the unique use of a P-loop-like nucleotide binding sequence in dUTPase among nucleotide pyrophosphatases is functional adaptation to high dUTP specificity likely related to the development of uracil-DNA repair. The structural and functional similarity to P-loops is also discussed.
Results and Discussion
Mutations in the P-Loop-Like Sequence Disable dUTPase’s Ability to Effectively Discriminate Between the Nucleoside Di- and Triphosphate Ligands.
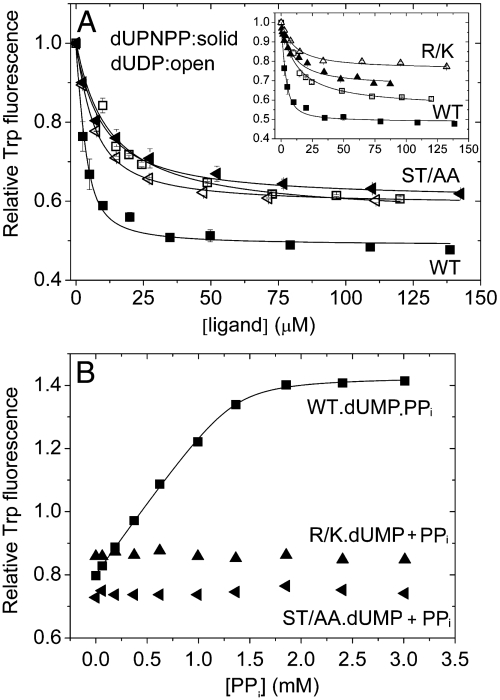
We took advantage of the discriminative power of the tryptophan (Trp) sensor built into the active site in place of F158 (shown in Fig. 1 and ref. 17) to investigate the interaction of dUTPase C-terminal motif V mutants with physiological ligands. The fluorescence of W158 is characteristically quenched in the dUTPase-ligand complexes compared with the apoenzyme (13) due to an aromatic stacking interaction between the uracil ring and the conserved aromatic side chain of the C-terminal motif V (18). The effect of the Trp substitution on the enzymatic and ligand binding properties of dUTPase is minor [kcat = 5.8 ± 0.5 and 6.8 ± 2 s-1, Kd,dUPNPP = 1.5 ± 1 and 2.0 ± 1 μM for the WT and Trp mutant, respectively when measured for direct comparison (18)]. In the apoenzyme, the C-terminal motif V displays increased flexibility in solution (5, 6) and is disordered in the crystal structure [PDB ID: 1Q5U (8)] but the C-terminal arm stays in proximity to the active site through nucleotide-independent intersubunit interactions according to our previous spectroscopy results (13). Fig. 2A shows fluorescence intensity titrations upon dUTPase binding to dUDP or to the nonhydrolysable substrate analog α-β-imidodUTP (dUPNPP). Both dUDP and dUPNPP binding curves of the hDUTST/AA mutant run close to the dUDP binding curve of the WT enzyme indicating that the binding mode of dUPNPP in this mutant is similar to the dUDP-bound conformation of the WT. The hDUTR/K mutant also displays a reduction in the fluorescence quench brought about by ligand binding (Fig. 2A inset). We also applied circular dichroism (CD) spectroscopy to measure ligand binding as the hDUTarmless mutant lacks the fluorescence signal (Fig. S4). The CD measurements are in line with the fluorescence-based ones in cases where both techniques were applied (Table 1). Dissociation constants obtained from data fitting are collected in Table 1 and show that while the WT binds dUPNPP stronger than dUDP, the complexes of the mutants with dUPNPP or dUDP are almost equally strong. The hDUTarmless mutant exhibits smaller binding constants than the other enzyme constructs in agreement with the fewest contacts it has with the ligands. Binding of the product dUMP to the mutants is not significantly affected as compared to the WT (Fig. S5, Table 1) and is one order of magnitude weaker than that of the substrate.
Fig. 2.
Effect of mutations in the P-loop-like sequence on nucleotide and PPi binding. Fluorescence intensity titration of the loop Trp is shown upon dUTPase binding to (A), dUPNPP (solid symbols) and dUDP (open symbols) (hDUTR/K data shown in the inset separately for clarity) or (B), upon dUMP saturated dUTPase binding to PPi. Signal intensities are normalized to the nucleotide-free state. Smooth lines through the data are hyperbolic fits yielding Kd values listed in Table 1. WT (squares), hDUTR/K (triangles), and hDUTST/AA (left-pointing triangles) were used at 4 μM concentration. Error bars indicate SD for n = 3. Further evidences on nucleotide binding properties of the mutants are shown in the SI Results and Discussion.
Table 1.
Dissociation constants of WT and P-loop mutant hDUT complexes with deoxyuridin mono-, di- and triphosphate ligands determined by Trp fluorescence or by CD
| Kd (μM) | |||
| Enzyme | dUMP | dUDP | dUPNPP |
| WT | 78 ± 4 | 12 ± 1 | 5.0 ± 3 3.4 ± 1* |
| hDUTR/K | 96 ± 14 | 12 ± 1 | 11 ± 4 10 ± 4 * |
| hDUTST/AA | 74 ± 6 | 5.6 ± 0.6 | 6.5 ± 1 6.8 ± 3 * |
| hDUTarmless | ND | 9.5 ± 3 * | 14 ± 0.4 * |
*determined by differential CD spectroscopy
ND: not determined
A strong proof of compromised γ-P binding by the C-terminal motif V mutants is shown in Fig. 2B. We could not observe any sign of ternary product complex (enzyme.dUMP.PPi) formation even at high PPi concentrations in contrast to results obtained with the WT. The lack of a binding curve here may indicate a highly elevated Kd or an altered binding mode that is not followed by the characteristic fluorescence signal change (13).
In summary, the similar degree of fluorescence quench and similar dissociation constants for the dUDP- and dUPNPP-mutant complexes together with an undetectable PPi binding to the mutant enzymes indicate that the P-loop-like nucleotide binding motif is the γ-P “detector” of dUTPase.
Mutations of the P-Loop-Like Sequence Result in a Large Decrease in the Enzymatic Activity and Specifically in the Hydrolysis Rate Constant.
The catalytic activity and efficiency of dUTPase is severely affected in these mutants in the order of hDUTarmless > hDUTR/K > hDUTST/AA≫WT (Fig. S6, Table 2). These results are in line with the expectations as only some H-bonds are disrupted in the hDUTST/AA and hDUTR/K mutants, while in the hDUTarmless, the entire γ-P coordination is abolished. Our results also agree with earlier data showing the near inactivity of armless mutants (5, 7, 10, 19) and of R/K mutants (7, 10, 20).
Table 2.
Kinetic parameters of WT and P-loop mutant dUTPases
| Enzyme | Vmax (s-1) | KM (μM) | kcat/KM (M-1 s-1) | kH (s-1) | kdUTP,on (μM-1 s-1) | kdUTP,off (s-1) | kISO (s-1) |
| WT | 6.5 ± 0.2 | 1.3 ± 0.5 | 5.0 × 106 | 5.5 ± 3 * | 120 * | 100 * | 24 ± 6 |
| hDUTR/K | 0.078 ± 0.002 | 6.1 ± 1.7 | 1.3 × 104 | 0.048 ± 0.001 | ND | ND | ND |
| hDUTST/AA | 0.26 ± 0.1 | 12 ± 2 | 2.2 × 105 | 0.16 ± 0.01 | 84 ± 8 | 250 ± 40 | 29 ± 5 |
| hDUTarmless | 0.010 ± 0.0007 | ND | 7.1 × 103† | 0.014 ± 0.0004 | NA | NA | NA |
*data from Toth and coworkers (1)
†Kd for the hDUTarmless.dUPNPP complex was used instead of KM
ND: not determined
NA: not applicable
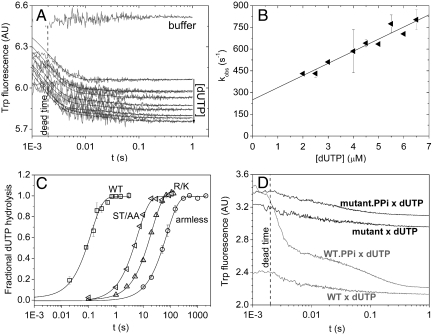
To determine the mechanism underlying the steady-state behavior of the mutant enzymes, transient kinetics experiments investigating the previously described kinetic steps (13) were performed on all or selected mutants. The kinetic parameters of dUTP binding to and dissociating from the hDUTST/AA were determined via stopped-flow using the Trp signal. Fig. 3A shows the time courses upon mixing hDUTST/AA with increasing concentrations of dUTP. The curves were fitted with double exponential functions. As in the case of the WT, a concentration dependent fast and a first-order slow phase (representing 5–8% of the total amplitude) could be distinguished. The concentration dependence of the fast process (Fig. 3B) yielded kdUTP,on = 84 μM-1 s-1 and kdUTP,off = 250 s-1. These rate constants report somewhat faster dUTP binding than to the WT enzyme [(13), Table 2], while the rate constants of the second, isomerization, process are similar in both enzymes (kISO = 29 s-1 vs. 24 s-1 in the WT).
Fig. 3.
Transient kinetic analysis of the mutants, (A and B) show stopped-flow experiments of hDUTAST/AA binding to dUTP. (A), Fluorescence time courses recorded upon the initial binding phase of the reaction of various concentrations of dUTP with 0.8 μM hDUTST/AA. Smooth lines are the best double exponential fits to the experimental curves. The first exponential (fast phase with 95–98% of the total amplitude) is analyzed in (B). The second exponential of small amplitude did not depend on concentration and yielded kISO = 29 ± 5 s-1 (SD for n = 10). (B), Concentration dependence of the observed rate constant of the fast phase. The linear fit yielded a second-order binding rate constant of 84 ± 8 μM-1 s-1 and a dissociation rate constant of 250 ± 36 s-1. Errors represent SD for n = 20. (C), Single turnover γ32P-dUTP hydrolysis by the WT and mutant dUTPases measured using the quench-flow technique. 25 μM protein was mixed with 12.5 μM γ32P-dUTP and the reaction was followed till completion. Each curve was fitted with single exponentials yielding hydrolysis rate constants of 5.5 ± 2.5 s-1 for WT (squares), 0.16 ± 0.01 s-1 for hDUTST/AA (left-pointing triangles), 0.048 ± 0.001 s-1 for hDUTR/K (triangles), and 0.01 ± 0.001 s-1 for hDUTarmless (circles). Errors represent SD for n = 3. (D), Time courses of PPi dissociation from the WT and hDUTST/AA was measured by dUTP chasing in the stopped-flow. 4 μM enzyme or its preequilibrated complex with 2 mM PPi was mixed with 1 mM dUTP. The fast phase of the dUTP binding reaction is lost in the dead-time as expected (13). In the chasing reaction, PPi dissociation limits the rate of dUTP binding which can be followed in case of the WT enzyme (gray lines) while no PPi dissociation can be observed in the mutant (black lines). WT and hDUTST/AA curves are shifted on the y-axis compared to each other for better viewing.
We took the next kinetic step, hydrolysis, under investigation by using the quenched-flow method. As shown in Fig. 3C (note the logarithmic time scale) the single turnover hydrolysis curves of the mutants report a large decrease in the hydrolysis rate constants compared to the WT. Single exponential functions fitted well to the curves and yielded rate constants (kH) comparable to those of the steady-state rate (Table 2).
Our fluorescence (Fig. 2A) and quenched-flow (Fig. 3C) data together indicate that perturbation of the secondary interactions between the C-terminal motif V and the γ-P results in a reduced ability of the enzyme-substrate complex to adopt a catalytically competent conformation. Hydrolysis still occurs at a reduced rate, the reduction being proportional to the expected severity of the perturbation (WT≫hDUTST/AA > hDUTR/K > hDUTarmless, Table 2). The C-terminal motif V is the major but not the only structural element of the enzyme that coordinates the γ-P. Arg85 of another conserved motif is also important for activity probably via the coordination and charge neutralization of the β- and γ-P (Fig. 1C, Fig. S1B). This interaction may explain the residual activity of the mutant enzyme lacking the P-loop-like sequence (hDUTarmless).
We attempted to measure the kinetics of product release from the enzyme-product(s) complex by dUTP chasing (Fig. 3D). The WT enzyme exhibited slower apparent dUTP binding when mixed with the enzyme.PPi complex as dissociation of PPi from the enzyme limited the dUTP binding process (13). Conversely, we observed no such effect when repeating the experiment using the hDUTST/AA mutant. The reason PPi dissociation was not observed may be that (i) PPi dissociation is as fast as, or faster than dUTP binding at the applied dUTP concentration, or (ii) no quantitative enzyme.PPi complex was formed. Both possibilities imply that the PPi off-rate from the P-loop mutant is probably higher than that from the WT (kp = 740 s-1 in ref. 13) and thus does not alter the steady-state rate.
This transient kinetic analysis indicates that the only kinetic step that affects the observed steady-state rate in the mutants is hydrolysis itself (i.e., hydrolysis or an indistinguishable conformational transition coupled to it). Other changes in the substrate binding and product release kinetics are not manifested in the steady-state rate because these processes remained orders of magnitude faster. We may conclude that the interaction of the γ-P with the C-terminal P-loop-like sequence in dUTPase promotes the entry into a hydrolysis competent state and therefore the frequency of the hydrolytic events. This phenomenon may be brought about by optimization of the geometry of the prehydrolysis enzyme.substrate complex or by electrostatics, i.e., partially distributing the excess negative charges through secondary interactions.
It is important to point out that the conserved aromatic residue within the P-loop-like sequence (Fig. 1A) makes a significant contribution to rate acceleration in dUTPase via its stacking interaction with the uracil ring of the substrate as we reported recently in the Mycobacterium tuberculosis and human enzymes (18). The lack of this interaction in the hDUTarmless mutant acts together with the lack of γ-P coordination. Interestingly, the P-loop-like motif in dUTPase forms catalytically important interactions with two distinct parts of the nucleotide substrate at both sides of the scissile bond.
The Presence of γ-P Is Required for Hydrolysis of the α-β Phosphoanhydride Bond by dUTPase.
As dUTPase removes PPi instead of only the terminal phosphate group of the nucleotide, it is possible to investigate the effect of γ-P substitution using nucleotide analogs. The ADP.BeFx complex has been reported to act as ATP analog (21, 22), we therefore assayed the binding (Fig. S7A) and hydrolysis (Fig. S7B) of the dUDP.BeFx complex by the WT enzyme. Using fluorescence spectroscopy we detected binding of BeFx to the enzyme.dUDP complex characterized by a fluorescence quench similar to that of dUPNPP binding. Hydrolysis, nevertheless, did not occur in the enzyme.dUDP.BeFx complex (see SI Results and Discussion). If dUDP.BeFx is truly a nucleoside triphosphate analog in dUTPase as indicated by our spectroscopy result, then it suggests that the γ-P specifically is required for hydrolysis of the α-β phosphoanhydride bond.
The hydrolysis of dUDP by the WT and motif V mutant enzymes was also investigated using thin layer chromatography (TLC). Although this method allowed the use of high enzyme and dUDP concentrations, dUDP hydrolysis was not detected in agreement with previous reports (Fig. S8) (23).
Because the presence of an intact C-terminal motif V enhances the catalytic efficiency of dUTPase 720-fold (kcat/KM values in Table 2), the γ-P—motif V interaction probably is the major factor in discriminating the nonsubstrate dUDP from the substrate dUTP in accordance with previous suggestions from E. coli (3, 5).
Comparison to Functionally Related Enzymes Suggests That the P-Loop-Like Motif in dUTPase Is Evolutionary Adaptation to High dUTP Specificity and dUDP Discrimination.
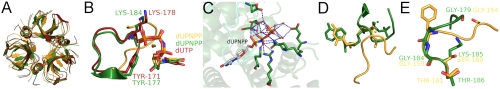
The use of a P-loop-like sequence to promote nucleotide pyrophosphatase activity and NDP/NTP discrimination is unique in dUTPase (for details see SI Results and Discussion). The most relevant structural and functional comparison to make is therefore with the homologous dUTPase superfamily enzymes dCTP deaminase (DCD) and dCTP deaminase/dUTPase (DCD-DUT), as well as with the nonhomologous but functionally comparable bifunctional dUDP/dUTPase. dUTPase, DCD, and DCD-DUT share a common fold and the first four motifs responsible for nucleotide and Mg2+ binding (Fig. 4A and in stereo in Fig. S9A). Importantly, however, only dUTPase possesses a P-loop-like sequence as its motif V. The C terminus of the other members of the superfamily also goes through a disordered-to-ordered transition upon substrate binding (24). This transition, however, results in relatively few interactions with the γ-P (Fig. 4B and in stereo in Fig. S9B) (25). These enzymes are significantly worse catalysts of dUTP hydrolysis than dUTPase (kcat/KM = 4.5 × 103 s-1 M-1) for DCD-DUT from M. tuberculosis at 37 °C (26) and 3.5 × 104 s-1 M-1 from Methanococcus jannaschii at 60 °C (27) to be compared to 5 × 106 s-1 M-1 at 25 °C for hDUT) even though their active site architecture including the position of the catalytic Asp and water molecule are similar. This observation implies that it is probably the presence of the C-terminal P-loop-like sequence that makes dUTPase the most efficient dUTP hydrolase within the superfamily. We therefore propose that the use of a P-loop-like sequence is an evolutionary adaptation of current dUTPases that allows for efficient catalysis of dUTP hydrolysis. The uracil world being more ancient than that including the methylated uracil, thymine, the action of dUTPase in preventing uracil incorporation into DNA must have gained importance only with the appearance of thymine and the uracil-specific DNA repair systems.
Fig. 4.
γ-P-protein interactions in dUTPase-related and in P-loop enzymes, (A), Identical fold of protein cores of M. tuberculosis dUTPase (PDB ID: 2PY4, orange), M. tuberculosis DCD-DUT (PDB ID: 2QPL, green), and E. coli DCD (PDB ID: 1XS1, red). The enzymes were superimposed with UCSF Chimera. (B), Superimposition of the actives sites of the above structures. The nucleotides and the C terminus contacting the phosphate chain are highlighted. Amino acids contacting the γ-P in DCD and DCD-DUT, and nucleotides are presented as sticks with atomic coloring. Mg2+ ions are presented as spheres. The γ-P interactions in dUTPase already shown in Fig. 1C are omitted here for clarity. (C), The active site of Campylobacter jejuni dUDP/dUTPase [PDB ID: 2CIC (30)]. β- and γ-P coordinating groups and their contacts to dUPNPP (atomic coloring with gray carbons) are highlighted. While the β-P has many contacts to the protein, the γ-P is only coordinated by water molecules (red spheres). Dashed blue lines depict hydrogen bonds. The protein is colored by subunits, Mg2+ ions as purple spheres. (D), Comparison of the relative positions of the C-terminal motif V in dUTPase (PDB ID: 2HQU, orange) and the P-loop in Dictyostelium discoideum myosin II [PDB ID: 1MMN (34), green]. dUPNPP and ATP are presented as sticks, Mg2+ ions as spheres. (E), Superimposition of the two loops presented in (D), aligned for the loop and not for the nucleotide. Walker A amino acids are highlighted by stick representation (atomic coloring). These structural images are also shown in stereo mode in Fig. S9.
The bifunctional dUDP/dUTPase enzymes generate dUMP from both dUDP and dUTP with comparable kcat/KM values in the order of 106 s-1 M-1 by a mechanism similar to that of DNA polymerases (28, 29). We analyzed the available structures of these enzymes and discovered that the γ-P of bound dUTP is only coordinated by water molecules and is largely exposed to the bulk solvent (Fig. 4C and in stereo in Fig. S9C). In the bifunctional enzyme, it is the β-P that makes extensive contacts with the enzyme residues and Mg2+ also necessary for catalysis (compare to Fig. 5A in ref. 30, Fig. 4C). The bifunctional dUDP/dUTPase activity in this case coincides with the lack of a P-loop-like sequence or other γ-P coordinating structural element. This phenomenon further supports our suggestion that a P-loop-like motif may be responsible for structural discrimination between (d)NDP and (d)NTP when no downstream coupled reaction occurs.
Discrimination against dUDP hydrolysis is probably not a major force of selection in the functional adaptation of trimeric dUTPases. It is more likely an incidental phenomenon that accompanies the increased catalytic efficiency dUTPases gained with the presence of a P-loop-like sequence.
Structural and Functional Comparison of the P-Loop-Like Motif V in dUTPase and the P-Loop in NTPases Indicates Convergent Evolution.
Comparing ATPases and dUTPase, we observe that while the P-loops and the phosphate chains can be perfectly superimposed within ATPases (shown in Fig. 5 in ref. 31), the P-loop-like motif in dUTPase adopts a different conformation relative to the nucleotide substrate (Fig. 4D). The superposition of the two loops, however, shows that there is structural similarity between their local conformation particularly around the single amino acid that makes the sequential difference between P-loop and dUTPase motif V (Fig. 4E and in stereo in Fig. S9D). The main difference between these conformations is that the P-loop in myosin (example for ATPase) surrounds both the β- and the γ-P, whereas the P-loop-like motif in dUTPase only contacts the γ-P (Fig. 1C and 4D). This difference, likely arising from the C-terminal position of the dUTPase loop and an eclipsed dUTP conformation, also supports the role of this motif in γ-P discrimination in dUTPase. The discriminatory role of the P-loop has also been shown in GTPases, in which most of the P-loop interactions target the β-P. Consequently, P-loop mutations may cause decreased dUDP affinity and preference for GTP binding finally resulting in overstimulation of certain signaling pathways (32).
The P-loop acts in different ways to achieve catalysis in NTPases and is therefore thought to have independently evolved in multiple instances (31). In G-proteins and myosins, its most likely role in catalysis is to orient the nucleotide and the attacking water molecule for efficient nucleophilic attack on the γ-P as well as charge stabilization (31, 33). In dUTPase, the γ-P coordination of the P-loop-like motif is also necessary for orientation of the catalytic apparatus and stabilization of the associative type transition state (17, 18). The completely different topology and the partly similar function of these loops indicates convergent evolution of phosphate binding motifs in a larger group of nucleotide hydrolases including PPi generating diphosphatases.
Methods
Proteins were expressed and purified as described previously (17). Site-directed mutagenesis was performed by the QuikChange method (Stratagene) and verified by sequencing of both strands. The following mutants where created: Ser160 and Thr161 to Ala (hDUTST/AA), Arg153 to Lys (hDUTR/K) while the truncated mutant missing the entire P-loop was created by generating a stop codon in place of the Thr151 residue (hDUTarmless). The enzyme conferring a single Trp in the active site (hDUTF158W) was used as WT (cf. ref. 13) thus all mutations were created within this construct. Protein concentration was measured using the Bradford method (Bio-Rad Protein Assay) and by UV absorbance (λ280 = 10,555 M-1 cm-1 for WT and hDUTarmless, λ280 = 16,055 M-1 cm-1 for hDUTF158W, hDUTAR/K, and hDUTST/AA) and is given in monomers. dUMP, dUDP, dUTP, and α,β-imido-dUTP (dUPNPP) where purchased from Jena Bioscience, other chemicals where from Sigma Aldrich. All measurements were carried out in a buffer comprising 20 mM Hepes pH 7.5, 100 mM NaCl, 2 mM MgCl2 and 1 mM DTT if not stated otherwise.
Stopped-Flow Experiments.
Measurements were made using an SX-20 (Applied Photophysics) stopped-flow apparatus. Measurement was carried out exactly as described in ref. 13. Time courses were analyzed using the curve fitting software provided with the stopped-flow apparatus or by Origin 7.5.
Quench flow experiments were carried out in an RQF-3 (KinTek Corp.) quench-flow apparatus using [γ-32P]dUTP as described earlier (13).
Fluorescence Intensity Titrations.
Fluorescence was measured in a Jobin Yvon Spex Fluoromax-3 spectrofluorometer at 20 °C, with excitation at 295 nm (slit 1 nm) and emission between 320–400 nm (slit 5 nm), or at 347 nm. 4 μM protein was titrated by addition of 1–2 μL aliquots from concentrated nucleotide solutions. Because large concentrations of nucleotides were used, care was taken to correct for any additional fluorescence or inner filter effect imposed on the measured intensities by the nucleotide stock solutions.
Statistical Analysis.
All measurements were carried out at least three times. Error bars represent standard deviations. Where no error bars are shown, a representative curve is displayed while the summary table shows the relevant standard deviations of a certain parameter obtained from several different measurements. In the stopped-flow experiments, typically 5–8 traces were collected and averaged.
Supplementary Material
Acknowledgments.
This work was supported by the US National Institutes of Health (Grant number 1R01TW008130-01); the János Bolyai Research Scholarship of the Hungarian Academy of Sciences, the Howard Hughes Medical Institutes (Grant number 55000342); the Hungarian Scientific Research Funds (Grant numbers PD72008, CK-78646, K68229, K72973, and NI68466); the National Office for Research and Technology, Hungary (Grant number JÁP_TSZ_071128_TB_INTER) and the EU FP6 (Grant numbers SPINE2c LSHG-CT-2006-031220, TEACH-SG LSSG-CT-2007-037198) and the Alexander von Humboldt Foundation.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013872108/-/DCSupplemental.
References
- 1.Vertessy BG, Toth J. Keeping uracil out of DNA: physiological role, structure and catalytic mechanism of dUTPases. Acc Chem Res. 2009;42:97–106. doi: 10.1021/ar800114w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Persson R, Cedergren-Zeppezauer ES, Wilson KS. Homotrimeric dUTPases; structural solutions for specific recognition and hydrolysis of dUTP. Curr Protein Pept Sc. 2001;2:287–300. doi: 10.2174/1389203013381035. [DOI] [PubMed] [Google Scholar]
- 3.Vertessy BG. Flexible glycine rich motif of Escherichia coli deoxyuridine triphosphate nucleotidohydrolase is important for functional but not for structural integrity of the enzyme. Proteins. 1997;28:568–579. [PubMed] [Google Scholar]
- 4.Prasad GS. Glycine rich P-loop motif in deoxyuridine pyrophosphatase. Curr Protein Pept Sc. 2001;2:301–311. doi: 10.2174/1389203013381017. [DOI] [PubMed] [Google Scholar]
- 5.Vertessy BG, et al. The complete triphosphate moiety of non-hydrolyzable substrate analogues is required for a conformational shift of the flexible C-terminus in E. coli dUTP pyrophosphatase. FEBS Lett. 1998;421:83–88. doi: 10.1016/s0014-5793(97)01545-7. [DOI] [PubMed] [Google Scholar]
- 6.Nord J, Nyman P, Larsson G, Drakenberg T. The C-terminus of dUTPase: observation on flexibility using NMR. FEBS Lett. 2001;492:228–232. doi: 10.1016/s0014-5793(01)02257-8. [DOI] [PubMed] [Google Scholar]
- 7.Freeman L, et al. The flexible motif V of Epstein-Barr virus deoxyuridine 5′-triphosphate pyrophosphatase is essential for catalysis. J Biol Chem. 2009;284:25280–25289. doi: 10.1074/jbc.M109.019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mol CD, Harris JM, McIntosh EM, Tainer JA. Human dUTP pyrophosphatase: uracil recognition by a beta hairpin and active sites formed by three separate subunits. Structure. 1996;4:1077–1092. doi: 10.1016/s0969-2126(96)00114-1. [DOI] [PubMed] [Google Scholar]
- 9.Chan S, et al. Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism. J Mol Biol. 2004;341:503–517. doi: 10.1016/j.jmb.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 10.Nemeth-Pongracz V, et al. Flexible segments modulate co-folding of dUTPase and nucleocapsid proteins. Nucleic Acids Res. 2007;35:495–505. doi: 10.1093/nar/gkl1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varga B, et al. Active site of mycobacterial dUTPase: structural characteristics and a built-in sensor. Biochem Biophys Res Commun. 2008;373:8–13. doi: 10.1016/j.bbrc.2008.05.130. [DOI] [PubMed] [Google Scholar]
- 12.Takacs E, Barabas O, Petoukhov MV, Svergun DI, Vertessy BG. Molecular shape and prominent role of beta-strand swapping in organization of dUTPase oligomers. FEBS Lett. 2009;583:865–871. doi: 10.1016/j.febslet.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Toth J, Varga B, Kovacs M, Malnasi-Csizmadia A, Vertessy BG. Kinetic mechanism of human dUTPase, an essential nucleotide pyrophosphatase enzyme. J Biol Chem. 2007;282:33572–33582. doi: 10.1074/jbc.M706230200. [DOI] [PubMed] [Google Scholar]
- 14.Kovari J, et al. Methylene substitution at the alpha-beta bridging position within the phosphate chain of dUDP profoundly perturbs ligand accommodation into the dUTPase active site. Proteins. 2008;71:308–319. doi: 10.1002/prot.21757. [DOI] [PubMed] [Google Scholar]
- 15.Barabas O, Pongracz V, Kovari J, Wilmanns M, Vertessy BG. Structural insights into the catalytic mechanism of phosphate ester hydrolysis by dUT ase. J Biol Chem. 2004;279:42907–42915. doi: 10.1074/jbc.M406135200. [DOI] [PubMed] [Google Scholar]
- 16.Tarbouriech N, Buisson M, Seigneurin JM, Cusack S, Burmeister WP. The monomeric dUTPase from Epstein-Barr virus mimics trimeric dUTPases. Structure. 2005;13:1299–1310. doi: 10.1016/j.str.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 17.Varga B, et al. Active site closure facilitates juxtaposition of reactant atoms for initiation of catalysis by human dUTPase. FEBS Lett. 2007;581:4783–4788. doi: 10.1016/j.febslet.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 18.Pecsi I, Leveles I, Harmat V, Vertessy BG, Toth J. Aromatic stacking between nucleobase and enzyme promotes phosphate ester hydrolysis in dUTPase. Nucleic Acids Res. 2010;38:7179–7186. doi: 10.1093/nar/gkq584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nord J, Kiefer M, Adolph HW, Zeppezauer MM, Nyman PO. Transient kinetics of ligand binding and role of the C-terminus in the dUTPase from equine infectious anemia virus. FEBS Lett. 2000;472:312–316. doi: 10.1016/s0014-5793(00)01453-8. [DOI] [PubMed] [Google Scholar]
- 20.Shao H, et al. Characterization and mutational studies of equine infectious anemia virus dUTPase. Biochim Biophys Acta. 1997;1339:181–191. doi: 10.1016/s0167-4838(96)00229-4. [DOI] [PubMed] [Google Scholar]
- 21.Ponomarev MA, Timofeev VP, Levitsky DI. The difference between ADP-beryllium fluoride and ADP-aluminum fluoride complexes of the spin-labeled myosin subfragment 1. FEBS Lett. 1995;371:261–263. doi: 10.1016/0014-5793(95)00898-j. [DOI] [PubMed] [Google Scholar]
- 22.Fisher AJ, et al. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP.BeFx and MgADP.AlF4. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 23.Larsson G, Nyman PO, Kvassman JO. Kinetic characterization of dUTPase from Escherichia coli. J Biol Chem. 1996;271:24010–24016. doi: 10.1074/jbc.271.39.24010. [DOI] [PubMed] [Google Scholar]
- 24.Huffman JL, Li H, White RH, Tainer JA. Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii. J Mol Biol. 2003;331:885–896. doi: 10.1016/s0022-2836(03)00789-7. [DOI] [PubMed] [Google Scholar]
- 25.Johansson E, et al. Structures of dCTP deaminase from Escherichia coli with bound substrate and product: reaction mechanism and determinants of mono- and bifunctionality for a family of enzymes. J Biol Chem. 2005;280:3051–3059. doi: 10.1074/jbc.M409534200. [DOI] [PubMed] [Google Scholar]
- 26.Helt SS, et al. Mechanism of dTTP inhibition of the bifunctional dCTP deaminase:dUTPase encoded by Mycobacterium tuberculosis. J Mol Biol. 2008;376:554–569. doi: 10.1016/j.jmb.2007.11.099. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Xu H, Graham DE, White RH. The Methanococcus jannaschii dCTP deaminase is a bifunctional deaminase and diphosphatase. J Biol Chem. 2003;278:11100–11106. doi: 10.1074/jbc.M212460200. [DOI] [PubMed] [Google Scholar]
- 28.Hidalgo-Zarco F, et al. Kinetic properties and inhibition of the dimeric dUTPase-dUDPase from Leishmania major. Protein Sci. 2001;10:1426–1433. doi: 10.1110/ps.48801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Musso-Buendia JA, et al. Kinetic properties and inhibition of the dimeric dUTPase-dUDPase from Campylobacter jejuni. J Enzym Inhib Med Ch. 2009;24:111–116. doi: 10.1080/14756360801915476. [DOI] [PubMed] [Google Scholar]
- 30.Moroz OV, et al. The crystal structure of a complex of Campylobacter jejuni dUTPase with substrate analogue sheds light on the mechanism and suggests the “basic module” for dimeric d(C/U)TPases. J Mol Biol. 2004;342:1583–1597. doi: 10.1016/j.jmb.2004.07.050. [DOI] [PubMed] [Google Scholar]
- 31.Smith CA, Rayment I. Active site comparisons highlight structural similarities between myosin and other P-loop proteins. Biophys J. 1996;70:1590–1602. doi: 10.1016/S0006-3495(96)79745-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klockow B, Ahmadian MR, Block C, Wittinghofer A. Oncogenic insertional mutations in the P-loop of Ras are overactive in MAP kinase signaling. Oncogene. 2000;19:5367–5376. doi: 10.1038/sj.onc.1203909. [DOI] [PubMed] [Google Scholar]
- 33.Pai EF, et al. Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis. Embo J. 1990;9:2351–2359. doi: 10.1002/j.1460-2075.1990.tb07409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gulick AM, Bauer CB, Thoden JB, Rayment I. X-ray structures of the MgADP, MgATPgammaS, and MgAMPPNP complexes of the Dictyostelium discoideum myosin motor domain. Biochemistry. 1997;36:11619–11628. doi: 10.1021/bi9712596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.