Abstract
Current theory predicts that a shift to a new habitat would increase the rate of diversification, while as lineages evolve into multiple species, intensified competition would decrease the rate of diversification. We used Holarctic amphipods of the genus Gammarus to test this hypothesis. We sequenced four genes (5,088 bp) for 289 samples representing 115 Gammarus species. A phylogenetic analysis showed that Gammarus originated from the Tethyan region with a saline ancestry in the Paleocene, and later colonized the freshwater habitat in the Middle Eocene. Ancestral range reconstruction and diversification mode analysis combined with paleogeological and paleoclimatic evidence suggested that the habitat shift from saline to freshwater led to an increased diversification rate. The saline lineage of Gammarus dispersed to both sides of the Atlantic at 55 million years ago (Ma), because of the few barriers between the Tethys and the Atlantic, and diversified throughout its evolutionary history with a constant diversification rate [0.04 species per million years (sp/My)]. The freshwater Gammarus, however, underwent a rapid diversification phase (0.11 sp/My) until the Middle Miocene, and lineages successively diversified across Eurasia via vicariance process likely driven by changes of the Tethys and landmass. In particular, the freshwater Gammarus lacustris and Gammarus balcanicus lineages had a relatively high diversification shift, corresponding to the regression of the Paratethys Sea and the continentalization of Eurasian lands during the Miocene period. Subsequently (14 Ma), the diversification rate of the freshwater Gammarus decreased to 0.05 and again to 0.01 sp/My. The genus Gammarus provides an excellent aquatic case supporting the hypothesis that ecological opportunities promote diversification.
Keywords: evolution, molecular dating, range expansion
Colonization of a new habitat often opens up new ecological opportunities and thus promotes lineage diversification (1–3). This is a major tenet in modern evolutionary biology and there are numerous successful case studies in terrestrial systems, particularly in oceanic archipelagos. Darwin's finches on the Galápagos Islands and the drosophilid flies of the Hawaiian Islands are perhaps the best known cases (4, 5). In aquatic systems, habitat shifts from marine to freshwater have been frequently documented (6); however, whether such habitat shift promotes diversification has not been vigorously tested. Marine and freshwater are very different ecosystems; in addition to a significant difference in salinity, locally adapted coinhabitants provide different resources and competitions (7). To investigate the evolutionary implications of such habitat shift, an appropriate choice of organism group is essential. A speciose group with limited long-distance dispersal ability, occurring in both freshwater and saline water and inhabiting an area with well-known geological history, would be ideal (8).
Amphipod crustaceans of the genus Gammarus seem to satisfy these criteria. The genus includes over 200 described species and is distributed across the entire Holarctic. Although the majority inhabit freshwater, a large number of species occur in amphi-Atlantic, Mediterranean, and Ponto-Caspian saline water systems (9). Early phylogenetic work suggested a possible habitat shift from marine to freshwater (10). Amphipods lack an independent larval dispersal stage, and all freshwater species except Gammarus lacustris are endemic to restricted areas. The current disjunctive distribution of Gammarus indicates that the evolutionary history of this group may bear a clear signature of past geological events, such as the plate tectonic effects and the Tethyan events described below.
In the Late Eocene [37–34 million years ago (Ma)], the collision between India and Asia promoted the uplift of the Tibetan plateau, which disrupted the exchange between the Tethys Ocean and the Eastern Asian water systems (8, 11). The convergence between Africa and Europe formed an active Alpine mountain belt along the southern border of the Eurasian continent, which isolated the Paratethys Sea from the Mediterranean Sea (11). The Paratethys existed as far east as Tajikistan and was connected with the Arctic Ocean and the North Sea by the Turgai Strait and the Danish-Polish trough (12, 13). These two seaways divided the Eurasian land into West Europe, East Europe, and Asia. During the Early Oligocene (34–32 Ma), the Paratethyan basin began to regress and the Turgai Strait became dry land, which opened up a migration pathway for European and Asian continental animals (11). In the Early Miocene (23–19 Ma), a rapid regression process in the eastern Paratethys enlarged the land areas. The Messinian salinity crisis (5.96–5.33 Ma) was a dramatic drying event of the Mediterranean (8), which might have had a significant impact on endemic aquatic organisms. The subsequent flooding from the Atlantic promoted many surviving species to recolonize the area (13). These geological events and climatic changes in the Tethyan regions are likely associated with the evolutionary history of the genus Gammarus.
Current theory predicts that a shift to a new habitat frees species from the competition with closely related species and would increase the rate of diversification followed by adaptive radiations. As lineages diversify into multiple species, competition between newly formed species would intensify. Consequently, the diversification rate would decrease as species accumulate in the limited space (3, 14). In this study, we use DNA sequence data and a variety of analytical methods to test whether the habitat shift of Gammarus promoted diversification rates. Phylogenetic inference is used to construct the history of Gammarus, to estimate the divergence times of its major lineages, and to determine when the shift from saline to freshwater occurred. We further conduct a biogeographic analysis to explore where Gammarus first colonized freshwater habitats and a diversification analysis to assess the temporal diversification mode associated with the habitat shift.
Results
Phylogenetic Inference Identifies an Eocene Habitat Shift from Saline to Freshwater.
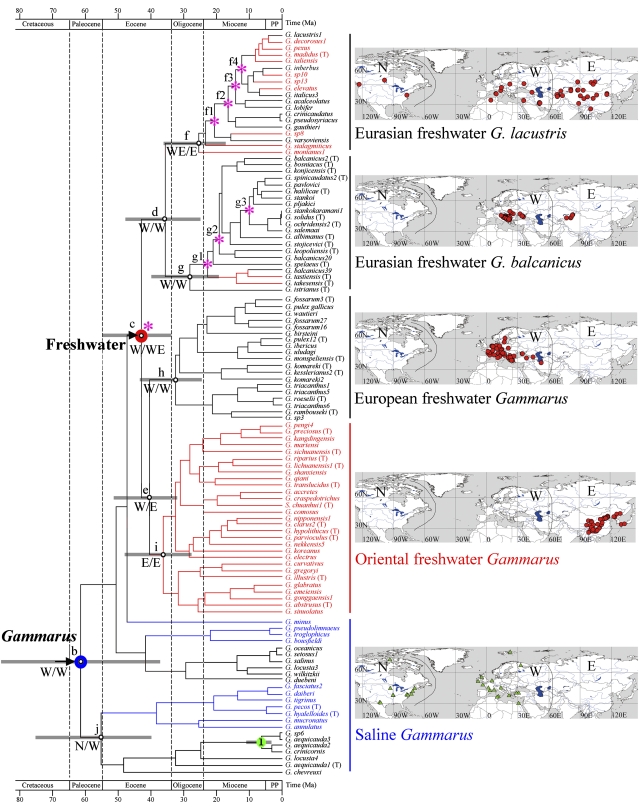
We sequenced four genes of 28S, 18S, cytochrome oxidase I (COI), and elongation factor 1α (EF1α) from 289 samples representing 115 Gammarus species, three Baikal amphipods, and 13 outgroup taxa. Parsimony and likelihood analyses revealed similar topologies, and the likelihood tree is presented in Fig. S1. The genus Gammarus, including Baikal amphipods and Sinogammarus, which was previously thought to be a junior synonym of Gammarus (10), formed a monophyletic group. At the base of the group, there were three saline lineages that are endemic to the Mediterranean–Atlantic area. A freshwater lineage, which is widely distributed in the Palearctic, was nested deeply in the tree. The North American freshwater species formed two groups inside the saline lineages, and they likely had saline common ancestors and colonized freshwater recently. We therefore included them in the saline lineages in subsequent analyses. The freshwater lineage was further divided into four morphologically and geographically distinct clades: the European Gammarus clade, which included the Gammarus fossarum, Gammarus pulex, Gammarus roeselii, and Gammarus komareki species complexes; the Oriental Gammarus clade, which contained all of the Eastern Asian species; the Gammarus lacustris lineage and the Gammarus balcanicus lineage. These last two clades are primarily located in Europe and Central Asia (Fig. 1).
Fig. 1.
Maximum clade credibility chronogram inferred from a BEAST dating analysis. Nodes b–j are nodes of interest with gray horizontal bars indicating the 95% highest posterior density interval; node 1 in green circle is used for calibration. Major ancestral range reconstructions are shown below the branches (N, Nearctic; W, West Palearctic; E, East Palearctic), and arrows show main dispersal events. Species names and terminal branches are in red for East Palearctic species, blue for Nearctic species, and black for West Palearctic species. Pink asterisks are nodes (c, f1–f4, and g1–g3) that showed an increase in diversification rate. The blue circle (node b) indicates the saline origin of the genus Gammarus and the red circle (node c) indicates the origin of freshwater species. Outgroups and Baikal amphipods are not shown (Figs. S1 and S2). (Right) Maps correspond to the sampling sites for the adjacent clade.
Divergence time estimates were conducted with a reduced dataset, in which all major clades defined by the phylogenetic analysis were evenly represented to avoid bias. The ages for nodes of interest estimated by BEAST are shown in Fig. 1, and more details are provided in Fig. S2 and Table S2. The genus Gammarus originated at ∼61 Ma, and started to radiate to freshwater habitat at 43 Ma during the Middle Eocene. The successive freshwater colonizations occurred in the late Eocene, at 36 Ma for the Eurasian G. lacustris and G. balcanicus clades, at 33 Ma for the European clade, and at 37 Ma for the Oriental clade (Fig. 1). All these freshwater colonization events are older than the earliest Gammarus fossil records, which are in the Lower Oligocene.
Biogeographic Analysis Indicates Two Major Range Shifts.
We used likelihood and parsimony methods to reconstruct the historical distribution ranges of the hypothetical ancestors. Both methods produced similar results and details are provided in Table S3. Two major range expansion events were detected in the evolutionary history of the genus Gammarus (Fig. 1). Ancestral saline Gammarus was distributed in the West Palearctic (node b), and subsequently dispersed to the Nearctic at 55 Ma (node j). The second dispersal event occurred within freshwater Gammarus (node c), which expanded from the West Palearctic to the East Palearctic at 40.6 Ma (node e).
Diversification Modes Associated with Habitat Shift.
We conducted a birth–death analysis to find a model that could best represent the diversification patterns for Gammarus overall, freshwater Gammarus, and saline Gammarus. For Gammarus overall, the Monte Carlo constant-rate (MCCR) test rejected the null hypothesis of constant rate (critical value = −3.33, P = 0.003), and the gamma (γ) statistic (γ = −4.3) indicated a decrease in the diversification rate over time. Of the six models tested, the variable-rate yule2rate model with one shift in diversification was selected as the best fit model for Gammarus overall, suggesting that the initial diversification rate of 0.07 species per million years (sp/My) shifted to 0.03 sp/My at 25 Ma. Similarly, the γ statistic and MCCR test significantly rejected the constant-rate model for the freshwater Gammarus, and the yule3rate model was chosen as the best fit model with two diversification decreases at 14 Ma (from 0.11 to 0.05 sp/My) and 6 Ma (from 0.05 to 0.01 sp/My). For saline Gammarus, the pure birth model with diversification rate of 0.04 sp/My was selected as the best model, which was supported by the γ and MCCR tests. Detailed results are provided in Table S4.
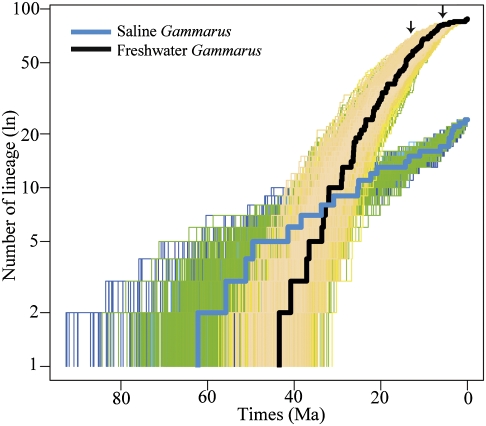
The semilogarithmic lineage-through-time (LTT) plots for Gammarus overall, freshwater, and saline Gammarus are shown in Fig. 2 and Fig. S3. The saline Gammarus showed a fairly slow but constant rate of lineage accumulation. On the contrary, freshwater Gammarus maintained a high diversification rate at the early stages of the colonization of freshwater habitat until ∼14 Ma. After that, two independent events of diversification decrease were identified, with 6 Ma being a major shift point.
Fig. 2.
Lineage-through-time plot estimated from 1,000 Bayesian trees for freshwater and saline Gammarus. The lines in bold correspond to the maximum credibility tree from the BEAST dating analysis. Arrows indicate changes in diversification rates that are estimated to have occurred according to the best fit model of lineage diversification.
Absolute net diversification rate within the genus Gammarus varied from 0.08 to 0.05 sp/My under two extremes of the relative extinction rate (0 and 0.9). The absolute net diversification rate for freshwater Gammarus ranged from 0.10 to 0.06 sp/My, whereas the rates for saline Gammarus ranged from 0.05 to 0.03 sp/My. The relative cladogenesis (RC) statistic indicated a significant difference in cladogenesis rate between freshwater and saline Gammarus (Fig. 1). Rapid shifts in diversification rate were also located within the freshwater G. lacustris and G. balcanicus lineages during the Miocene period (Table S5).
Discussion
Saline Origin, Eocene Habitat Shift, and Diversification in Freshwater.
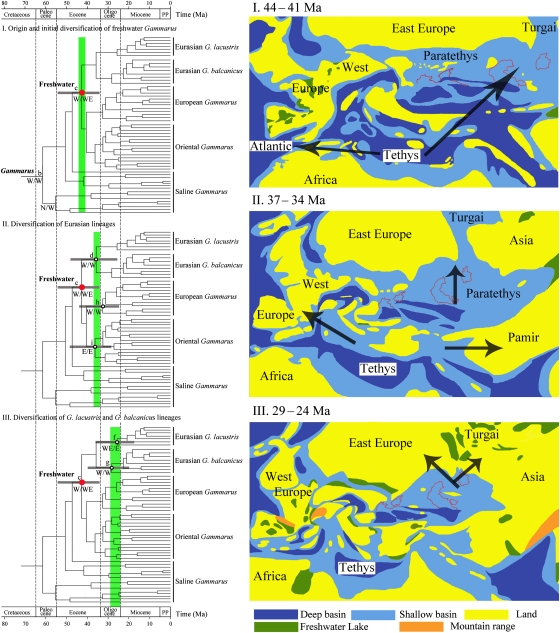
The genus Gammarus originated in the West Palearctic with a saline habitat at ∼61 Ma in the Paleocene and subsequently colonized a freshwater habitat and radiated across Eurasia starting at ∼43 Ma in the Middle Eocene (Figs. 1 and 3, I). During the Paleocene period, the Tethys Ocean stretched out from west to east across Southern Europe and Central Asia, connecting the Atlantic Ocean and the Pacific Ocean (11), where saline Gammarus dominated (Fig. 1). When Gammarus started to diversify, the exchange between Tethys and Atlantic was free, which may have led to the first range expansion from the Tethys Ocean to both sides of the Atlantic. Along with the progressive crust movements and Tethyan regression during Eocene, the accretion of landmass formed a prominent new freshwater aquatic ecosystem. A euryhaline marine ancestor of Gammarus colonized freshwater habitats of the growing Eurasian land, and this habitat shift may have played a significant role in the diversification evolution of Gammarus. Our analysis clearly suggests a Tethyan saline origin for Gammarus, which contradicts the traditional view that Holarctic freshwater Gammarus are of a continental origin and dispersed to coastal seas secondarily (9, 15).
Fig. 3.
Biogeographic scenarios for freshwater Gammarus. Each time frame as in Fig. 1 is shown to the Left of the respective paleomap, and corresponding nodes are shown. Vertical green bars on Left phylogeny correspond to the times of Right paleomaps. (I) Origin and initial diversification of freshwater Gammarus (node c); map redrawn after Meulenkamp and Sissingh (12). (II) Diversification of Eurasian lineages (nodes d, h, and i); map redrawn after Popov et al. (13). (III) Diversification of G. lacustris and G. balcanicus lineages (nodes f and g); map redrawn after Popov et al. (13).
After the successful colonization of freshwater habitats, Gammarus underwent a second range expansion from the West Palearctic to the East Palearctic at about 40 Ma and rapidly diverged allopatrically across Eurasian freshwater habitats in the late Eocene. The Oriental Gammarus (node i; Figs. 1 and 3, II) separated from its common ancestor with the European freshwater Gammarus at 37 Ma, followed by the separation of the Eurasian lineages G. lacustris and G. balcanicus clade at 36 Ma (node d; Figs. 1 and 3, II). The European freshwater endemic species complexes G. fossarum, G. pulex, G. roeselii, and G. komareki diversified at 33 Ma (node h; Figs. 1 and 3, II). Several paleogeological events are likely responsible for these hypothetical radiation events of freshwater Gammarus. The collision between the Indian and Asian plates during the Eocene (37–34 Ma) forced the uplift of the Tibetan Plateau, resulting in the interruption of the Tethys Ocean and causing the formation of a freshwater environment in the Eastern Asia. The Turgai Strait, as a shallow seaway, supplied a route for the Paratethyan Gammarus to spread northwards and colonize the Eurasian freshwater habitats. The convergence between the African and Eurasian plates during the Oligocene (34–30 Ma) and the subsequent retreat of the Tethys Ocean contributed to the growing European landmass available for Gammarus species to inhabit.
Most speciation events that produced the extant species occurred in the Miocene (Fig. 1). The diversification of the G. lacustris and G. balcanicus clades were probably driven by the closing of the Turgai Strait at ∼29 Ma, which made it possible for terrestrial and freshwater animals present in Eurasia to expand their ranges (11). The two lineages were formed at the same time as the closing, and diversified afterward (nodes f and g; Figs. 1 and 3, III). Together, drying of the Turgai Strait, shrinking of the Paratethyan basin, and Eurasian continentalization created available ecological niches for freshwater Gammarus. The majority of the freshwater species have a restricted distribution and demonstrate a strong vicariant pattern (9, 10). This is not surprising considering their history and a lack of long-distance dispersal ability. G. lacustris is an exception and has a Holarctic distribution. It usually inhabits mountain and glacial lakes and presumably dispersed to northern Europe and North America during glaciations (9).
Habitat Shift Promoted Freshwater Gammarus Diversification.
Our diversification analyses have revealed that the saline Gammarus diversified with a constant rate of 0.04 sp/My throughout its evolutionary history. This is partially expected; as saline Gammarus originated from the Tethyan region, they could freely disperse to both sides of the Atlantic without physical barriers. On the other hand, freshwater Gammarus initially had a high diversification rate (0.11 sp/My) from the Middle Eocene to Middle Miocene (43–14 Ma), after which the diversification rate decreased to 0.05 sp/My until the Miocene/Pliocene boundary and again declined to 0.01 sp/My toward the modern descendants (Fig. 2 and Table S4). The RC test also suggested that freshwater Gammarus went through a rapid diversification within the G. lacustris and G. balcanicus lineages during the Early and Middle Miocene (Fig. 1). This diversification scenario for freshwater Gammarus implies that they experienced a rapid adaptive diversification when they first shifted to a new habitat and that their diversification rate slowed down with the species accumulation, as a model has predicted (3). This finding suggests that freshwater Gammarus diversification was driven by ecological opportunities created by the emergence of largely unoccupied habitats and few competitors (2, 16).
The absolute net diversification rate of freshwater Gammarus (0.10–0.06 sp/My) is about twice that of saline Gammarus (0.05–0.03 sp/My). The rate of freshwater Gammarus is comparable with fastest-diversifying bivalves 0.09 sp/My (17) and ground beetles 0.10 sp/My (18). However, it is slower than that of Mediterranean plants, such as Dianthus carnations, which have a rate of 2.2–7.6 sp/My (19), and Brassicaceae mustard 0.18–0.22 sp/My (20). The ancient Tethyan region is one of the hotspots of global biodiversity for both terrestrial and aquatic ecosystems. This likely attributes to its complex geological history and heterogeneous topography. This region will continue to provide an excellent setting for studying evolutionary processes.
The genus Gammarus is a taxonomically challenging group. Our data unequivocally placed Sinogammarus and the Baikal amphipods as part of Gammarus (10, 15). The genus “Sinogammarus” consists exclusively of cave dwellers, and the Baikal amphipods are endemic to Lake Baikal. Both have highly specialized morphology. We suggest that new synapomorphies should be examined to redefine the genus Gammarus and its species groups on the basis of a solid phylogenetic framework.
The hypothesis that ecological opportunities promote lineage diversification is well studied in island terrestrial systems. The genus Gammarus presents a strong case in support of this hypothesis in aquatic ecosystems. A habitat shift from saline to freshwater, coupled with increased landmass and thus increased available bodies of freshwater, led to a rapid radiation of freshwater Gammarus species. Habitat shift is no rare event in aquatic life, and therefore more studies along these lines are likely to shed more light on the formation of biodiversity.
Materials and Methods
Taxon Sampling.
Our sampling covered all major geographic regions across the Northern Hemisphere (Table S1). When possible, samples from the type localities of nominal taxa were included and multiple samples were used to represent widespread species. In total, we used 289 samples of 115 Gammarus species inhabiting both freshwater and saline habitats, including two samples of Sinogammarus, which is believed to be a junior synonym of Gammarus (10). Three Baikal amphipods, which are closely related to Gammarus (15), were also included. Another 13 individuals from different genera of the family Gammaridae and several distantly related families were chosen as outgroups. For molecular dating, biogeographic reconstruction, and diversity analysis, unequal taxon sampling could induce bias toward better represented clades (16). Therefore, several taxa were pruned to ensure that all of the major clades recovered in the broader analysis were evenly represented. The reduced matrix consisted of 129 taxa.
Laboratory Protocols and Phylogenetic Analyses.
Genomic DNA extraction, amplification, and sequence alignment were performed as in Hou et al. (10). Primers are listed in Table S6. The complete dataset totaled 305 taxa with 5,088 bp, including 1,526 bp for 28S, 656 bp for COI, 2,305 bp for 18S, and 601 bp for EF1α (Table S1). Nucleotide polymorphisms caused by heterozygosity in EF1α were limited (less than 10 bp) and were coded as missing data.
Phylogenetic analyses were performed using maximum parsimony (MP) and maximum likelihood (ML) criteria. MP analysis and bootstrap support evaluation were conducted using PAUP* 4.0b10 (21). Separated MP analyses of individual genes found no conflict among well-supported nodes (bootstrap index >70%); therefore, the four genes were combined for all other analyses. ML analysis was carried out using GARLI 1.0 (22). To confirm the optimal topology, analyses were repeated until two independently derived topologies of the highest likelihood were identical. A rapid bootstrap analysis with 1,000 replicates was performed under partitioned data mode with RAxML 7.0.3 (23) and a thorough ML search was used to compare with the result of GARLI.
To infer the habitat preference of the hypothetical ancestors, a ML approach was used to map the habitat type onto the Gammarus phylogeny (Figs. S1 and S2) with Mesquite 2.74 (24). Freshwater and saline states were coded as binary characters. The proportional likelihoods of ancestral states were calculated for each node under asymmetrical two-parameter Markov k-state model.
Molecular Dating Analysis.
Fossil records of amphipod crustaceans are rare and have no detailed affinities with extant taxa (25). Instead, we used three geological events for molecular clock calibration. First, the origin of Mediterranean saline Gammarus (node 1, Fig. 1 and Fig. S2) was set at 5 Ma. This is based on the assumption that all gammarids in the region went extinct during the Messinian salinity crisis (8), and that the current lineage was established after the crisis as a consequence of a recolonization of the Mediterranean. Second, the most recent common ancestor of the Baikal clade (node 2, Fig. S2) was set to 30 Ma. The permanent Lake Baikal originated at ∼35–30 Ma, and the main Baikal group of Acanthogammaridae amphipods were estimated to occur no later than 30 Ma, on the basis of molecular data (26). Third, we constrained 37 Ma to the ancestral node of Sarothrogammarus and Rhipidogammarus (node 3, Fig. S2). The Paratethys covered Tajikistan until the late Eocene (11, 13), and the regression of the Paratethys likely resulted in the split of present disjunctive distribution of the Central Asian Sarothrogammarus and the Mediterranean Rhipidogammarus species. Freshwater Sarothrogammarus in Tajikistan was stranded to be a relict group. Different combinations of calibration schemes were tested to determine how well calibration nodes and divergence dates correlated with each other (Table S2).
The divergence times were obtained by applying a Bayesian method implemented in BEAST 1.6.1 (27). We used the uncorrelated lognormal molecular clock model with a Yule process for the speciation model, GTR+I+G for the substitution model, and a normal distribution with SD of 1 as priors on the calibration nodes to accommodate for calibration uncertainty. The Markov chain Monte Carlo was run for 50 million generations and sampled every 1,000 generations. Two independent runs were performed to confirm the convergence of the analysis. The stationary of each run was examined using the effective sampling size of each parameter (>200). The last 40 million generations were used to construct the maximum clade credibility tree and the associated 95% highest posterior density distributions around the estimated node ages. For the purpose of comparison and confirmation, we also estimated divergence time using a penalized likelihood method implemented in r8s 1.7.1 (28). We specified an optimal smoothing value of 1,000 in the analysis, as obtained by a cross-validation test. The confidence intervals were calculated for each node following the r8s bootstrap kit.
The estimates from the penalized likelihood method and the Bayesian method agreed with each other; all of the mean nodal ages estimated by the former were within the confidence intervals calculated by the latter. Calibrations with all three nodes corresponded well with calibration using nodes 1 and 2, whereas calibration using nodes 1 and 3 produced younger estimations. The BEAST conservative estimation with calibration nodes 1 and 2 was therefore reported and used for biogeographic and diversification analyses.
Biogeographic Analysis.
To reconstruct the historical distribution range of the hypothetical ancestors, we used a likelihood method implemented in the program Lagrange 20101113 (29). The major areas of Gammarus distribution were broadly categorized into three areas: the Nearctic (N), the West Palearctic (W), and the East Palearctic (E) (9). Widespread species were assigned to more than one area (Table S1). The chronogram produced by BEAST was used (Fig. 1), with the outgroups and the Baikal amphipods removed from the tree. The Baikal lineage survived a particularly strong morphological diversification and evolved separately endemic to Lake Baikal (15). Dispersal probabilities were modified to 0.1 between the Nearctic and the East Palearctic or 0.1–1.0 between areas on the basis of paleogeographic reconstructions of area position through time (Table S3) (30).
A parsimony-based analysis was performed for comparison. The same tree topology was used and area assignments with the “maxareas” option unlimited or constrained to two. To estimate the support for ancestral range reconstructions, we randomly sampled 1,000 trees from the BEAST output and conducted a Bayes-DIVA analysis using the program S-DIVA 1.5c (31).
Diversification Pattern.
We assigned a conservative estimate of 204 species calculated only for described undisputed species, including 160 freshwater and 44 saline species (32). The diversification pattern was assessed for freshwater, saline Gammarus lineages, and Gammarus overall chronogram (Fig. 1). To examine the diversification rate change over time, semilogarithmic LTT plots were constructed in APE 2.5–1 (33). The confidence intervals of the LTT plots were generated on 1,000 trees sampled from the converged BEAST trees.
A birth–death likelihood method was used to test several variable-rate models against the null hypothesis of constant diversification rate with the package LASER 2.3 (34). Akaike information criterion (AIC) scores were computed for the constant-rate and the variable-rate models, including the pure birth, birth–death, yule2rate, yule3rate, variable speciation rate and constant extinction rate (SPVAR), and variable extinction rate and constant speciation rate (EXVAR) models. The AIC difference between best constant-rate model and each of the variable-rate models was computed to select the best fit model. In addition, the MCCR test was used by comparing the empirical γ statistic with the distribution of γ statistics of 1,000 simulated incomplete phylogenies under a pure birth model to test whether the diversification has decelerated through time (35).
The absolute net diversification rates were calculated by using the BEAST chronogram under two extremes of the relative extinction rate (speciation rate/extinction rate = 0 and 0.9) following the whole-clade method (17) implemented in LASER. The RC statistic was calculated with GEIGER 1.3–1 (36) to detect rapid shifts in diversification rates at any specified time. Lineages with more or fewer descendents than expected under the constant-rate model were hypothesized as a diversification rate shift.
Supplementary Material
Acknowledgments
We thank Professor J. Fu and two anonymous reviewers for improvements to the manuscript. This research was supported by National Natural Sciences Foundation of China (NSFC-30870473 and -30970336), China National Funds for Distinguished Young Scientists (NSFC-31025023), and by the state budget from the Slovenian Research Agency (Program no. P1-0184, Zoology and Speleobiology).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. are given in Table S1).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104636108/-/DCSupplemental.
References
- 1.Schluter D. The Ecology of Adaptive Radiation. Oxford: Oxford Univ Press; 2000. [Google Scholar]
- 2.Hughes C, Eastwood R. Island radiation on a continental scale: Exceptional rates of plant diversification after uplift of the Andes. Proc Natl Acad Sci USA. 2006;103:10334–10339. doi: 10.1073/pnas.0601928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gavrilets S, Losos JB. Adaptive radiation: Contrasting theory with data. Science. 2009;323:732–737. doi: 10.1126/science.1157966. [DOI] [PubMed] [Google Scholar]
- 4.Grant PR, Grant BR. How and Why Species Multiply: The Radiation of Darwin's Finches. Princeton, NJ: Princeton Univ Press; 2008. [Google Scholar]
- 5.Carson HL, Kaneshiro KY. Drosophila of Hawaii: Systematics and ecological genetics. Annu Rev Ecol Syst. 1976;7:311–345. [Google Scholar]
- 6.Lovejoy NR, Bermingham E, Martin AP. Marine incursion into South America. Nature. 1998;396:421–422. [Google Scholar]
- 7.Logares R, et al. Infrequent marine-freshwater transitions in the microbial world. Trends Microbiol. 2009;17:414–422. doi: 10.1016/j.tim.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 8.Salvo G, Ho SYW, Rosenbaum G, Ree R, Conti E. Tracing the temporal and spatial origins of island endemics in the Mediterranean region: A case study from the citrus family (Ruta L., Rutaceae) Syst Biol. 2010;59:705–722. doi: 10.1093/sysbio/syq046. [DOI] [PubMed] [Google Scholar]
- 9.Väinölä R, et al. Global diversity of amphipods (Amphipoda; Crustacea) in freshwater. Hydrobiologia. 2008;595:241–255. [Google Scholar]
- 10.Hou Z, Fu J, Li S. A molecular phylogeny of the genus Gammarus (Crustacea: Amphipoda) based on mitochondrial and nuclear gene sequences. Mol Phylogenet Evol. 2007;45:596–611. doi: 10.1016/j.ympev.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Rögl F. Palaeogeographic considerations for Mediterranean and Paratethys seaways (Oligocene to Miocene) Ann Naturhis Mus Wien. 1998;99A:279–310. [Google Scholar]
- 12.Meulenkamp JE, Sissingh W. Tertiary palaeogeography and tectonostratigraphic evolution of the Northern and Southern Peri-Tethys platforms and the intermediate domains of the African–Eurasian convergent plate boundary zone. Palaeogeogr Palaeocl. 2003;196:209–228. [Google Scholar]
- 13.Popov SV, et al. Lithological-Paleogeographic maps of Paratethys: 10 maps Late Eocene to Pliocene. Cour Forsch Senck. 2004;250:1–46. [Google Scholar]
- 14.Parent CE, Crespi BJ. Ecological opportunity in adaptive radiation of Galápagos endemic land snails. Am Nat. 2009;174:898–905. doi: 10.1086/646604. [DOI] [PubMed] [Google Scholar]
- 15.MacDonald KS, 3rd, Yampolsky L, Duffy JE. Molecular and morphological evolution of the amphipod radiation of Lake Baikal. Mol Phylogenet Evol. 2005;35:323–343. doi: 10.1016/j.ympev.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 16.Hines HM. Historical biogeography, divergence times, and diversification patterns of bumble bees (Hymenoptera: Apidae: Bombus) Syst Biol. 2008;57:58–75. doi: 10.1080/10635150801898912. [DOI] [PubMed] [Google Scholar]
- 17.Magallón S, Sanderson MJ. Absolute diversification rates in angiosperm clades. Evolution. 2001;55:1762–1780. doi: 10.1111/j.0014-3820.2001.tb00826.x. [DOI] [PubMed] [Google Scholar]
- 18.Ober KA, Heider TN. Phylogenetic diversification patterns and divergence times in ground beetles (Coleoptera: Carabidae: Harpalinae) BMC Evol Biol. 2010;10:262. doi: 10.1186/1471-2148-10-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valente LM, Savolainen V, Vargas P. Unparalleled rates of species diversification in Europe. Proc R Soc B. 2010;277:1489–1496. doi: 10.1098/rspb.2009.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couvreur TLP, et al. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae) Mol Biol Evol. 2010;27:55–71. doi: 10.1093/molbev/msp202. [DOI] [PubMed] [Google Scholar]
- 21.Swofford DL. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sunderland, MA: Sinauer; 2002. [Google Scholar]
- 22.Zwickl DJ. Austin, TX: University of Texas; 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. PhD dissertation. [Google Scholar]
- 23.Stamatakis A, Hoover PJ, Rougemont J. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 24.Maddison WP, Maddison DR. Mesquite: A molecular system for evolutionary analysis. Version 2.74. 2010. Available at http://mesquiteproject.org. Accessed October 3, 2010.
- 25.Karaman GC. Critical remarks to the fossil Amphipoda with description of some new taxa. Poljoprivreda I Šumarstvo. 1984;30:87–104. [Google Scholar]
- 26.Ogarkov OB, Kamaltynov RM, Belikov SI, Sherbakov DY. Phylogenetic relatedness of the Baikal Lake endemical amphipods (Crustacea, Amphipoda) deduced from partial nucleotide sequences of the cytochrome oxidase subunit III genes. Mol Biol. 1997;31:24–29. [PubMed] [Google Scholar]
- 27.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanderson MJ. r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19:301–302. doi: 10.1093/bioinformatics/19.2.301. [DOI] [PubMed] [Google Scholar]
- 29.Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 2008;57:4–14. doi: 10.1080/10635150701883881. [DOI] [PubMed] [Google Scholar]
- 30.Clayton JW, Soltis PS, Soltis DE. Recent long-distance dispersal overshadows ancient biogeographical patterns in a pantropical angiosperm family (Simaroubaceae, Sapindales) Syst Biol. 2009;58:395–410. doi: 10.1093/sysbio/syp041. [DOI] [PubMed] [Google Scholar]
- 31.Yu Y, Harris AJ, He X. S-DIVA (Statistical Dispersal-Vicariance Analysis): A tool for inferring biogeographic histories. Mol Phylogenet Evol. 2010;56:848–850. doi: 10.1016/j.ympev.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 32.Holsinger JR, Shafer J, Fong DW, Culver DC. Gammarus cohabitus, a new species of subterranean amphipod crustacean (Gammaridae) from groundwater habitats in central Pennsylvania, USA. Subterranean Biol. 2008;6:31–41. [Google Scholar]
- 33.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 34.Rabosky DL. LASER: A maximum likelihood toolkit for detecting temporal shifts in diversification rates from molecular phylogenies. Evol Bioinform Online. 2006;2:273–276. [PMC free article] [PubMed] [Google Scholar]
- 35.Pybus OG, Harvey PH. Testing macro-evolutionary models using incomplete molecular phylogenies. Proc R Soc B. 2000;267:2267–2272. doi: 10.1098/rspb.2000.1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harmon LJ, Weir JT, Brock CD, Glor RE, Challenger W. GEIGER: Investigating evolutionary radiations. Bioinformatics. 2008;24:129–131. doi: 10.1093/bioinformatics/btm538. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.