Abstract
Domestication of plants and animals promoted humanity's transition from nomadic to sedentary lifestyles, demographic expansion, and the emergence of civilizations. In contrast to the well-documented successes of crop and livestock breeding, processes of microbe domestication remain obscure, despite the importance of microbes to the production of food, beverages, and biofuels. Lager-beer, first brewed in the 15th century, employs an allotetraploid hybrid yeast, Saccharomyces pastorianus (syn. Saccharomyces carlsbergensis), a domesticated species created by the fusion of a Saccharomyces cerevisiae ale-yeast with an unknown cryotolerant Saccharomyces species. We report the isolation of that species and designate it Saccharomyces eubayanus sp. nov. because of its resemblance to Saccharomyces bayanus (a complex hybrid of S. eubayanus, Saccharomyces uvarum, and S. cerevisiae found only in the brewing environment). Individuals from populations of S. eubayanus and its sister species, S. uvarum, exist in apparent sympatry in Nothofagus (Southern beech) forests in Patagonia, but are isolated genetically through intrinsic postzygotic barriers, and ecologically through host-preference. The draft genome sequence of S. eubayanus is 99.5% identical to the non-S. cerevisiae portion of the S. pastorianus genome sequence and suggests specific changes in sugar and sulfite metabolism that were crucial for domestication in the lager-brewing environment. This study shows that combining microbial ecology with comparative genomics facilitates the discovery and preservation of wild genetic stocks of domesticated microbes to trace their history, identify genetic changes, and suggest paths to further industrial improvement.
Keywords: beer yeast, next-generation sequencing, yeast ecology, yeast taxonomy
The beginning of agriculture and the domestication of plants and animals are among the most decisive events in human history because they triggered the rise of civilizations and the attendant demographic, technological, and cultural developments (1). The domestication of barley in the Fertile Crescent (2) led to the emergence of the forebear of modern beer in Sumeria 6,000 y ago (3). Beer and other alcoholic beverages may have played a pivotal role in cementing human societies through the social act and rituals of drinking (4) and by providing a source of nutrition, medicine, and uncontaminated water (5). Since the emergence of fermented beverages roughly matches the domestication of plants and animals, it is likely that some yeast lineages with favored traits were also unwittingly domesticated.
In Europe, brewing gradually evolved during the Middle Ages to produce ale-type beer, a process conducted by Saccharomyces cerevisiae, the same species involved in producing wine and leavened bread. Lager-brewing arose in 15th century Bavaria, gained broad acceptance by the late 19th century (6), and has since become the most popular technique for producing alcoholic beverages, with over 250 billion dollars of global sales in 2008 (7). Unlike most ales and wines, lagers require slow, low-temperature fermentations that are carried out by cryotolerant Saccharomyces pastorianus (syn. Saccharomyces carlsbergensis) strains (8); two other cryotolerant Saccharomyces spp. have been associated with beer as contaminants (Saccharomyces bayanus) and with cider or wine fermented at low temperatures (Saccharomyces uvarum) (9). S. pastorianus has never been isolated from the wild, depends on humans for its propagation, and appears to be an allotetraploid hybrid species of S. cerevisiae and an unidentified species (10, 11). Several hypotheses have been advanced for the source of the non-S. cerevisiae genome present in S. pastorianus, including the taxonomically and genetically complex species S. bayanus (12–14) and an unknown “lager” lineage distinct both from S. bayanus and S. uvarum (11, 15). Identifying the wild genetic stock of the cryotolerant subgenome of S. pastorianus is necessary for resolving the taxonomy and systematics of this important species complex, and for understanding the key events that led to the domestication of lager yeast.
In contrast to extensive investigation into domestication of crops and livestock (2, 16–19), studies of domestication of eukaryotic microbes have been limited (20–24), perhaps because of the inability to conduct direct field studies. Identifying the genetic basis of traits under selection during domestication may clarify the emergence of new traits and show the way toward further improvement. Because domesticated lineages derive from a subset of the original populations, a genetic bottleneck is likely to have caused the disappearance of some alleles (17), especially in microbes, which are often propagated clonally. In an age of accelerated habitat destruction and diminishing biodiversity, discovery of wild genetic stocks of domesticated microbes will facilitate preservation of their genetic resources for strain improvement.
Results and Discussion
Discovery of Wild Populations of Cryotolerant Saccharomyces.
Saccharomyces spp. are associated with oak trees (Fagaceae) in the Northern Hemisphere (25, 26). Because species of the genus Nothofagus (Southern beeches, also members of the Fagales) occupy the oak niche in temperate regions of the Southern Hemisphere (27), our survey in Northwestern Patagonia for Saccharomyces focused on woodlands containing populations of Nothofagus antarctica, Nothofagus dombeyi, and Nothofagus pumilio, within and near Lanin and Nahuel Huapi National Parks (Argentina) (Fig. S1). We also surveyed stromata of Cyttaria hariotii (an obligate ascomycete parasite of Nothofagus spp.) because these fruiting structures are rich in simple sugars and provide a favorable yeast habitat (28). A total of 133 samples of Nothofagus bark, soil from underneath the trees, and Cyttaria stromata, collected from 2006 to 2008, yielded 123 isolates of cryotolerant Saccharomyces and two isolates of S. cerevisiae (Table S1).
For a preliminary identification of the cryotolerant Saccharomyces isolates, we determined the DNA sequence of individual genes, performed PCR-fingerprinting, and examined restriction fragment length polymorphisms (RFLPs), using S. bayanus CBS 380T, S. uvarum CBS 395T, and S. uvarum CBS 7001 (referred to as S. bayanus in genomics literature) as references (Fig. S2). The isolates discretely fall into two groups: group A appears related to S. bayanus (78 isolates); group B is closely related to S. uvarum (45 isolates). The almost complete occupancy of the Nothofagus niche by cryotolerant species contrasts with our ongoing survey of Saccharomyces biogeography in the Northern Hemisphere oak niche (North America, Mediterranean, Central Europe, and Japan), where we have isolated ∼240 Saccharomyces strains from more than 500 oak samples and observed that sympatric species tend to have different growth temperature preferences. For example, S. cerevisiae (thermotolerant) and Saccharomyces kudriavzevii (cryotolerant) co-occur in Mediterranean regions, but Saccharomyces paradoxus (thermotolerant) and S. uvarum (cryotolerant) co-occur in temperate Europe and North America (26). Therefore, the detection of a pair of cryotolerant species in Patagonia and the near absence of thermotolerant species suggest the Patagonian ecosystem supporting Saccharomyces spp. may be unusual. One potential explanation is that the relatively low annual average temperatures in our Patagonian isolation sites [6/8 °C with mean low temperatures of −1/−2 °C and mean high temperatures of 22/23 °C (29)] may favor cryotolerant over thermotolerant species.
Ecological and Genetic Isolation of Two Cryotolerant Species.
The unanticipated detection of two closely related sympatric cryotolerant populations prompted us to investigate the degree of genetic isolation between them by measuring the meiotic sterility of hybrids of the two populations. Spore viability within populations was 89% to 91%, but hybrids produced only 7.3% viable spores. Thus, the two populations exhibit considerable intrinsic postzygotic isolation and can be considered to be two different biological species, although they are phenotypically indistinguishable (Fig. S3). Moreover, population A was found in association with N. antarctica and N. pumilio, whereas population B was associated with N. dombeyi (P < 10−7; Fisher's exact test) (Table 1). The evergreen N. dombeyi prevails at mesic midelevation sites, but N. antarctica and N. pumilio are deciduous and tend to replace N. dombeyi at high-elevation and xeric sites (27), suggesting that local niche-partitioning at least partially explains the apparent coexistence of these two species. Detailed field and laboratory studies into the causes of isolation are called for.
Table 1.
Isolation of cryotolerant Saccharomyces in Patagonia
| Isolates |
|||
| No. of samples | Population A | Population B | |
| N. antarctica | 47 (9C, 23B, 15S) | 29 (7C, 17B, 5S) | 4 (2C, 2B) |
| N. pumilio | 32 (2C, 15B, 15S) | 25 (1C, 10B, 14S) | 5 (1C, 3B, 1S) |
| N. dombeyi | 54 (12C, 27B, 15S) | 8 (5C, 2B, 1S) | 38 (8C, 21B, 9S) |
| Total | 133 (23C, 65B, 45S) | 62 | 47 |
The samples used in the isolations correspond to Cyttaria hariotii (C), Nothofagus bark (B), and soil collected underneath the trees (S). Numerals refer to number of samples or number of isolates. Statistically significantly associations are in boldface (P < 10−7).
Genome Sequences Resolve Saccharomyces Taxonomy and Systematics.
The identification and taxonomy of S. bayanus, S. pastorianus, and S. uvarum is problematic and controversial because S. bayanus and S. pastorianus have only been isolated from human-associated fermentations. Indeed, all known representatives of these two species have been suspected (S. bayanus) (13) or confirmed (S. pastorianus) (10) to be interspecies hybrids. One potential exception is the brewing contaminant NBRC 1948, which was asserted to be a pure strain of S. bayanus based primarily on RFLP evidence from all 16 chromosomes (15). A broad survey of several strains previously assigned to S. bayanus, S. pastorianus, and S. uvarum led to the conclusion that an additional “lager” lineage or species exists that contributed its genome to S. pastorianus (15). In contrast, the genome sequence of S. uvarum CBS 7001 lacks any signs of hybridization, introgression, or horizontal gene transfer events from other Saccharomyces spp. (30).
To resolve these taxonomic and systematic issues and conclusively identify our Patagonian strains, we generated draft genome sequences of a representative from each Patagonian species and several key brewing isolates by assembling millions of 36-bp sequences, using the S. uvarum genome sequence as a reference. Comparison of these genome sequences allowed us to conclusively test: (i) whether our wild Patagonian populations A and B are comprised of pure lineages; (ii) whether NBRC 1948 is a pure line or a hybrid; (iii) the composition of the type strains of S. uvarum (CBS 395T) and S. bayanus (CBS 380T); and (iv) the identity of the non-S. cerevisiae moiety of the S. pastorianus genome.
S. eubayanus sp. nov. Is the Missing Wild Genetic Stock of S. pastorianus.
Comparison of the draft genome sequences revealed that the two Patagonian species differ at ∼6% to 8% of nucleotides across the genomes of surveyed strains [Fig. 1 (explained in detail in the legend), and Datasets S1 and S2] (average divergence 6.89%). The uniformity of sequence divergence suggests that there has been little, if any, recent introgression between the Patagonian species, consistent with the low spore viability of the hybrid cross. (Introgression of regions that are small, subtelomeric, difficult to assemble, or missing from some strains cannot be excluded.) The species B strain is indeed closely related to the reference strain of S. uvarum (average divergence 0.52%) and to its type strain CBS 395T, but the species A strain is closely related to the non-S. cerevisiae moiety of the S. pastorianus lager yeast genome (average divergence 0.44%). In contrast to these essentially pure strains, the main component of the genome of the type strain of S. bayanus (CBS 380T) is S. uvarum (67%), although there are substantial contributions from species A (33%), including several large heterozygous regions (19% of the genome). Similarly, large portions of the genome of NBRC 1948 are derived from S. uvarum (37%), but the majority of these portions are derived from species A (63%).
Fig. 1.
S. eubayanus sp. nov. is distinct from S. uvarum and donated its genome to S. pastorianus. All 16 S. uvarum chromosomes are shown as x axes, with hash marks every 100 kbp; divergence (uncorrected) relative to S. uvarum CBS 7001 (30) is plotted above each chromosome; divergence relative to S. eubayanus is plotted (inverted) below each chromosome. Strains are color-coded according to the key. Each strain appears above and below the chromosomes to represent both comparisons. S. uvarum CBS 395T is only shown in Dataset S1; the Patagonian S. eubayanus reference strain is only shown above the chromosomes. To avoid overlaps of the comparisons, some strains are offset from the x axis as follows: S. eubayanus, 0.3%; NBRC 1948, 0.2%; CBS 380T, 0.1%. CBS 380T contains both S. uvarum and S. eubayanus alleles in several regions, so its percentage heterozygosity (of base pairs) is also shown in thin gray above the chromosomes. The divergence plots for a strain generally move in parallel above and below the chromosomes because nearly all regions of each strain are closely related to either the S. uvarum or the S. eubayanus reference (within 2% of the axis on one side and 6–8% on the other side). Quantitative differences reflect regions with different evolutionary rates, heterozygous regions (CBS 380T), or regions that may harbor older alleles because of incomplete lineage sorting. For example, on the left arm of chromosome VI, CBS 380T is heterozygous, but NBRC 1948 contains only S. uvarum alleles; moving right, CBS 380T becomes homozygous S. uvarum, and NBRC 1948 contains a large section from S. eubayanus before returning to S. uvarum characteristics toward the telomere. Across the filtered draft genome sequences (∼80% of 1-kb windows), S. pastorianus is comprised of S. eubayanus alleles (>99.9%), and the Patagonian S. uvarum is closely related to the S. uvarum reference (>99.9%).
We also screened the short sequence reads for evidence of introgression, hybridization, or horizontal gene transfer using two different methods and the available Saccharomyces spp. reference genomes (see SI Materials and Methods). We found no evidence of foreign genes in either Patagonian strain or in the S. uvarum type strain. Although these analyses cannot exclude foreign contributions among genes missing from the Saccharomyces spp. ortholog set, they are sensitive enough to readily detect multiple subtelomeric contributions from S. cerevisiae in the two strains of S. bayanus (CBS 380T and NBRC 1948) (Dataset S3).
Given the clear differences in ecological background and in genomic constitution between the hybrid species S. bayanus and the essentially pure species A, we propose regarding the wild Patagonian lineage as a distinct species: S. eubayanus sp. nov. Moreover, these genome sequence analyses firmly establish S. eubayanus as the donor of the non-S. cerevisiae subgenome of S. pastorianus, and exclude the contribution by an unknown “lager” species substantially divergent from S. cerevisiae and S. eubayanus (Dataset S4). Instead, a broad survey of strains (Fig. S2) and the genome sequences of both of our representative strains of S. bayanus (Fig. 1 and Datasets S1 and S3) suggest that the diversity of this hybrid species can be explained by the contribution of mixtures of alleles from S. uvarum and S. eubayanus, along with some genes contributed by S. cerevisiae.
Evidence of Domestication.
Domestication of crops and livestock selects for desirable characteristics through directional breeding so that domesticated lineages become genetically distinct from their wild ancestors in ways that make them more useful to humans (16, 17). To determine which genetic changes might have been favored in brewing, we searched for differences between the genome of S. eubayanus and three domesticated strains associated with brewing (S. pastorianus and the triple hybrids CBS 380T and NBRC 1948).
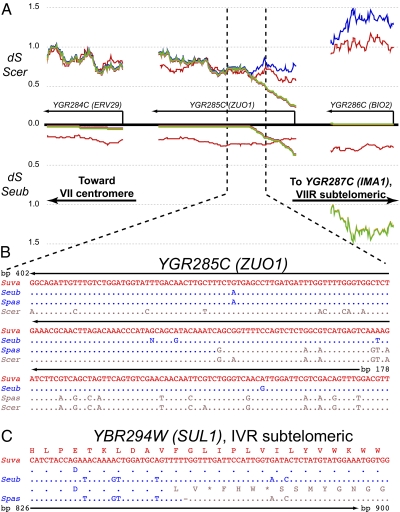
The disaccharide maltose is one of the most abundant sugars in wort, so its utilization is a strongly selected trait in the brewing environment (11, 31). Like S. pastorianus, the triple hybrid strains associated with brewing contain subtelomeric maltose (MAL) gene clusters horizontally transferred from S. cerevisiae, as well as the S. cerevisiae SUC4 gene for processing the disaccharide sucrose (Dataset S3). Surprisingly, we discovered that these three strains share an identical chromosome translocation breakpoint within ZUO1 (YGR285C) that fuses the right arm of S. eubayanus chromosome VII to subtelomeric sequences on the right arm of S. cerevisiae chromosome VII (Fig. 2 A and B). The transferred fragment carries the S. cerevisiae IMA1 (YGR287C) gene that encodes isomaltase, which catalyzes cleavage of the disaccharide isomaltose (32). The identical breakpoints indicate a common origin for this S. cerevisiae sugar-processing gene in all three hybrid strains and suggest strong selection for optimal sugar utilization during brewing.
Fig. 2.
S. pastorianus and two triple hybrid strains associated with brewing share identical domestication alleles. (A) Sliding window analysis of synonymous site divergence (dS, Jukes-Cantor corrected) for subtelomeric coding regions with arrows marking direction of transcription and with the color scheme and y axis as in Fig. 1, except the dS shown is to S. cerevisiae (Scer), rather than to S. uvarum (Suva); the y-value offsets necessary for visualization are 10-fold larger than in Fig. 1 because of the different scale; 2,000 sites are plotted with a 100-site window, a step of one, and an arbitrary intergenic spacer of 200 sites. (B) Close-up of partial coding sequences with dots denoting identity. Note the breakpoint fusing the 3′ portion of S. eubayanus (Seub) ZUO1 (blue) to the 5′ portion of ScerZUO1 (brown) and distal genes, including ScerBIO2 and ScerIMA1. [S. pastorianus clearly possesses ScerIMA1 and the ZUO1 breakpoint, but there is a gap across ScerBIO2 in the published assembly (11)]. (C) Partial coding sequence and translation of SUL1 highlighting a 1-bp deletion in SpasSUL1 that causes an inactivating frame-shift mutation. S. pastorianus (Spas) also represents NBRC 1948 and CBS 380T in B and C because all called base pairs are identical in the coding sequences of ERV29, ZUO1, BIO2, and SUL1 in those strains.
Sulfite formation is also important in lager-brewing because sulfite is an antioxidant and flavor stabilizer (31). S. pastorianus Weihenstephan 34/70 was previously noted to carry inactive copies of both the S.cerevisiae and S. eubayanus SUL1 genes, while retaining functional versions of both SUL2 genes (11), which encode the high affinity transporters of sulfate, the metabolic precursor of sulfite (33). Surprisingly, we found that both triple hybrid strains contain the same frame-shift mutation as S. pastorianus in their copies of S. eubayanus SUL1 (Fig. 2C). Interestingly, selective expression of SUL2, especially the S. eubayanus allele, has been shown to improve sulfite production (34). Because we found that the SUL1 gene is intact in S. eubayanus, it is likely that the inactivation of SUL1 is a consequence of artificial selection in the brewing environment. The losses of SUL1 and some MAL transporters suggest a tendency to selectively discard less-efficient nutrient transport systems and retain or acquire others that are more efficient under brewing conditions, a trade-off possibly resulting from the 2D space constraints of the plasma membrane.
Evolution of S. pastorianus and S. bayanus Under Domestication.
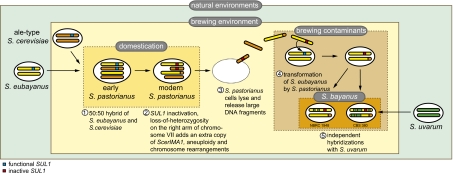
Based on our ecological and comparative genomic analyses, S. bayanus encompasses a set of hybrid strains known only from the industrial brewing environment, whereas S. eubayanus exists as an essentially pure lineage in natural conditions in Patagonia. According to our model (Fig. 3), in the lager-brewery environment of the 15th century (6) wild S. eubayanus genomes began fusing with ale-type S. cerevisiae genomes to give rise to allotetraploid hybrids, rare events that seem to have happened at least twice (10). In these ancestors of modern-day S. pastorianus, mitotic crossovers and other DNA repair mechanisms fused some S. cerevisiae chromosomes to S. eubayanus chromosomes, making some portions of the genome homozygous and other regions aneuploid (10, 11). The S. eubayanus portions of these chromosomal fusions that occurred in S. pastorianus would have provided ample nearly identical sequence to seed the recombination needed to introduce S. cerevisiae DNA into other S. eubayanus and S. uvarum strains arriving in the brewing environment, thus gradually giving rise to the complex S. bayanus genome. Alternatively, the tremendous population sizes achieved in the brewing environment may have allowed for the recovery of extremely rare viable hybrid spores or for strains to return to euploidy via a parasexual cycle. Because all S. cerevisiae genes detected in the triple hybrids are subtelomeric in the S. uvarum or S. cerevisiae reference genomes, we believe transformation is the more likely mechanism because it requires only one crossover proximal to the subtelomeric gene under selection, and it explains the absence of widespread transfer of S. cerevisiae proximal genes that would be expected under the other models. Regardless of the mechanism of gene transfer, the identical frame-shift mutations (SUL1), chromosome breakpoints (IMA1), and identical hitchhiking sequences suggest that strong positive selection was imposed by the brewers choosing the best strains or by the competitive brewing environment itself to spread these alleles to S. pastorianus and both S. bayanus triple hybrids.
Fig. 3.
A model of the formation of S. pastorianus and the hybrid strains of S. bayanus. First, wild S. eubayanus and ale-type S. cerevisiae hybridized to form an allotetraploid that gave rise to S. pastorianus. Second, domestication imposed strong selective pressure for strains with the most desirable brewing properties. Third, in the brewing vats with high densities of S. pastorianus, cell lysis releases large DNA fragments that occasionally transform, fourth, contaminating wild strains of S. eubayanus because of the lack of pure culture techniques. Fifth, multiple hybridization events with wild strains of S. uvarum gave rise to CBS 380T and NBRC 1948. This model does not exclude prior or parallel involvement of S. uvarum in brewing or contamination.
It is surprising that European isolates of S. eubayanus have never been found, despite records of yeast isolation since the late 19th century, including a recent emphasis on sampling more natural arboreal environments (26). The facile recovery of this species from Patagonia suggests that S. eubayanus may have been absent in Europe until it was imported from overseas after the advent of trans-Atlantic trade. In sharp contrast, its sister species, S. uvarum, has been repeatedly isolated in Europe from both artificial (e.g., brewing) and natural substrates (e.g., insects, bark). Although additional environmental sampling is called for worldwide, it seems likely that any natural European niches suitable for S. eubayanus are occupied by other species, but that it found great success through hybridization in the new artificial environment of lager breweries.
The ∼7% genome-wide sequence divergence between S. eubayanus and S. uvarum is the lowest level observed within the Saccharomyces genus to result in genetic and ecological isolation (Table S2). The geographic distribution and apparent niche differentiation of these sister species make them a genetically tractable system to study fungal speciation in allopatry with secondary contact, or in sympatry because of ecological factors.
Taxonomy of S. eubayanus, S. bayanus, and S. uvarum.
The nomenclature of S. bayanus and S. uvarum has been confusing and controversial for decades. High DNA-DNA reassociation values between the type strains of these two species (35) led Naumov to defend their merger, with S. uvarum initially considered a synonym of S. bayanus (36), and later a variety (12). Other studies employing molecular methods documented two well-separated groups (37, 38), leading to proposals for the reinstatement of S. uvarum as a separate species (13, 14). Unfortunately, the absence of a clear resolution of these taxonomic issues caused CBS 7001, a strain selected for genome-sequencing (30), to be referred to as S. bayanus (instead of S. uvarum) in most of the ensuing genomics literature and databases. However, all known strains of S. bayanus (including the type strain CBS 380T) appear to be hybrids of S. eubayanus and S. uvarum that contain contributions from S. cerevisiae in at least some cases (Fig. S2). Therefore, like S. pastorianus, S. bayanus is not a “species” in the ecological and evolutionary sense and is best viewed as a product of the artificial brewing environment with no occurrence in nature. Because it is now evident that the varietal and other designations adopted by some researchers lack scientific support, we propose that “S. uvarum” and “S. eubayanus” be used as descriptors of biologically meaningful species, whereas “S. bayanus” and “S. pastorianus” should be restricted to the domesticated and hybrid lineages.
Standard description.
Standard description of Saccharomyces eubayanus Sampaio, Libkind, Hittinger, P. Gonçalves, Valério, C. Gonçalves, Dover et Johnston sp. nov.
Etymol.: The epithet is chosen to refer to the pure and natural lineage that is related to the hybrid species Saccharomyces bayanus Saccardo.
Cells globose to ovoidal (2.5–5 × 5–7 μm). Pseudomycelium absent. Asci oval and persistent containing two to four round ascospores. The main diagnosis characteristic is the genome sequence deposited in NCBI database. Salient physiological properties are listed in Dataset S5. Strain CBS 12357 T (CRUB 1568T, PYCC 6148T) is designated as the type strain.
Latin description.
Latin description of Saccharomyces eubayanus Sampaio, Libkind, Hittinger, P. Gonçalves, Valério, C. Gonçalves, Dover et Johnston sp. nov.
Species generis Saccharomycetis. Cellulae globosae ad ovoideae. Pseudomycelium nullum. Asci ovales, persistentes, 2–4 ascosporis globosis. Sequentia genomae totae in collectione sequentiarum acidi nucleici NCBI numero SRP006155 in SRA030851 deposita. Cultura typica CBS 12357T.
Domestication Processes Across Taxonomic Kingdoms.
Domestication is an inherently evolutionary process, the understanding of which requires the inference of ancestral states, either by comparison with wild progenitor populations or by archaeological investigations. The characteristics of the previously undescribed species S. eubayanus explain the instantaneous formation of S. pastorianus by hybridization with S. cerevisiae. Selection imposed by the brewing environment further refined lager yeast, and we pinpointed several genetic changes attendant to lager production. Given the diverse mechanistic natures of genetic changes that occurred during crop and livestock domestication (2, 17, 18), we anticipate that additional genetic changes that are not obvious from sequence comparisons alone will be uncovered in the future, such as changes in gene regulation and mechanisms to integrate two genomes separated by millions of years of evolution. Identification of these evolutionary changes and access to the previously unknown wild stock promise to illuminate the role that fermented beverages have played in human civilization and provide new strategies for improving yeasts for brewing and biofuel production.
Materials and Methods
Yeast Isolation, Preliminary Characterization, and Crosses.
The selective protocol used for Saccharomyces isolations was previously described (26). Preliminary identification was based on confirmation of Saccharomyces-type ascospore production; DNA-sequencing of FSY1, FUN14, HIS3, MET2, RIP1 (Dataset S6), and the ITS region of rDNA; PCR-fingerprinting with primers (GTG)5 and M13 (39); and PCR-RFLP using FUN14 digested with BanII, RIP1 digested with PstI, and HIS3 digested with RsaI (15) (Fig. S2). Isolated ascospores of strains S. eubayanus CRUB 1568 and S. uvarum CRUB 1595 were crossed to obtain interspecies hybrids. Hybridization was confirmed with PCR-RFLP as above. Interspecific spore viability was determined by examining 362 ascospores produced by three independent hybrid strains. For the assessment of intraspecific spore viability, ∼100 ascospores of each of the two species (strains CRUB 1568, CRUB 1595) were studied.
Genome Sequencing and Analyses.
The draft genome sequences of monosporic-derivatives of CRUB 1568 (FM1318), CRUB 1595 (FM1317), and NBRC 1948 (FM1309) were determined by assembling single-pass 36-bp Illumina sequence reads to the S. uvarum CBS 7001 reference genome sequence (30), as described previously (40) with modifications (SI Materials and Methods). Viable spores of CBS 380T and CBS 395T were not recovered, so the genomes of these strains were sequenced without further manipulation and shown to be aneuploid, providing a likely explanation for their sterility. Genome sequence analyses were performed using custom PERL scripts and standard software.
Supplementary Material
Acknowledgments
We thank the Argentinean National Parks Administration for collecting permits. We thank the NITE Biological Resource Center of Japan for providing strain NBRC 1948, R. Ulloa for help during preliminary field work, M. Weiss for preparing the Latin diagnosis, and R. A. Sclafani for critical reading of the manuscript; and S. Fontenla and M. R. van Broock for assistance and advice. This work was supported by Fundação para a Ciência e a Tecnologia Grants PTDC/BIA-BDE/71734/2006 (to P.G. and J.P.S.), SFRH/BPD/46471/2008 (to E.V.), UNComahue Grant CRUB B143 (to D.L.), National Institutes of Health Grant GM032540 (to M.J.), and the James S. McDonnell Foundation (C.T.H. and M.J.). C.T.H. is the Maclyn McCarty Fellow of the Helen Hay Whitney Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Genome data have been deposited in the National Center for Biotechnology Information's Sequence Read Archive (www.ncbi.nlm.nih.gov/sra) (sequence nos. SRP006155 of SRA030851); individual genes have been deposited in GenBank (accession nos. JF786614–JF786710).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105430108/-/DCSupplemental.
References
- 1.Bar-Yosef O. The Natufian culture of the Levant, threshold to the origins of agriculture. Evol Anthropol. 1998;6:159–177. [Google Scholar]
- 2.Salamini F, Özkan H, Brandolini A, Schäfer-Pregl R, Martin W. Genetics and geography of wild cereal domestication in the near east. Nat Rev Genet. 2002;3:429–441. doi: 10.1038/nrg817. [DOI] [PubMed] [Google Scholar]
- 3.Hornesey I. A History of Beer and Brewing. Cambridge, UK: RSC paperbacks; 2003. [Google Scholar]
- 4.Joffe AH, et al. Alcohol and social complexity in ancient western Asia. Curr Anthropol. 1998;39:297–322. [Google Scholar]
- 5.McGovern PE. Uncorking the Past: The Quest for Wine, Beer, and Other Alcoholic Beverages. Berkeley, CA: University of California Press; 2009. [Google Scholar]
- 6.Corran HS. A History of Brewing. London, UK: David and Charles; 1975. [Google Scholar]
- 7.Beer: Global industry guide. http://www.researchandmarkets.com/reports/53577 (accessed 15 January 2011)
- 8.Bond U. The genomes of lager yeasts. In: Laskin AI, Sariaslani S, Gadd GM, editors. Advances in Applied Microbiology. Vol 69. Burlington: Academic Press; 2009. pp. 159–182. [DOI] [PubMed] [Google Scholar]
- 9.Rainieri S, Zambonelli C, Kaneko Y. Saccharomyces sensu stricto: Systematics, genetic diversity and evolution. J Biosci Bioeng. 2003;96:1–9. [PubMed] [Google Scholar]
- 10.Dunn B, Sherlock G. Reconstruction of the genome origins and evolution of the hybrid lager yeast Saccharomyces pastorianus. Genome Res. 2008;18:1610–1623. doi: 10.1101/gr.076075.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakao Y, et al. Genome sequence of the lager brewing yeast, an interspecies hybrid. DNA Res. 2009;16:115–129. doi: 10.1093/dnares/dsp003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naumov G. Saccharomyces bayanus var. uvarum comb. nov., a new variety established by genetic analysis. (Translated from Russian) Mikrobiologiia. 2000;69:410–414. [PubMed] [Google Scholar]
- 13.Nguyen H-V, Gaillardin C. Evolutionary relationships between the former species Saccharomyces uvarum and the hybrids Sacharomyces bayanus and Saccharomyces pastorianus; reinstatement of Sacccharomyces uvarum (Beijerinck) as a distinct species. FEM Yeast Res. 2005;5:471–483. doi: 10.1016/j.femsyr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Pulvirenti A, et al. Saccharomyces uvarum, a proper species within Saccharomyces sensu stricto. FEMS Microbiol Lett. 2000;192:191–196. doi: 10.1111/j.1574-6968.2000.tb09381.x. [DOI] [PubMed] [Google Scholar]
- 15.Rainieri S, et al. Pure and mixed genetic lines of Saccharomyces bayanus and Saccharomyces pastorianus and their contribution to the lager brewing strain genome. Appl Environ Microbiol. 2006;72:3968–3974. doi: 10.1128/AEM.02769-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–707. doi: 10.1038/nature01019. [DOI] [PubMed] [Google Scholar]
- 17.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Larson G, et al. Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc Natl Acad Sci USA. 2010;107:7686–7691. doi: 10.1073/pnas.0912264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeder MA. Domestication and early agriculture in the Mediterranean Basin: Origins, diffusion, and impact. Proc Natl Acad Sci USA. 2008;105:11597–11604. doi: 10.1073/pnas.0801317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Legras J-L, Merdinoglu D, Cornuet J-M, Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- 22.Liti G, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rokas A. The effect of domestication on the fungal proteome. Trends Genet. 2009;25:60–63. doi: 10.1016/j.tig.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 24.Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sniegowski PD, Dombrowski PG, Fingerman E. Saccharomyces cerevisiae and Saccharomyces paradoxus coexist in a natural woodland site in North America and display different levels of reproductive isolation from European conspecifics. FEM Yeast Res. 2002;1:299–306. doi: 10.1111/j.1567-1364.2002.tb00048.x. [DOI] [PubMed] [Google Scholar]
- 26.Sampaio JP, Gonçalves P. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl Environ Microbiol. 2008;74:2144–2152. doi: 10.1128/AEM.02396-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Veblen TT, Donoso C, Kitzberger T, Rebertus AJ. Ecology of southern Chilean and Argentinean Nothofagus forests. In: Veblen TT, Hill RS, Read J, editors. The Ecology and Biogeography of Nothofagus forests. New Haven, CT: Yale University Press; 1996. pp. 293–351. [Google Scholar]
- 28.Libkind D, Ruffini A, van Broock M, Alves L, Sampaio JP. Biogeography, host specificity, and molecular phylogeny of the basidiomycetous yeast Phaffia rhodozyma and its sexual form, Xanthophyllomyces dendrorhous. Appl Environ Microbiol. 2007;73:1120–1125. doi: 10.1128/AEM.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paruelo JM, Beltrán A, Jobbágy E, Sala OE, Golluscio RA. The climate of Patagonia: General patterns and controls on biotic processes. Ecologia Austral. 1998;8:85–101. [Google Scholar]
- 30.Cliften PF, Fulton RS, Wilson RK, Johnston M. After the duplication: Gene loss and adaptation in Saccharomyces genomes. Genetics. 2006;172:863–872. doi: 10.1534/genetics.105.048900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Donalies UE, Nguyen HTT, Stahl U, Nevoigt E. Improvement of Saccharomyces yeast strains used in brewing, wine making and baking. Adv Biochem Eng Biotechnol. 2008;111:67–98. doi: 10.1007/10_2008_099. [DOI] [PubMed] [Google Scholar]
- 32.Brown CA, Murray AW, Verstrepen KJ. Rapid expansion and functional divergence of subtelomeric gene families in yeasts. Curr Biol. 2010;20:895–903. doi: 10.1016/j.cub.2010.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cherest H, et al. Molecular characterization of two high affinity sulfate transporters in Saccharomyces cerevisiae. Genetics. 1997;145:627–635. doi: 10.1093/genetics/145.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakao Y, Kodama Y, Shimonaga T. Sulfate ion transporter gene and use thereof. US Patent Appl No. 11/922,219. 2006 [Google Scholar]
- 35.Rosini G, Federici F, Vaughan AE, Martini A. Systematics of the species of the yeast genus Saccharomyces associated with the fermentation industry. Appl Microbiol Biotechnol. 1982;15:188–193. [Google Scholar]
- 36.Naumov GI. Genetic identification of biological species in the Saccharomyces sensu stricto complex. J Ind Microbiol. 1996;17:295–302. [Google Scholar]
- 37.Nguyen HV, Gaillardin C. Two subgroups within the Saccharomyces bayanus species evidenced by PCR amplificatiom and restriction polymorphism of the non-transcribed spacer 2 in the ribosomal DNA unit. Syst Appl Microbiol. 1997;20:286–294. [Google Scholar]
- 38.Rainieri S, Zambonelli C, Hallsworth JE, Pulvirenti A, Giudici P. Saccharomyces uvarum, a distinct group within Saccharomyces sensu stricto. FEMS Microbiol Lett. 1999;177:177–185. doi: 10.1111/j.1574-6968.1999.tb13729.x. [DOI] [PubMed] [Google Scholar]
- 39.Sampaio JP, et al. Polyphasic taxonomy of the basidiomycetous yeast genus Rhodosporidium: Rhodosporidium kratochvilovae and related anamorphic species. Int J Syst Evol Microbiol. 2001;51:687–697. doi: 10.1099/00207713-51-2-687. [DOI] [PubMed] [Google Scholar]
- 40.Hittinger CT, et al. Remarkably ancient balanced polymorphisms in a multi-locus gene network. Nature. 2010;464:54–58. doi: 10.1038/nature08791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.