Abstract
Regeneration of peripheral differentiated tissue in mammals is rare, and regulators of this process are largely unknown. We carried out a forward genetic screen in mice using N-ethyl-N-nitrosourea mutagenesis to identify genetic mutations that affect regenerative healing in vivo. More than 400 pedigrees were screened for closure of a through-and-through punch wound in the mouse ear. This led to the identification of a single pedigree with a heritable, fast, and regenerative wound-healing phenotype. Within 5 wk after ear-punch, a threefold decrease in the diameter of the wound was observed in the mutant mice compared with the wild-type mice. At 22 wk, new cartilage, hair follicles, and sebaceous glands were observed in the newly generated tissue. This trait was mapped to a point mutation in a receptor for TGF-β, TGFBR1. Mouse embryonic fibroblasts from the affected mice had increased expression of a subset of TGF-β target genes, suggesting that the mutation caused partial activation of the receptor. Further, bone marrow stromal cells from the mutant mice more readily differentiated to chondrogenic precursors, providing a plausible explanation for the enhanced development of cartilage islands in the regenerated ears. This mutant mouse strain provides a unique model to further explore regeneration in mammals and, in particular, the role of TGFBR1 in chondrogenesis and regenerative wound healing.
Keywords: skin, epimorphic, appendage
Wound healing and regeneration are the two major responses to injury that exist in higher organisms (1). In amphibians, epimorphic regeneration is common, leading to replacement of entire appendages after amputation (1–3). However, although studies from axolotls to zebrafish have implicated known developmental or tyrosine receptor kinase pathways in this process, the exact mechanisms underlying regeneration are not well understood (2–6).
In mammals, wounding of peripheral appendages typically results in a patch of fibroblasts and a collagen-rich scar, rather than regeneration of the differentiated structures of normal tissue (1). One of the few known examples of mammalian regeneration is the healing of through-and-through ear punch wounds in the MRL/MpJ mouse strain (7). Healing of an ear punch in this strain exhibits some of the hallmarks of regeneration, including generation of a blastema-like structure (or a mesenchymal cell mass from which new tissue is created) expressing the early chondrocyte marker aggrecan, and the de novo generation of cartilaginous islands and hair follicles (8). Genetic loci that control this healing trait have been mapped through intercrosses between MRL/MpJ and other mouse strains that heal ear punch wounds more slowly (9–12). However, to date, none of these loci have been resolved to the point of identification of the causative genetic variant (9–12), a difficult process given the low mapping resolution of the quantitative trait loci approach.
To identify and establish mouse lines with genetically tractable regenerative healing traits, we carried out a forward genetic screen after N-ethyl-N-nitrosourea (ENU) mutagenesis to identify strains with a fast healing phenotype upon through-and-through ear hole punch. The genetic mutation underlying the regeneration phenotype was successfully identified through SNP mapping and candidate gene sequencing. This is a genetically defined mouse model with regenerative traits and should provide a valuable tool to study regeneration and its underlying molecular mechanisms in mammals.
Results
Identification of a Heritable, Fast-Healing Phenotype.
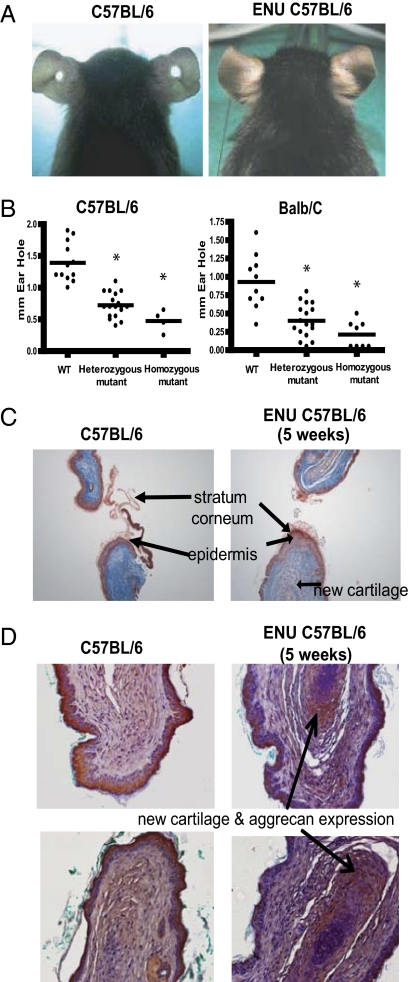
In an effort to further elucidate pathways that control regeneration in mammals, we used ENU mutagenesis to introduce random mutations into the germline of a slow-healing mouse strain, C57BL/6, and screened families of progeny mice for fast-healing mutant strains. A 2-mm hole was punched in the ears of 5-wk-old animals. At 5 wk after wounding, the average hole diameter for wild-type mice or mice from the ENU families was 1.4 mm. However, one pedigree from the ENU screen had a heritable wound-healing phenotype that mapped to a single genetic locus—affected animals from this strain had an average wound diameter of only 0.4–0.6 mm (Fig. 1 A and B).
Fig. 1.
Accelerated wound-healing trait in an ENU mutant pedigree. A 2-mm through-and-through hole was punched in the ears of 5-wk-old C57BL/6 wild-type mice and ENU-induced mutant mice to screen for acceleration of wound healing. (A) Example of incomplete ear hole closing in unaffected colony control mice (Left) and complete ear hole closing in the affected ENU strain (Right). Images were taken 4 wk after wounding. (B) Wild-type, heterozygous, and homozygous mice (C57BL/6 and BALB/c strains) were given a centrally located ear hole punch at 5 wk of age. A significant healing trait was demonstrated in both strains compared with wild type 5 wk later (*P < 0.01, n ≥ 5 per group). (C) Masson’s trichrome-stained ear sections taken 5 wk after wounding in control and heterozygous mutant mice. Initial thickening of the epidermis and new cartilage formation was observed in the heterozygous mutant mice. Representative images shown from five mice studied of each genotype. (D) Ear sections from wild-type and homozygous mutant mice, taken 5 wk after wounding, were stained with anti-aggrecan antibody overnight at 4 °C. The increase red/brown staining signifies aggrecan expression, which was increased in the homozygous mutant mice. Representative images shown from five mice studied of each genotype.
We bred affected mice to two slow-healing strains, C57BL/6 and BALB/c. Fast healers were selected at each generation for further breeding. Intercrossing and phenotyping of the progeny of these crosses, coupled with a comparison of the phenotypes with genotyping of the causative allele on archived DNA (see below) demonstrated that the fast-healing trait was semidominant on two backgrounds (Fig. 1B). Animals carrying two copies of the mutation had an average hole diameter of only 0.2 mm on the BALB/c background.
A thickened wound epidermis was apparent in both the heterozygous mutant and wild-type mice at 7 d after wounding (Fig. S1A), with no apparent difference in cellular proliferation when assessed by staining for the mitotic marker proliferating cell nuclear antigen (PCNA) (Fig. S1B). In the heterozygous mutant mice, cartilage-like formation was clearly apparent in the healing wound at 5 wk after ear punch (Fig. 1C), although some of the wild-type mice also appeared to have some cartilage formation during the healing process. Moreover, immunohistochemical staining for the cartilage matrix protein aggrecan revealed a consistent and increased expression in the mice expressing the mutation compared with wild-type mice (Fig. 1D). Twenty-two weeks after the ear punch, the wound was mostly repaired macroscopically, and histological analysis of the new tissue showed sebaceous glands, hair follicles, and islands of cartilage (Fig. S1C). These histological data suggest that the wound healing response in this strain is more akin to a regenerative response, reminiscent of the response to ear punch described for the MRL/MpJ mice. Additionally, the accelerated cartilage development suggests a role for mesenchymal stem cells in the healing process.
Identification of an ENU-induced mutation in the strain with the fast healing trait.
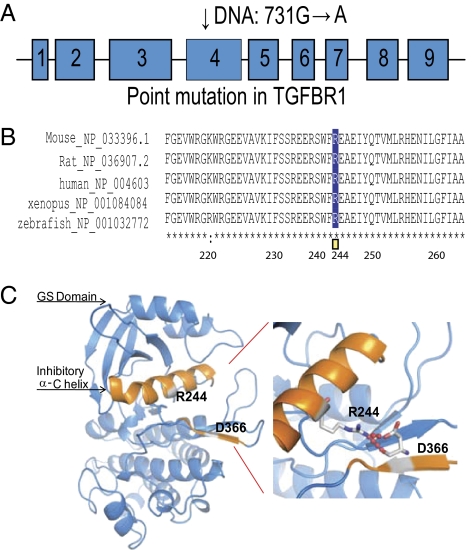
To identify the ENU mutation responsible for the fast-healing trait, we crossed affected mice on a C57BL/6 background to wild-type mice from the BALB/c strain. Offspring from this cross were back-crossed once again to BALB/c, and the resulting F1N1 animals were phenotyped and genotyped with a panel of 329 SNPs between the C57BL/6 and BALB/c strains. Most affected animals were heterozygous across a 4.6-Mb interval on chromosome 4 with base position from 46.6 to 51.1 Mb (Ensembl build 59), whereas unaffected controls were homozygous for BALB/c alleles in this region (Table S1). Sequencing of candidate genes in this region from phenotypically wild-type and from fast-wound-healing mice identified an ENU-induced mutation in the TGFBR1 gene. This gene encodes TGF-β type I receptor, TGFBR1 (also known as Alk5). The ENU-induced mutation was a G to A transition at nucleotide 731, predicted to cause substitution of a conserved R (arginine) to Q (glutamine) at residue 244 of TGFBR1. To rule out additional functional mutations in this 4.5-Mb region, the entire region was targeted for sequencing. A custom Nimblegen array was used to enrich the genomic DNA in this genomic region from a pool of five affected mice. DNA was further paired-end sequenced on an Illumina Genome Analyzer GAIIx. The highest-quality homozygous SNPs were confirmed with Sanger sequencing and compared with a wild-type C57BL/6 mouse. The only nonsynonymous SNP that was confirmed by Sanger sequencing was the one described above in TGFBR1.
Signaling function of R244Q TGFBR1.
R244 in the mouse TGFBR1 lies in a highly conserved intracellular regulatory region between the GS domain (glycine and serine-rich sequence) and the kinase domain, suggesting a critical functional role (Fig. 2 A and B). In the model of TGFBR1 activation proposed by Huse et al. (13), ligand binding to the TGF-β type II receptor type (TGFBR2) receptor leads to recruitment of TGFBR1 and phosphorylation of the GS sequence of TGFBR1 by the kinase domain of TGFBR2. Before activation, the unphosphorylated GS domain “locks” TGFBR1 in an inactive conformation. According to the structure of human TGFBR1 (13, 14), R244 in the regulatory α-C helix and D366 in the kinase domain form a salt bridge that helps to hold the regulatory α-C helix in an inhibitory position (Fig. 2C). We hypothesize that the R244Q mutation weakens this salt bridge, destabilizing the inactive conformation and resulting in hyperresponsiveness of the mutated receptor with or without ligand stimulation.
Fig. 2.
An R244Q mutation in TGFBR1 in the ENU-induced mutant mice with accelerated would healing. (A) The R244 mutation (731G→A) is located in exon 4. (B) R244 is evolutionarily conserved in TGFBR1 across vertebrate species. (C) Modeling of the intracellular region of TGFBR1 is shown here to illustrate the effect of R244Q mutation. The colored helix is the α-C helix, which has an inhibitory function on kinase activity. R244 is located between the GS domain and the kinase domain of the human TGFBR1 protein (13, 14). R244 in the α-C helix and D366 in the kinase domain form a salt bridge that is thought to help to hold the regulatory α-C helix in an inhibitory position.
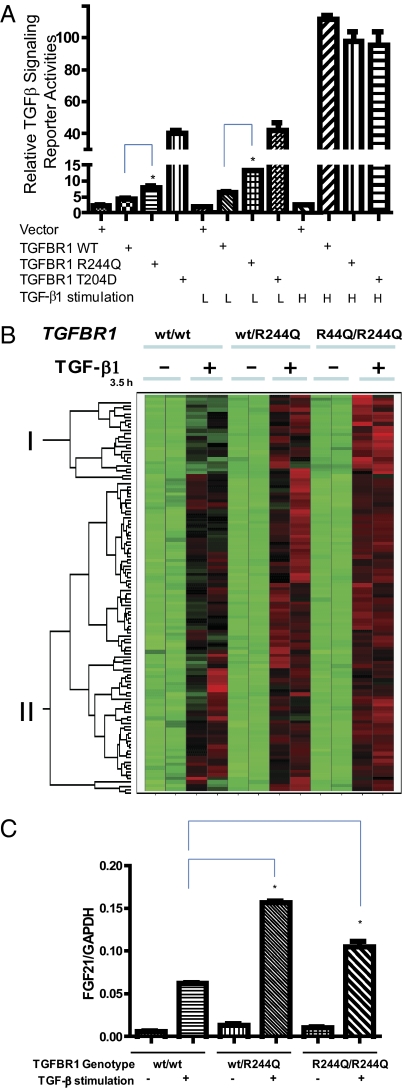
To test this hypothesis, we generated TGFBR1 expression cDNA constructs encoding the following variants of TGFBR1: the wild-type sequence, the R244Q mutant, and the constitutively active mutant T204D, which mimics the activating phosphorylation event. We transfected these into the TGFBR1-deficient cell line L17 (mink lung epithelial cells) (11) and monitored activity of the receptor using a destabilized luciferase reporter construct driven by TGF-β/Smad-responsive elements in a promoter from the TGF-β target gene, Plasminogen Activator Inhibitor Type 1 (PAI-1) (15). This experiment demonstrated that the R244Q variant receptor had a twofold increase in reporter gene activity compared with the wild-type receptor in the absence of TGF-β1 stimulation (Fig. 3A) but was fivefold less active than the T204D mutant, a constitutively active mutant. A similar result was observed upon suboptimal stimulation of TGF-β1 but not upon treatment of a high dose of TGF-β1 in L17 cells transfected with different TGFBR1 constructs. The variant R244Q receptor remained dependent on its kinase activity, because introduction of the kinase-dead K322R mutation prevented induction of the reporter gene.
Fig. 3.
TGFBR1 R244Q mutant is a weak partial activation mutant. (A) The TGFBR1 mutant activates a TGF-β–dependent reporter assay in a TGFBR1 defective cell line, L17. Activity of the mutant receptor is greater than the WT TGFBR1 but less than the constitutively active mutant T204D, under basal conditions and during suboptimal stimulation of TGF-β1 (8 pg/mL). All receptor constructs activated the reporter gene to equivalent levels at a high dose of TGF-β1 (1 ng/mL). *Signaling activity difference between the wild-type and R244Q mutant is statistically significant (P < 0.05 by t test). (B) MEFs with three different genotypes (TGFBR1wt/wt, TGFBR1wt/R244Q, and TGFBR1R244Q/R244Q) were treated with or without 5 ng/mL TGF-β1 for the indicated times. RNAs were harvested with the RNeasy RNA extraction kit (Qiagen). Hierarchical clustering was used to compare the expression levels of the TGF-β response genes (up-regulated by TGFβ1 in wild-type MEFs) among individual MEFs to determine the expression pattern and correlation between each individual MEF donor. Cluster I and Cluster II are shown as indicated. TGF-β response genes in Cluster I were increased by the mutant allele and were largely unchanged in Cluster II. (C) Quantitative RT-PCR (TaqMan) assay of FGF21 mRNA in pooled MEF cells with or without TGF-β1 stimulation. For each genotype, five separate MEF cells from independent embryos were pooled and plated in duplicates. *Difference in FGF21/GAPDH levels between the wild-type and WT/R244Q or the wild-type and R244Q/244Q MEF cells is statistically significant (P < 0.05 by t test) upon ligand stimulation.
We then tested whether Smad phosphorylation was increased by the R244Q variant in primary cells from the mutant mice. Mouse embryonic fibroblasts (MEFs) were generated from each of wild-type (wt/wt), heterozygous (TGFBR1wt/R244Q), and homozygous mutant (TGFBR1R244Q/R244Q) mice, and Smad2 phosphorylation was measured after stimulation of the cells with TGF-β1. MEF cells from heterozygous TGFBR1wt/R244Q mice showed a slight increase in Smad 2 phosphorylation after ligand binding, relative to the wild-type cells. However, this increase in proximal signaling associated with the R244Q mutation was not statistically significant.
To further characterize gene expression changes affected by the R244Q TGFBR1 mutation, we examined MEFs before and after stimulation with TGF-β1. Approximately one fifth of the TGF-β response genes were hyperinduced in TGFBR1wt/R244Q and TGFBR1R244Q/R244Q MEFs after stimulation (Cluster I, Fig. 3B, and Table S2), such as FGF21 (Fibroblast Growth Factor 21). Quantitative PCR (TaqMan assays) confirmed the microarray result for FGF21 with separate MEF cells derived from independent mouse embryos (Fig. 3C). We also found that the expression levels of several known TGF-β target genes, such as HAS2 (Hyaluronan Synthase 2) and PLAUR (Plasminogen Activator, Urokinase Receptor), as well as around three-quarters of the TGF-β response genes identified by microarray (Cluster II, Fig. 3B, and Table S3) were not appreciably affected by the TGFBR1 R244Q mutation in MEF cells. This result indicates that the R244Q mutation seems to partially activate only a small subset of TGF-β target genes. Overall, the partial activation by the R244Q mutation is relatively weak. Microarray analysis of MEF cells from all three genotypes did not uncover modulation of any additional pathway, other than TGF-β signaling, under either unstimulated or TGF-β–stimulated conditions.
Role of immune response in the fast-healing trait in TGFBR1 R244Q mice.
TGF-β is an immunosuppressive cytokine, and the immune response to wounding has been linked with regenerative wound healing in lower vertebrates (16–20). Moreover, in MRL/MpJ mice, hematopoietic cells produce more TGF-β1 than in control mice and have an impaired inflammatory response to lipopolysaccharide challenge (21) and lower white blood cell counts (22). We therefore asked whether accelerated wound healing in our strain might be secondary to a dampened immune response at the wound site caused by partially increased signaling through the mutant receptor. Blood cell counts did not differ significantly between the TGFBR1wt/R244Q and wild-type mice (Fig. S2A); TGF-β1 production levels in splenocytes were equivalent between TGFBR1wt/R244Q and wild-type mice (Fig. S2B). We then tested whether the wound-healing phenotype was affected by bone marrow transplantation from either wild-type or TGFBR1R244Q/R244Q mice into irradiated wild-type recipients. The wound-healing phenotypes of recipient mice were equivalent regardless of whether bone marrow came from wild-type or mutant donors (Fig. S3). Taken together, these data do not provide evidence that the TGFBR1 R244Q mutation is affecting the wound-healing process through an immunomodulatory mechanism.
Accelerated Chondrogenic Differentiation in TGFBR1 R244Q Mutant Mice.
Three lines of evidence prompted us to ask whether the accelerated healing phenotype in our mutant mice might be affected by increased chondrogenesis. First, TGF-β family ligands are effective inducers of chondrogenesis in cultures of bone marrow-derived mesenchymal stromal cells (BMSCs) (23, 24). Second, chondrocyte progenitors and aggrecan expression have been reported in the blastema-like structures at the ear punch wound margins in the MRL/MpJ strain (8). Third, the healing response in the ear includes histological evidence of additional aggrecan-expressing chondrocytes and cartilage deposition in the TGFBR1 mutant mice compared with wild-type mice (Fig. 1 C and D and Fig. S1B).
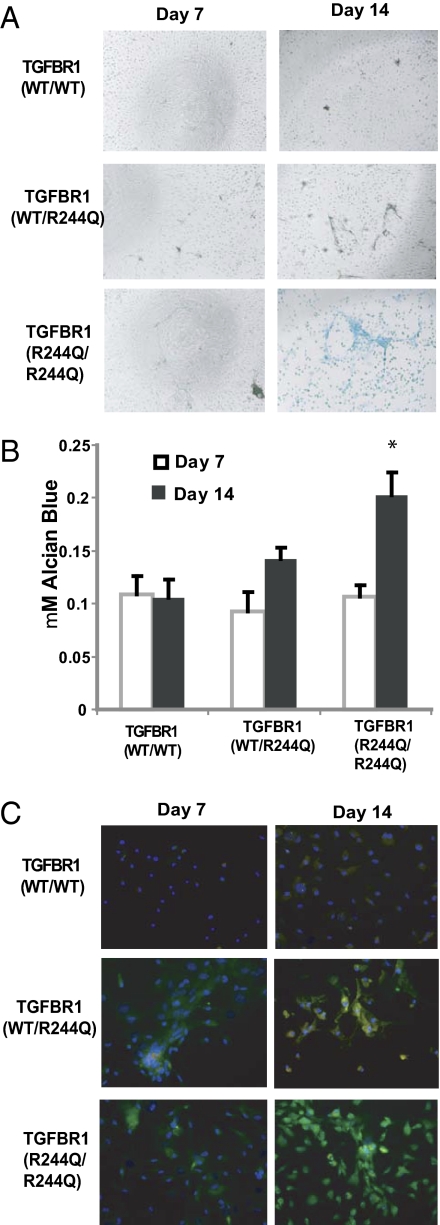
We isolated BMSCs from mice of all three genotypes and examined their ability to undergo spontaneous chondrogenesis under low serum conditions. Although no Alcian blue staining was observed after 7 d, small nodules, similar to the cartilage islands in the ear, were seen in BMSCs from heterozygous TGFBR1wt/R244Q mice and more substantially in BMSCs from TGFBR1R244Q/R244Q mice (Fig. 4A). By day 14, these structures had increased in size and showed significantly more Alcian blue staining in the homozygous mutants than in the wild-type controls (Fig. 4B). At both time points there were no significant differences in the cell numbers or the number of colony-forming units in the two cell populations. We confirmed an increase in two chondrocyte-specific genes by high content imaging, type II collagen (2.2-fold) and Sox9 (3.4-fold), in the BMSC cultures from the mutant TGFBR1 mice (Fig. 4C). Taken together these data suggest that the accelerated wound-healing response in the mutant mouse strain could be due in part to an increase in chondrogenesis from resident mesenchymal stem or progenitor cells.
Fig. 4.
Mutant TGFBR1 mice BMSC have accelerated chondrogenesis in vitro. (A) Alcian blue staining (indicative of cartilage proteoglycans) demonstrated condensing and nodule formation in the mutant TGFBR1 mice by day 7. At day 14 substantially increased Alcian blue staining was further observed in cells from the mutant TGFBR1 mice compared with wild-type mice. (B) Spectophometric quantification of Alcian blue staining. *P < 0.05, data pooled from six wells per genotype. (C) BMSC stained with anti-type II collagen (green), anti-sox9 (red), and DAPI demonstrate an increase in cartilage-specific protein expression in the mutant TGFBR1 BMSC. The data were quantified by high content imaging platform (Perkin-Elmer) to determine a 2.2-fold increase in type II collagen staining and a 3.4-fold increase in Sox9 expression in the TGFBR1 mutant cells compared with the control.
Discussion
Ear hole wound healing is a well-established model to assess soft tissue healing and regeneration (25). Our data show that a single base change in TGFBR1 can accelerate wound healing in slow-healing strains, exemplified here by C57BL6 and BALB/c. Importantly, fast healing in the TGFBR1R244Q strain is associated with recapitulation of tissue structures, including cartilage, hair follicles, and sebaceous glands (instead of fibrosis), which is akin to regeneration in amphibians. In support of our findings, genetic deletion of Smad3, a signal mediator of the TGF-β signaling network, delays ear hole wound healing in the mouse (26), whereas ectopic overexpression of Smad3 accelerates ear hole wound healing in the rabbit (27, 28). In axolotls, TGF-β1 is up-regulated in regenerating limbs, and treatment of the amputated limb with a TGFBR1 inhibitor, SB-431542, blocks limb regeneration (29). In a zebrafish fin regeneration study, SB-431542 also inhibited blastema formation (30), which suggests that TGF-β signaling could at least be partially required for the process. It would be of interest to examine whether the MRL/MpJ mice have altered regulation of the TGF-β pathway compared with strains with slower-healing phenotypes.
Wound healing in mammals is a complex process involving the surrounding matrix, tissue architecture, resident cell types, growth factors, and secreted proteins (1). The TGF-β superfamily includes a variety of members that are involved in these diverse processes (31–33). For example, TGF-β family members can act as attractants and/or inhibitors of fibroblast/keratinocyte migration or immune cell infiltration, increase collagen production, and modulate matrix turnover (31–33). Depending on the specific context, for example, wounding methods and host immune status, TGF-β family members have been described as both positive and negative regulators of the wound-healing process (31–33).
Furthermore, each TGF-β isoform has specific roles in wound healing. For example, in the embryo, wound healing takes place in the absence of a scar, in part owing to the precise regulation of TGF-β1, -2, and -3 secretion and localization (32), which have opposing effects. It is likely that the anti-scarring effects of TGF-β3 are mediated by TGFBR1 and subsequent feedback regulation or cross-talk with other signaling pathways. Preliminary studies using our TGFBR1 mutant mice in a dorsal split wound-healing model show a small but significant acceleration in closure of the wounded site, on the basis of the total wound size after 7 d of healing. In contrast with our data in the ear, the healing architecture and histological appearance in this model do not dramatically differ between mutant and wild-type controls. This might reflect the different healing processes that predominate in wounds of the back skin compared with the ear, which in turn might require diverse signaling pathways or different signaling thresholds.
This is a unique genetic mouse model to demonstrate that a monogenic mutation can affect regenerative traits. During the preparation of this manuscript, Dr. Heber-Katz’s group also published a similar regenerative phenotype in p21−/− mice (34). In our model the partial activation of TGFBR1 signaling may lead to accelerated chondrogenesis in vitro, the development of cartilage islands in vivo, and regenerative healing of ear wounding in mice. However, other potential R244Q-dependent molecular and cellular changes may also contribute to this trait. Further studies of this TGFBR1 mutant mouse line may help to elucidate the regulation of wound healing and lead to improved treatments for burns, skin ulcers, or surgical scars through specific regulation of TGFBR1 (31–33). In addition, ear hole wound healing has been correlated with other soft tissue regeneration traits in MRL/MpJ mice, such as heart, cartilage, cornea, or retinal regeneration (35–38); thus this TGFBR1 mutant mouse strain may help to understand the balance between remodeling, scar formation, and regeneration in these healing processes.
Materials and Methods
Generation of ENU Mice, Husbandry, and Phenotyping.
C57BL/6 ENU mice were generated as previously described (39). All animals were housed and bred at the Genomics Institute of the Novartis Research Foundation (GNF). The experimental protocols were in compliance with animal welfare regulations and approved by the Institutional Animal Care and Use Committee at GNF. Five-week-old animals were wounded with a 2-mm through-and-through hole in the central cartilaginous part of each ear. The hole diameter was measured 5 wk later.
Mapping, Sequencing of TGFBR1, and Genotyping of Mice.
All exons of TGFBR1, plus an average of 100 bp into surrounding introns, were PCR amplified from mouse genomic DNA and sequenced on an Applied Biosystems 3730xl using standard reagents and protocols (Applied Biosystems). Genotyping SNPs (n = 329) identified between C57BL/6 and BALB/c were typed using matrix-assisted laser desorption ionization-time-of-flight single-base extension reactions (iPLEX; Sequenom).
Genomic DNA Deep Sequencing.
A custom sequence capture array (Nimblegen) was designed from base positions 46,553,370–51,099,633 on mouse chromosome 4 (University of California, Santa Cruz build mm9). Repetitive regions of the genome were masked, resulting in coverage of 3,974,040 bases, or 87.4%. The July 2010 version of the Nimblegen protocol for Paired-End Illumina Libraries was used to enrich for this genomic region in genomic DNA from a pool of five affected mice. DNA was paired-end sequenced for 60 cycles on a Genome Analyzer GAIIx (Illumina) using the TruSeq Clust Gen Kit v5 (Illumina) and the TruSeq Kit v5 (Illumina) for cluster generation and sequencing, respectively. Each sequencing read was separately aligned against the mouse genome with BWA (version 0.5.8a) (40). At least five sequencing reads covered 99.9% of the targeted bases in the genomic interval. SAMTools (version 1.12a) (41) was used to call sequence variants. Homozygous SNPs were annotated with ANNOVAR (42). Sanger sequencing was performed on the highest-quality SNPs and compared with a wild-type C57BL/6 mouse from our colony.
Ear Histology and Immunohistochemistry.
Mouse ears were fixed in 10% phosphate-buffered formalin for 24 h, then embedded in paraffin and cut into 5 μm sections. The slides were stained with hematoxylin and eosin or Masson’s trichrome blue. The Masson’s trichrome blue method stains cell cytoplasm in red, nuclei in black, and collagen in blue. For immunohistochemistry, sections were deparaffinized, hydrated, and antigen retrieved with the Decloaking Chamber (Biocare), blocked for endogenous peroxidase, and incubated with a primary antibody against PCNA (BD Transduction Laboratories) or aggrecan (Millipore), followed by an enhanced secondary antibody detection system, the envision kit (Dako), and enhanced diaminobenzidine (Dako), and counterstained with hematoxylin (Vector Laboratories), according to the manufacturer’s instructions.
Additional methods describing constructs, cell culture, transfection, Western blot, TaqMan, MicroArray, TGF-β1 measurement, and bone marrow transplantation are included in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Joan Massagué for providing L17 cells and p3TP-luc plasmid, Dr. Michael Cooke and Susan Sutton for bone marrow transplantation, Dr. James Watson for histology studies, Dr. Jianwei Che for generating Fig. 2C, Ying Sun, Guorong Guo, and Dr. Allen Li for technical assistance, Carie Jackson for FACS analysis of blood cells, Glenn Federe and Whitney Barnes for deep sequencing, and Richard Eddins and Alejandra Rocha for animal handling. This work was supported by the Novartis Research Foundation and the Skaggs Institute for Chemical Biology (P.G.S.). This is manuscript 19998 of the Scripps Research Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111056108/-/DCSupplemental.
References
- 1.Gurtner GC, Werner S, Barrandon Y, Longaker MT. Wound repair and regeneration. Nature. 2008;453:314–321. doi: 10.1038/nature07039. [DOI] [PubMed] [Google Scholar]
- 2.Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. 2007;21:1292–1315. doi: 10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- 3.Sánchez Alvarado A, Tsonis PA. Bridging the regeneration gap: Genetic insights from diverse animal models. Nat Rev Genet. 2006;7:873–884. doi: 10.1038/nrg1923. [DOI] [PubMed] [Google Scholar]
- 4.Christen B, Robles V, Raya M, Paramonov I, Belmonte JC. Regeneration and reprogramming compared. BMC Biol. 2010;8:5. doi: 10.1186/1741-7007-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rojas-Muñoz A, et al. ErbB2 and ErbB3 regulate amputation-induced proliferation and migration during vertebrate regeneration. Dev Biol. 2009;327:177–190. doi: 10.1016/j.ydbio.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 6.Whitehead GG, Makino S, Lien CL, Keating MT. fgf20 is essential for initiating zebrafish fin regeneration. Science. 2005;310:1957–1960. doi: 10.1126/science.1117637. [DOI] [PubMed] [Google Scholar]
- 7.Heber-Katz E, Leferovich JM, Bedelbaeva K, Gourevitch D. Spallanzani’s mouse: A model of restoration and regeneration. Curr Top Microbiol Immunol. 2004;280:165–189. doi: 10.1007/978-3-642-18846-6_5. [DOI] [PubMed] [Google Scholar]
- 8.Metcalfe AD, Willis H, Beare A, Ferguson MW. Characterizing regeneration in the vertebrate ear. J Anat. 2006;209:439–446. doi: 10.1111/j.1469-7580.2006.00632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heber-Katz E, et al. Regeneration in MRL mice: Further genetic loci controlling the ear hole closure trait using MRL and M.m. Castaneus mice. Wound Repair Regen. 2004;12:384–392. doi: 10.1111/j.1067-1927.2004.012308.x. [DOI] [PubMed] [Google Scholar]
- 10.Masinde GL, et al. Identification of wound healing/regeneration quantitative trait loci (QTL) at multiple time points that explain seventy percent of variance in (MRL/MpJ and SJL/J) mice F2 population. Genome Res. 2001;11:2027–2033. doi: 10.1101/gr.203701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McBrearty BA, Clark LD, Zhang XM, Blankenhorn EP, Heber-Katz E. Genetic analysis of a mammalian wound-healing trait. Proc Natl Acad Sci USA. 1998;95:11792–11797. doi: 10.1073/pnas.95.20.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yu H, Mohan S, Masinde GL, Baylink DJ. Mapping the dominant wound healing and soft tissue regeneration QTL in MRL x CAST. Mamm Genome. 2005;16:918–924. doi: 10.1007/s00335-005-0077-0. [DOI] [PubMed] [Google Scholar]
- 13.Huse M, et al. The TGF beta receptor activation process: An inhibitor- to substrate-binding switch. Mol Cell. 2001;8:671–682. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- 14.Huse M, Chen YG, Massagué J, Kuriyan J. Crystal structure of the cytoplasmic domain of the type I TGF beta receptor in complex with FKBP12. Cell. 1999;96:425–436. doi: 10.1016/s0092-8674(00)80555-3. [DOI] [PubMed] [Google Scholar]
- 15.Cárcamo J, Zentella A, Massagué J. Disruption of transforming growth factor beta signaling by a mutation that prevents transphosphorylation within the receptor complex. Mol Cell Biol. 1995;15:1573–1581. doi: 10.1128/mcb.15.3.1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 17.Gorelik L, Flavell RA. Transforming growth factor-beta in T-cell biology. Nat Rev Immunol. 2002;2:46–53. doi: 10.1038/nri704. [DOI] [PubMed] [Google Scholar]
- 18.Wan YY, Flavell RA. Regulatory T cells, transforming growth factor-beta, and immune suppression. Proc Am Thorac Soc. 2007;4:271–276. doi: 10.1513/pats.200701-020AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan YY, Flavell RA. ‘Yin-Yang’ functions of transforming growth factor-beta and T regulatory cells in immune regulation. Immunol Rev. 2007;220:199–213. doi: 10.1111/j.1600-065X.2007.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harty M, Neff AW, King MW, Mescher AL. Regeneration or scarring: An immunologic perspective. Dev Dyn. 2003;226:268–279. doi: 10.1002/dvdy.10239. [DOI] [PubMed] [Google Scholar]
- 21.Kench JA, et al. Aberrant wound healing and TGF-beta production in the autoimmune-prone MRL/+ mouse. Clin Immunol. 1999;92:300–310. doi: 10.1006/clim.1999.4754. [DOI] [PubMed] [Google Scholar]
- 22.Davis TA, Lennon G. Mice with a regenerative wound healing capacity and an SLE autoimmune phenotype contain elevated numbers of circulating and marrow-derived macrophage progenitor cells. Blood Cells Mol Dis. 2005;34:17–25. doi: 10.1016/j.bcmd.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 23.Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res. 1998;238:265–272. doi: 10.1006/excr.1997.3858. [DOI] [PubMed] [Google Scholar]
- 24.Mackay AM, et al. Chondrogenic differentiation of cultured human mesenchymal stem cells from marrow. Tissue Eng. 1998;4:415–428. doi: 10.1089/ten.1998.4.415. [DOI] [PubMed] [Google Scholar]
- 25.Clark LD, Clark RK, Heber-Katz E. A new murine model for mammalian wound repair and regeneration. Clin Immunol Immunopathol. 1998;88:35–45. doi: 10.1006/clin.1998.4519. [DOI] [PubMed] [Google Scholar]
- 26.Arany PR, et al. Smad3 deficiency alters key structural elements of the extracellular matrix and mechanotransduction of wound closure. Proc Natl Acad Sci USA. 2006;103:9250–9255. doi: 10.1073/pnas.0602473103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kloeters O, et al. Alteration of Smad3 signaling in ischemic rabbit dermal ulcer wounds. Wound Repair Regen. 2007;15:341–349. doi: 10.1111/j.1524-475X.2007.00236.x. [DOI] [PubMed] [Google Scholar]
- 28.Sumiyoshi K, Nakao A, Setoguchi Y, Okumura K, Ogawa H. Exogenous Smad3 accelerates wound healing in a rabbit dermal ulcer model. J Invest Dermatol. 2004;123:229–236. doi: 10.1111/j.0022-202X.2004.22730.x. [DOI] [PubMed] [Google Scholar]
- 29.Lévesque M, et al. Transforming growth factor: Beta signaling is essential for limb regeneration in axolotls. PLoS ONE. 2007;2:e1227. doi: 10.1371/journal.pone.0001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jaźwińska A, Badakov R, Keating MT. Activin-betaA signaling is required for zebrafish fin regeneration. Curr Biol. 2007;17:1390–1395. doi: 10.1016/j.cub.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 31.Faler BJ, Macsata RA, Plummer D, Mishra L, Sidawy AN. Transforming growth factor-beta and wound healing. Perspect Vasc Surg Endovasc Ther. 2006;18:55–62. doi: 10.1177/153100350601800123. [DOI] [PubMed] [Google Scholar]
- 32.O’Kane S, Ferguson MW. Transforming growth factor beta s and wound healing. Int J Biochem Cell Biol. 1997;29:63–78. doi: 10.1016/s1357-2725(96)00120-3. [DOI] [PubMed] [Google Scholar]
- 33.Roberts AB. Transforming growth factor-beta: Activity and efficacy in animal models of wound healing. Wound Repair Regen. 1995;3:408–418. doi: 10.1046/j.1524-475X.1995.30405.x. [DOI] [PubMed] [Google Scholar]
- 34.Bedelbaeva K, et al. Lack of p21 expression links cell cycle control and appendage regeneration in mice. Proc Natl Acad Sci USA. 2010;107:5845–5850. doi: 10.1073/pnas.1000830107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueno M, et al. Accelerated wound healing of alkali-burned corneas in MRL mice is associated with a reduced inflammatory signature. Invest Ophthalmol Vis Sci. 2005;46:4097–4106. doi: 10.1167/iovs.05-0548. [DOI] [PubMed] [Google Scholar]
- 36.Leferovich JM, et al. Heart regeneration in adult MRL mice. Proc Natl Acad Sci USA. 2001;98:9830–9835. doi: 10.1073/pnas.181329398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fitzgerald J, et al. Evidence for articular cartilage regeneration in MRL/MpJ mice. Osteoarthritis Cartilage. 2008;16:1319–1326. doi: 10.1016/j.joca.2008.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Tucker B, Klassen H, Yang L, Chen DF, Young MJ. Elevated MMP expression in the MRL mouse retina creates a permissive environment for retinal regeneration. Invest Ophthalmol Vis Sci. 2008;49:1686–1695. doi: 10.1167/iovs.07-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miosge LA, Blasioli J, Blery M, Goodnow CC. Analysis of an ethylnitrosourea-generated mouse mutation defines a cell intrinsic role of nuclear factor kappaB2 in regulating circulating B cell numbers. J Exp Med. 2002;196:1113–1119. doi: 10.1084/jem.20020959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. 1000 Genome Project Data Processing Subgroup: The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.