Abstract
The protease activity of the paracaspase Malt1 contributes to antigen receptor-mediated lymphocyte activation and lymphomagenesis. Malt1 activity is required for optimal NF-κB activation, but little is known about the responsible substrate(s). Here we report that Malt1 cleaved the NF-κB family member RelB after Arg-85. RelB cleavage induced its proteasomal degradation and specifically controlled DNA binding of RelA- or c-Rel–containing NF-κB complexes. Overexpression of RelB inhibited expression of canonical NF-κB target genes and led to impaired survival of diffuse large B-cell lymphoma cell lines characterized by constitutive Malt1 activity. These findings identify a central role for Malt1-dependent RelB cleavage in canonical NF-κB activation and thereby provide a rationale for the targeting of Malt1 in immunomodulation and cancer treatment.
Keywords: signal transduction, T-cell receptor
The antigen receptor-driven activation of the nuclear factor-kappa B (NF-κB) signaling pathway has recently gained considerable interest because genetic deficiencies in this pathway are linked to immune deficiencies, whereas aberrant constitutive NF-κB activation is associated with the development of autoimmune disease and neoplastic disorders (1–4).
The NF-κB family of transcription factors comprises five transcription factors that share a Rel homology domain (RHD) required for DNA binding and homo- or heterodimerization (1, 5). The transcriptionally active form of NF-κB is a heterodimer containing a member with an RHD (p50 or p52) and one with an RHD and an additional transcription activation domain (RelA, RelB, or c-Rel). NF-κB family members are present in the cytoplasm in an inactive form that can be mobilized by either the classical (canonical) or alternative (noncanonical) pathway. The classical pathway is thought to control the activation of p50–RelA and p50–c-Rel complexes by stimulus-dependent degradation of inhibitor of κB (IκB) proteins that bind these complexes and prevent their nuclear translocation. The alternative pathway, on the other hand, controls the generation of transcriptionally active p52–RelB complexes through stimulation-induced processing of the p52 precursor p100 (5).
T-cell receptor (TCR)-induced NF-κB activation has been shown to rely on the activation of both RelA- and c-Rel–containing NF-κB complexes that occur in a timely staggered manner (6). However, the exact mechanism controlling persistent RelA–p50 and c-Rel–p50 activation in T cells remains not well characterized.
Biochemical and genetic studies have identified an essential role for proteins of the Carma1/Bcl-10/Malt1 (CBM) signaling module in TCR-induced NF-κB activation (2, 7–9). The current model of CBM-dependent NF-κB activation suggests that antigen triggering leads to the phosphorylation of Carma1 by PKC family and probably additional kinases, inducing a conformational change in Carma1 that allows it to recruit preformed Bcl-10/Malt1 complexes (7, 10). Malt1 is thought to control the activation of the IKK complex by binding to the ubiquitin ligase TRAF6 and subsequent TRAF6-mediated ubiquitination of Bcl-10, Malt1, and TRAF6 itself (11). This may then lead to physical recruitment of the IKK complex, whose catalytically inactive IKKγ subunit contains a ubiquitin binding motif. Subsequently, TRAF6-dependent activation of the Ser/Thr kinase TAK1 is thought to promote activation of the IKKβ kinase subunit through its phosphorylation (11).
Recently, we and others have demonstrated a protease activity for Malt1, which is required for optimal NF-κB activation in B and T cells (12, 13) and for the NF-κB–dependent growth of cell lines derived from diffuse large B-cell lymphoma (DLBCL) of the activated B-cell (ABC) subtype (14, 15). Interestingly, Malt1 inhibition has no influence on IKK-dependent IκBα phosphorylation (13, 16), suggesting that another, IKK-independent mechanism of NF-κB activation must contribute to canonical NF-κB activation in lymphocytes.
Here, we have investigated the mechanism by which the Malt1 protease activity controls NF-κB activation. We identify RelB as a Malt1 substrate and show that Malt1-dependent RelB cleavage led to its degradation, which resulted in enhanced RelA- and c-Rel–dependent DNA binding in activated lymphocytes and ABC DLBCL cell lines. RelB overexpression inhibited expression of NF-κB target genes, like IL-2 and Bcl-xL, and impaired survival of ABC DLBCL cell lines. These findings identify a new role for the Malt1 protease and its substrate RelB in lymphocyte activation and lymphoma development.
Results
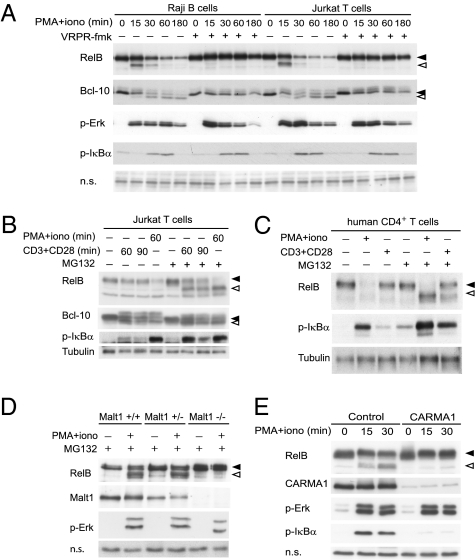
Inhibition of Malt1 activity affects NF-κB activation without noticeable effect on IκBα or IκBβ phosphorylation or degradation (13, 16) (Fig. S1), suggesting that the protease activity of Malt1 controls NF-κB activation independently of the IKK complex. To start to investigate the molecular mechanism underlying Malt1 protease-dependent NF-κB activation, we initially assessed the possibility of a direct, Malt1-dependent cleavage of NF-κB subunits as a mechanism to regulate their activity in antigen receptor- or phorbol myristate acetate (PMA)- and ionomycin-stimulated lymphocytes. Initially, we stimulated human T- and B-cell lines with PMA and ionomycin, which mimic many aspects of lymphocyte activation and potently activate Malt1, leading to cleavage of the Malt1 substrate Bcl-10 (Fig. 1A). This treatment also led to the appearance of a smaller migrating isoform of RelB of ∼55 kDa and subsequent RelB degradation (Fig. 1A). Pretreatment of the cells with the Malt1 inhibitor, under conditions that efficiently block Bcl-10 cleavage, prevented both the stimulation-induced RelB cleavage and its degradation (Fig. 1A). RelB degradation was dependent on the proteasome, because pretreatment of cells with the proteasome inhibitor MG132 led to accumulation of the faster-migrating RelB isoform in both Jurkat T cells and primary human CD4+ T cells, which were stimulated using either PMA and ionomycin or agonistic anti-CD3 and anti-CD28 antibodies (Fig. 1 B and C). Taken together, this suggests that RelB is first cleaved by Malt1 and the resulting cleavage fragment is subsequently degraded by the proteasome. Similar results were obtained using primary mouse splenocytes (Fig. S2). Cleavage of RelB was strictly dependent on Malt1, because splenocytes of Malt1-deficient mice (17) showed no evidence of RelB cleavage (Fig. 1D). RelB cleavage and degradation were inhibited by the PKC inhibitor bisindoleylmaleimide VIII (BIM VIII) (Fig. S3) and by Carma1 silencing (Fig. 1E), which both efficiently block the formation of the CBM complex and thus Malt1 activation (13). IKK activity on the other hand was not essential for the induction of RelB cleavage, because RelB was still efficiently cleaved in Nemo-deficient Jurkat cells (Fig. S3).
Fig. 1.
Malt1 cleaves RelB. (A–E) Indicated lymphocyte cell lines or primary lymphocytes were stimulated using PMA and ionomycin (PMA+iono) or cross-linked anti-CD3 and anti-CD28 antibodies (CD3/CD28) for 30 min or the indicated times, and postnuclear lysates were analyzed by Western blotting (WB). Where indicated, cells were treated with the Malt1 inhibitor VRPR-fmk or solvent control before and during stimulation. In some experiments (B–E), cells were pretreated with the proteasome inhibitor MG132 or solvent alone (DMSO) before stimulation. (D) Splenocytes from Malt1 wild-type, heterozygous, or knockout mice were pretreated and stimulated as indicated. (E) Jurkat T cells were transduced with either control or Carma1 shRNA and pretreated with MG132 before stimulation. In all figure parts, black and open arrowheads indicate uncleaved and cleaved forms, respectively, of the Malt1 substrates RelB and Bcl-10. Data are representative of at least three (A–C) or two (D and E) independent experiments.
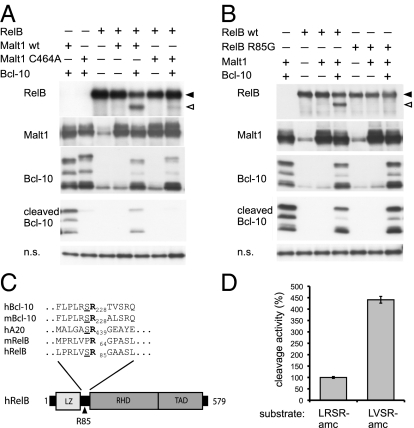
To identify the potential cleavage site in RelB and confirm the requirement of active Malt1 for RelB cleavage, we next assessed the effect of wild-type (WT) Malt1 and a catalytically inactive version of Malt1 on RelB cleavage in 293T cells. In this system, the presence of Bcl-10 is required for optimal Malt1 activation (13). Coexpression of RelB with WT Malt1 and Bcl-10 induced the formation of a C-terminal RelB cleavage fragment of 55 kDa (Fig. 2A). Cleavage was strongly impaired when a catalytically inactive form of Malt1 was used, in which the active site residue Cys-464 is mutated into Ala (C464A) (Fig. 2A). Under these conditions, a small degree of residual RelB cleavage could be observed, which is most likely due to the activation of endogenous Malt1 by Bcl-10 overexpression. Because Malt1 cleaves its two known substrates, Bcl-10 and A20, after an Arg residue that is preceded by a Ser residue (12, 13), we next mutated two Arg residues in the N-terminal part of RelB, Arg-49 and Arg-85 in the sequence motifs AVSR49 and LVSR85, respectively, and assessed the capacity of Malt1 to cleave these constructs. Mutation of Arg-85, but not Arg-49, abolished the capacity of Malt1 to cleave RelB in 293T cells and in Raji cells (Fig. 2B and Fig. S4).
Fig. 2.
Malt1 cleaves human RelB after Arg-85. (A and B) Expression constructs for Malt1 (WT or catalytically inactive C464A mutant), Bcl-10, and RelB (WT or noncleavable R85G mutant) were cotransfected in the indicated combinations into 293T cells and postnuclear lysates were analyzed by Western blotting as indicated. (C) Alignment of Malt1-dependent cleavage sites in human and mouse Bcl-10 (hBcl-10 and mBcl-10), human A20 (hA20), and human RelB (hRelB). Cleavage occurs after the conserved Arg residue (R) shown in bold, the preceding Ser residue (S) is underlined. (D) The in vitro proteolytic activity of Strep–Malt1 bound to Streptactin beads was assessed by incubation with either LRSR-amc or LVSR-amc (corresponding to the Bcl-10 or RelB cleavage site, respectively) in cleavage assay buffer at 30 °C. Graph shows cleavage activity using LVSR-amc relative to LRSR-amc. Results are expressed as means ± SD (n = 3). Data are representative of at least three (A and B) or two (D) independent experiments. Black and open arrowheads indicate uncleaved and cleaved proteins, respectively.
These findings identify RelB as a Malt1 substrate and suggest that Malt1 preferentially cleaves its substrates after a conserved amino acid sequence motif (hydrophobic-variable-Ser-Arg), which is present in three identified human Malt1 substrates (Fig. 2C). Of note, cleavage of both human and mouse RelB occurs C terminal to the leucine zipper motif, a predicted protein–protein interaction motif, whose exact functional relevance for RelB is unknown, and which is not present in other Rel family members (18, 19). To further assess the specificity of Malt1-mediated RelB cleavage, we made use of a previously developed in vitro cleavage assay, in which recombinant oligomeric Malt1 is incubated with fluorogenic tetrapeptide substrates (13). When comparing identical molar concentrations of the two peptides, Malt1 cleaved the RelB peptide, LVSR-amc, with more than fourfold higher efficiency than the Bcl-10 peptide, LRSR-amc (Fig. 2D). None of the two peptides was cleaved by the corresponding C464A mutant of Malt1.
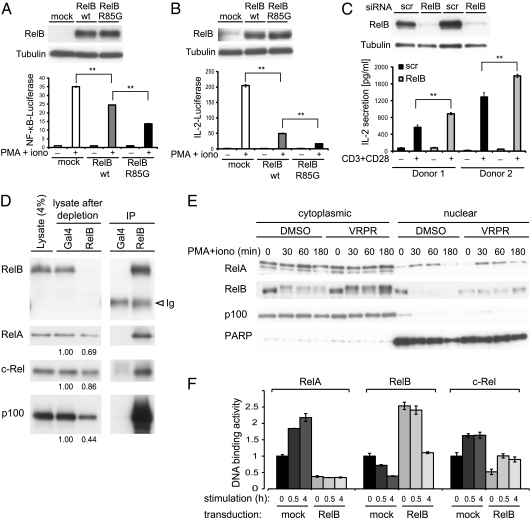
So far, our data suggested that Malt1 activation leads to lowered RelB expression in activated lymphocytes, but it was unclear how this would affect Malt1-dependent NF-κB activation. To assess the effect of varying RelB levels on the activity of NF-κB in lymphocytes, we first expressed WT or noncleavable (R85G) forms of RelB in Jurkat T cells. RelB overexpression clearly inhibited NF-κB induction by PMA and ionomycin stimulation (Fig. 3A), which depends on Malt1 activity (13). The inhibitory effect was more pronounced upon overexpression of noncleavable RelB, which is fully resistant to Malt1-dependent cleavage (Fig. 3A). A similar inhibitory effect of RelB was observed using a Jurkat T-cell line stably transfected with a reporter gene construct containing the promotor of the NF-κB target gene IL-2 (Fig. 3B). To confirm the inhibitory role of RelB in antigen receptor-mediated IL-2 production, we silenced its expression in human primary naïve T cells by siRNA. Stimulation of the RelB-silenced cells with agonistic anti-CD3 and anti-CD28 antibodies (Fig. 3C) or PMA and ionomycin (Fig. S5) resulted in increased IL-2 levels. To gain insight into how RelB affects classical, RelA-, and c-Rel–dependent NF-κB activation, we initially tested the effect of increasing amounts of RelB on Malt1- and Bcl-10–induced NF-κB activation in reporter assays using 293T cells. In this experimental system, RelB did not promote activation of canonical NF-κB, but clearly inhibited Malt1- and Bcl-10–induced NF-κB activation (Fig. S6). Interestingly, RelB expression also inhibited NF-κB activation by RelA itself (Fig. S6), suggesting that inhibition occurs directly at the level of the transcription factors. Consistent with this idea, we detected constitutive binding of endogenous or Flag-tagged RelB to a significant proportion of RelA and c-Rel in Jurkat T cells (Fig. 3D and Fig. S7). RelB stabilization by the Malt1 inhibitor did not interfere with RelA translocation into the nucleus after stimulation with PMA and ionomycin, but led to higher RelB levels in the nuclear fractions (Fig. 3E). We therefore assessed the effect of RelB on the DNA-binding capacity of individual NF-κB subunits using an ELISA-based assay. Interestingly, RelB overexpression clearly correlated with impaired DNA binding of both RelA and c-Rel in unstimulated and stimulated Jurkat T cells (Fig. 3F). Collectively, these findings suggest that RelB does not prevent nuclear translocation of RelA, but rather inhibits the binding of RelA and c-Rel to NF-κB binding sites, both by sequestering RelA and c-Rel in RelB-containing complexes and by competition for DNA binding.
Fig. 3.
RelB inhibits expression of NF-κB gene targets in T cells. (A and B) Jurkat T cells (A) or Jurkat T cells stably expressing an IL-2 reporter construct (B) were transfected with the indicated expression constructs, treated with or without PMA and ionomycin for 16 h, and assessed for NF-κB activation (A) or IL-2 gene activation (B) by luciferase reporter assay. The corresponding expression level of the constructs is shown by Western blot. Data are representative of at least three independent experiments. (C) Naïve human primary T cells were transfected with either scrambled (scr) or RelB siRNA. Forty-eight hours after transfection, cells were stimulated with agonistic anti-CD3 and anti-CD28 antibodies and IL-2 secretion was quantified 16 h later by ELISA. (D) Jurkat cells were lysed and assessed for the binding of endogenous RelB to the indicated proteins. Position of the Ig heavy chain (Ig) is indicated. (E) Jurkat T cells were pretreated for 30 min with the Malt1 inhibitor VRPR-fmk and stimulated as indicated. (F) Jurkat cells transduced with RelB or control vector were stimulated for the indicated times with PMA and ionomycin and DNA binding of indicated Rel subunits was assessed by TransAM DNA-binding ELISA. In all figure parts, results are expressed as means ± SD from three independent experiments (**P ≤ 0.005).
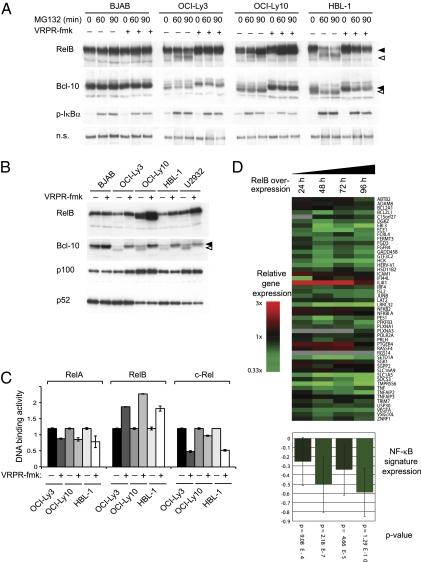
Next, we assessed RelB cleavage and expression levels in lysates from DLBCL cell lines that were incubated in the presence or absence of the Malt1 inhibitor. In the ABC DLBCL lines OCI-Ly3, OCI-Ly10, HBL-1, and U2932, which are characterized by constitutive Malt1 activity and cleavage of the Malt1 substrate Bcl-10 (14, 15), treatment of the cells with MG132 led to spontaneous accumulation of the cleaved isoform of RelB, which could be prevented by pretreatment with the Malt1 inhibitor (Fig. 4A). Malt1 inhibition for 48 h resulted in a strong increase in total RelB levels in the ABC DLBCL lines OCI-Ly3, OCI-Ly10, HBL-1, and U2932, whereas this was not the case for the germinal centre B-cell like (GCB) DLBCL cell line BJAB, in which Malt1 is not active (Fig. 4B). RelB stabilization by the Malt1 inhibitor led to an increase in both total and nuclear RelB levels (Fig. S8). Noticeably, the processing of p100 to the active form p52 was not at all affected by Malt1 inhibition in all GCB and ABC DLBCL cell lines tested (Fig. 4B). Higher RelB levels in the cells correlated with an increase in DNA binding and a decrease in RelA and c-Rel recruitment (Fig. 4C). These findings suggest that RelB specifically controls the transcriptional activity of RelA and c-Rel at the level of their DNA-binding capacity, independently of IκBα or IκBβ. To confirm an inhibitory role on the classical NF-κB signaling pathway by RelB stabilization, we quantified the NF-κB target gene expression using gene arrays. After retroviral transduction, we inducibly expressed FLAG-tagged RelB in the ABC DLBCL line OCI-Ly10, which is characterized by constitutive MALT1 activity (14, 15). RelB expression for 24, 48, 72, and 96 h led to a significantly reduced expression of the majority of target genes that are also down-regulated upon treatment of the cells with the IKK inhibitor MLN120b (Fig. 4D). NF-κB target genes that were affected by RelB overexpression include the prosurvival gene BCL2L1 (also known as Bcl-xL), whose down-regulation could also be confirmed at the protein level in three different ABC DLBCL lines (Fig. S9).
Fig. 4.
RelB is constitutively cleaved in ABC DLBCL cell lines. (A) DLBCL cell lines of the GCB (BJAB) or ABC type (all others) were treated with the proteasome inhibitor MG132 or solvent control for the indicated times, in the presence or absence of the Malt1 inhibitor VRPR-fmk. Postnuclear lysates were analyzed for expression and cleavage of RelB. Bcl-10 and IκBα blots serve as control for efficient Malt1 and proteasome inhibition, respectively. A nonspecific band (n.s.) of the anti–p-IκBα blot serves as a loading control (B) DLBCL cell lines of the GCB (BJAB), or ABC subtype (all others) were pretreated with 75 μM of the Malt1 inhibitor VRPR-fmk or solvent control for 48 h, and RelB expression was assessed by Western blotting of postnuclear cell extracts. Black and open arrowheads indicate uncleaved and cleaved proteins, respectively. Data in A and B are representative of at least three independent experiments. (C) OCI-Ly3, OCI-Ly10, and HBL-1 cells were treated with 50 μM of the Malt1 inhibitor VRPR-fmk or solvent control for 16 h, and DNA binding of individual NF-κB subunits was assessed by TransAM assay. Data are expressed as means ± SD (n = 3). (D) Gene expression profiling of the ABC DLBCL cell line OCI-Ly10 was performed following induction of RelB expression at the indicated time points (24, 48, 72, and 96 h). Gene expression changes are depicted according to the color scale shown. (Lower) Significance of the RelB-mediated down-regulation of the NF-κB signature. Gene expression measurements for the component genes in the NF-κB signature (Upper) were averaged for each of the indicated time points.
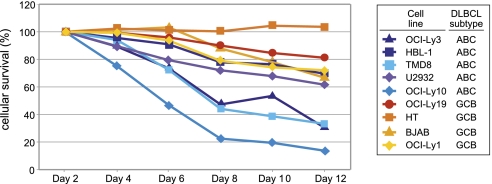
Collectively, these findings suggest that RelB inhibits the expression of canonical NF-κB target genes not only in activated lymphocytes but also in ABC DLBCL lines. The Malt1-dependent reduction of RelB levels in ABC DLBCL lines (Fig. 4B), together with the observed inhibition of NF-κB target genes by RelB overexpression (Fig. 4D and Fig. S9), suggested that Malt1-dependent down-modulation of RelB levels might be essential for the survival of cells derived from ABC DLBCL, which critically depend on the CBM pathway (14, 15, 20). Consistent with this hypothesis, RelB overexpression clearly led to reduced survival in four out of five cell lines derived from ABC DLBCL, while showing little or no effect on the survival of four cell lines derived from GCB DLBCL, which do not depend on CBM-dependent signaling for their growth (Fig. 5) (14, 15, 20).
Fig. 5.
RelB overexpression impairs the survival of ABC DLBCL lines. Cell lines derived from the indicated DLBCL subtypes were retrovirally transduced with an expression vector for human RelB together with green fluorescent protein (GFP). Live GFP+ cells were enumerated by flow cytometry on the indicated days postretroviral transduction and normalized to the value at day 2 following retroviral transduction. Data are representative of three independent experiments.
Discussion
Here, we provide several lines of evidence for a negative regulatory role of RelB in lymphocyte activation that is controlled by the Malt1-dependent proteolytic degradation of RelB. First, Malt1 cleaved RelB at a consensus site that resembled previously identified Malt1 cleavage sites in Bcl-10 and A20. Second, RelB cleavage induced the subsequent, proteasome-dependent degradation of RelB within a time frame that correlated with persistent nuclear translocation of NF-κB. Third, continuous RelB degradation was observed in ABC DLBCL cell lines that were characterized by constitutive Malt1 activity. Fourth, RelB overexpression reduced the DNA binding of RelA and c-Rel, most likely via the formation of transcriptionally inactive RelB/RelA and RelB/c-Rel complexes and/or competition for nuclear NF-κB binding sites and so led to impaired expression of NF-κB target genes, which correlates with impaired survival of cell lines derived from ABC DLBCL.
The identification of RelB as a negative regulator of RelA- and c-Rel–dependent target genes may seem initially surprising, but is indeed consistent with a number of previously published findings. A negative regulatory role of RelB is clearly supported by the phenotype of RelB knockout mice, which show multiorgan inflammation with increased IL-2 levels (21). This inflammatory phenotype is reduced when RelB-deficient mice are crossed to T-cell–deficient mice, strongly suggesting a pathogenic role for hyperactive T cells in the RelB-deficient mice (22). Indeed, RelB-deficient mice show increased proliferation of naïve T cells in response to stimulation with antigen-presenting cells (23).
Our findings also correlate with an earlier report that had identified RelB as i-Rel (inhibitor of Rel) and attributed a negative regulatory role to RelB in NF-κB activation (24). Initial experiments using cell lines had suggested that RelB represses the transcription of p50/RelA-dependent reporter genes in activated Jurkat cells (24). Subsequently, both, positive and negative regulatory effects of RelB have been reported in a variety of different studies assessing differing target or reporter genes (25–29). Therefore, the nature of the NF-κB target genes suppressed by RelB seems to strongly depend on the individual promotor studied and the cellular trigger involved.
How exactly does RelB inhibit classical NF-κB activation in lymphocytes? Our data strongly suggest that Malt1 activity does not control IKK-mediated phosphorylation of IκBα (a rapid, Malt1 activity-independent event that takes only a few minutes) and its subsequent proteasomal degradation, but most likely controls the DNA-binding capacity of a subset of NF-κB1 complexes through degradation of RelB. We propose that Malt1 scaffold-dependent activation of the IKK complex controls initial nuclear translocation of NF-κB1 via degradation of IκB proteins, while Malt1 protease activity controls the prolonged DNA-binding capacity of a subset of RelA- and c-Rel-containing NF-κB1 complexes via degradation of RelB. The molecular mechanisms by which RelB inhibits classical NF-kB1 complexes is not well understood but our experiments together with previously reported findings (25, 28, 30) support the idea of competition on the DNA-binding side or direct physical interaction between RelB and RelA or RelB and c-Rel.
How T-cell activation triggers RelB cleavage by Malt1 is going to be an interesting aspect of future work. Some insight into this question is provided by an earlier study that had reported a signal-induced phosphorylation of RelB, which was proposed to precede RelB degradation by the proteasome (30). However, mutation of the respective phosphorylation site did not prevent RelB cleavage by Malt1 in 293T cells, indicating that RelB phosphorylation at this site is not absolutely required for its Malt1-dependent cleavage (Fig. S10).
In conclusion, our study identifies the Malt1-dependent RelB cleavage as a key event in the antigen receptor-mediated activation of the canonical NF-κB pathway and the associated immune responses. Moreover, our data support the idea of targeting the protease activity of Malt1 as a rational strategy for immunomodulation and the treatment of specific forms of lymphoma.
Materials and Methods
Antibodies.
Primary antibodies used in this study include rabbit monoclonal anti-RelB and rabbit polyclonal anti–c-Rel (Cell Signaling), monoclonal mouse anti-p100 (Upstate), mouse anti-phospho ERK (Sigma), rabbit anti-phospho-IκBα (5A5; Cell Signaling), rabbit anti-IκBα (Sigma), mouse anti-tubulin (Sigma), rabbit anti–Bcl-10 (H-197; Santa Cruz), rabbit anti-Carma1 (Cell Signaling), mouse anti-p100 (Upstate), rabbit anti-PARP (Cell Signaling), and an affinity-purified anti-Malt1 antibody generated against a GST–Malt1 fusion protein comprising amino acids 1–824 of human Malt1 (13). An antibody specific for cleaved Bcl-10 was generated as described (14). Western blots were revealed using HRP-coupled goat anti-mouse or anti-rabbit antibodies (Jackson Immunoresearch).
Plasmids.
Malt1 constructs (WT and C464A) have been described (13). RelB expression constructs (WT, R49G, and R85G) were generated by standard PCR amplification approaches on human RelB cDNA (kindly provided by G. Natoli, IEO, Milano, Italy) and cloned into vectors derived from pCR3 (Invitrogen) with an N-terminal tag.
Cells, Transfection, and Transduction.
The human DLBCL cell lines SUDHL-4, SUDHL-6, HBL-1, and U2932 were grown as described (14). TMD-8, OCI-Ly1, and OCI-Ly19 were cultured in Iscove's modified Dulbecco medium with 10% FCS. The cell lines Jurkat, BJAB, Raji, Ramos, and HT were grown at 37 °C in RPMI 1640 supplemented with 10% FCS and antibiotics. 293T cells were grown in DMEM supplemented with 10% FCS and antibiotics. A Jurkat clone stably expressing an IL-2 reporter plasmid was used for IL-2 reporter assays (kindly provided by G. Zenke, Novartis, Basel, Switzerland). Jurkat cells expressing a Flag-tagged construct encompassing wild-type or R85G mutant human RelB were generated by lentiviral transduction and puromycin selection as described (14). The lentiviral silencing vector specific for Carma1 has been described (13). Expression of RelB in DLBCL lines was achieved by retroviral transduction as described previously (20), using cell lines engineered to express the murine ecotropic retroviral receptor for efficient transduction and the bacterial tetracycline repressor for doxycycline-inducible expression. RelB cDNA was either inserted into a modified version of the inducible pRetroSUPER dual-promoter vector or in a noninducible pMSCV vector. Primary human CD4+ T cells were isolated from blood samples of healthy volunteers, using anti–CD4-coated MACS beads according to the manufacturer's description (Miltenyi). Naïve T cells were isolated with the help of anti–CD45RA-coated MACS beads, after prior removal of B cells with anti–CD19-coated MACS beads (Miltenyi). Transfections with siRNA (ON-TARGETplus SMARTpool; Thermo Scientific) were performed using Human T-Cell Nucleofector kit (Amaxa) according to the manufacturer's protocol. Mouse total splenocytes were obtained from C57BL/6 mice that were bred in the animal facilities of the University of Lausanne according to institutional guidelines for animal care. Splenocytes were isolated by mechanical tissue separation and subsequent red blood cell lysis in hypotonic Tris-NH4Cl buffer. Cells were washed with PBS and resuspended in prewarmed RPMI 1640 supplemented with 10% FCS for 10 min at 37 °C before stimulation. Transient transfection of 293T cells was performed as described before (13). Transient transfection of Jurkat T cells was achieved by electroporation of 107 cells in DPBS (Gibco) at 220 V and 950 μF (GenePulser Xcell; Biorad).
Cell Activation and Lysis.
Lymphocyte stimulation was initiated by addition of PMA (10 ng/mL; Alexis) and ionomycin (1 μM; Calbiochem). T cells were stimulated with a combination of anti-human CD3ε (10 μg/mL of OKT3; Apotech) and anti-CD28 (10 μg/mL of CD28.2; Immunotech) antibodies, immediately followed by addition of 5 μg/mL of cross-linking goat anti-mouse antibody (The Jackson Laboratory). In some stimulation experiments, cells were preincubated with 75 μM of z-VRPR-fmk (Alexis Biochemicals), 5 μM MG132 (Calbiochem), or with corresponding volumes of solvent for the indicated times at 37 °C. Pelleted cells were lysed in Tris-NaCl lysis buffer containing 1% Nonidet P-40, protease inhibitors (Complete; Roche) and phosphatase inhibitors (mixtures I and II; Sigma). Transfected 293T cells were washed in cold Tris-NaCl buffer and lysed in Tris-NaCl lysis buffer. Postnuclear cell lysates were boiled with reducing SDS sample buffer and analyzed by SDS/PAGE. For optimal resolution, samples were analyzed using Anderson gels as described (13).
Cell Fractionation.
After washing the cells with cold PBS, cells were incubated for 5 min on ice with a hypotonic lysis buffer (10 mM Hepes, 1.5 mM MgCl2, 300 mM sucrose, 0.5% Nonidet P-40, 10 mM KCl, and 0.5 mM DTT). Cells were centrifuged at 9,000 rpm for 1 min and supernatants were transferred in a new cup (cytosolic fraction). The pellets were resuspended in a buffer containing 20 mM Hepes, 100 mM NaCl, 0.2 mM EDTA, 20% glycerol, 100 mM KCl, and 0.5 mM DTT) and exposed to three freeze/thaw cycles and 10 s of sonication. Nuclear fractions were collected after centrifugation for 10 min at 13,000 rpm.
Gene Reporter, DNA Binding, and ELISAs.
Activation of gene transcription was assessed by transient transfection of cells with a NF-κB firefly luciferase reporter construct together with a Renilla luciferase vector (phRL-TK). Cells were lysed in passive lysis buffer (Promega) and luciferase activity assessed using dual luciferase assay (Promega) on a TD-20/20 luminometer (Turner Design). DNA binding of Rel subunits was determined using TransAM Assay (Active Motif) according to the manufacturer's instructions. Measurement of IL-2 concentrations by ELISA was done as described (13).
Gene Expression Profiling.
Gene expression profiling was performed in the ABC DLBCL cell line OCI-Ly10 after retroviral transduction with RelB cDNA after 24, 48, 72, and 96 h. RNA was prepared using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Gene expression was measured using whole-genome Agilent 4 × 44K gene expression arrays (Agilent Technologies), following the manufacturer's protocol. Signals from RelB cDNA expressing OCI-Ly10 cells (labeled with Cy5) were compared with signals from uninduced OCI-Ly10 cells (labeled with Cy3). A previously developed NF-κB target gene signature was applied to the gene expression data to determine the effect of RelB overexpression on the NF-κB signaling pathway (31). Genes that were significantly down-regulated were determined by paired t test (P < 0.05).
In Vitro Protease Activity Assay.
N-terminally Strep-tagged Malt1 was transfected into 293T cells. Cells were lysed in Tris-NaCl lysis buffer, postnuclear lysates were precleared for 30–60 min on Sepharose 6B beads, and Strep–Malt1 was then precipitated for 20 min using Streptactin beads (IBA BioTAGnology). Malt1 protease activity in the precipitates was determined upon addition of 25 μM Ac-LRSR-amc or Ac-LVSR-amc (Peptides International) in cleavage assay buffer (12) and incubation at 30 °C for 4 h, using a Synergy microplate reader (BioTek).
Supplementary Material
Acknowledgments
We thank Fabio Martinon, Etienne Meylan, and Ben Marsland for critical reading of the manuscript; Vishva Dixit for Malt1-deficient mice; Fabien Rebeaud, Myriam Tapernoux, Stephan Duss, Richard Iggo, and Gioacchino Natoli for plasmids; Robert Weil for cell lines; and Leonhard Heinz for technical advice. This work was supported by grants from the Swiss National Science Foundation, the Swiss Cancer League, the foundations Leenaards, Pierre Mercier, and Emma Muschamp (to M.T.), and by a fellowship of the Faculty of Biology and Medicine of the University of Lausanne (to J.E.C.). G.L. was supported by grants from the German Research Foundation, the Deutsche Krebshilfe, and the Else Kröner-Fresenius Stiftung.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105020108/-/DCSupplemental.
References
- 1.Li Q, Verma IM. NF-kappaB regulation in the immune system. Nat Rev Immunol. 2002;2:725–734. doi: 10.1038/nri910. [DOI] [PubMed] [Google Scholar]
- 2.Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4:348–359. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 3.Staudt LM, Dave S. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv Immunol. 2005;87:163–208. doi: 10.1016/S0065-2776(05)87005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baud V, Karin M. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov. 2009;8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vallabhapurapu S, Karin M. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol. 2009;27:693–733. doi: 10.1146/annurev.immunol.021908.132641. [DOI] [PubMed] [Google Scholar]
- 6.Kane LP, Lin J, Weiss A. It's all Rel-ative: NF-kappaB and CD28 costimulation of T-cell activation. Trends Immunol. 2002;23:413–420. doi: 10.1016/s1471-4906(02)02264-0. [DOI] [PubMed] [Google Scholar]
- 7.Rawlings DJ, Sommer K, Moreno-García ME. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol. 2006;6:799–812. doi: 10.1038/nri1944. [DOI] [PubMed] [Google Scholar]
- 8.Schulze-Luehrmann J, Ghosh S. Antigen-receptor signaling to nuclear factor kappa B. Immunity. 2006;25:701–715. doi: 10.1016/j.immuni.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 9.Lin X, Wang D. The roles of CARMA1, Bcl10, and MALT1 in antigen receptor signaling. Semin Immunol. 2004;16:429–435. doi: 10.1016/j.smim.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 10.Rueda D, Thome M. Phosphorylation of CARMA1: The link(er) to NF-kappaB activation. Immunity. 2005;23:551–553. doi: 10.1016/j.immuni.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 11.Thome M. Multifunctional roles for MALT1 in T-cell activation. Nat Rev Immunol. 2008;8:495–500. doi: 10.1038/nri2338. [DOI] [PubMed] [Google Scholar]
- 12.Coornaert B, et al. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat Immunol. 2008;9:263–271. doi: 10.1038/ni1561. [DOI] [PubMed] [Google Scholar]
- 13.Rebeaud F, et al. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol. 2008;9:272–281. doi: 10.1038/ni1568. [DOI] [PubMed] [Google Scholar]
- 14.Hailfinger S, et al. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci USA. 2009;106:19946–19951. doi: 10.1073/pnas.0907511106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferch U, et al. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J Exp Med. 2009;206:2313–2320. doi: 10.1084/jem.20091167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Düwel M, et al. A20 negatively regulates T cell receptor signaling to NF-kappaB by cleaving Malt1 ubiquitin chains. J Immunol. 2009;182:7718–7728. doi: 10.4049/jimmunol.0803313. [DOI] [PubMed] [Google Scholar]
- 17.Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302:1581–1584. doi: 10.1126/science.1090769. [DOI] [PubMed] [Google Scholar]
- 18.Dobrzanski P, Ryseck RP, Bravo R. Both N- and C-terminal domains of RelB are required for full transactivation: Role of the N-terminal leucine zipper-like motif. Mol Cell Biol. 1993;13:1572–1582. doi: 10.1128/mcb.13.3.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 20.Ngo VN, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 21.Weih F, et al. Multiorgan inflammation and hematopoietic abnormalities in mice with a targeted disruption of RelB, a member of the NF-kappa B/Rel family. Cell. 1995;80:331–340. doi: 10.1016/0092-8674(95)90416-6. [DOI] [PubMed] [Google Scholar]
- 22.Weih F, et al. Both multiorgan inflammation and myeloid hyperplasia in RelB-deficient mice are T cell dependent. J Immunol. 1996;157:3974–3979. [PubMed] [Google Scholar]
- 23.Ishimaru N, Kishimoto H, Hayashi Y, Sprent J. Regulation of naive T cell function by the NF-kappaB2 pathway. Nat Immunol. 2006;7:763–772. doi: 10.1038/ni1351. [DOI] [PubMed] [Google Scholar]
- 24.Ruben SM, et al. I-Rel: A novel rel-related protein that inhibits NF-kappa B transcriptional activity. Genes Dev. 1992;6:745–760. doi: 10.1101/gad.6.5.745. [DOI] [PubMed] [Google Scholar]
- 25.Marienfeld R, et al. RelB forms transcriptionally inactive complexes with RelA/p65. J Biol Chem. 2003;278:19852–19860. doi: 10.1074/jbc.M301945200. [DOI] [PubMed] [Google Scholar]
- 26.Xia Y, Pauza ME, Feng L, Lo D. RelB regulation of chemokine expression modulates local inflammation. Am J Pathol. 1997;151:375–387. [PMC free article] [PubMed] [Google Scholar]
- 27.Xia Y, et al. RelB modulation of IkappaBalpha stability as a mechanism of transcription suppression of interleukin-1alpha (IL-1alpha), IL-1beta, and tumor necrosis factor alpha in fibroblasts. Mol Cell Biol. 1999;19:7688–7696. doi: 10.1128/mcb.19.11.7688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saccani S, Pantano S, Natoli G. Modulation of NF-kappaB activity by exchange of dimers. Mol Cell. 2003;11:1563–1574. doi: 10.1016/s1097-2765(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 29.Wright CW, Duckett CS. The aryl hydrocarbon nuclear translocator alters CD30-mediated NF-kappaB-dependent transcription. Science. 2009;323:251–255. doi: 10.1126/science.1162818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marienfeld R, et al. Signal-specific and phosphorylation-dependent RelB degradation: A potential mechanism of NF-kappaB control. Oncogene. 2001;20:8142–8147. doi: 10.1038/sj.onc.1204884. [DOI] [PubMed] [Google Scholar]
- 31.Kloo B, et al. Critical role of PI3K signaling for NF-kappaB-dependent survival in a subset of activated B-cell-like diffuse large B-cell lymphoma cells. Proc Natl Acad Sci USA. 2011;108:272–277. doi: 10.1073/pnas.1008969108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.