Abstract
The nucleotide-binding domain and leucine-rich repeats containing proteins (NLRs) serve as immune receptors in both plants and animals. Overaccumulation of NLRs often leads to autoimmune responses, suggesting that the levels of these immune receptors must be tightly controlled. However, the mechanism by which NLR protein levels are regulated is unknown. Here we report that the F-box protein CPR1 controls the stability of plant NLR resistance proteins. Loss-of-function mutations in CPR1 lead to higher accumulation of the NLR proteins SNC1 and RPS2, as well as autoactivation of immune responses. The autoimmune responses in cpr1 mutant plants can be largely suppressed by knocking out SNC1. Furthermore, CPR1 interacts with SNC1 and RPS2 in vivo, and overexpressing CPR1 results in reduced accumulation of SNC1 and RPS2, as well as suppression of immunity mediated by these two NLR proteins. Our data suggest that SKP1-CULLIN1-F-box (SCF) complex-mediated stability control of plant NLR proteins plays an important role in regulating their protein levels and preventing autoimmunity.
Plants and animals rely on innate immunity to defend against microbial pathogen infections. Although plant surface-residing receptors often recognize common features of the microbes, intracellular receptors detect specific effectors from pathogens to initiate a downstream defense cascade (1). Remarkably, plants and animals use immune sensors with similar structural features, such as nucleotide-binding (NB) and leucine-rich repeats (LRR) domains (2). These immune receptors are commonly named Nod-like Receptors (NLRs; or nucleotide-binding and leucine-rich repeat-containing) after the human innate immunity receptor Nod1 and Nod2 (3). In plants, they are designated as NB-LRR resistance proteins (NLR R proteins). Upon detection of specific pathogen effectors, R proteins mount a quick and robust reaction, usually culminating in a hypersensitive response, a type of programmed cell death, to defend against and restrict further spread of the pathogen (1). Because of the detrimental effects of R protein activation on plant cell growth and development, normally R protein-mediated immunity has to be under multiple levels of tight negative control. Overaccumulation of R proteins often leads to autoimmunity, implying the importance of stability control of R protein levels. In addition, gain-of-function mutants of NLRs, such as snc1 (suppressor of npr1-1, constitutive 1) and ssi4 (suppressor of salicylic acid insensitivity of npr1-5, 4), can render the NLR proteins constitutively active without pathogen interaction (4, 5). These mutations are speculated to enhance the stability or activities of the NLRs. Constitutive expression of defense marker Pathogenesis Related (PR) genes, enhanced pathogen resistance, and altered plant development, such as dwarfism, are general features of plant autoimmunity.
The Arabidopsis genome contains about 170 genes encoding NLRs (6), with either Toll/IL-1 receptor (TIR) or coiled-coil (CC) domains at their N terminus. The detailed activation mechanism of NLR R proteins is unclear. During the past decade, intensive studies on RAR1, SGT1, and HSP90 using genetic and biochemical approaches established these components as members of a protein complex required for chaperone activities to properly fold and stabilize NLR R proteins (7, 8). Interestingly, mammalian SGT1 and HSP90 were also shown to be required for NLR-mediated immune responses. Some NLRs, including Nod1 and Nod2, form complexes with HSP90 and SGT1 (9).
Aside from the positive roles SGT1 plays in R protein folding, it is also involved in the negative regulation of R protein stability. A loss-of-function mutation in SGT1b restores reduced accumulation of CC-type NLR RPS5 in rar1 mutant plants (10). In addition, accumulation of SNC1 is increased in sgt1b mutant plants. Recently, the evolutionarily conserved SRFR1 was shown to interact directly with SGT1 (11). Loss-of-function of SRFR1 results in increased accumulation of SNC1 and activation of SNC1-mediated defense responses (11, 12). However, the mechanism on how SGT1 and SRFR1 negatively regulate the accumulation of R proteins is unclear.
Besides interactions with HSP90 and RAR1, SGT1 was also shown to associate with SKP1 and CULLIN1 (CUL1) (7, 13), members of the SCF (SKP1-CUL1-F-box protein) E3 ubiquitin ligase complex that targets specific substrate proteins for ubiquitination and most often subsequent protein degradation. The F-box protein usually interacts directly with the protein substrate and serves as the substrate determinant of SCF. The association between SGT1 and SCF suggests potential connections between SCF and R protein-mediated immunity. Here we provide experimental evidence that Arabidopsis F-box protein CPR30/CPR1 targets NLR proteins SNC1 and RPS2 for degradation, revealing how some NLR R protein levels are controlled mechanistically.
Results
snc1 Mutation Affects the Stability of SNC1.
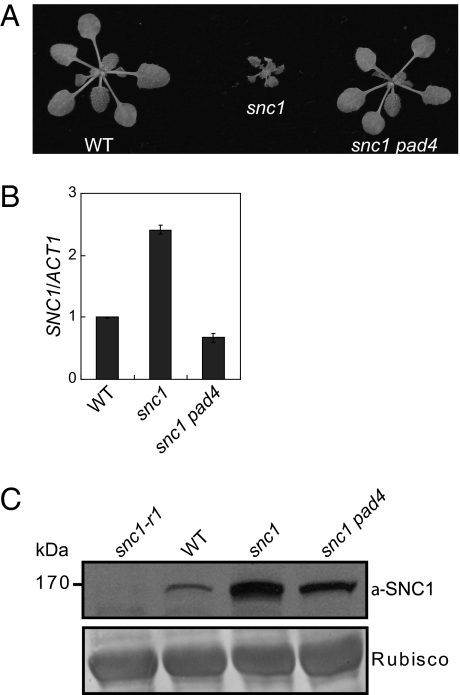
The gain-of-function mutant snc1 carries a mutation in SNC1, an NLR R-like gene that leads to constitutive activation of defense responses in Arabidopsis (4, 14). Like other TIR-type R proteins, snc1 signals through PAD4 (15). Mutations in PAD4 can completely suppress the dwarfism of snc1 caused by autoimmunity (Fig. 1A). To investigate how the E552 to K552 change in the linker region causes constitutive activation of SNC1, we first examined the snc1 transcript level in the mutant and found only a moderate increase (Fig. 1B). This increase is fully suppressed when PAD4, an essential signaling component downstream of SNC1, is mutated (Fig. 1B), indicating that the increased snc1 transcription is caused by feedback up-regulation from downstream defense signaling. With the availability of an antibody specific to the endogenous SNC1 protein (11), we analyzed snc1 protein levels in snc1 and snc1 pad4 plants. To our surprise, snc1 protein levels are still considerably higher in the snc1 pad4 double-mutant plants than that in the wild-type (Fig. 1C), suggesting that the snc1 mutation renders the R protein more stable.
Fig. 1.
Increased snc1 protein levels in snc1 and snc1 pad4 double mutant. (A) Morphology of wild-type Col (WT), snc1 (gain-of-function allele of SNC1), and snc1 pad4 double-mutant plants (14). The picture was taken with 4-wk-old soil-grown plants. (B) qRT-PCR analysis of SNC1 transcript levels in WT, snc1, and snc1 pad4 plants. (C) Western blot analysis of SNC1/snc1 protein levels in snc1-r1 (a loss-of-function deletion allele of SNC1), WT, snc1, and snc1 pad4 plants (14). Rubisco levels serve as loading control.
cul1-7 Exhibits Increased SNC1 Level and Constitutive Defense Responses.
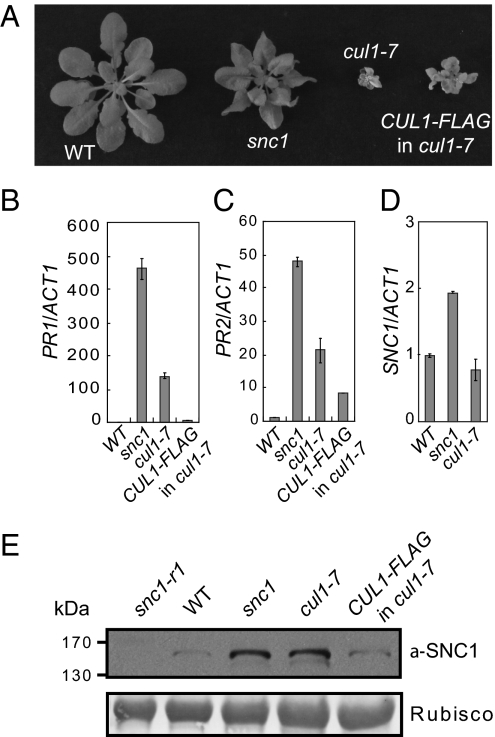
Because a mutation in SGT1b also results in increased SNC1 protein level and SGT1 associates with SKP1 and CUL1 (7, 11, 13), two shared members of the SCF E3 ubiquitin ligase complex, we asked whether the stability of SNC1 could be controlled by SCF-mediated protein degradation. In Arabidopsis, null mutations in CUL1 are lethal (16). A partial loss-of-function allele of CUL1, cul1-7, was found to exhibit a dwarf phenotype similar to snc1 (17) (Fig. 2A). In cul1-7, the T510 to I510 substitution seems to affects the C terminus of the protein, as well as the stability of CUL1, thus resulting in the misregulation of SCFs. The mutant exhibited accumulation of target proteins of many known SCF complexes, including AUS/IAA1 and RGA1 for auxin and gibberellic acid signaling, respectively (17). Additionally, cul1-7 plants express high levels of defense-marker PR genes, PR1 and PR2 (Fig. 2 B and C), suggesting that the mutation causes activation of defense responses. The dwarfism of cul1-7 is more severe than that of snc1 and can be partially complemented by a transgene expressing the CUL1-FLAG fusion protein (17) (Fig. 2A). Although the transcript level of SNC1 in cul1-7 is not significantly different from that of wild-type (Fig. 2D), Western blot analysis showed that the SNC1 protein level in cul1-7 was much higher than that in wild-type and this increased accumulation of SNC1 can be partially complemented by the CUL1-FLAG transgene (Fig. 2E), indicating that CUL1 contributes to the control of SNC1 levels. Furthermore, quantitative RT-PCR analysis of a variety of known R genes indicates that transcript levels of these R genes are not significantly increased in cul1-7 (Fig. S1), suggesting that SCF-mediated protein ubiquitination has little effect on R gene transcription.
Fig. 2.
Increased accumulation of SNC1 protein levels in cul1-7 mutant. (A) Morphology of WT, snc1, cul1-7, and cul1-7 carrying the partially complementing transgene CUL1-FLAG (CUL1-FLAG in cul1-7) (17). The picture was taken with 5-wk-old soil-grown plants. (B–D) qRT-PCR analysis of the expression of PR1 (B), PR2 (C), and SNC1 (D) in the indicated genotypes. (E) Western blot analysis of SNC1/snc1 protein levels in snc1-r1 (a loss-of-function deletion allele of SNC1), WT, snc1, cul1-7, and CUL1-FLAG in cul1-7.
SNC1 Protein Levels Are Increased in cpr1 and cpr30 Mutants.
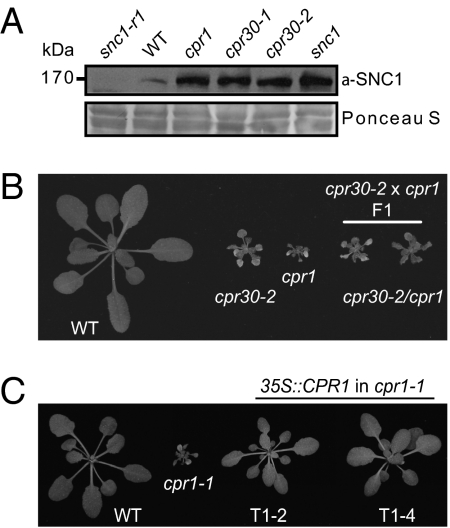
The substrate specificity of the SCF complexes is most often determined by the F-box proteins. Previously it was shown that mutations in the F-box protein, CPR30 [At4g12560; Constitutive PR gene expression, 30 (cpr30)], lead to constitutive activation of PR genes, accumulation of the defense hormone salicylic acid, and a dwarf phenotype strikingly similar to that of snc1 (18) (Fig. S2). Western blot analysis revealed that SNC1 overaccumulates in the cpr30 mutant plants (Fig. 3A). Another well-known mutant with constitutive defense responses and snc1-like morphology is cpr1 (19) (Fig. 3B and Fig. S2). Although cpr1 was isolated over 15 y ago, its identity was unknown. Interestingly, similar to the cpr30 alleles, the SNC1 protein level in cpr1 was much higher than that in wild-type (Fig. 3A). The levels of SNC1 in cpr1 and two alleles of cpr30 are very similar. To exclude the possibility that CPR1 negatively controls SNC1 expression, we checked the SNC1 expression level in cpr1 and cpr1 pad4 mutants. As shown in Fig. S3A, SNC1 transcript level is only slightly increased in cpr1 and this increase is completely reverted by pad4 in the cpr1 pad4 double mutant, suggesting that the small increase of SNC1 transcription is caused by feedback up-regulation from downstream defense signaling. In contrast, SNC1 levels in the cpr1 pad4 double mutants remain much higher than in wild-type and not drastically reduced from that in cpr1 (Fig. S3B), confirming that CPR1 negatively regulates SNC1 protein level. Interestingly, SNC1 levels in cpr30 and cpr30 pad4 are also much higher than that in wild-type (Fig. S3B). Furthermore, quantitative RT-PCR analysis of several other R genes indicates that their transcript levels are not drastically increased in cpr1 or cpr1 pad4 (Fig. S3 C–I), suggesting that CPR1 has little effect on R gene transcription.
Fig. 3.
Both cpr1 and cpr30 are alleles of At4g12560. (A) Western blot analysis of SNC1 protein levels in cpr1 (renamed cpr1-1), cpr30-1 (renamed cpr1-2), and cpr30-2 (renamed cpr1-3). snc1-r1 (a loss-of-function deletion allele of SNC1), WT, and snc1 plants were used as controls. (B) Allelism test between cpr30-2 and cpr1. Morphology of two F1 plants from the cross between cpr30-2 and cpr1, with WT, cpr30-2, and cpr1 plants as controls. (C) Complementation of cpr1-1 by the CPR1 transgene under 35S promoter. Morphologies of two independent transgenic lines (T1-2 and T1-4) are shown.
cpr1 and cpr30 Are Allelic Mutations.
Because cpr1 and cpr30 are both recessive mutations mapped to chromosome 4 (18, 19), we crossed cpr1 and cpr30-2 to test for allelism. As shown in Fig. 3B, cpr1 and cpr30-2 failed to complement in F1, indicating that they are allelic to each other. Because cpr1 was identified as the founding member of the cpr-type mutants, for simplicity and to avoid confusion in the literature, we renamed cpr1 as cpr1-1, the allele obtained by Gou et al. (18) (previously named cpr30-1) as cpr1-2, and the T-DNA allele (SALK_045148; previously named cpr30-2) as cpr1-3. Further sequence analysis of cpr1-1 revealed a G to A point mutation in At4g12560. This mutation is located at an intron-exon junction (Fig. S4), leading to a shift of the splicing site which results in a reading frame change in the gene. When a construct expressing At4g12560 under the control of a 35S promoter was transformed into cpr1-1, all transgenic plants exhibited wild-type morphology (Fig. 3C); this confirms that the cpr1-1 mutant phenotype was caused by the mutation in At4g12560. Phylogenetic analysis of CPR1 and its homologs indicates that they are also present in other higher plants (Fig. S5).
Constitutive Defense Responses in cpr1-3 Are Largely Suppressed by Knocking Out SNC1.
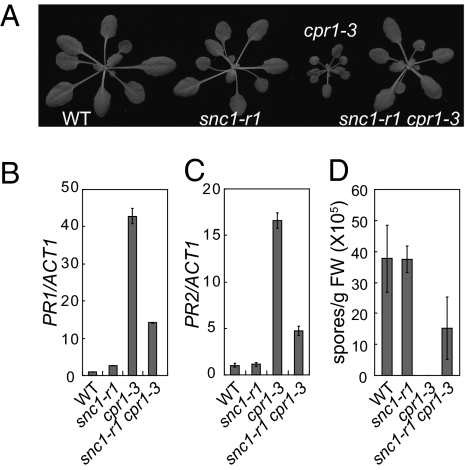
To test whether the increased SNC1 protein accumulation contributes to the activation of defense responses in the cpr1 mutants, we crossed snc1-r1, a loss-of-function deletion allele of SNC1 (14), into cpr1-3. As shown in Fig. 4A, the snc1-r1 mutation reverted cpr1-3 to wild-type morphology. Analysis of PR gene expression showed that constitutive expression of both PR1 and PR2 was reduced in snc1-r1 cpr1-3 (Fig. 4 B and C). In addition, enhanced resistance against the virulent oomycete pathogen Hyaloperonospora arabidopsidis Noco2 in cpr1-3 was also attenuated by the snc1-r1 mutation (Fig. 4D). These data suggest that overaccumulation of SNC1 in cpr1-3 is one of the main factors leading to the cpr1 autoimmune mutant phenotypes.
Fig. 4.
CPR1 regulates SNC1-mediated defense responses. (A) Morphology of WT, snc1-r1 (a loss-of-function deletion allele of SNC1), cpr1-3, and snc1-r1 cpr1-3 mutant plants. (B and C) qRT-PCR analysis of PR1 (B) and PR2 (C) expression in snc1-r1, cpr1-3, and snc1-r1 cpr1-3. (D) Growth of H. arabidopsidis Noco2 spores on the indicated genotypes.
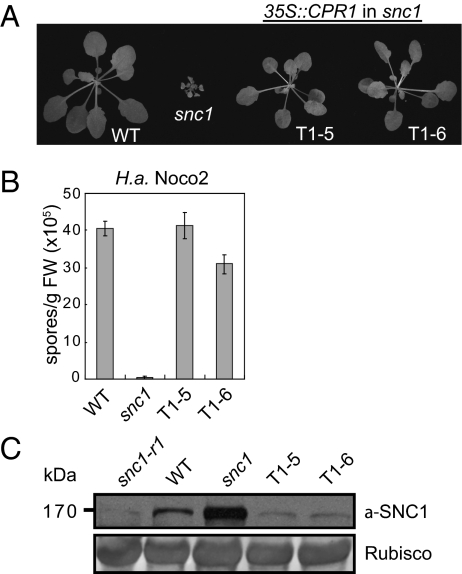
Overexpression of CPR1 Results in Reduced snc1 Protein Levels and Suppression of snc1 Mutant Phenotypes.
Increased accumulation of SNC1 protein in cpr1 mutants and suppression of cpr1-3 phenotypes by snc1-r1 suggest that CPR1 might target SNC1 for degradation. Because the F-box proteins are usually the limiting factor in SCF-mediated target protein degradation, we tested whether overexpression of CPR1 would reduce the accumulation of SNC1 by overexpressing CPR1 in the snc1 mutant background. As shown in Fig. 5A, snc1 transgenic lines T1-5 and T1-6 carrying 35S::CPR1 exhibit wild type-like morphology. Both lines expressed high levels of CPR1 (Fig. S6A) and modestly reduced level of SNC1 (Fig. S6B) because of reduced feedback up-regulation in snc1. In addition, enhanced resistance against H. arabidopsidis Noco2 (Fig. 5B) and constitutive PR gene expression (Fig. S6 C and D) in snc1 were completely suppressed in the CPR1 overexpression lines. Western blot analysis revealed that snc1 protein accumulation was clearly reduced even below the wild-type level in the two CPR1 overexpression lines (Fig. 5C). These data support that CPR1 is the F-box protein targeting SNC1 for degradation.
Fig. 5.
Overexpression of CPR1 in snc1 alleviates the enhanced disease resistance phenotypes of snc1 and reduces the protein levels of snc1. (A) Morphology of WT, snc1, and two independent transgenic lines (T1-5 and T1-6) overexpressing CPR1 in the snc1 mutant background. (B) Growth of H. arabidopsidis Noco2 spores on the indicated genotypes. (C) Western blot analysis of snc1/SNC1 protein levels in the transgenic lines over-expressing CPR1. snc1-r1, WT, and snc1 plants were used as controls.
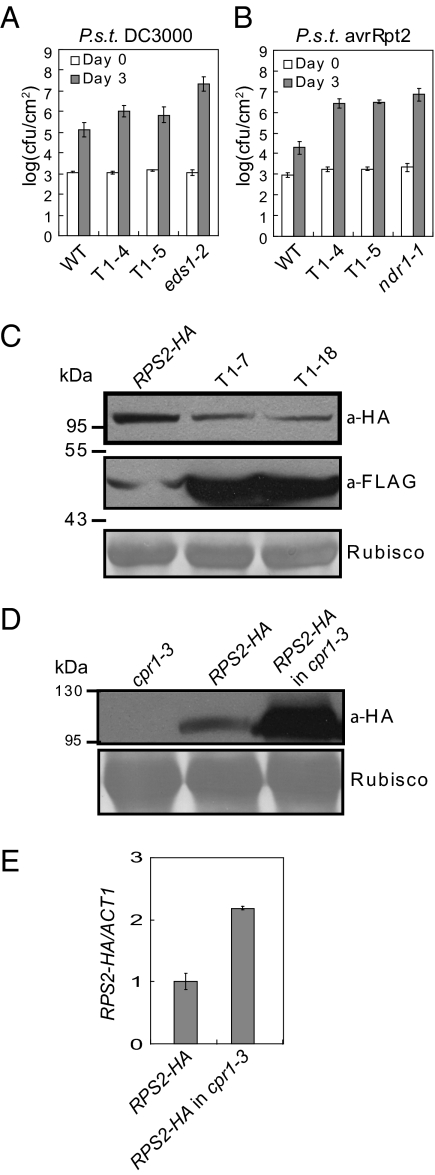
Overexpression of CPR1 Affects R Protein-Mediated Resistance.
Although snc1-r1 completely suppresses the dwarfism of cpr1-3, constitutive PR gene expression and enhanced resistance against H. arabidopsidis Noco2 in cpr1-3 is only partially attenuated by snc1-r1 (Fig. 4 B–D), suggesting that CPR1 may also target R proteins other than SNC1 for degradation. The residual enhanced resistance in snc1-r1 cpr1-3 is probably caused by increased accumulation of the other R proteins. To test this hypothesis, we generated transgenic lines overexpressing CPR1 in the wild-type Columbia (WT) background (Fig. S7A) and challenged the transgenic plants with pathogens carrying different effectors that activate specific R protein-mediated resistance. The two CPR1 overexpression lines were slightly more susceptible to the virulent bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Fig. 6A). In contrast, resistance mediated by RPS2 and RPM1 is severely compromised when CPR1 is expressed at high levels (Fig. 6B and Fig. S7B), suggesting that CPR1 may also target R proteins, such as RPS2 and RPM1, for degradation. Because the expression levels of RPS2 and RPM1 are not significantly affected in these transgenic plants, the defects in RPS2- and RPM1-mediated resistance in CRP1 overexpressing lines are not likely caused by reduced RPS2 and RPM1 transcription (Fig. S8 G and H). Overexpression of CPR1 also has modest effects on resistance mediated by RPS5, RPS4, RPP2, and RPP4 (Fig. S7 C–F). No drastic change of transcript levels of these R genes were observed (Fig. S8 B–I).
Fig. 6.
Regulation of RPS2-mediated resistance and RPS2 protein accumulation by CPR1. (A and B) Growth of P. syringae pv. tomato DC3000 (A) and P. syringae pv. tomato DC3000 carrying avrRpt2 that is recognized by RPS2 (B) in two independent transgenic lines (T1-4 and T1-5) overexpressing CPR1 in wild-type Col background. WT, eds1-2, and ndr1-1 plants were used as controls. (C) Western blot analysis of RPS2-HA protein levels in two CPR1-FLAG transgenic lines (T1-7 and T1-18) expressing CPR1-FLAG under its own promoter in a previously described RPS2-HA transgenic background (20). The weak band in the RPS2-HA lane is the result of nonspecific hybridization from anti-FLAG antibody. (D) Western blot analysis of RSP2-HA protein levels in cpr1-3. The RPS2-HA transgene was crossed into cpr1-3. (E) Expression levels of the RPS2-HA transgene in RPS2-HA and cpr1-3 crossed into RPS2-HA.
CPR1 Regulates the Stability of RPS2.
RPS2 and RPM1 are both CC-type NLR R proteins. RPS2 was chosen for further testing because the RPS2-HA transgenic line was available to us (20) (a kind gift of B. Staskawicz, University of California at Berkeley). To test whether over-expression of CPR1 affects the accumulation of RPS2, we transformed plants expressing the RPS2-HA fusion protein with a construct expressing CPR1 with a C-terminal 3×FLAG tag, under its own promoter. As shown in Fig. 6C, increased expression of CPR1-FLAG protein correlated with reduced accumulation of RPS2-HA. Next we tested whether loss of CPR1 function leads to increased accumulation of RPS2 by crossing cpr1-3 into the RPS2-HA transgenic line. Western blot analysis showed that RPS2-HA protein level dramatically increased in cpr1-3 (Fig. 6D). Because RPS2 transcript level in cpr1-3 was only moderately higher than in wild-type (Fig. 6E), the increase in RPS2-HA protein level is most likely due to increased stability of RPS2-HA in cpr1-3.
A similar approach was used to test for the correlation between CPR1 and RPS4, where minor effect was observed for RPS4-mediated immunity in CPR1 overexpression lines (Fig. S7D). When we transformed the RPS4-HA transgenic line (21) (a gift of Jane Parker, MPI, Köln) with the same CPR1-3×FLAG construct. As shown in Fig. S9, the RPS4-HA level remains the same in cpr1 and CPR1 overexpression backgrounds as in wild-type. In contrast, when the same samples were probed with anti-SNC1 antibody, reverse correlation between CPR1 and SNC1 levels was observed (Fig. S9). Thus CPR1 does not seem to regulate RPS4 stability.
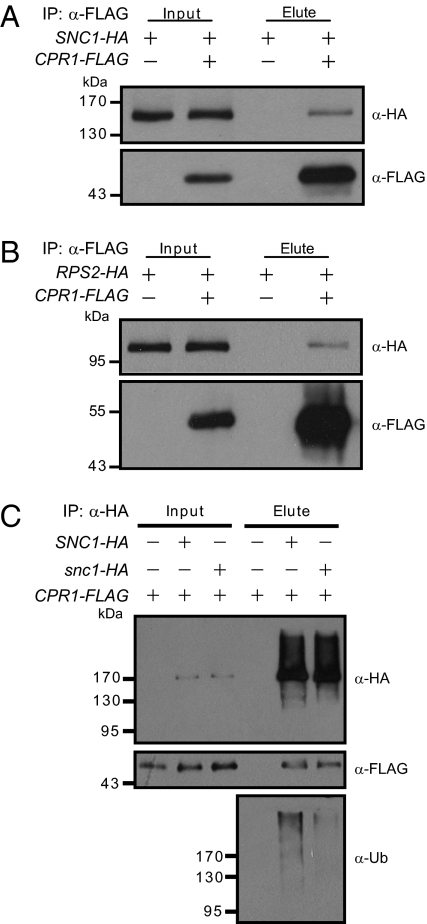
CPR1 Interacts with SNC1 and RPS2 in Vivo.
F-box proteins are known to form SCF complexes with their substrates. The genetic interaction between CPR1 and SNC1, plus the negative correlations between CPR1 protein level and accumulation of SNC1 and RPS2, prompted us to test whether CPR1 associates with SNC1 and RPS2 in vivo. Constructs expressing SNC1-HA and CPR1-FLAG fusion proteins were cotransformed into Arabidopsis mesophyll protoplasts and immunoprecipitation was subsequently performed on the protein extracts using anti-FLAG agarose beads. As shown in Fig. 7A, SNC1-HA coimmunoprecipitated with the CPR1-FLAG protein. When RPS2-HA and CPR1-FLAG were coexpressed in Arabidopsis mesophyll protoplasts, RPS2-HA also coimmunoprecipitated with CPR1-FLAG (Fig. 7B). These data suggest that CPR1 does associate with SNC1 and RPS2 in vivo, probably in SCF complexes, to target SNC1 and RPS2 for degradation.
Fig. 7.
Interactions between CPR1 and SNC1 (A) or RPS2 (B). (A) In vivo pull-down of SNC1-HA by CPR1-FLAG. (B) In vivo pull-down of RPS2-HA by CPR1-FLAG. (C) Immunoprecipitation of SNC1-HA or snc1-HA and ubiquitination levels of SNC1-HA or snc1-HA proteins. Arabidopsis mesophyll protoplasts were transfected with the indicated constructs. Total protein extracts were subjected to immunoprecipitation with anti-FLAG agarose beads (A and B) or anti-HA microbeads (C). Crude lysates (Input) and immunoprecipitated proteins (Elute) were detected with anti-FLAG, anti-HA, or anti-Ub antibodies.
Because the point mutation in snc1 stabilizes the SNC1 protein, we asked whether it caused a reduction of affinity with CPR1. As shown in Fig. 7C, in Arabidopsis mesophyll protoplasts coexpressing CPR1-FLAG and SNC1-HA or snc1-HA, when immunoprecipitation was performed on the protein extracts using anti-HA microbeads, SNC1-HA and snc1-HA are both able to pull down CPR1. The snc1 mutation does not seem to have any obvious effect on interactions between CPR1 and SNC1 in this assay. However, when the immunoprecipitated samples were probed with an anti-ubiquitin (Ub) antibody, much less ubiquitination was observed in the immunoprecipitated snc1-HA, suggesting that SNC1 is a better substrate for ubiquitination than snc1.
Discussion
In plants, SCF-mediated protein degradation is involved in the regulation of diverse biological processes. The most well-studied examples are from auxin signaling. SCFTIR1 and its homologous F-box proteins serve as auxin receptors and modulate Aux/IAA proteins for degradation (22). There is evidence that SCFs are also involved in the regulation of plant immunity. In tobacco, N-mediated resistance response to TMV was compromised when SKP1 was silenced (23). In addition, the tobacco F-box protein ACIF1 is required for the Cf-9– and Cf-4–mediated hypersensitive response, whereas the Arabidopsis F-box protein SON1 plays a negative role in defense (24, 25).
Recently it was shown that mutations in the F-box protein CPR30/CPR1 leads to constitutive expression of PR genes and enhanced pathogen resistance (18), but the mechanism of how CPR1 regulates plant defense responses is unclear. In this study, we provide strong evidence that SCFCPR1 targets the NLR R-like protein SNC1 for degradation to prevent overaccumulation of SNC1 and autoimmunity. Loss-of-function mutations in CPR1 result in increased SNC1 accumulation, and the constitutive defense responses in cpr1 are largely dependent on SNC1. In addition, overexpression of CPR1 reduces the accumulation of SNC1 protein and suppresses the autoimmunity phenotype in snc1. Previously it was shown that SRFR1 and SGT1b are also involved in the negative regulation of SNC1 accumulation (11). Because SRFR1 interacts with SGT1 and SGT1 associates with SKP1 and CUL1 in vivo (7, 11, 13), SRFR1 and SGT1b may regulate the stability of SNC1 through modulating the activity of SCFCPR1. In support of this hypothesis, association of SNC1 and SRFR1 was detected from coimmunoprecipitation experiments in transient expression system in tobacco (12).
In addition to SNC1, SCFCPR1 also negatively regulates the stability of RPS2. Loss of CPR1 function leads to increased RPS2 levels, whereas overexpression of CPR1 reduces the accumulation of RPS2 and severely compromises RPS2-mediated immunity. Furthermore, overexpression of CPR1 abolishes immunity mediated by RPM1 and slightly compromises resistance specified by several other R proteins, such as RPP2 and RPP4, suggesting that SCFCPR1 may also target these R proteins for degradation. In contrast, overexpression of CPR1 hardly affects the resistance mediated by RPS4 and RPS5. Whether the stability of these R proteins is controlled by other F-box proteins remains to be determined. It was surprising to us that CPR1 targets SNC1 and RPS2, two quite different NLR proteins, yet doesn't target RPS4, which is more closely related to SNC1. It is possible that CPR1 may recognize a common feature shared between SNC1 and RPS2, but not with RPS4.
Previous studies on RAR1-SGT1-HSP90 demonstrated that correct folding and maintaining NLR R proteins above a threshold level are required for quick induction of defense responses when under pathogen attack. Meanwhile, the activities and protein levels of NLR R proteins need to be tightly controlled to prevent activation of defense responses without the presence of pathogens because constitutive activation of defense responses is detrimental to growth and development (7). Our study identified SCF-mediated protein degradation as a critical mechanism for regulating the stability of plant NLR R proteins, suggesting that NLR R proteins are maintained at proper levels by balanced activities from the RAR1-SGT1-HSP90–mediated assembly and proteasome-mediated degradation. Because overexpression of NLR immune receptors also results in autoactivation of immune responses in animals, and plants and animal NLRs are structurally similar, it will be interesting to test whether the stability of NLR proteins in animals is also controlled by a similar SCF-mediated mechanism.
Materials and Methods
Plant Growth Conditions and Mutant Phenotypic Characterization.
All plants were grown at 22 °C under a long-day (16-h light/8-h night) regime. Gene-expression analysis was carried out by extracting total RNA from 2-wk-old plate-grown plants. The extracted RNA was then reverse-transcribed to obtain cDNA. Real-time PCR was performed and the expression levels of Actin1, PR1, and PR2 were determined as described previously (14). Infection experiments with P. syringae and H. arabidopsidis (previously Hyaloperonospora parasitica) were performed as previously described (4). H. arabidopsidis infection details were visualized under a light microscope after staining leaves with lactophenol Trypan blue (26).
Construction of Plasmids and Arabidopsis Transformation.
The coding sequence of At4g12560 (CPR1/CPR30) was PCR-amplified using primers 5′ cggGGTACCATGGCGACGATTCCAATGGA 3′ and 5′ CGCggatccTTATAAGACCAGCTTGAATC 3′ from wild-type Col cDNA. The amplified fragment was then digested with KpnI and BamHI and cloned into pHAN-35S to generate pHAN-35S::CPR1. For the construction of pCAMBIA1305-pCPR1::CPR1-3×FLAG, the fragment containing 1,914 bp upstream of the start codon of CPR1 and the CPR1 genomic sequence was amplified by using primers 5′ cggGGTACCaaatcacaagtcacctgacc 3′ and 5′ CGCggatccTAAGACCAGCTTGAATCCTTTGG 3′ from wild-type Col genomic DNA. The fragment was digested with KpnI and BamHI and cloned into pCAMBIA-3×FLAG to generate pCAMBIA1305-pCPR1::CPR1-3×FLAG. The above plasmids were electroporated into Agrobacterium and subsequently transformed into the appropriate Arabidopsis genotypes by floral dipping method (27).
Transient expression vectors pUC19-35S-FLAG-RBS and pUC19-35S-HA-RBS (kindly provided by Jian-Min Zhou, National Institute of Biological Sciences, Beijing, People's Republic of China) containing the cauliflower mosaic virus 35S promoter, 3×FLAG or 3×HA, and a Rubisco Small Subunit terminator were used for transient expression of CPR1 and R protein-coding genes in protoplasts. CPR1, SNC1, and RPS2 coding sequences were PCR-amplified using primers listed in Table S1. The amplified fragments were digested with restriction enzymes indicated in the same table and ligated into pUC19-35S-FLAG-RBS or pUC19-35S-HA-RBS. The resulting constructs were used in the Arabidopsis mesophyll protoplasts transient expression system (28).
Generation of Arabidopsis Protoplasts.
Arabidopsis mesophyll protoplasts were generated by following the protocol from Yoo et al. (28), with minor modifications. Leaf strips were digested in enzyme solution (0.4 M mannitol, 20 mM KCl, 20 mM Mes pH 5.7, 10 mM CaCl2 and 0.1% BSA) with 1.3% cellulose R10 (Yakult Pharmaceutical Ind. Co., Ltd.) and 0.3% Macerozyme R10 (Yakult Pharmaceutical Ind. Co., Ltd.) for 2.5 h with gentle shaking. The protoplast solution was filtered through a 100-μm nylon mesh and washed once with W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM Mes pH 5.7). Isolated protoplasts were resuspended in W5 solution and incubated on ice for 30 min. After incubation, protoplasts were pelleted down and resuspended in MMG solution (0.4 M mannitol, 15 mM MgCl2, and 4 mM Mes pH 5.7). For coimmunoprecipitation, we used 2 mL of protoplasts (in MMG solution), 100 μL of plasmid A (1 μg/μL), 100 μL of plasmid B (1 μg/μL; or H2O for controls), and 2.2 mL PEG solution [40% PEG4000 (Fluka; cat. no. 81240), 0.2 M mannitol and 100 mM CaCl2]. The resulting transfection mix was well mixed and allowed to react for 10 min. The mix was then diluted with 8 mL W5 solution to stop the reaction.
Plant Total Protein Extraction, Protein Immunoprecipitation, and Western Blot Analyses.
Plant total protein was extracted from 100 mg of 12-d-old plate-grown plants using extraction buffer (100 mM Tris-HCl pH 8.0, 0.2% SDS and 2% β-mercaptoethanol). Laemmli buffer (4×) was added to each protein sample and boiled for 5 to 10 min. The resulting protein samples were subjected to Western blot analyses.
For protein immunoprecipitation, proteins in the tranfected protoplasts were extracted using 1.5 mL grinding buffer [50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 150 mM NaCl, 0.1% Nonidet P-40, 1 mM PMSF, 1× Protease Inhibitor Coctail (Roche; Cat. #11873580001), and 100 μM MG132]. The sample was spun at 16,000 × g for 10 min at 4 °C to remove cellular debris. Forty microliters of the supernatant was saved as input. The rest of the supernatant was transferred to a tube containing 35 μL anti-FLAG M2 beads (Sigma; Cat. #A2220) and incubated for 2 h at 4 °C with gentle rotation. After incubation, the beads were spun down at 1,500 × g for 30 s at 4 °C. The beads were washed thoroughly with 1 mL of grinding buffer three times before immunoprecipitated proteins were eluted with 60 μL 3 × FLAG peptide (150 μg/mL; Sigma, Cat. #F4799).
The anti-SNC1 antibody was generated against a SNC1-specific peptide in rabbit (11). The anti-HA antibody was from Roche (Cat. #11867423001). The anti-FLAG antibody and the anti-Ubiquitin antibody were both from Sigma (Cat. #F1804 and U0508, respectively).
Supplementary Material
Acknowledgments
We thank our colleagues around the globe who kindly provided us with materials: Dr. Judy Callis for seeds of the cul1-7 and transgenic CUL1-FLAG in cul1-7; Dr. Jian-Min Zhou for pUC19-35S-FLAG-RBS and pUC19-35S-HA-RBS; Dr. Brian Staskawitz for seeds of the RPS2-HA transgenic line; Dr. Jane Parker for seeds of the RPS4-HA transgenic line; Dr. Guoying Wang for seeds of cpr1-2 (previously cpr30-1) and cpr1-2 pad4; Dr. Mary Berbee and Dr. Sean Graham for their help with phylogenetic analysis of CPR1 and its homologs; Kaeli Johnson and Virginia Woloshen for careful reading of the manuscript; and Yan Li for assistance with making figures. Work described is supported by funds from Natural Sciences and Engineering Research Council of Canada (to X.L.) and the Chinese Ministry of Science and Technology (to Y.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105685108/-/DCSupplemental.
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Eitas TK, Dangl JL. NB-LRR proteins: Pairs, pieces, perception, partners, and pathways. Curr Opin Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magalhaes JG, Sorbara MT, Girardin SE, Philpott DJ. What is new with Nods? Curr Opin Immunol. 2011;23(1):29–34. doi: 10.1016/j.coi.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Clarke JD, Zhang Y, Dong X. Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact. 2001;14:1131–1139. doi: 10.1094/MPMI.2001.14.10.1131. [DOI] [PubMed] [Google Scholar]
- 5.Shirano Y, Kachroo P, Shah J, Klessig DF. A gain-of-function mutation in an Arabidopsis Toll Interleukin1 receptor-nucleotide binding site-leucine-rich repeat type R gene triggers defense responses and results in enhanced disease resistance. Plant Cell. 2002;14:3149–3162. doi: 10.1105/tpc.005348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tan X, et al. Global expression analysis of nucleotide binding site-leucine rich repeat-encoding and related genes in Arabidopsis. BMC Plant Biol. 2007;7:56. doi: 10.1186/1471-2229-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shirasu K. The HSP90-SGT1 chaperone complex for NLR immune sensors. Annu Rev Plant Biol. 2009;60:139–164. doi: 10.1146/annurev.arplant.59.032607.092906. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Kadota Y, Prodromou C, Shirasu K, Pearl LH. Structural basis for assembly of Hsp90-Sgt1-CHORD protein complexes: Implications for chaperoning of NLR innate immunity receptors. Mol Cell. 2010;39:269–281. doi: 10.1016/j.molcel.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mayor A, Martinon F, De Smedt T, Pétrilli V, Tschopp J. A crucial function of SGT1 and HSP90 in inflammasome activity links mammalian and plant innate immune responses. Nat Immunol. 2007;8:497–503. doi: 10.1038/ni1459. [DOI] [PubMed] [Google Scholar]
- 10.Holt BF, 3rd, Belkhadir Y, Dangl JL. Antagonistic control of disease resistance protein stability in the plant immune system. Science. 2005;309:929–932. doi: 10.1126/science.1109977. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, et al. SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog. 2010;6:e1001111. doi: 10.1371/journal.ppat.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SH, et al. The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog. 2010;6:e1001172. doi: 10.1371/journal.ppat.1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azevedo C, et al. The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science. 2002;295:2073–2076. doi: 10.1126/science.1067554. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Goritschnig S, Dong X, Li X. A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell. 2003;15:2636–2646. doi: 10.1105/tpc.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wiermer M, Feys BJ, Parker JE. Plant immunity: The EDS1 regulatory node. Curr Opin Plant Biol. 2005;8:383–389. doi: 10.1016/j.pbi.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 16.Shen WH, et al. Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol Biol Cell. 2002;13:1916–1928. doi: 10.1091/mbc.E02-02-0077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilkerson J, et al. Isolation and characterization of cul1-7, a recessive allele of CULLIN1 that disrupts SCF function at the C terminus of CUL1 in Arabidopsis thaliana. Genetics. 2009;181:945–963. doi: 10.1534/genetics.108.097675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gou M, et al. An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J. 2009;60:757–770. doi: 10.1111/j.1365-313X.2009.03995.x. [DOI] [PubMed] [Google Scholar]
- 19.Bowling SA, et al. A mutation in Arabidopsis that leads to constitutive expression of systemic acquired resistance. Plant Cell. 1994;6:1845–1857. doi: 10.1105/tpc.6.12.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 21.Wirthmueller L, Zhang Y, Jones JDG, Parker JE. Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol. 2007;17:2023–2029. doi: 10.1016/j.cub.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 22.Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Schiff M, Serino G, Deng XW, Dinesh-Kumar SP. Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell. 2002;14:1483–1496. doi: 10.1105/tpc.002493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van den Burg HA, et al. The F-box protein ACRE189/ACIF1 regulates cell death and defense responses activated during pathogen recognition in tobacco and tomato. Plant Cell. 2008;20:697–719. doi: 10.1105/tpc.107.056978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HS, Delaney TP. Arabidopsis SON1 is an F-box protein that regulates a novel induced defense response independent of both salicylic acid and systemic acquired resistance. Plant Cell. 2002;14:1469–1482. doi: 10.1105/tpc.001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koch E, Slusarenko A. Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell. 1990;2:437–445. doi: 10.1105/tpc.2.5.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 28.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.