Abstract
Recent epigenome-wide mapping studies describe nucleosome-depleted regions (NDRs) at transcription start sites and enhancers. However, these static maps do not address causality or the roles of NDRs in gene control, and their relationship to transcription factors and DNA methylation is not well understood. Using a high-resolution single-molecule mapping approach to simultaneously investigate endogenous DNA methylation and nucleosome occupancies on individual DNA molecules, we show that the unmethylated OCT4 distal enhancer has an NDR, whereas NANOG has a clear NDR at its proximal promoter. These NDRs are maintained by binding of OCT4 and are required for OCT4 and NANOG expression. Differentiation causes a rapid loss of both NDRs accompanied by nucleosome occupancy, which precedes de novo DNA methylation. NDRs can be restored by forced expression of OCT4 in somatic cells but only when there is no cytosine methylation. These data show the central role of the NDRs, established by OCT4, in ensuring the autoregulatory loop of pluripotency and, furthermore, that de novo methylation follows the loss of NDRs and stabilizes the suppressed state.
During development, each cell acquires its own epigenetic signature that provides guidelines to its cellular identity (1, 2). This epigenetic signature is accomplished by multiple epigenetic mechanisms, including DNA methylation, histone modifications, nucleosome positioning, and noncoding RNAs (3, 4). Pluripotent cells have a distinctive signature that is more dynamic compared with differentiated cells and allows for self-renewal and pluripotency. Developmentally important genes are bivalent in embryonic stem cells, containing both active and repressive histone modifications (5, 6). The transcription factors OCT4, SOX2, and NANOG are known as core regulators of the transcription circuitry in pluripotent cells. The transcription autoregulatory loop ensures high levels of expression of these key stemness genes; they bind to their own regulatory regions, thereby maintaining expression patterns necessary for establishing and preserving pluripotent states (7).
The nucleosome is the basic unit of chromatin and consists of DNA wound around a histone octamer protein core to achieve high compaction. Besides its role in packing the genome, pioneering studies have shown that nucleosome occupancy at gene promoters inhibits transcription initiation (8) and plays a critical role in epigenetic regulation (9). Genome-wide studies have shown that nucleosome-depleted regions (NDRs) are present at the transcription start sites of active genes and enhancers (10–14). More recently, genome-wide studies have begun to focus on the relationship between nucleosome positioning and gene expression (15, 16). Despite these observations, the role of dynamic nucleosome occupancy at gene regulatory regions and the relationship with transcription factors has not been well characterized at high resolution, particularly during the initial steps when embryonic stem cells lose their pluripotency.
In this study, we used our high-resolution single-molecule nucleosome occupancy and methylome sequencing (NOMe-seq) approach to investigate endogenous DNA methylation as well as the distribution of nucleosomes on the same DNA strand (Fig. 1A). NOMe-seq is based on the accessibility of DNA to the GpC methyltransferase (M.CviPI) to provide a digital footprint of nucleosome occupancies and DNA methylation within the same DNA molecule, regardless of CpG density or methylation state (17, 18). Using NOMe-seq to assess nucleosome occupancy and DNA methylation at genomic regulatory elements underlying pluripotency, we show that NDRs at the OCT4 distal enhancer and the NANOG proximal promoter play key roles in the expression of these genes. The OCT4 protein physically binds to the DNA regulatory regions of OCT4 and NANOG, a process that is crucial for establishing and maintaining NDRs. Furthermore, nucleosome occupancy precedes de novo DNA methylation, which can act as a major epigenetic barrier to gene reactivation during differentiation. These data provide a potential mechanism for de novo DNA methylation after nucleosome insertion at regulatory regions of pluripotent genes during differentiation, thereby ensuring gene silencing.
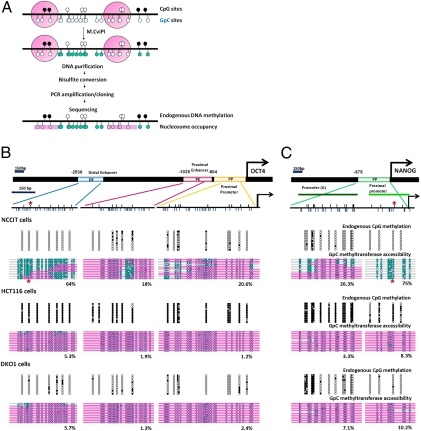
Fig. 1.
OCT4 and NANOG show distinct nucleosome configurations at DNA regulatory regions that correlate with transcriptional activities. (A) Scheme of the NOMe-seq assay. Upper black circles represent CpG sites (white circles represent unmethylated CpG sites, and black circles represent methylated CpG sites). Lower blue circles represent GpC sites (unfilled blue circles represent GpC sites that are inaccessible to GpC methyltransferase, and green-filled blue circles represent sites that are accessible to GpC methyltransferase). Pink bars represent regions of inaccessibility large enough to accommodate a nucleosome. (B and C) Schematic diagrams of the human OCT4 distal enhancer (DE), proximal enhancer (PE), and proximal promoter (PP) and the NANOG promoter. OCT4 recognizes specific octamer sequences at the OCT4 distal enhancer and NANOG proximal promoter (denoted by asterisks). Arrows indicate the transcription start sites. Small upper black lines indicate CpG sites, and lower blue lines indicate GpC sites of those regions. Endogenous DNA methylation and nucleosome occupancy were analyzed by the NOMe-seq assay. A 150-bp marker gives the approximate size that would accommodate a nucleosome. Each horizontal line represents individual OCT4 enhancers and promoter or NANOG promoter modules.
Results
Nucleosome Occupancy at DNA Regulatory Regions Is a Key Player in Pluripotent Gene Expression.
To investigate the roles of distinct epigenetic mechanisms in silencing pluripotency genes, we used embryonic carcinoma cell line NCCIT (19, 20), colon cancer cell line HCT116, and cell line DKO1, the hypomethylated derivative of HCT116 that exhibits 95% reduced DNA methylation because of the genetic knockdown of DNA methyltransferases DNMT3B and DNMT1 (21, 22). These cell lines were selected because they represent distinct transcriptional contexts of pluripotency genes in the presence and absence of DNA methylation. NCCIT cells are easy to culture and manipulate, yet they have a very similar transcriptional profile and comparable pluripotency potential to embryonic stem cells. To rule out cancer cell-specific effects, we confirmed our results with human embryonic stem cells and other normal cell lines. As expected, only NCCIT cells strongly expressed OCT4 (Fig. S1A). Three OCT4 regulatory elements—the distal enhancer, proximal enhancer, and proximal promoter (23)—were largely unmethylated in both NCCIT and DKO1 cells but were extensively methylated in HCT116 cells (Fig. 1B). Similar results were obtained for the NANOG promoter (Fig. 1C) (24), which is expressed in NCCIT cells but not in HCT116 or DKO1 cells (Fig. 1C and Fig. S1B). These results showed that OCT4 and NANOG expression was associated with low levels of DNA methylation at regulatory elements. However, these genes can also be silenced in the absence of DNA methylation because neither gene was expressed in DKO1 cells, which contained little methylation.
A large NDR was present in the majority of OCT4 distal enhancer modules, although some modules contained nucleosomes in the expressing NCCIT cells, indicating the presence of an NDR in which the modules were alternatively occupied or unoccupied by nucleosomes (Fig. 1B). In contrast, the proximal enhancer of OCT4 showed a highly defined NDR (<150 bp), and, although the proximal promoter showed some accessibility, it did not show a clear NDR (Fig. 1B). Interestingly, the nonexpressing HCT116 and DKO1 cells showed a complete loss of the NDR throughout the distal and proximal enhancers, suggesting that nucleosome occupancy at enhancers is associated with gene suppression in the two cell types and that endogenous DNA methylation is not required for silencing. A similar situation was observed at the proximal promoter of the NANOG gene (Fig. 1C). ChIP analysis using a histone 3 (H3) antibody generally confirmed the nucleosome occupancies we observed in OCT4 and NANOG regulatory regions in each of the three cell lines (Fig. S1B). However, it did not show the more detailed configuration of nucleosome occupancy provided by NOMe-seq. Once again, a clear NDR was present in the expressing cells but was absent in the nonexpressing cell types. These observations were confirmed with a human embryonic stem cell line (H1), human fibroblasts (LD419), and keratinocytes (HaCaT) (Fig. S1C). We also performed the NOMe-seq assay after high salt concentration (400 nM) washings, which would be expected to dissociate other transcription factors and chromatin factors in both NCCIT and H1 cells (Fig. S1D). The increased salt resulted in only a slight increase in GpC enzyme accessibility; however, nucleosome positions remain unaltered. These data suggest that the chromatin configuration we saw initially (Fig. 1B and Fig. S1C) is caused by the presence of nucleosomes rather than by tightly binding transcription factors or chromatin remodelers.
Nucleosome Occupancy Precedes DNA Methylation During Differentiation.
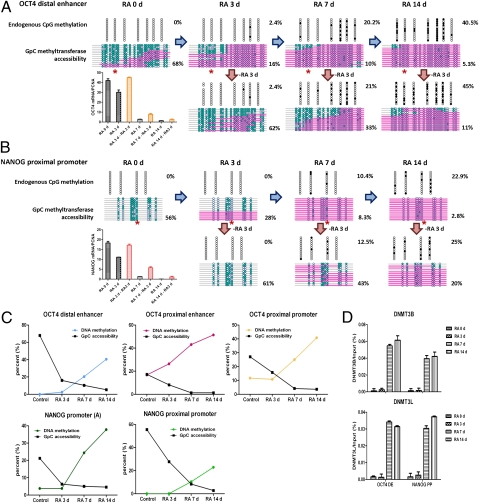
NCCIT cells, like human embryonic stem cells, can be induced to differentiate in response to retinoic acid (RA) (19, 20). Treatment of NCCIT cells with RA reduced OCT4 and NANOG mRNA expression at 3 d after treatment, and expression had decreased to baseline at 7 and 14 d after treatment (Fig. 2A). We next performed NOMe-seq experiments to measure changes in endogenous CpG methylation and nucleosome occupancy of the OCT4 distal enhancer and the NANOG proximal promoter (Fig. 2 A and B). Endogenous DNA methylation progressively increased, reaching 41% at the OCT4 distal enhancer and 23% at the NANOG proximal promoter after 14 d of treatment. Loss of the NDRs at the OCT4 distal enhancer and the NANOG proximal promoter could be observed as early as 3 d after RA treatment (Fig. 2 A and B), with the NDR completely obscured by 14 d after induction of differentiation. We confirmed the change in nucleosome occupancy with ChIP for H3 (Fig. S2A). Other OCT4 and NANOG regulatory regions showed similar nucleosome occupancy kinetics during differentiation (Fig. 2C and Fig. S2B). Importantly, gene expression decreased and regulatory region NDRs were lost, resulting in nucleosome occupancy before changes in endogenous DNA methylation (Fig. 2C). Enrichment of DNMT3B and DNMT3L at days 7 and 14 further support the finding that changes in DNA methylation occur after changes in nucleosome occupancy and decreased gene expression (Fig. 2D).
Fig. 2.
DNA methylation follows nucleosome insertion during cell differentiation and inhibits reactivation of OCT4 and NANOG gene expression. NCCIT cells were exposed to 10 μM RA for the indicated days. (A and B) Endogenous DNA methylation and nucleosome occupancy at the OCT4 distal enhancer and NANOG proximal promoter were analyzed by the NOMe-seq assay during differentiation. OCT4 recognizes specific octamer sequences at the OCT4 distal enhancer and NANOG proximal promoter that do not include CpG sites (denoted by asterisks). For reactivation experiments, RA was removed and cells were cultured for an additional 3 d with fresh growth media. The expression levels of OCT4 and NANOG were determined by quantitative PCR at each indicated time point. (C) Kinetics of the change in DNA methylation and nucleosome occupancy at the OCT4 and NANOG DNA regulatory regions. (D) Chromatin was immunoprecipitated with anti-DNMT3B and anti-DNMT3L antibodies at each time point during the differentiation, and their binding to the OCT4 distal enhancer and NANOG proximal promoter was analyzed by quantitative PCR.
To examine the role of DNA methylation in stabilizing gene repression, we removed RA after 3, 7, and 14 d of exposure. Cells exposed to RA for 3 d before the onset of DNA methylation reexpressed OCT4 and NANOG upon RA withdrawal (Fig. 2 A and B, Lower Left). However, in cells exposed to RA for 14 d after DNA methylation was established, little OCT4 and NANOG reexpression was detected after RA withdrawal (Fig. 2 A and B, Lower Right). These data demonstrate the role of DNA methylation in stabilizing, rather than initiating, gene repression.
OCT4 Is Required for Maintaining NDRs, Which Ensures the Autoregulatory Loop of Pluripotency.
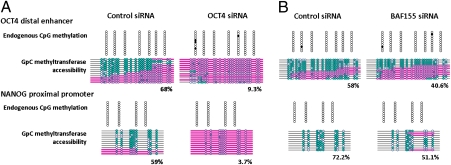
Interestingly, prominent NDRs detected at both the distal enhancer of OCT4 and the proximal promoter of NANOG have OCT4/SOX2 binding motifs (25, 26). We performed ChIP assays to examine whether physical OCT4 binding is important for the presence of NDRs at these regions. These data showed that RA-mediated differentiation resulted in decreased occupancy of OCT4 and SOX2 at both regulatory regions (Fig. S3A). Disruption of the autoregulatory loop by downregulating OCT4 via siRNA allowed us to determine whether the OCT4 gene product itself was required for the presence of the NDR. We first verified OCT4 knockdown and also found reduced NANOG and SOX2 expression (Fig. S3B). Next, we examined the OCT4 distal enhancer and found that it was inaccessible after OCT4 knockdown, compared with 68% of the enhancer modules that contained an NDR in cells transfected with the control siRNA (Fig. 3A). Likewise, the NANOG proximal promoter showed a decrease in GpC methyltransferase accessibility from 59% to 4% after OCT4 knockdown (Fig. 3A). Altogether, these data suggest that active expression of OCT4 is required for maintaining NDRs by directly binding to DNA within the NDRs, which likely ensures that the autoregulatory loop is functional.
Fig. 3.
OCT4 maintains NDRs at regulatory regions of OCT4 and NANOG, which ensures the autoregulatory loop of pluripotency. (A) At 72 h posttransfection with OCT4 and control siRNAs (100 nmol/L), endogenous DNA methylation and nucleosome occupancy at the OCT4 distal enhancer and NANOG proximal promoter were analyzed by the NOMe-seq assay. The data are representative of three biological experiments. (B) At 72 h posttransfection with BAF155 or OCT4 siRNA in NCCIT cells, the NOMe-seq assay was performed. The data are representative of two biological experiments.
Recent studies have shown that OCT4 can physically bind to BAF155, which is a unique subunit of the Brg1/Brm-associated factor (BAF) complex in embryonic stem cells (27–29). We found a decrease of BAF155 binding during RA differentiation and OCT4 knockdown experiments (Fig. S3C). Western blot analysis showed that changes in the expression level of BAF155 correlate with changes in occupancy (Fig. S3D). However, BAF155 knockdown experiments showed the BAF155 enrichment only plays a minor role in maintaining NDRs and OCT4 expression (Fig. 3B and Fig. S3E), suggesting that residual OCT4 protein can recruit other factors that maintain the NDRs and ensure OCT4 and NANOG expression.
OCT4 Can Establish NDRs at Unmethylated but Not Methylated DNA Regulatory Regions of OCT4 and NANOG.
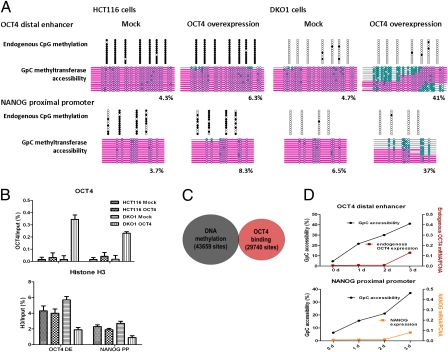
Because OCT4 controls the expression of many transcription factors in addition to itself, it was important to exclude the possibility that these other factors could be directly involved in setting up the NDRs. To examine the more direct role of OCT4 in establishing and maintaining NDRs, we introduced exogenous OCT4 into HCT116 and DKO1 cells, which exhibit different DNA methylation contexts. Exogenous OCT4 established NDRs at OCT4 and NANOG regulatory regions in unmethylated DKO1 cells, whereas HCT116 cells with a high level of DNA methylation did not undergo these changes (Fig. 4A). Transfected DKO1 cells showed a small amount of endogenous OCT4 and NANOG expression (determined by using specific primers that detect 3′ untranslated regions), whereas HCT116 cells did not, consistent with nucleosome occupancy as determined by NOMe-seq (Fig. 4A and Fig. S4A).
Fig. 4.
Exogenous OCT4 expression initiates nucleosome depletion at DNA regulatory regions of OCT4 and NANOG, which are inhibited by DNA methylation. (A) Transfection of HCT116 and DKO1 cells with exogenous OCT4 and mock vectors was carried out with Lipofectamine LTX. At 72 h posttransfection, endogenous DNA methylation and nucleosome occupancy at the OCT4 distal enhancer and NANOG proximal promoter were analyzed by the NOMe-seq assay. (B) At the same time points, ChIP assays were performed with OCT4 and histone H3. (C) To understand the relationship between OCT4 binding and DNA methylated regions, genome-wide studies in human embryonic stem cells (H1) using ENCODE and GEO data (wgEncodeHudsonalphaMethylSeqRegionsRep1H1hesc for DNA methylation and GSM518373 for OCT4 ChIP-Seq) were performed. The data comprised 100-bp windows of OCT4 binding regions (29,740 sites) and DNA methylation (43,659 sites). (D) DKO1 cells were transfected with exogenous OCT4 and mock vectors. The NOMe-seq assay was performed at the indicated time after transfection.
OCT4 is known to be a sequence-specific binding transcription factor with no CpG sites in its binding motif. Therefore, the methylation effects are most likely indirect. We next investigated changes in OCT4 binding with ChIP assays using OCT4 antibodies, which addresses the question of DNA methylation and OCT4 binding. The results showed that OCT4 does not bind in HCT116 cells in which the flanking CpG sites of its consensus motifs are methylated but does bind to unmethylated DNA in DKO cells (Fig. 4B). We also analyzed the relationship between OCT4 binding regions and DNA methylated regions genome-wide in human embryonic stem cells (H1) by using Encyclopedia of DNA Elements (ENCODE) and Gene Expression Omnibus (GEO) data (wgEncodeHudsonalphaMethylSeqRegionsRep1H1hesc for DNA methylation and GSM518373 for OCT4 ChIP-Seq). These data show that 100-bp windows of OCT4 binding regions (29,740 sites) and DNA methylation (43,659 sites) are mutually exclusive (Fig. 4C). This evidence suggests two possibilities: (i) OCT4 cannot bind its recognition sites when they have flanking methylated DNA or (ii) DNA methylation stabilizes nucleosomes that cannot be dissociated by OCT4. Recent work by Choy et al. (30), showing that CpG methylation increases nucleosome compaction and rigidity, lends more weight to the latter idea. In summary, OCT4 competes with relatively unstable nucleosomes that do not have DNA methylation, and it establishes NDRs, which are crucial for target gene expression.
Although we could detect endogenous OCT4 and NANOG expression in DKO1 cells at 3 d posttransfection (Fig. S4A), it was very low compared with pluripotent NCCIT cells. Notably, there was little change in the expression of OCT4 downstream target genes, such as other pluripotent genes (SOX2, KLF4, C-MYC, LIN28, AP, and REST) and master regulatory genes involved in lineage commitment (CDX2, PAX6, and NEUROG1) (Fig. S4B), indicating that the changes we see are because of a change in OCT4 expression and subsequent binding rather than an indirect response to a change in pluripotency. Furthermore, the time course of exogenous OCT4 introduction shows that nucleosome depletion could be detected before endogenous OCT4 and NANOG expression (Fig. 4D), demonstrating that OCT4 can directly reorganize the chromatin structure at the DNA regulatory regions of OCT4 and NANOG without changing the expression of other downstream genes.
To examine which domain of OCT4 is involved in this process, we performed deletion mutant experiments. The N-terminal transactivation domain of OCT4 is unique to pluripotent cells, whereas the POU and C-terminal domains are shared with other isoforms that are also expressed in HCT116, DKO1, and NCCIT cells (31). The pluripotent cell specificity of the N terminus of OCT4 suggests that the N terminus is necessary for this chromatin-remodeling activity. To determine the role of other domains, we overexpressed three deletion mutants (N terminus alone, N terminus and POU domain, and N and C termini) in DKO cells (Fig. S4C). The presence of the N terminus alone did not result in remodeling, whereas some remodeling occurred when either the POU or C-terminal domain was present. These results suggest that, although both regions are capable of remodeling activity, complete chromatin remodeling requires all three domains. These data confirm that the OCT4 transcription factor has a key role in establishing and maintaining NDRs, which are essential for OCT4 and NANOG expression, and that DNA methylation acts as an epigenetic barrier preventing reexpression.
Discussion
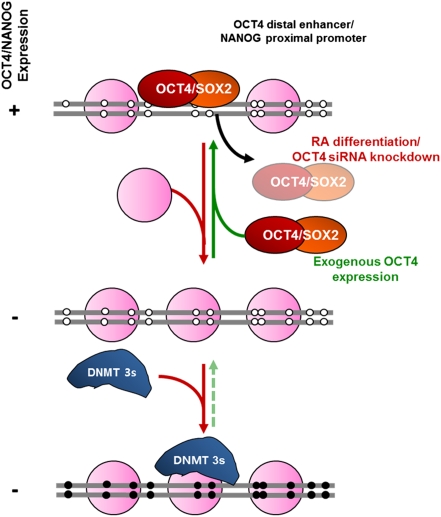
In this study, we used a nucleosome-positioning analysis technique (NOMe-seq) to show endogenous DNA methylation and nucleosome occupancy within the same DNA molecule. Our results are summarized in the model outlined in Fig. 5. The key regulatory regions, which contain OCT4 binding sites (OCT4 distal enhancer and NANOG proximal promoter), show nucleosome depletion and OCT4/SOX2 occupancy in pluripotent states. When cells receive a differentiation signal, the NDRs are lost first, followed by de novo DNA methylation, which acts as a major barrier to reactivation by locking in nucleosome occupancy. NANOG shows the canonical nucleosome configuration of an open promoter with an NDR upstream of the transcription start site when the gene is transcriptionally active (16, 32, 33). OCT4, on the other hand, shows a very interesting footprint: some OCT4 distal enhancer modules contain nucleosomes, whereas others show a striking NDR that contains regions in which two or more nucleosomes are missing. In contrast, the OCT4 proximal enhancer shows a highly defined NDR. The distinct enhancer chromatin structures are intriguing given that the proximal enhancer is required for epiblast expression in vivo, whereas the distal enhancer is required for preimplantation and germ-cell lineage expression of Oct4 in the mouse (34). Interestingly, the NDR within the OCT4 proximal enhancer has various transcription factor binding motifs (Table S1). The specific transcription factor that maintains the highly defined NDR within the proximal enhancer remains to be elucidated. Recently, it has been shown that the transcription factor PRDM14 binds to the OCT4 proximal enhancer and has crucial functions in maintaining pluripotency in human embryonic stem cells, making it a possible candidate (35). Unlike the promoter of NANOG, the OCT4 proximal promoter does not show a clear NDR in our results. It is possible that the enhancer, which does contain an NDR, is close enough to the transcription start site that it can recruit the transcription machinery and initiate transcription.
Fig. 5.
A proposed model of the mechanism of repression and de novo DNA methylation of OCT4 and NANOG during cell differentiation. (Refer to Discussion for a detailed explanation.)
The mechanisms regulating nucleosome positioning are of great interest and have been the subject of several recent outstanding reviews (15, 16, 32). Based on the Segal laboratory's prediction program and/or the NuPoP program, the NANOG proximal promoter showed a lower nucleosome occupancy score compared with other regions. However, in vivo, nucleosome positioning is affected by competition between transcription factors and nucleosomes for a given piece of DNA. The NDR formation and loss described here are likely caused by the competition between OCT4 and nucleosomes. This result is supported by a “thermodynamic equilibrium model,” which describes the dynamic repositioning of nucleosomes that occurs as the relative input balance between nucleosomes and specific transcription factors changes (15). Specifically, the cells receive a signal that down-regulates OCT4 protein levels, shifting the balance in the favor of nucleosomes; as a result, the NDRs begin to lose, amplifying the negative autoregulatory loop. In corroboration, Zhang et al. report a packing mechanism that can override DNA intrinsic positioning (36).
It has been known for some time that silencing precedes DNA methylation (37), and some studies have investigated the roles of histone marks, such as H3K9me, in this process (38–40). However, previous works have overlooked the role of nucleosome occupancy in silencing by not, for example, correcting ChIP data for histone occupancy at key regulatory regions. The follow-up study by Bergman and colleagues showed that G9a binding is more crucial rather than the repressive histone mark (41). In addition, our H3K9me3/H3 data showed that the changes of nucleosome occupancy were more significant and occurred before repressive histone modification changes (Fig. S2A). The high-resolution approach that we used shows nucleosome occupancy and endogenous DNA methylation on the same single molecule of DNA by identifying changes at individual CpG and GpC sites, which is not possible with ChIP/DNase I. Decreased levels of OCT4, initiated by either RA-induced differentiation or siRNA knockdown, result in the collapse of the NDRs by nucleosome insertion into these regions. These data demonstrate the critical importance of OCT4 occupancy accompanying nucleosome depletion at key regulatory elements in ensuring the activity of the autoregulatory network. Furthermore, exogenous OCT4 overexpression shows that OCT4 has chromatin reorganizing activity that establishes NDRs at the unmethylated endogenous OCT4 distal enhancer and NANOG proximal promoter in somatic cells.
Both the differentiation experiments in pluripotent cells and the experiments with somatic cell lines show that the presence of nucleosomes at the OCT4 distal enhancer and NANOG proximal promoter is associated with transcriptional repression, even in the absence of DNA methylation. The presence of nucleosomes provides a substrate leading to the recruitment of the DNA methyltransferases, which apply DNA methylation to the region. This result is consistent with a seminal study showing that DNMT3A, DNMT3B, and DNMT3L complexes require nucleosomes to initiate DNA methylation (42). It has also recently been shown that de novo DNA methyltransferases selectively anchor to nucleosomes and that nucleosomes containing methylated DNA stabilize de novo DNA methyltransferases (43, 44), emphasizing the important role of nucleosomes not only in gene silencing but also in initiating de novo DNA methylation. These data support the idea that DNA methylation may be required not to initiate gene repression but to maintain silencing over a protracted time period in differentiated cell types (45). These data correlate with previous studies showing that the efficiency of induced pluripotent stem cell formation can be increased by using DNA methylation inhibitors (46, 47).
Our work, using the precise digital-mapping approach that integrates the analysis of two fundamental epigenetic control mechanisms, establishes the critical role of the NDRs in transcriptional competency and de novo DNA methylation. Our results have implications for cell biology, stem cell differentiation, and reprogramming. These data show that OCT4 is a master structural regulator with the central role of establishing and maintaining NDRs in its distal enhancer and the NANOG proximal promoter; these NDRs are crucial in the control of pluripotency gene regulation. Our results also show that nucleosomal occupancy precedes de novo DNA methylation and that DNA methylation is required not to initiate silencing of genes but to keep them silenced over a protracted time period in differentiated cell types.
Methods
Details of the methods used in this article, including cell culture, NOMe-seq, reagents and antibodies, RNA extraction and RT-PCR, Western blot analysis, ChIP, siRNA transfection, and introduction of exogenous OCT4 experiments, are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Dr. Jeung-Whan Han, Dr. C. Andreu-Vieyra, and K. Pandiyan for discussions about the manuscript. Funding for this work was provided by National Institutes of Health Grant R37 CA-082422-12 (to P.A.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111309108/-/DCSupplemental.
References
- 1.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 2.Hochedlinger K, Plath K. Epigenetic reprogramming and induced pluripotency. Development. 2009;136:509–523. doi: 10.1242/dev.020867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christophersen NS, Helin K. Epigenetic control of embryonic stem cell fate. J Exp Med. 2010;207:2287–2295. doi: 10.1084/jem.20101438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 6.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 7.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorch Y, LaPointe JW, Kornberg RD. Nucleosomes inhibit the initiation of transcription but allow chain elongation with the displacement of histones. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 9.Mellor J. The dynamics of chromatin remodeling at promoters. Mol Cell. 2005;19:147–157. doi: 10.1016/j.molcel.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Schones DE, et al. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 12.Jiang C, Pugh BF. Nucleosome positioning and gene regulation: Advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin C, et al. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–945. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He HH, et al. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–347. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bai L, Morozov AV. Gene regulation by nucleosome positioning. Trends Genet. 2010;26:476–483. doi: 10.1016/j.tig.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 17.Wolff EM, et al. Hypomethylation of a LINE-1 promoter activates an alternate transcript of the MET oncogene in bladders with cancer. PLoS Genet. 2010;6:e1000917. doi: 10.1371/journal.pgen.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kelly TK, et al. H2A.Z maintenance during mitosis reveals nucleosome shifting on mitotically silenced genes. Mol Cell. 2010;39:901–911. doi: 10.1016/j.molcel.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Damjanov I, Horvat B, Gibas Z. Retinoic acid-induced differentiation of the developmentally pluripotent human germ cell tumor-derived cell line, NCCIT. Lab Invest. 1993;68:220–232. [PubMed] [Google Scholar]
- 20.You JS, et al. Depletion of embryonic stem cell signature by histone deacetylase inhibitor in NCCIT cells: Involvement of Nanog suppression. Cancer Res. 2009;69:5716–5725. doi: 10.1158/0008-5472.CAN-08-4953. [DOI] [PubMed] [Google Scholar]
- 21.Egger G, et al. Identification of DNMT1 (DNA methyltransferase 1) hypomorphs in somatic knockouts suggests an essential role for DNMT1 in cell survival. Proc Natl Acad Sci USA. 2006;103:14080–14085. doi: 10.1073/pnas.0604602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhee I, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 23.Freberg CT, Dahl JA, Timoskainen S, Collas P. Epigenetic reprogramming of OCT4 and NANOG regulatory regions by embryonal carcinoma cell extract. Mol Biol Cell. 2007;18:1543–1553. doi: 10.1091/mbc.E07-01-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chan KK, et al. KLF4 and PBX1 directly regulate NANOG expression in human embryonic stem cells. Stem Cells. 2009;27:2114–2125. doi: 10.1002/stem.143. [DOI] [PubMed] [Google Scholar]
- 25.Chew JL, et al. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 2005;25:6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda T, et al. Octamer and Sox elements are required for transcriptional cis regulation of Nanog gene expression. Mol Cell Biol. 2005;25:2475–2485. doi: 10.1128/MCB.25.6.2475-2485.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van den Berg DL, et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 2010;6:369–381. doi: 10.1016/j.stem.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pardo M, et al. An expanded Oct4 interaction network: Implications for stem cell biology, development, and disease. Cell Stem Cell. 2010;6:382–395. doi: 10.1016/j.stem.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Choy JS, et al. DNA methylation increases nucleosome compaction and rigidity. J Am Chem Soc. 2010;132:1782–1783. doi: 10.1021/ja910264z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang X, Dai J. Concise review: Isoforms of OCT4 contribute to the confusing diversity in stem cell biology. Stem Cells. 2010;28:885–893. doi: 10.1002/stem.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arya G, Maitra A, Grigoryev SA. A structural perspective on the where, how, why, and what of nucleosome positioning. J Biomol Struct Dyn. 2010;27:803–820. doi: 10.1080/07391102.2010.10508585. [DOI] [PubMed] [Google Scholar]
- 33.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 34.Ng JH, Heng JC, Loh YH, Ng HH. Transcriptional and epigenetic regulations of embryonic stem cells. Mutat Res. 2008;647:52–58. doi: 10.1016/j.mrfmmm.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Chia NY, et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature. 2010;468:316–320. doi: 10.1038/nature09531. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, et al. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lock LF, Takagi N, Martin GR. Methylation of the Hprt gene on the inactive X occurs after chromosome inactivation. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- 38.Feldman N, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 39.Strunnikova M, et al. Chromatin inactivation precedes de novo DNA methylation during the progressive epigenetic silencing of the RASSF1A promoter. Mol Cell Biol. 2005;25:3923–3933. doi: 10.1128/MCB.25.10.3923-3933.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mutskov V, Felsenfeld G. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J. 2004;23:138–149. doi: 10.1038/sj.emboj.7600013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Epsztejn-Litman S, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ooi SKT, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jeong S, et al. Selective anchoring of DNA methyltransferases 3A and 3B to nucleosomes containing methylated DNA. Mol Cell Biol. 2009;29:5366–5376. doi: 10.1128/MCB.00484-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sharma S, De Carvalho DD, Jeong S, Jones PA, Liang G. Nucleosomes containing methylated DNA stabilize DNA methyltransferases 3A/3B and ensure faithful epigenetic inheritance. PLoS Genet. 2011;7:e1001286. doi: 10.1371/journal.pgen.1001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol. 2010;20:609–617. doi: 10.1016/j.tcb.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.