Abstract
The Ten-Eleven-Translocation 2 (TET2) gene encodes a member of TET family enzymes that alters the epigenetic status of DNA by oxidizing 5-methylcytosine to 5-hydroxymethylcytosine (5hmC). Somatic loss-of-function mutations of TET2 are frequently observed in patients with diverse myeloid malignancies, including myelodysplastic syndromes, myeloproliferative neoplasms, and chronic myelomonocytic leukemia. By analyzing mice with targeted disruption of the Tet2 catalytic domain, we show here that Tet2 is a critical regulator of self-renewal and differentiation of hematopoietic stem cells (HSCs). Tet2 deficiency led to decreased genomic levels of 5hmC and augmented the size of the hematopoietic stem/progenitor cell pool in a cell-autonomous manner. In competitive transplantation assays, Tet2-deficient HSCs were capable of multilineage reconstitution and possessed a competitive advantage over wild-type HSCs, resulting in enhanced hematopoiesis into both lymphoid and myeloid lineages. In vitro, Tet2 deficiency delayed HSC differentiation and skewed development toward the monocyte/macrophage lineage. Our data indicate that Tet2 has a critical role in regulating the expansion and function of HSCs, presumably by controlling 5hmC levels at genes important for the self-renewal, proliferation, and differentiation of HSCs.
DNA methylation is highly aberrant in cancer (1, 2). Cancer cells show global hypomethylation associated with genomic instability as well as promoter hypermethylation associated with inactivation of tumor suppressor, cell cycle, or repair-related genes (3–5). In the hematopoietic system, whole-genome sequencing and other genetic analyses have identified recurrent somatic alterations that contribute to the pathogenesis of a variety of myeloid malignancies (1, 2); many of these mutations involve proteins that regulate the epigenetic landscape of cancer cells by altering DNA methylation and histone modification patterns (3, 4, 6, 7). Thus, mutations in genes encoding the polycomb proteins EZH2 and ASXL1, and the DNA methyltransferase DNMT3A, are frequently observed in patients with acute myeloid leukemia (AML), myelodysplastic syndromes (MDS), and myeloproliferative neoplasms (MPN) (8–10). In mice, the maintenance DNA methyltransferase Dnmt1 is critical for self-renewal and the differentiation potential of mouse hematopoietic stem cells (HSCs) (11, 12); similarly, Dnmt3a and Dnmt3b, de novo DNA methyltransferases that also maintain methylation in certain contexts (1), are individually dispensable for lymphoid and myeloid differentiation, but their combined loss results in impaired self-renewal of HSCs (13). Collectively, these results implicate DNA methylation in the regulation of self-renewal, differentiation, and oncogenic transformation of HSCs.
The Ten-Eleven-Translocation (TET) proteins TET1, TET2, and TET3 are α-ketoglutarate and Fe2+-dependent enzymes capable of modifying DNA methylation status by converting 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC) (14–18). The chromosome 4q24 region containing the TET2 gene undergoes frequent microdeletions and uniparental disomy in patients with myeloid malignancies (19, 20), and TET2 mutations—including base substitutions, out-of-frame insertions/deletions, and splice site mutations—are observed in a diverse spectrum of myeloid malignancies classified as MDS (19∼26% of cases), MPN (12∼37% of cases), chronic myelomonocytic leukemia (CMML; 20∼50% of cases), AML, and secondary AML (sAML) (20–27).
We showed that leukemia-associated missense mutations impair the enzymatic activity of TET2 (17), and that TET2 mutational status correlates well with decreased genomic levels of 5hmC in bone marrow samples from patients with myeloid malignancies (17, 27). Moreover, small hairpin RNA (shRNA)-mediated knockdown of Tet2 in mouse hematopoietic stem/progenitor cells (HSPCs) revealed a potential function of Tet2 in controlling myeloid differentiation and the homeostasis of HSPCs (17, 28). Tet2 mRNA is broadly expressed in hematopoietic cell subsets including stem/progenitor and mature cells, and 5hmC is present at clearly detectable levels in their genomes (17, 29). Collectively, these results suggest that loss-of-function mutations of TET2 perturb normal hematopoiesis by impairing the conversion of 5mC into 5hmC at specific genetic loci that are critical for the maintenance and function of HSCs.
In this study, we report the generation of conditional gene-disrupted mice to clarify the role of Tet2 in hematopoietic development. We find that Tet2 deficiency decreases 5hmC levels as expected, increases the pool of HSPCs in the bone marrow in a cell-intrinsic manner, enhances the multilineage repopulation capacity of HSCs, and regulates myeloid differentiation potential.
Results
Targeted Disruption of the Tet2 Gene in Mice.
We demonstrated that conversion of 5mC to 5hmC via TET family members requires a signature HxD motif (14, 17). The orthologous residues H1302 and D1304 are encoded by exon 9 in murine Tet2. To conditionally ablate Tet2 enzymatic activity in vivo, we inserted LoxP sites into the endogenous Tet2 locus flanking exons 8–10 (Fig. S1A). Embryonic stem (ES) cells were electroporated with the targeting construct containing a neomycin resistance cassette, and correct integration of the targeting construct in resistant clones was ascertained by Southern blot analyses with probes flanking the upstream and downstream homology arms (Fig. S1B, Upper Left). The integrity of the incorporated LoxP sites was confirmed by in vitro treatment of targeted clones with recombinant TAT-CRE fusion proteins (Fig. S1B, Lower Left). Correctly targeted ES cells were injected into blastocysts, and progenies bearing the targeted allele were bred to FLP-E deletor and CMV-CRE deletor mice to delete the neomycin cassette and generate Tet2+/− mice, respectively. F1 crosses of Tet2+/− mice produced Tet2−/− mice at a Mendelian ratio without any evidence for embryonic lethality.
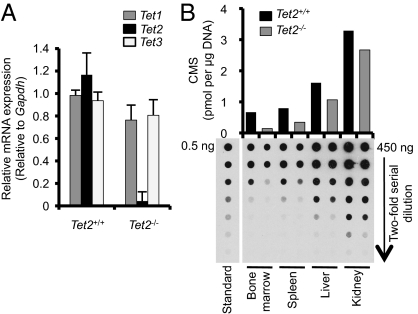
Naïve CD4 T cells express all Tet family members at high levels (17). Thus, we used these cells to assess targeting efficiency. Real-time PCR analysis showed that Tet2 mRNA is highly expressed in WT CD4 T cells but barely detected in Tet2−/− T cells (Fig. 1A). There was no compensatory up-regulation of Tet1 or Tet3 in the absence of Tet2. Genomic DNAs isolated from several organs of Tet2+/+ or Tet2−/− mice (bone marrow, spleen, liver, kidney) were treated with sodium bisulfite to convert 5hmC into cytosine 5-methylenesulfonate (CMS), and 5hmC was quantified by dot blot using anti-CMS antibody (17, 30). 5hmC levels were substantially decreased in bone marrow and spleen of Tet2−/− mice compared with Tet2+/+ controls, but less severely affected in liver and kidney (Fig. 1B), most likely reflecting differences in relative expression of Tet2 versus other Tet family proteins in hematopoietic versus nonhematopoietic organs.
Fig. 1.
Tet mRNA expression and 5hmC levels after Tet2 deletion in mice. (A) Expression of Tet1, Tet2, and Tet3 in sorted naïve CD4+ (CD4+CD8−CD44loCD62L+) T cells derived from Tet2+/+ or Tet2−/− mice. Cells were isolated from pooled spleen and lymph nodes by flow cytometry, and quantitative RT-PCR analysis was performed. The relative levels of Tet1-3 after normalization to the level of Gapdh mRNA in the same cell population are shown, with the amount in the Tet2+/+ naïve CD4+ population arbitrarily set to 1. Error bars show SD, n = 4. (B) Quantification of 5hmC by anti-CMS dot blot. 5hmC levels in bone marrow, spleen, liver, and kidney in the Tet2+/+ or Tet2−/− mice were assessed by dot blot assay with anti-CMS antibody after treatment of genomic DNA with bisulfite. A synthetic oligonucleotide with a known amount of CMS was used as standard. Data are representative of two independent experiments.
Tet2 Restricts Expansion of HSPCs in the Bone Marrow.
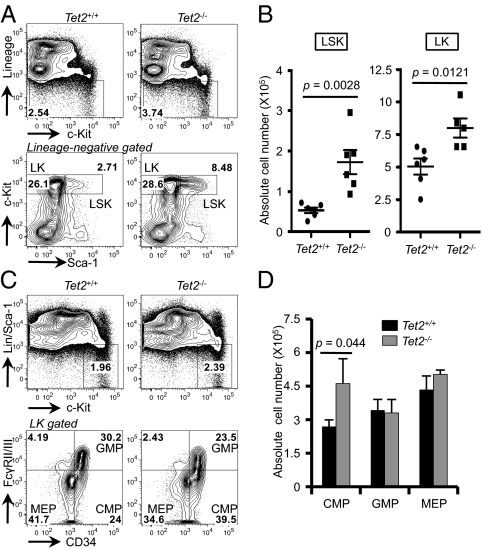
To investigate whether Tet2 deficiency affects the development of HSCs in the bone marrow, we analyzed the major hematopoietic lineages by flow cytometry. We were unable to detect any phenotypic differences in the bone marrow of 5- to 6-wk-old mice; however, Tet2 deficiency led to an increase in total cell numbers in the bone marrow and spleen as early as 8–12 wk of age (Fig. S2A). In these Tet2−/− mice, the development of myeloid (Gr-1/Mac-1), B lymphoid (B220+CD19+), and erythroid (Ter119+) cells was largely unchanged (Fig. S2 B and C), and T-cell development in the thymus and periphery was normal (Fig. S3). Tet2−/− bone marrow displayed a significantly greater frequency of HSCs, defined as lineage-negative (Lin−: B220−Gr-1−Mac-1−Ter119−CD3ε−), c-Kit+Sca-1+ (LSK) cells and a slightly increased frequency of myeloid progenitors, Lin−c-Kit+Sca-1− (LK) cells (Fig. 2A). The absolute number of LSK and LK was greater in Tet2−/− mice compared with controls (Fig. 2B). Within the LK compartment, the absolute number of common myeloid progenitors (CMP, LK CD34+FcγRII/IIIlo) was increased in Tet2−/− mice compared with WT controls (Fig. 2 C and D). Taken together, these data suggest that Tet2 restrains the expansion of the HSPC compartment in the bone marrow.
Fig. 2.
Tet2 regulates the size of the hematopoietic stem/progenitor pool. (A) Flow cytometric analysis of LSK (Lin−c-Kit+Sca-1+) and LK (Lin−c-Kit+Sca-1−) subsets in the bone marrow of 8- to 12-wk-old Tet2+/+ or Tet2−/− mice. The frequency of LSK and LK was assessed by discriminating lineage-negative cells (Lineage: Gr-1, Mac-1, Ter-119, CD3ε, and B220) based on the expression of c-Kit and Sca-1. n = 5–6 per group. P, Student's t test. (B) Absolute number of LSK and LK cells calculated in A. Error bars show SD, n = 5–6 per group. Bone marrow cells were flushed from two femurs and two tibias from each 8- to 12-wk-old mouse. (C) Distribution of myeloid progenitor cell subsets was assessed by the expression of FcγRII/III and CD34 within LK population (n = 3 per group). (D) Absolute number of myeloid progenitor cells calculated in C. CMP, common myeloid progenitor, GMP, granulocyte-monocyte progenitor, MEP, megakaryocyte-erythroid progenitor. Error bars show SD, n = 3 per group. P, Student t test.
Cell-Autonomous Effect of Tet2 Deficiency.
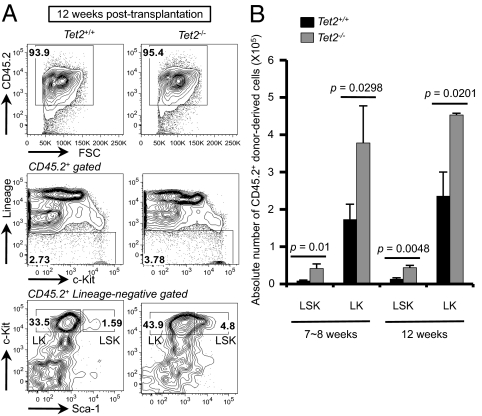
Tet2 was systemically deleted in our system, thus the increased numbers of HSPCs in Tet2−/− mice could reflect a cell-intrinsic effect of Tet2 deficiency in hematopoietic cells or could be due to loss of Tet2 in bone marrow stromal cells. To explore this issue, we transplanted red cell-depleted bone marrow cells from CD45.2+ Tet2+/+ or Tet2−/− mice into lethally irradiated CD45.1+ congenic recipients. At 7–8 or 12 wk after transplantation, the chimeric mice reconstituted with Tet2−/− bone marrow displayed an increase in the frequency and absolute number of LSK and LK cells (Fig. 3 A and B and Fig. S4A). Tet2 deficiency did not significantly alter the development of B cells and myeloid cells in this experimental setting (Fig. S4B), and splenomegaly was observed in some mice reconstituted with Tet2−/−, but not Tet2+/+, bone marrow (Fig. S4C). Collectively, these results demonstrate a cell-autonomous effect of Tet2 deficiency on the magnitude of HSPC pool in the bone marrow.
Fig. 3.
Cell-intrinsic effect of Tet2 deficiency on the size of the hematopoietic stem/progenitor compartment. (A) Flow cytometric analysis of LSK or LK cells in chimeric mice reconstituted with Tet2+/+ or Tet2−/− bone marrow cells. CD45.2+ donor-derived (Top), lineage-negative (Middle) cells in the bone marrow of chimeric mice were further subclassified based on expression of c-Kit and Sca-1 (Bottom). A representative result at 12 wk after transplantation is shown (n = 3 per group). (B) Absolute number of cells in CD45.2+ donor-derived LSK and LK subsets calculated in Fig. 3A and Fig. S4A. Error bars show SD (for 7–8-wk samples) or range of duplicate (for 12-wk samples). n = 3 for 7–8-wk samples; n = 2 for 12-wk samples. P, Student t test.
Tet2 Deficiency Enhances the Repopulating Capacity of HSPCs in Vivo.
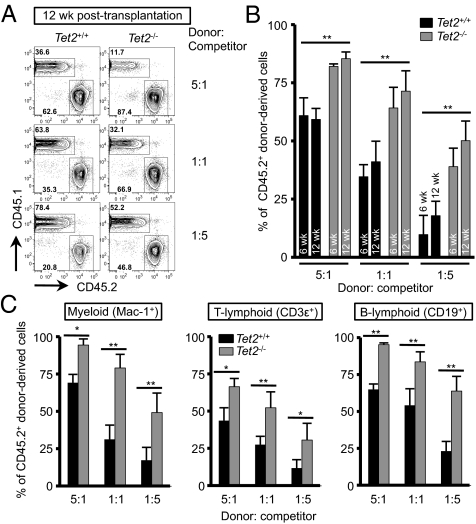
To explore whether Tet2 deficiency alters the function of HSPCs, we performed competitive repopulation assays. Total bone marrow cells from CD45.2+ Tet2+/+ or Tet2−/− mice were depleted of red blood cells, mixed at ratios of 5:1, 1:1, or 1:5 with CD45.1+ Tet2+/+ competitor bone marrow cells, and injected into lethally irradiated CD45.1+ recipients. Six and 12 weeks after transplantation, we collected peripheral blood samples from each recipient and assessed the relative proportion of donor (CD45.2+) or competitor (CD45.1+) cells. The extent of hematopoietic reconstitution by Tet2−/− cells was significantly greater than that by Tet2+/+ donor cells in every condition, and Tet2−/− chimerism was higher than Tet2+/+ chimerism in every hematopoietic lineage: myeloid (Mac-1+), T lymphoid (CD3ε+), and B lymphoid (B220+) cells (Fig. 4 and Fig. S5). Collectively, these data indicate that Tet2 deficiency increases the self-renewal capacity of HSCs, conferring a competitive advantage on Tet2−/− cells relative to Tet2+/+ cells in their ability to give rise to multiple hematopoietic lineages in mice.
Fig. 4.
Augmented hematopoietic repopulation capacity upon Tet2 deficiency. (A) Competitive repopulation assay. CD45.2+ bone marrow cells from Tet2+/+ or Tet2−/− mice were mixed with CD45.1+ competitor cells at different ratios as shown and transplanted into lethally irradiated CD45.1+ congenic recipients. At 6 or 12 wk after transplantation, peripheral blood was examined for donor/competitor chimerism. A representative result at 12 wk after transplantation is shown. n = 2–4 per group. (B) Percentage of CD45.2+ donor chimerism at 6 or 12 wk after transplantation calculated in Fig. 4A and Fig. S5A. n = 2 for Tet2+/+ in 5:1 group; n = 4 for the other samples. *P < 0.05, **P < 0.005 (Student's t test.) (C) Percentage of CD45.2+ donor chimerism in hematopoietic lineages in peripheral blood at 12 wk after transplantation. The percentage of CD45.2+ cells was calculated after gating on myeloid (Mac-1+) (Left), T-cell (CD3ε+) (Center) and B-cell (CD19+) (Right) populations. n = 2 for Tet2+/+ in 5:1 group; n = 4 for the other samples. *P < 0.05, **P < 0.005 (Student's t test).
Sustained Progenitor Properties and Premature Initiation of Myeloid Developmental Program in Tet2-Deficient HSPCs in Vitro.
shRNA-mediated depletion of Tet2 has been shown to alter differentiation of HSPCs toward myeloid lineages and contribute to prolonged maintenance of stem/progenitor cell markers in vitro (17, 28). To confirm these observations, LSK cells isolated from the bone marrow of Tet2+/+ or Tet2−/− mice were cultured in vitro in the presence of two different combinations of cytokines: stem cell factor (SCF), thrombopoietin (TPO), interleukin (IL)-3, and IL-6 or SCF, TPO, and granulocyte-macrophage colony stimulating factor (GM-CSF). The majority of Tet2−/− LSK cells maintained progenitor properties with lower expression of lineage markers, whereas most of the Tet2+/+ LSK cells underwent differentiation in vitro (Fig. 5A, Upper). As reported (28), the fraction of c-Kit+ cells and cells displaying phenotypes similar to LSK were slightly greater in Tet2−/− cell culture than in Tet2+/+ cell culture (Fig. S6 A and B). A significant proportion of Tet2−/− cells expressed lower levels of the myeloid markers Gr-1 and Mac-1, relative to Tet2+/+ cells, which expressed them at relatively high levels (Fig. 5A, Lower), suggesting that Tet2 is a negative regulator of HSC differentiation toward the myeloid lineage. Notably, loss of Tet2 induced developmental skewing with increased production of CD115+F4/80+ monocyte/macrophage cells (Fig. 5B, Upper). Analysis of Gr-1 and Mac-1 expression within CD115+F4/80+ cells suggested that Tet2 deficiency induced premature differentiation of immature progenitor cells expressing low levels of myeloid markers toward the monocyte/macrophage lineage (Fig. 5B, Lower). Similar data were obtained when lineage-negative bone marrow cells were used. Tet2 deficiency did not significantly affect the extent of myeloid differentiation of the LK myeloid progenitor compartment as assessed by Gr-1 and Mac-1 expression, but it still promoted monocyte/macrophage differentiation as assessed by expression of CD115 and F4/80 (Fig. S6C). The increased number of CD115+F4/80+ cells was due at least in part to increased proliferation: CD115+ cells in Tet2−/− cultures incorporated significantly more 5-bromo-2-deoxyuridine (BrdU), a thymidine analog, during an acute pulse in vitro compared with CD115+ cells in WT cultures (Fig. S7). Collectively, these results suggest that Tet2 negatively regulates differentiation of HSCs in vitro, but it also inhibits premature initiation of the monocyte/macrophage differentiation program in progenitor cells.
Fig. 5.
Tet2-deficient hematopoietic stem cells retain sustained progenitor properties in vitro but preferentially undergo monocyte/macrophage differentiation. (A) Effect of Tet2 deficiency on myeloid differentiation. LSK cells isolated from bone marrow of Tet2+/+ or Tet2−/− mice were grown in the presence of cytokine mixtures as shown. After 5 d, flow cytometric analysis was performed for the expression of lineage markers (Upper) and Gr-1 and Mac-1, myeloid markers (Lower). Data are representative of at least two independent experiments (n = 3–5). (B) The cells in A were analyzed for their expression of CD115 (M-CSFR) and F4/80, monocyte/macrophage markers (Upper). The CD115+F4/80+ cells were further assessed for their expression of myeloid markers (Lower). Data are representative of at least two independent experiments (n = 3–5).
Discussion
To clarify the function of Tet2 in hematopoiesis, we generated mice with a systemic deletion of the Tet2 catalytic domain. We find that ablation of Tet2 diminishes genomic 5hmC levels in all organs tested (Fig. 1B) and causes an increase in cellularity in the bone marrow and in the frequency and number of HSPCs (Fig. 2). Transplantation of Tet2+/+ and Tet2−/− bone marrow cells into congenic WT mice confirmed a cell-intrinsic function for Tet2 in stem cells (Fig. 3), and competitive repopulation experiments demonstrated the enhanced ability of Tet2−/− HSCs, relative to WT HSCs, to reconstitute hematopoiesis in vivo (Fig. 4). Finally, Tet2 deficiency restrains HSCs from undergoing differentiation in vitro, as assessed by expression of lineage markers upon differentiation (Fig. 5). Despite this antagonizing effect, however, the number of monocyte/macrophage cells was greater in Tet2−/− cultures compared with controls, and immature Tet2−/− progenitor cells differentiated prematurely into cells of the monocyte/macrophage lineage (Fig. 5 and Fig. S6). These results indicate that Tet2 deficiency alters stem/progenitor cell properties to induce developmental skewing toward the monocyte/macrophage lineage. Although Tet1 and Tet3 are expressed in hematopoietic stem/progenitor cells (17) and share similar catalytic function (14, 16–18), they are not up-regulated in Tet2-deficient cells and apparently do not compensate sufficiently for the loss-of-function of Tet2 (Fig. 1).
Our data are consistent with recent characterizations of three other strains of Tet2 gene-disrupted mice (29, 31): hypomorphic gene trap mice and conditional gene-disrupted mice in which either exon 3 or exon 11 in the Tet2 locus was targeted. Tet2 deficiency gave rise to similar phenotypes in all studies. Loss of Tet2 enhanced the serial replating ability of HSPCs in vitro but antagonized HSPC differentiation; Tet2-deficient mice had enlarged HSPC compartments and the cells possessed a competitive advantage upon hematopoietic reconstitution in vivo; and haploinsufficiency of Tet2 sufficed to induce aberrant hematopoiesis. The data point to a role for Tet2 in homeostatic regulation, in which Tet2 restrains aberrant self-renewal and expansion of HSCs. In contrast in ES cells, 5hmC and Tet1/Tet2 expression levels correlate with pluripotent status (16, 18), and depletion of Tet1 and Tet2 impaired self-renewal and lineage specification (18). Thus, the role of Tet proteins in controlling the function of stem cells is divergent depending on the cellular context.
Loss-of-function mutations in TET2 represent one of the most common acquired mutations in patients with myeloid malignancies (17, 20–25, 32). The data suggest that decreased TET2 activity in stem/progenitor cells potentiates myeloid transformation by influencing not only HSCs, but also cells such as myeloid progenitors that are further along in the differentiation pathway compared with HSCs (33). TET2-mutated human CD34+ stem cells isolated from MPD patients reconstituted hematopoiesis in NOD-SCID mice and displayed skewed differentiation toward myeloid lineages at the expense of lymphoid lineages (22); similarly, TET2 depletion in human cord blood CD34+ progenitor cells impaired lymphoid and erythroid differentiation but promoted monocyte differentiation at the expense of granulocyte development (27). In the mouse hematopoietic system, Tet2−/− progenitor cells expanded in vitro were shown to share gene expression signatures with CMPs, with increased expression of genes related to self-renewal (29). Our own studies show that Tet2-depleted HSPCs (17) as well as sorted Tet2-deficient LSK and LK cells (Fig. 5, Fig. S6, and Fig. S7) are preferentially committed to differentiate into cells of the monocyte/macrophage lineage; in another study, however, Tet2 depletion led to an increase in HSPCs but impaired myelopoiesis in vitro (28). Interestingly, ectopic expression of recurrently mutated forms of the metabolic enzymes isocitrate dehydrogenase (IDH) 1 and IDH2 had similar effects; these mutant enzymes act dominantly to produce the “oncometabolite” 2-hydroxyglutarate, which impairs the catalytic activity of TET2 and other Fe2+/α-ketoglutarate–dependent dioxygenases, apparently by competing with the substrate α-ketoglutarate (28, 34). Overall, a plausible hypothesis is that the functions and cell fate decisions of HSPC are affected by mutations in TET2 or IDH1/IDH2 that decrease TET2 activity, and that this phenomenon has a crucial role in the pathogenesis of diverse myeloid malignancies (26, 32, 35).
It is well established that TET2 mutations are most frequent in CMML (20, 21). Ablation of Tet2 in mice was sufficient to induce a CMML-like disease, featuring persistent monocytosis and splenomegaly associated with myeloproliferation (29, 31), and Tet2 deficiency in LK cells induced developmental skewing toward the monocytic lineage (Fig. S6C). In patients with CMML, however, TET2 mutations frequently coexist with somatic mutations in genes encoding other signaling and epigenetic regulators such as JAK2, ASXL1, EZH2, CBL, and CBLB (36, 37). Presumably, full-blown myeloid malignancies in humans involve additional genetic alterations that occur before or after TET2 mutations (32, 35, 38). Notably, loss-of-function mutations in TET1 or TET3 have not been reported in most TET2-mutated cancers (17, 21). If TET2 loss-of-function contributes to myeloid transformation because of impaired 5hmC production, it might be beneficial from the perspective of cancer therapies to develop strategies to activate the enzymatic activity of other TET proteins.
Is the tumor suppressor function of TET2 related to its ability to alter DNA methylation patterns by consuming 5mC? The available data on the effect of TET2 loss-of-function on CpG methylation are contradictory. In our own studies, we examined DNA methylation status of bone marrow samples from patients with diverse myeloid malignancies (MDS and CMML, with a few MPN and sAML; TET2 mutant patients largely had CMML). We grouped patients by high versus low 5hmC, rather than WT and mutant TET2, and analyzed DNA methylation patterns by using the Illumina Infinium Methylation array (17). In contrast, Figueroa et al. analyzed only patients with de novo AML, classified as expressing WT or mutant TET2 (28), and used the HELP assay (39) to profile methylation status. We found global hypomethylation at most differentially methylated CpG sites in 5hmC low versus high samples (17), whereas Figueroa et al. found localized hypermethylation at several HpaII sites in de novo AML (28). There are several explanations for the discrepancy, including cancer subtype; the use of different platforms for profiling, which results in almost complete lack of overlap in the sites examined; and the use of different statistical methods for analysis of the data. Thus, the effects of TET2 and DNMT3A mutations on DNA methylation status remain to be elucidated.
Recent genome-wide analysis of Tet1 and 5hmC localization in ES cells revealed that Tet1 and 5hmC are enriched in regions with specific histone modifications related to unique transcriptional regulation (40, 41). Therefore, TET2 mutations may alter gene expression in a more complex way that involves not only changes in DNA methylation status, but also deregulated histone modifications. Our studies show that in ES cells, 5hmC-containing regions contain dual H3K4 and H3K27 trimethylation, a “bivalent” mark characteristic of genes that are “poised” to be up-regulated upon ES cell differentiation (41). Similarly, Tet2 expression and/or 5hmC might be markers for genes whose expression changes in HSCs in response to self-renewal signals. Future studies should focus on how TET2 loss-of-function alters DNA methylation and/or histone modifications at specific genetic loci implicated in controlling the function, self-renewal, and differentiation of HSCs.
Materials and Methods
Generation of Tet2-Deficient mice.
We targeted the endogenous Tet2 locus to generate a conditional allele that would enable deletion of exons 8, 9, and 10 (Fig. S1A). These exons encode the bulk of the double-stranded beta-helix (DSBH) domain, which contains the catalytic triad (HxD…R) required for the enzymatic functions of Tet2 (15). Site-specific insertion of LoxP sites into introns upstream of exon 8 and downstream of exon 10 was achieved by standard gene targeting protocols. Briefly, a targeting vector comprising upstream and downstream homology arms, an FRT-flanked neomycin resistance cassette, and Loxp-flanked central region was generated as depicted in Fig. S1A. The targeting vector was linearized and electroporated into ART B6-3 (genetic background: C57BL/6 NTac) ES cells. Twenty-four hours after transfection, the ES cells were selected by using G418, and colonies of resistant clones were isolated and expanded. Correct integration of the targeted construct at the endogenous Tet2 locus via homologous recombination was ascertained by Southern blot analysis of EcoRI or EcoRV digested genomic DNA as shown in Fig. S1B. Two correctly targeted clones were injected into blastocysts, which were subsequently implanted into B6-albino females. The current studies were conducted from mice derived from one germ-line transmitted clone, H8. The transmission of the targeted locus was guided by coat color of the progeny and confirmed by PCR analyses of DNA obtained from ear clips. Progenies bearing the targeted floxed allele were mated with FLP-E deletor mice to delete the neomycin resistance cassette, and subsequent progenies were bred to CMV-CRE deletor mice to globally delete Tet2 exons 8–10 in all tissues. Tet2+/− mice were viable and fertile and transmitted a germ-line deleted allele at a Mendelian ratio to their progeny. F1 crosses of Tet2+/− mice produced Tet2−/− mice at a Mendelian ratio without any evidence for embryonic lethality.
Bone Marrow Transplantation and Competitive Repopulation Assays.
Total bone marrow cells were isolated from donor (CD45.2+) Tet2+/+ or Tet2−/− mice, and red blood cells were depleted by incubation with ACK lysis buffer (0.15 M NH4Cl, 10 mM KHCO3, and 0.1 mM EDTA) at room temperature for 1 min. Cells (2 × 106) were injected i.v. into lethally irradiated (10 Gy) WT congenic (CD45.1+) B6.SJL mice. For competitive repopulation assays, 106 total bone marrow cells derived from donor (CD45.2+) Tet2+/+ or Tet2−/− mice were mixed at ratios of 5:1, 1:1, or 1:5 with competitor (CD45.1+) bone marrow cells, and the mixtures were injected into lethally irradiated (10 Gy) congenic (CD45.1+) B6.SJL mice. To assess donor/competitor chimerism in hematopoietic subsets, peripheral blood was collected from chimeric mice at 6–7 or 12 wk after transplantation, depleted of red blood cells, and stained with combinations of antibodies against CD45.1, CD45.2, CD3ε, CD19, and Mac-1 (CD11b).
Flow Cytometry and Cell Sorting.
Bone marrow cells were flushed out of two femurs and two tibias of 8- to 12-wk-old mouse, and red blood cells were depleted. Thymocytes and splenocytes were prepared by mincing thymus and spleen, respectively, onto a 70-μm cell strainer (BD Biosciences). Cells were stained with monoclonal antibodies in PBS containing 1% heat-inactivated FBS and 0.1% (wt/vol) sodium azide. Biotinylated antibodies were revealed by PerCP-Cy5.5–conjugated streptavidin (BD Biosciences). Flow cytometric analyses and cell sorting were performed by using FACS Canto II or FACSAria II flow cytometers (BD Biosciences), and data were analyzed by using FlowJo software. LSK (Lin−c-Kit+Sca-1+) and LK (Lin−c-Kit+Sca-1−) cells were isolated by cell sorting.
In Vitro Differentiation Assays.
Upon cell sorting by flow cytometry, LSK, LK, or Lin− BM cells were cultured in complete RPMI 1640 medium [20% FBS, 100 U/mL penicillin, 100 U/mL streptomycin, 50 μg/mL β-mercaptoethanol, 1% Glutamax (GIBCO), 1% nonessential amino acid (GIBCO), and 1% sodium pyruvate (GIBCO)] supplemented with 50 ng/mL SCF and 10 ng/mL TPO with either 6 ng/mL IL-3 and 10 ng/mL IL-6 or 10 ng/mL GM-CSF (all from R&D Systems). After 4–5 d, cell were harvested and analyzed for expression of stem/progenitor markers and myeloid markers by flow cytometry. For BrdU incorporation assays to assess cell proliferation, cells at day 6 were pulse-labeled with 10 μM BrdU (BD Biosciences) for 1 h and stained to evaluate cell surface CD115 (M-CSFR) expression, followed by intracellular staining for BrdU and for DNA content by staining cells with 7-aminoactinomycin D (7-AAD). BrdU incorporation was assessed after gating on CD115+ (M-CSFR+) monocyte/macrophage cells.
Dot Blot Analysis.
Dot blot assays were performed as described (17). Briefly, cells were incubated with 200 μg/mL proteinase K (Roche) overnight at 55 °C, and genomic DNA was isolated by phenol-chloroform extraction. Then, DNA samples were treated with sodium bisulfite by using the EpiTect Bisulfite kit (QIAGEN) and denatured in 0.4 M NaOH, 10 mM EDTA at 95 °C for 10 min, followed by neutralization with cold 2 M ammonium acetate (pH 7.0). Twofold serial dilutions of the denatured DNA samples were spotted on a nitrocellulose membrane in an assembled Bio-Dot apparatus (Bio-Rad) per manufacturer's instructions. The membrane was washed with 2× SSC buffer, air-dried, and vacuum-baked at 80 °C for 2 h, then blocked with 5% nonfat milk for 1 h and incubated with anti-CMS antiserum (1:1,000) overnight at 4 °C. After incubating with horseradish peroxidase-conjugated anti-rabbit IgG secondary antibody, the membrane was visualized by enhanced chemiluminescence.
Real-Time RT-PCR.
Total RNA was prepared with TRIzol reagent (Invitrogen) per manufacturer's instructions, followed by reverse transcription using SuperScript III (Invitrogen). Diluted cDNAs were analyzed by real-time PCR using StepONE plus real-time PCR system (Applied Biosystems) and FastStart Universal SYBR Green Master kit (Roche). Data were analyzed by StepONE plus real-time PCR software. Cycling parameters and primer sequences are described (17).
ACKNOWLEDGMENTS.
We thank members of the A.R. laboratory for valuable discussions. This work was funded by National Institutes of Health (NIH) Grants AI44432, RC1 DA028422, and HD065812 (to A.R.) and R37 AI054636 (to K. R.), a Special Fellow award from the Leukemia and Lymphoma Society (to M.K.), and postdoctoral fellowships from the Lady Tata Memorial Trust and from the GlaxoSmithKline-Immune Disease Institute Alliance (to H.S.B.).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112317108/-/DCSupplemental.
References
- 1.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: Modifications, screening, and therapy. Annu Rev Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 2.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 3.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 4.Gaudet F, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300:489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 5.Ehrlich M. DNA hypomethylation in cancer cells. Epigenomics. 2009;1:239–259. doi: 10.2217/epi.09.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ernst T, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42:722–726. doi: 10.1038/ng.621. [DOI] [PubMed] [Google Scholar]
- 7.van Haaften G, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Walter MJ, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25:1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan XJ, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 11.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5:442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bröske AM, et al. DNA methylation protects hematopoietic stem cell multipotency from myeloerythroid restriction. Nat Genet. 2009;41:1207–1215. doi: 10.1038/ng.463. [DOI] [PubMed] [Google Scholar]
- 13.Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med. 2007;204:715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer LM, Tahiliani M, Rao A, Aravind L. Prediction of novel families of enzymes involved in oxidative and other complex modifications of bases in nucleic acids. Cell Cycle. 2009;8:1698–1710. doi: 10.4161/cc.8.11.8580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito S, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ko M, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koh KP, et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viguié F, et al. Common 4q24 deletion in four cases of hematopoietic malignancy: Early stem cell involvement? Leukemia. 2005;19:1411–1415. doi: 10.1038/sj.leu.2403818. [DOI] [PubMed] [Google Scholar]
- 20.Jankowska AM, et al. Loss of heterozygosity 4q24 and TET2 mutations associated with myelodysplastic/myeloproliferative neoplasms. Blood. 2009;113:6403–6410. doi: 10.1182/blood-2009-02-205690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdel-Wahab O, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood. 2009;114:144–147. doi: 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delhommeau F, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 23.Langemeijer SM, et al. Acquired mutations in TET2 are common in myelodysplastic syndromes. Nat Genet. 2009;41:838–842. doi: 10.1038/ng.391. [DOI] [PubMed] [Google Scholar]
- 24.Tefferi A, et al. Frequent TET2 mutations in systemic mastocytosis: Clinical, KITD816V and FIP1L1-PDGFRA correlates. Leukemia. 2009;23:900–904. doi: 10.1038/leu.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tefferi A, et al. Detection of mutant TET2 in myeloid malignancies other than myeloproliferative neoplasms: CMML, MDS, MDS/MPN and AML. Leukemia. 2009;23:1343–1345. doi: 10.1038/leu.2009.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abdel-Wahab O. Genetics of the myeloproliferative neoplasms. Curr Opin Hematol. 2011;18:117–123. doi: 10.1097/MOH.0b013e328343998e. [DOI] [PubMed] [Google Scholar]
- 27.Pronier E, et al. Inhibition of TET2-mediated conversion of 5-methylcytosine to 5-hydroxymethylcytosine disturbs erythroid and granulo-monocytic differentiation of human hematopoietic progenitors. Blood. 2011 doi: 10.1182/blood-2010-12-324707. 10.1182/blood-2010-12-324707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moran-Crusio K, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell. 2011;20:11–24. doi: 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang YPW, et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS ONE. 2010;5:e8888. doi: 10.1371/journal.pone.0008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quivoron C, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell. 2011;20:25–38. doi: 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Tefferi A. Novel mutations and their functional and clinical relevance in myeloproliferative neoplasms: JAK2, MPL, TET2, ASXL1, CBL, IDH and IKZF1. Leukemia. 2010;24:1128–1138. doi: 10.1038/leu.2010.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaub FX, et al. Clonal analysis of TET2 and JAK2 mutations suggests that TET2 can be a late event in the progression of myeloproliferative neoplasms. Blood. 2010;115:2003–2007. doi: 10.1182/blood-2009-09-245381. [DOI] [PubMed] [Google Scholar]
- 34.Xu W, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Delhommeau F, Jeziorowska D, Marzac C, Casadevall N. Molecular aspects of myeloproliferative neoplasms. Int J Hematol. 2010;91:165–173. doi: 10.1007/s12185-010-0530-z. [DOI] [PubMed] [Google Scholar]
- 36.Grossmann V, et al. Molecular profiling of chronic myelomonocytic leukemia reveals diverse mutations in >80% of patients with TET2 and EZH2 being of high prognostic relevance. Leukemia. 2011;25:877–879. doi: 10.1038/leu.2011.10. [DOI] [PubMed] [Google Scholar]
- 37.Makishima H, et al. CBL, CBLB, TET2, ASXL1, and IDH1/2 mutations and additional chromosomal aberrations constitute molecular events in chronic myelogenous leukemia. Blood. 2011;117:e198–e206. doi: 10.1182/blood-2010-06-292433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acquaviva C, Gelsi-Boyer V, Birnbaum D. Myelodysplastic syndromes: Lost between two states? Leukemia. 2010;24:1–5. doi: 10.1038/leu.2009.157. [DOI] [PubMed] [Google Scholar]
- 39.Figueroa ME, Melnick A, Greally JM. Genome-wide determination of DNA methylation by Hpa II tiny fragment enrichment by ligation-mediated PCR (HELP) for the study of acute leukemias. Methods Mol Biol. 2009;538:395–407. doi: 10.1007/978-1-59745-418-6_20. [DOI] [PubMed] [Google Scholar]
- 40.Wu H, et al. Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature. 2011;473:389–393. doi: 10.1038/nature09934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pastor WA, et al. Genome-wide mapping of 5-hydroxymethylcytosine in embryonic stem cells. Nature. 2011;473:394–397. doi: 10.1038/nature10102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.