Abstract
The WAVE regulatory complex (WRC) is a critical element in the control of actin polymerization at the eukaryotic cell membrane, but how WRC is activated remains uncertain. While Rho GTPase Rac1 can bind and activate WRC in vitro, this interaction is of low affinity, suggesting other factors may be important. By reconstituting WAVE-dependent actin assembly on membrane-coated beads in mammalian cell extracts, we found that Rac1 was not sufficient to engender bead motility, and we uncovered a key requirement for Arf GTPases. In vitro, Rac1 and Arf1 were individually able to bind weakly to recombinant WRC and activate it, but when both GTPases were bound at the membrane, recruitment and concomitant activation of WRC were dramatically enhanced. This cooperativity between the two GTPases was sufficient to induce WAVE-dependent bead motility in cell extracts. Our findings suggest that Arf GTPases may be central components in WAVE signalling, acting directly, alongside Rac1.
Dynamic assembly of the actin cytoskeleton is central to the architecture and movement of eukaryotic cells. Actin polymerization is nucleated by the ubiquitous Arp2/3 complex, which is activated by nucleation promoting factors (NPFs), most prominently N-WASP (neural Wiskott–Aldrich syndrome protein) and the WAVE (WASP family veroprolin homologue) regulatory complex (WRC), which comprises WAVE, Cyfip, Nap1, Abi1, and HSPC300 or their homologues (1). It has been established that purified N-WASP can be activated by the Rho GTPase Cdc42 and the lipid PIP2 (2), which trigger a conformational change in N-WASP, exposing its actin-polymerizing VCA domain (3). In contrast, the mechanism of WRC activation remains relatively unclear. Purified Rho GTPase Rac1 can bind and activate recombinant WRC in vitro (4), and the crystal structure of the WRC identified a potential binding site for Rac1 in Cyfip (5), prompting a proposal that, analogous to Cdc42 activation of N-WASP, binding of Rac1 leads to activation of the WRC by triggering exposure of the WAVE VCA domain. However, the Rac1 interaction with WRC in vitro is of very low affinity, about 8 μM (5), supporting the likelihood that additional factors may be important in WRC activation (6). This may be especially evident at the membrane. We aimed to establish which determinants could be key to this process by reconstituting WAVE-dependent actin polymerization at phospholipid membranes in a complex mammalian brain cell extract.
Results
Reconstitution of WAVE-Dependent Actin Assembly at the Membrane.
It has previously been established that Cdc42/N-WASP-dependent actin assembly can be reconstituted on PIP2-containing liposomes added to mammalian cell extract (7, 8). We used a similar approach to reconstitute Rac1/WAVE-dependent actin polymerization using silica beads coated with a lipid bilayer of phosphatidylcholine (PC), phosphatidylinositol (PI), and a low concentration (4%) of either PIP3 or, as a control, PIP2 (Fig. 1 and Fig. S1A). When added to mammalian cell extract, both beads recruited actin, and when the extract was activated (i.e., by adding GTPγS to stimulate endogenous small GTPases) comet tails were formed immediately on both PIP2 and PIP3 beads, which were propelled through the extract for an hour or longer (Fig. 1 A, i, and B, i, and Movies S1, S2, and S3).
Fig. 1.
Determinants of PIP2- and PIP3-based motility in cell extract. Actin-based motility of silica beads coated with phospholipid bilayers containing PC:PI (phosphatidylcholine:phosphatidylinositol) and either (A) PIP2 or (B) PIP3, depicted in the respective cartoons. Fluorescence microscopy of rhodamine-actin assembly on the PIP2 or PIP3 beads (i) in extract alone or supplemented with GTPγS, and (ii) in extract preincubated with the indicated inhibitors, initiated by adding GTPγS. Insets show magnifications of dotted areas. Scale bars, 10 μm. Fig. S2 shows comparable motility of PIP2 or PIP3 beads coated in bilayers containing phosphatidylcholine (PC), phosphatidyethanolamine (PE), phosphatidylserine (PS), and cholesterol (Ch). (iii) Proteins recruited by PIP2 and PIP3 beads in extract (+GTPγS) shown in (A i and B i). Isolated proteins were separated by consecutive anion and cation chromatography, from which a selected cation heparin fraction (H3) is shown following SDS/PAGE, Coomassie Blue staining and protein identification by mass spectrometry. Comprehensive results are presented in Fig. S3 and Dataset S1. Numbers correspond to protein band labeling retained in Fig. S3.
Actin recruitment and consequent movement by both PIP2 and PIP3 beads was inhibited by preincubation of extract with an inhibitor of Rac1 and Cdc42 (PBD, the GTPase-binding domain from PAK1) (9, 10), or with RhoGDI, which extracts membrane Rho GTPases into an inactive soluble complex (11) (Fig 1 A, ii, and B, ii), confirming reports that Rac1 and/or Cdc42 are required to initiate actin assembly (8). PIP2-induced actin comet tail formation was also prevented by preincubation of extract with a dominant-negative inhibitor of N-WASP (the derivative N-WASPΔVCA) but not by the Rac1 inhibitor NSC23766 (12) (Fig. 1A, ii), while PIP3-induced actin tail formation was inhibited only when both N-WASPΔVCA and NSC23766 inhibitors were added together (Fig. 1B, ii). Immunodepletion of WAVE from the extract efficiently inhibited PIP3 motility in the presence of N-WASPΔVCA (Fig. S1 B and C), confirming that actin assembly on these beads is WAVE-dependent. These assays demonstrate that whereas PIP2 actin-dependent movement is due solely to the Cdc42/N-WASP pathway, both this pathway and the Rac1/WAVE pathway contribute to PIP3 motility.
WAVE Complex Activators: Implicating Arf.
To determine the components driving actin assembly at the membrane, we scaled up the motility assays to isolate PIP2 and PIP3 beads on which comet tails had reached maximum (c. 15 min) and identified the recruited proteins by mass spectrometry. Key single fractions are shown in Fig. 1 A, iii, and B, iii; the full results are presented in Fig. S3 and summarized in Dataset S1. We identified 106 proteins on PIP3 beads and 87 on PIP2 beads; 43 were present on both. Significantly, the only known NPFs associated with either PIP2 or PIP3 were N-WASP and WAVE; neither bead recruited any Arp2/3-independent actin nucleators (e.g., formins). PIP2 and PIP3 beads both achieved N-WASP-dependent motility, and consistent with this Cdc42 and N-WASP were found on both of them (Fig. 1 A, iii, and B, iii). WRC components PIR121, Nap1, Abi1 and WAVE were, like Rac1, also prominent on both PIP2 and PIP3 beads (Fig. 1A, iii and B, iii). The GTPγS-dependent recruitment of these key proteins was reproducible and was confirmed by immunoblotting (Fig. S4), which also identified the smaller fifth WRC component, HSPC300, on both. A significant and surprising observation was that although the WRC was present on PIP2 and PIP3 beads, only PIP3 triggered Rac/WAVE-mediated motility (Fig. 1 A, ii and B, ii). The inability of PIP2 to activate the WRC was not due to Rac1 activation status (Fig. S5). Collectively, these data demonstrated that WRC recruitment and Rac1 activity are not sufficient to activate actin assembly in a complex extract and suggest an additional factor is crucial to WAVE activation at the membrane.
As we sought to identify this missing factor, we noticed that the Arf GAP (inactivator) GIT1 (13) (and its binding partners β-Pix and PAK) was recruited only by PIP2. In contrast, arfaptin, which specifically binds to active Arf GTPases (14), was only detected on PIP3. This suggested that a critical difference could be that the Arf GTPases found on PIP3 beads were active, while those on PIP2 beads were not. We therefore activated the Arfs recruited to PIP2 by prebinding the Arf GEF ARNO (15) [which contains a phosphatidylinositol-binding pleckstrin homology (PH) domain] to the beads before their addition to the extract. Actin comet tails were generated, even when the N-WASP pathway was inhibited (Fig. 2A and Movie S4). This strongly implicated Arf GTPase activity as the missing factor key to WAVE-dependent actin assembly. To confirm this, we initially preincubated extract with brefeldin A, a commonly used inhibitor of Arf. This had no effect on actin comet tail assembly, but this is perhaps not surprising because brefeldin A is not a direct inhibitor of Arf, per se, but actually inhibits a subset of Arf Gefs (16). Consequently, we utilized GAT, a domain of GGA1, which specifically binds and inhibits active, GTP-bound Arf GTPases (17, 18). Preincubation of extract with GAT had no effect on the actin-dependent motility of either PIP2 beads (i.e., N-WASP-dependent) or PIP3 beads (which activate both N-WASP and WAVE; Fig. 2B). However, in N-WASPΔVCA-inhibited extract, GAT abolished PIP3-induced comet tail formation (Fig. 2b). This demonstrated that active Rac is not enough for WRC activation. Arf is required.
Fig. 2.
Requirement for active Arf in WAVE-dependent motility. (A) Actin-based motility of PC:PI-coated beads with PIP2 alone (left) or PIP2 bound by Arf GEF (PIP2ArfGEF) initiated by GTPγS in extract containing N-WASP inhibitor (+N-WASPΔVCA). (B) Motility of PIP2 (left) or PIP3 (middle and right) beads, initiated by GTPγS in extract containing Arf inhibitor (GAT) and where indicated (right) N-WASPΔVCA. Fluorescence microscopy as in Fig. 1. Scale bars, 10 μm.
Arf Interaction with the WAVE Complex.
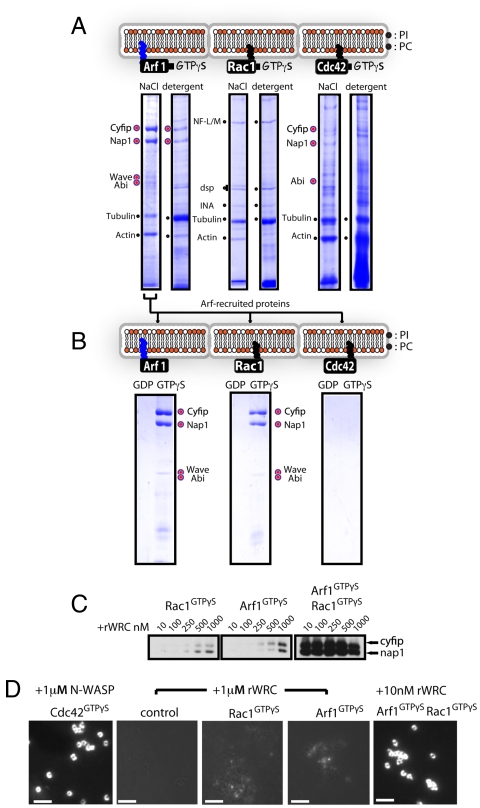
We next focused on the ability of Rac1 and Arf1 to recruit the WRC from extract to the membrane. To eliminate any contributions from the PIP2 and PIP3 phospholipids themselves, purified in vitro acylated Rac1 or Arf1 (or control Cdc42) were each anchored to beads coated with PC:PI alone and activated by loading with GTPγS, before adding to the extract. NaCl extraction of these Arf1GTPγS beads revealed distinct protein bands (Fig. 3A and Fig. S6A), which mass spectrometry identified as WRC components Cyfip, Nap1 (which are structurally homologous and together constitute a subcomplex (5), and in smaller amounts, WAVE and Abi. Detergent solubilization of the lipid bilayer coat to release membrane-associated GTPases and remaining high-affinity proteins again detected Cyfip and Nap1 (Fig. 3A), indicating their strong interaction with the Arf1GTPγS beads. In contrast, no WRC components were detected on Rac1GTPγS beads (Fig. 3A and Fig. S6B and confirmed by Western blotting; Fig. S4), which recruited small GTPases including Cdc42, and nonspecific proteins like tubulin and actin that were also found on control PC:PI beads (Fig. S6C). Control Cdc42GTPγS beads recruited detectable levels of Cyfip, Nap1, and Abi1 but not WAVE (Fig. 3A and Fig. S6D).
Fig. 3.
Binding and activation of WRC by membrane-anchored Arf1, Rac and Cdc42. (A) Proteins recruited from extract by active acylated Arf1GTPγS, Rac1GTPγS, and Cdc42GTPγS, each anchored to PC:PI bilayer-coated beads. Recruited proteins were sequentially extracted with 1 M NaCl, then detergent before SDS/PAGE and Coomassie Blue staining. Proteins identified from prominent bands are indicated, with WRC components marked with large colored circles. Comprehensive results are presented in Fig. S6 and Dataset S1. (B) Binding of WRC components by acylated membrane-anchored Arf1, Rac1, and Cdc42. The 1M NaCl extraction shown in Fig. 3A) was diluted fivefold prior to incubation with beads coated with GTPases loaded with either GDP or GTPγS. (C) Recombinant WRC recruitment from buffer by Arf1 and Rac1. Arf1GTPγS or Rac1GTPγS alone, or in combination, anchored to PC:PI-coated beads were preincubated with indicated concentrations of purified recombinant WAVE regulatory complex (WRC) then washed and isolated beads immunoblotted using antibodies against Cyfip and Nap. (D) Recombinant WRC activation at the membrane in buffer by Arf1 and Rac1. Arf1GTPγS, Rac1GTPγS, or Cdc42GTPγS, alone or in combination, anchored to PC:PI-coated beads were incubated with either purified recombinant N-WASP or the WAVE regulatory complex (rWRC) at the indicated concentrations, then washed prior to addition of Arp2/3 and rhodamine-actin to visualize actin recruitment. Scale bars, 5 μm.
To confirm that Arf1 can directly interact with one or more WRC components, the Cyfip/Nap1-enriched NaCl extraction from the Arf1GTPγS beads (Fig. 3A) was diluted and incubated with lipid-coated beads anchored with either active (GTPγS-bound) or inactive (GDP-bound) forms of each GTPase. Control Cdc42 bound neither Cyfip nor Nap1, nor did Arf1GDP, but Arf1GTPγS bound both (Fig. 3B). An unbiased search of the protein structure database revealed that the structural homologues Cyfip and Nap1 each contain a subdomain that shows a striking similarity to the Arf-binding GAT domain of GGA1 and GGA3 (Fig. S7 A and B), and consistent with this Arf1GTPγS (but not Arf1GDP) bound directly to purified, recombinant Cyfip, Nap1, and Cyfip-Nap1 subcomplex (Fig. S7C). Rac1GTPγS (but not RacGDP) also bound Cyfip and Nap1, compatible with active Rac1GTPγS interacting with purified WRC components (19). But as we showed (Fig. 3A), unlike Arf1, Rac is not sufficient to recruit the WRC in our motility assay. Rac1GTPγS beads showed only weak recruitment of purified recombinant WRC (rWRC), even at micromolar concentrations (Fig. 3C), consistent with the reported low affinity of Rac1 for the WRC in solution (5). Arf1GTPγS beads also recruited rWRC only at high concentrations (Fig. 3C), which was surprising because these beads recruited the complex from brain extract (Fig. 3A). Because Rac1 was present on ArfGTPγS beads isolated from extract (Fig. S6), it seemed the two GTPases might cooperate to bring the WRC efficiently to the membrane. Indeed, anchoring both Arf1GTPγS and Rac1GTPγS to beads allowed recruitment of rWRC when present at low nanomolar concentrations (Fig. 3C). Collectively, these results confirm that while Rac and Arf can interact with the WRC individually, both GTPases are required for efficient, stable recruitment to the membrane.
To determine whether Arf1 and Rac1 can activate purified rWRC at the membrane, acylated GTPγS-bound GTPases were each anchored in turn to PC:PI-coated beads prior to incubation with rWRC, Arp2/3 and fluorescently labeled actin, and WAVE activity monitored by accumulation of actin at the bead surface. In this assay, control Cdc42GTPγS beads preincubated with purified N-WASP efficiently induced actin polymerization at the surface of 95 ± 3.3% of beads, while empty PC:PI-coated beads did not activate rWRC (Fig. 3D). When incubated with 1 μM rWRC (conditions that support rWRC membrane recruitment; Fig. 3C), both Rac1GTPγS beads and Arf1GTPγS beads displayed weak activation, as indicated by a low level of actin localization (typified in Fig. 3D) around 11 ± 3.1% and 14 ± 2.6% of beads, respectively. In contrast, 95 ± 4.8% of beads anchored with both Rac1 and Arf1 were associated with pronounced actin accumulation, even when only 10 nM rWRC was present (Fig. 3D), indicating strong activation.
WAVE Complex Activation by Cooperating GTPases Arf and Rac1.
We next assessed Arf and Rac cooperativity in activating endogenous WRC in brain extract. Acylated GTPγS-bound Arf1GTPγS and Rac1GTPγS (and control Cdc42GTPγS) were again (as in Figs 1 and 2) anchored individually to PC:PI-coated beads prior to motility assay, but no GTPγS was added to the extract (i.e., endogenous GTPases were not activated). Cdc42GTPγS beads generated comet tails and moved in response to the N-WASP pathway, whereas beads anchored with Rac1GTPγS did not move (Fig. 4A), again consistent with the inability of Rac to recruit the WRC to the membrane (Fig. 3A). Arf1GTPγS beads induced formation of actin comet tails shorter than those induced by PIP3 but only after a 3–4 min lag (Fig. 4A). This initially promoted bead motility, but in contrast to PIP3-induced movement it was not sustained beyond 15 min. Arf1GTPγS beads, like PIP3 beads, also moved in N-WASP-inhibited extract (Fig. 4A and Movie S5). When we activated endogenous GTPases by adding GTPγS, the Arf1GTPγS beads formed long comet tails, allowing sustained motility (Fig. 4B and Movie S6). Arf1GTPγS bead motility was abolished in extract preincubated with the PBD inhibitor (Fig. 4B), suggesting that Rac1 enhanced the Arf1-induced movement. This was confirmed by coanchoring Rac1GTPγS and Arf1GTPγS to PC:PI-coated beads, which when added to extract immediately generated long actin tails that sustained movement comparable to that of PIP3 beads, even in the presence of N-WASPΔVCA (Fig. 4B and Movie S7). This motility was inhibited by addition of PBD or GAT, emphasizing that active GTP-bound Rac1 and Arf1 are both required to activate the WRC.
Fig. 4.
WRC activation by Arf family GTPases. Motility of PC:PI-coated beads anchored with one or two activated GTPase(s). (A) Arf1GTPγS, Rac1GTPγS, or Cdc42GTPγS anchored to beads in extract alone or with added N-WASP inhibitor (+N-WASPΔVCA). (B) Arf1GTPγS-anchored beads in extract with additional free GTPγS (left), or coanchored Rac1GTPγS (right). Extract contained N-WASPΔVCA alone (top), or inhibitors of Rac (PBD, middle) or Arf (GAT, bottom). (C) Arf5GTPγS or Arl1GTPγS anchored beads, coanchored with Rac1GTPγS, in the same extracts as B. Scale bars, 10 μm.
WAVE Activation by Other Arf Family GTPases.
To determine whether Arf1’s ability to recruit and activate the WRC is common to other Arf family GTPases we repeated the recruitment and motility experiments described in Figs. 3A and 4 with the closely related Arf5 or the more distant Arl1. Each Arf GTPase recruited the WRC to the membrane (Fig. S8) and triggered WAVE-dependent bead motility (i.e., in N-WASPΔVCA-inhibited extract), either alone or when coanchored with Rac1GTPγS (Fig. 4C). Motility of Arf5 and Arl1 beads was prevented by PBD, but only Arf5-dependent motility was abolished by GAT, which binds active Arfs but not Arls (Fig. 4C). Arl1, like Arf5 and Arf1, therefore acts directly and not via activation of other Arfs.
Discussion
How the WRC is activated to promote actin assembly is an important question in cell biology. A previous study (4) has reported in vitro activation of immunopurified WRC by weakly bound Rac1 (when present with acidic phospholipids). Here, by using a reconstitution system comprising lipid-coated beads added to a complex cell extract, we established that Rac1 is necessary but not sufficient to induce WAVE-dependent actin motility; another unknown factor was required. Extensive attempts at reconstitution assays and systematic mass spectrometry analysis of the protein platforms recruited to the lipid-coated beads implicated Arf as the missing factor. By using specific inhibitors of Rac1, Cdc42, and Arf GTPases, we showed that Arf activity is required for WAVE-mediated bead motility. In a second step, we uncovered the role of Arf by anchoring recombinant acylated Arf and Rac1 to lipid-coated beads, which enabled us to directly demonstrate synergy between Rac1 and Arf in recruitment and activation of the WRC. Acylated Rac1 alone promoted a low level of activation of recombinant WRC at the surface of lipid-coated beads. However, when Arf was also present on the membrane, WAVE activation was dramatically enhanced and could be achieved with much lower concentrations of WRC. We went on to show that Arf directly binds both Cyfip and Nap1, structurally homologous components of the WRC. We could not achieve saturatable binding to Rac1 or Arf beads even when WRC was present at 1 μM, suggesting that the apparent KD for both Arf and Rac1 is > 1 μM. However, when both GTPases were present, the apparent KD is much stronger, as efficient binding was seen even at low nM concentrations of WRC. This could be because the low affinity binding of one GTPase triggers a conformational change in the WRC that increases the affinity for the second GTPase, or it may simply be the result of increased avidity. The role of Arf binding does not seem to be limited to increasing the affinity of WRC binding as, like Rac1, Arf alone could also induce WRC activity. Our findings open the possibility that both these small GTPases, Arf and Rac1, play a direct cooperative role in triggering the conformational changes in the WRC that lead to VCA domain exposure and consequent actin assembly.
Although Arf GTPases are best known for their role in vesicular transport, they are increasingly recognized as key regulators of the actin cytoskeleton at the plasma membrane (20). Expression of dominant-negative Arf derivatives, or the depletion of Arfs by RNAi, has been shown to inhibit Rac-dependent breast cancer cell migration (21), membrane ruffling (22), neurite outgrowth (23), and phagocytosis (24), whereas expression of the Arf GEF ARNO induces the formation of lamellipodia and increases motility in epithelial cells (25). Evidence suggests that Arfs, especially Arf6, can act upstream of Rac1, promoting its activation and/or traffic to the membrane (25). However, forced targeting of a constitutively active Rac1 mutant to the plasma membrane could not relieve the inhibition of membrane ruffling caused by expression of a dominant-negative Arf6 construct (26). Our findings in the cell extract reveal a more direct function for Arfs in Rac1-mediated actin rearrangements, showing that Arf can act directly alongside Rac1 to play a central role in recruitment and activation of the WRC.
Our experiments demonstrated that recruitment and activation of the WRC at the membrane is not unique to Arf1 but can be similarly directed by Arf5 and Arl1, close and distant members of the Arf GTPase family, respectively. Indeed Arf family GTPases appear to have at least partially redundant or overlapping functions, because RNAi-mediated knockdown of indiviual Arfs had little impact on Golgi morphology, whereas knockdown of multiple Arfs had profound effects (27). We showed that Arf activity alone was not sufficient to activate WAVE in the cell extracts; Rac1 was also required, suggesting that Arf activation might only trigger WRC-dependent actin assembly if it is spatially and temporally coincident with Rac1 activation. Rac1 is best known for promoting WRC activity at the plasma membrane. Although Arf6 is the most prominent Arf family member found at the plasma membrane, Arfs 1–5 cycle through the cytosol when inactive and are thus available to bind to other membrane compartments and could therefore be available to activate WRC. Likewise Rac1, although predominantly found at the plasma membrane, has also been demonstrated to form complexes at other known sites of Arf action—e.g., the Golgi (28).
The specificity of Arf signaling seems to be determined by the large family of Arf GEFs, which activate Arfs at specific cellular locations to produce precise responses (16), so it is likely that GEFs will play a significant role in determining which Arfs promote WRC activation, and where this occurs, in vivo. There are multiple examples of the coordinate regulation of both Rac and Arf GTPases. For example, the Arf GEF ARNO forms a complex with the Rac GEF DOCK180 to promote lamellipodia formation and cell migration (29). Significantly, a mutant ARNO derivative that is unable to activate Arf but can still recruit DOCK180 (and therefore trigger Rac1 activity) fails to induce actin rearrangements (30), suggesting a direct role for Arf. A negative regulator of Arf signalling, the Arf GAP GIT1, also forms a complex with a Rac1 GEF, β-PIX, and during α4 integrin-mediated migration has been proposed to localize to the sides and rear of motile cells where it functions to inhibit Rac1-mediated actin assembly, thereby limiting protrusion to the cell’s leading edge (31). The Slit2-Robo4 complex promotes vascular stablity by recruiting GIT1 to inhibit Rac1-dependent protrusive activity (32).
Our results also appear to strengthen the possibility that WRC could function in the many cellular pathways known to be controlled by Arf GTPases. Arfs 1–5 localize predominantly to the Golgi where they are required for correct Golgi morpholgy (27). The WRC has also been shown to be required for maintainence of Golgi structure in Drosophila cells (33). Arf1 also acts to recruit the adaptor protein AP-1 to the trans-Golgi network (TGN) to mediate traffic between the TGN, endosomes, and the plasma membrane, and the WRC has been found colocalized with these AP-1 complexes (34). Additional work will be required to decipher the potential role of WRC away from the plasma membrane.
The in vivo importance and mechanistic details of our findings will, as is the case with Rac1, require further studies. Due to the redundancy of Arf family members and potential pleiotropic effects of their knockdown, it is likely that focusing on the regulators of Arf signalling (GEFs and GAPs) will be required to illuminate this pathway clearly. Deciphering the dual and seemingly cooperative actions of the two key GTPases will have a significant impact on our understanding of the regulation of cytoskeletal structure and function.
Methods
Assay of Actin-Based Motility by Phospholipid-Coated Beads.
A 60 μl motility-mix (extract) was prepared on ice in the following order: 40 μl brain extract, 3 μl 20x energy mix (300 mM creatine phosphate, 40 mM MgCl2, 40 mM ATP), 3 μl G-actin/rhodamine actin (140 μM, prepared as described) (35), 6 μl 10x salt buffer (600 mM KCL, 200 mM 3-phosphoglycerate), 6 μl 50 mM BAPTA (Merck) and 1 μl 300 mM DTT (Merck) and, when appropriate, 1 μl 30 mM GTPγS (Roche). Actin-dependent motility assays were initiated by adding 0.1 vol phospholipids-coated beads to 10 μl motility mix, then1 μl was applied to a microscope slide and sealed under a glass coverslip with Vaseline:lanolin:paraffin (1∶1∶1), before viewing immediately under a fluorescence microscope (Leica DM IRBE) at RT. Digital images were captured (CCD camera, Hamamatsu) and analyzed (Volocity, Improvision) then figures assembled using Adobe Photoshop and Illustrator CS3. Movies of actin-based motility were captured at three frames per min and shown at 12 frames per second. Comparable comet tails were observed with 0.1 μm to 3.0 μm beads (e.g., Movie S8).
When indicated, extract was preincubated with inhibitors before addition of beads. No inhibition was observed with any of the inhibitors once polymerization was initiated. Recombinant GDI and PAK-PBD were used at 0.5 mM final concentration, the Rac inhibitor NSC23766 (Merck) at 1 mM. N-WASPΔVCA was used at 5 μM, 10-fold higher than required to abolish PIP2 bead motility. Later assays of WAVE-dependent motility were performed in extract preincubated with 10 mM N-WASPΔVCA (“N-WASP-inhibited extract”). For complete inhibition, GAT was used at 30 μM (Kd ∼ 7 μM) for PIP2/PIP3 motility, and at 70 μM for assays with recombinant Arfs.
For protein isolation, actin-motility assays were scaled up to 30 mL then incubated (20 mins, RT) before phospholipid-coated beads were isolated by low-speed centrifugation (1,000 × g) and washed 10 times in HKS buffer supplemented with 1 mM MgCl2 (HKSM). Bead-associated proteins were extracted by rotation (5 mins) in HKSM supplemented with 1 M NaCl prior to extraction of remaining bound proteins with HKSM supplemented with 1% T × 100 detergent. Ionic strength in the extraction was lowered to 50 mM by dilution, then the detergent-extracted proteins were loaded consecutively onto 1ml Heparin and Q-sepharose ion-exchange chromatography columns (GE Healthcare), washed with 10-column volumes of HKSM buffer, then each column eluted over a 10-column volume linear 0–1 M NaCl gradient.
Assay of WAVE Complex Activation on Lipid-Coated Beads.
WAVE complex activation on lipid-coated beads was assayed essentially as previously (36). Briefly, acylated GTPases were anchored to PC:PI-coated beads and incubated with purified recombinant WAVE complex in HKSM buffer for 10 mins on ice. Beads were then washed and resuspended in HKSM buffer prior to sequential addition of 1 mM ATP, 2 mg/mL BSA, 50 nM Arp2/3 complex and 2 μM G-actin/rhodamine-actin. Actin polymerization was visualized by fluorescence.
Supplementary Material
Acknowledgments.
We thank Colin Hughes and Donald Tipper for advice and critical reading of the manuscript; Patrick Casey, John Collard, Shamshad Cockroft, David Owen, John Sondeck, and Martin Spiess for providing plasmids; and Michael Rosen for providing purified recombinant WRC proteins. This work was supported by the Wellcome Trust and the Cambridge Isaac Newton Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1107666108/-/DCSupplemental.
References
- 1.Campellone KG, Welch MD. A nucleator arms race: Cellular control of actin assembly. Nat Rev Mol Cell Biol. 2010;11:237–251. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rohatgi R, Ho HY, Kirschner MW. Mechanism of N-WASP activation by CDC42 and phosphatidylinositol 4, 5-bisphosphate. J Cell Biol. 2000;150:1299–1310. doi: 10.1083/jcb.150.6.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim AS, Kakalis LT, Abdul-Manan N, Liu GA, Rosen MK. Autoinhibition and activation mechanisms of the Wiskott–Aldrich syndrome protein. Nature. 2000;404:151–158. doi: 10.1038/35004513. [DOI] [PubMed] [Google Scholar]
- 4.Lebensohn AM, Kirschner MW. Activation of the WAVE complex by coincident signals controls actin assembly. Mol Cell. 2009;36:512–524. doi: 10.1016/j.molcel.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen Z, et al. Structure and control of the actin regulatory WAVE complex. Nature. 2010;468:533–538. doi: 10.1038/nature09623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davidson AJ, Insall RH. Actin-based motility: WAVE regulatory complex structure reopens old SCARs. Curr Biol. 2011;21:R66–68. doi: 10.1016/j.cub.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 7.Ho HY, et al. Toca-1 mediates Cdc42-dependent actin nucleation by activating the N-WASP-WIP complex. Cell. 2004;118:203–216. doi: 10.1016/j.cell.2004.06.027. [DOI] [PubMed] [Google Scholar]
- 8.Ma L, Cantley LC, Janmey PA, Kirschner MW. Corequirement of specific phosphoinositides and small GTP-binding protein Cdc42 in inducing actin assembly in Xenopus egg extracts. J Cell Biol. 1998;140:1125–1136. doi: 10.1083/jcb.140.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Benard V, Bohl BP, Bokoch GM. Characterization of rac and cdc42 activation in chemoattractant-stimulated human neutrophils using a novel assay for active GTPases. J Biol Chem. 1999;274:13198–13204. doi: 10.1074/jbc.274.19.13198. [DOI] [PubMed] [Google Scholar]
- 10.Knight-Krajewski S, et al. Deregulation of the Rho GTPase, Rac1, suppresses cyclin-dependent kinase inhibitor p21(CIP1) levels in androgen-independent human prostate cancer cells. Oncogene. 2004;23:5513–5522. doi: 10.1038/sj.onc.1207708. [DOI] [PubMed] [Google Scholar]
- 11.Fukumoto Y, et al. Molecular cloning and characterization of a novel type of regulatory protein (GDI) for the rho proteins, ras p21-like small GTP-binding proteins. Oncogene. 1990;5:1321–1328. [PubMed] [Google Scholar]
- 12.Gao Y, Dickerson JB, Guo F, Zheng J, Zheng Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc Natl Acad Sci USA. 2004;101:7618–7623. doi: 10.1073/pnas.0307512101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Premont RT, et al. beta2-Adrenergic receptor regulation by GIT1, a G protein-coupled receptor kinase-associated ADP ribosylation factor GTPase-activating protein. Proc Natl Acad Sci USA. 1998;95:14082–14087. doi: 10.1073/pnas.95.24.14082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanoh H, Williger BT, Exton JH. Arfaptin 1, a putative cytosolic target protein of ADP-ribosylation factor, is recruited to Golgi membranes. J Biol Chem. 1997;272:5421–5429. doi: 10.1074/jbc.272.9.5421. [DOI] [PubMed] [Google Scholar]
- 15.Chardin P, et al. A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature. 1996;384:481–484. doi: 10.1038/384481a0. [DOI] [PubMed] [Google Scholar]
- 16.Casanova JE. Regulation of Arf activation: The Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 17.Collins BM, Watson PJ, Owen DJ. The structure of the GGA1-GAT domain reveals the molecular basis for ARF binding and membrane association of GGAs. Dev Cell. 2003;4:321–332. doi: 10.1016/s1534-5807(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 18.Nakayama K, Takatsu H. Analysis of Arf interaction with GGAs in vitro and in vivo. Methods Enzymol. 2005;404:367–377. doi: 10.1016/S0076-6879(05)04032-2. [DOI] [PubMed] [Google Scholar]
- 19.Kobayashi K, et al. p140Sra-1 (specifically Rac1-associated protein) is a novel specific target for Rac1 small GTPase. J Biol Chem. 1998;273:291–295. doi: 10.1074/jbc.273.1.291. [DOI] [PubMed] [Google Scholar]
- 20.Myers KR, Casanova JE. Regulation of actin cytoskeleton dynamics by Arf-family GTPases. Trends Cell Biol. 2008;18:184–192. doi: 10.1016/j.tcb.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boulay PL, Cotton M, Melancon P, Claing A. ADP-ribosylation factor 1 controls the activation of the phosphatidylinositol 3-kinase pathway to regulate epidermal growth factor-dependent growth and migration of breast cancer cells. J Biol Chem. 2008;283:36425–36434. doi: 10.1074/jbc.M803603200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Radhakrishna H, Al-Awar O, Khachikian Z, Donaldson JG. ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J Cell Sci. 1999;112:855–866. doi: 10.1242/jcs.112.6.855. [DOI] [PubMed] [Google Scholar]
- 23.Albertinazzi C, Za L, Paris S, de Curtis I. ADP-ribosylation factor 6 and a functional PIX/p95-APP1 complex are required for Rac1B-mediated neurite outgrowth. Mol Biol Cell. 2003;14:1295–1307. doi: 10.1091/mbc.E02-07-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Q, Cox D, Tseng CC, Donaldson JG, Greenberg S. A requirement for ARF6 in Fcgamma receptor-mediated phagocytosis in macrophages. J Biol Chem. 1998;273:19977–19981. doi: 10.1074/jbc.273.32.19977. [DOI] [PubMed] [Google Scholar]
- 25.Santy LC, Casanova JE. Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J Cell Biol. 2001;154:599–610. doi: 10.1083/jcb.200104019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Calafat J, Janssen H, Greenberg S. ARF6 is required for growth factor- and rac-mediated membrane ruffling in macrophages at a stage distal to rac membrane targeting. Mol Cell Biol. 1999;19:8158–8168. doi: 10.1128/mcb.19.12.8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volpicelli-Daley LA, Li Y, Zhang CJ, Kahn RA. Isoform-selective effects of the depletion of ADP-ribosylation factors 1–5 on membrane traffic. Mol Biol Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anitei M, et al. Protein complexes containing CYFIP/Sra/PIR121 coordinate Arf1 and Rac1 signalling during clathrin-AP-1-coated carrier biogenesis at the TGN. Nat Cell Biol. 2010;12:330–340. doi: 10.1038/ncb2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Santy LC, Ravichandran KS, Casanova JE. The DOCK180/Elmo complex couples ARNO-mediated Arf6 activation to the downstream activation of Rac1. Curr Biol. 2005;15:1749–1754. doi: 10.1016/j.cub.2005.08.052. [DOI] [PubMed] [Google Scholar]
- 30.White DT, McShea KM, Attar MA, Santy LC. GRASP and IPCEF promote ARF-to-Rac signaling and cell migration by coordinating the association of ARNO/cytohesin 2 with Dock180. Mol Biol Cell. 2010;21:562–571. doi: 10.1091/mbc.E09-03-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishiya N, Kiosses WB, Han J, Ginsberg MH. An alpha4 integrin-paxillin-Arf-GAP complex restricts Rac activation to the leading edge of migrating cells. Nat Cell Biol. 2005;7:343–352. doi: 10.1038/ncb1234. [DOI] [PubMed] [Google Scholar]
- 32.Jones CA, et al. Slit2-Robo4 signalling promotes vascular stability by blocking Arf6 activity. Nat Cell Biol. 2009;11:1325–1331. doi: 10.1038/ncb1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondylis V, et al. The golgi comprises a paired stack that is separated at G2 by modulation of the actin cytoskeleton through Abi and Scar/WAVE. Dev Cell. 2007;12:901–915. doi: 10.1016/j.devcel.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Baust T, Czupalla C, Krause E, Bourel-Bonnet L, Hoflack B. Proteomic analysis of adaptor protein 1A coats selectively assembled on liposomes. Proc Natl Acad Sci USA. 2006;103:3159–3164. doi: 10.1073/pnas.0511062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGhie EJ, Hayward RD, Koronakis V. Control of actin turnover by a salmonella invasion protein. Mol Cell. 2004;13:497–510. doi: 10.1016/s1097-2765(04)00053-x. [DOI] [PubMed] [Google Scholar]
- 36.Suetsugu S, et al. Optimization of WAVE2 complex-induced actin polymerization by membrane-bound IRSp53, PIP(3), and Rac. J Cell Biol. 2006;173:571–585. doi: 10.1083/jcb.200509067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.