Abstract
Upon infection of human cells, the herpes simplex virus protein VP16 associates with the endogenous cell-proliferation factor HCF. VP16 can also associate with HCFs from invertebrates, suggesting that VP16 mimics a cellular protein whose interaction with HCF has been conserved. Here, we show that VP16 mimics the human basic leucine-zipper protein LZIP, which, through association with HCF, may control cell-cycle progression. VP16 and LZIP share a tetrapeptide motif—D/EHXY—used to associate with human HCF. The LZIP-related Drosophila protein BBF-2/dCREB-A contains this HCF-binding motif, indicating that the LZIP–HCF interaction has been conserved during metazoan evolution.
The initiation of herpes simplex virus (HSV) infection involves the interaction of viral and cellular proteins. The HSV virion carries the viral transactivator VP16 (also known as Vmw65 and αTIF), which is released into the cell upon infection and associates with two cellular factors—HCF and Oct-1—to initiate a cascade of viral gene expression (for review, see Thompson and McKnight 1992; O’Hare 1993). VP16 first associates with HCF (also known as C1, VCAF, and CFF), a protein involved in cell proliferation (Goto et al. 1997). HCF binding to VP16 promotes subsequent association with the POU DNA-binding domain of the transcription factor Oct-1 on the cis-regulatory target of VP16 activation: the TAATGARAT motif found in HSV immediate-early promoters. The interplay between viral and cellular elements afforded by this VP16-induced complex provides a mechanism for coordination of HSV infection with the state of the infected cell.
VP16 interacts with each of the other components in the VP16-induced complex, the DNA, Oct-1, and HCF. Two of these interactions, however, are not highly conserved: VP16 from different herpesviruses differs in its DNA-binding specificity (Huang and Herr 1996; Misra et al. 1996), and VP16 associates poorly with mouse Oct-1, which differs from human Oct-1 on the surface of the POU domain critical for association with VP16 (Cleary et al. 1993; Suzuki et al. 1993).
In contrast, the VP16–HCF interaction is highly conserved: HCF from organisms as different as vertebrates and invertebrates can associate with VP16 and stabilize the VP16-induced complex (Kristie et al. 1989; Wilson et al. 1993b). This conservation suggests that VP16 mimics a cellular protein whose interaction with HCF is essential for viability in metazoans. Here, we describe the isolation of such a candidate: the human basic leucine zipper (bZIP) protein LZIP. Strikingly, LZIP and a related Drosophila protein called BBF-2 (Abel et al. 1992) or dCREB-A (Smolik et al. 1992) share with VP16 a short tetrapeptide motif used to associate with HCF.
Results
HCF is synthesized as a large precursor protein of ∼2000 amino acids, which is subsequently processed by proteolysis at multiple sites into amino- and carboxy-terminal fragments, which remain noncovalently bound (Wilson et al. 1993a, 1995b; Kristie et al. 1995). VP16 associates with a 380-residue amino-terminal domain of HCF, called the HCFVIC domain (Wilson et al. 1997). To identify human proteins that bind the HCFVIC domain, we used the yeast two-hybrid assay (Fields and Song 1989) to screen a human HeLa cell cDNA library for proteins able to associate with a GAL4 DNA-binding domain (DBD) fusion to the first 450 residues of HCF (DBD ⋅ HCFN450).
We identified three (out of 1 × 106) clones that interact specifically with HCFN450. All three clones were derived from the same human gene, encoding a bZIP protein related to the mouse protein LZIP, a protein of little known biological function (Burbelo et al. 1994). We refer to the human protein as human LZIP or hLZIP. A contemporaneous study by Lu et al. (1997) also identified LZIP (referred to as Luman in that study) in a two-hybrid screen for human HCF-interacting proteins.
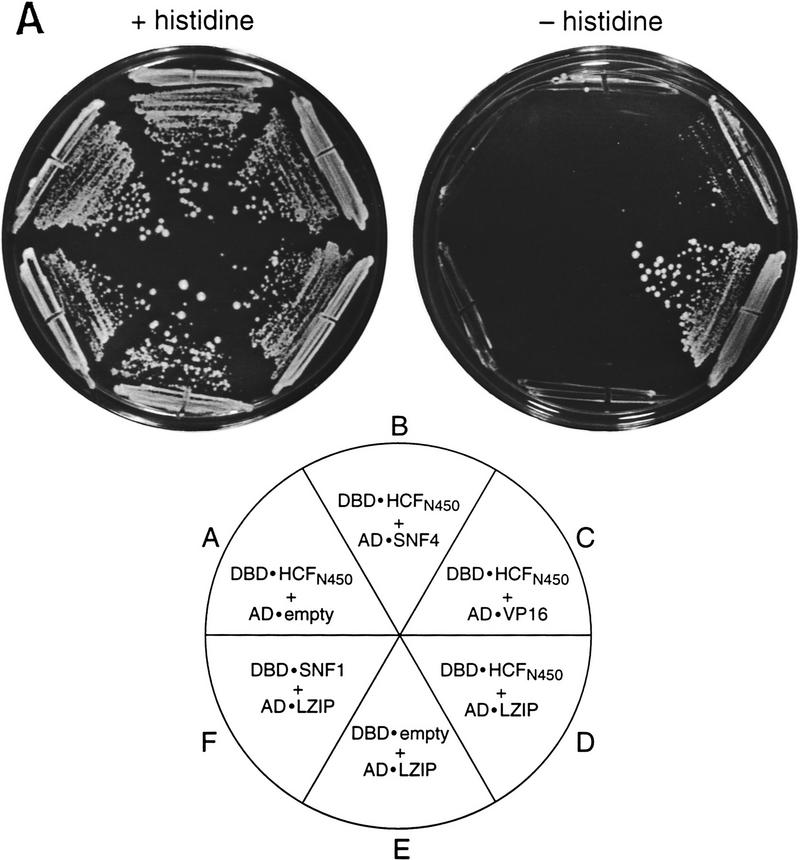
Figure 1A shows the specificity of the HCFN450–hLZIP interaction. Neither the DBD ⋅ HCFN450 fusion protein nor a GAL4 transcriptional activation domain hLZIP fusion (AD ⋅ hLZIP) activates the Gal1–His3 reporter on its own (sectors A and E, respectively) or together with an irrelevant partner (sectors B and F, respectively). DBD ⋅ HCFN450 does, however, activate the GAL1–HIS3 reporter in the presence of AD ⋅ hLZIP (sector D)—as does a VP16 activation domain fusion protein (AD ⋅ VP16; sector C)—demonstrating specific HCFN450 interaction with LZIP.
Figure 1.
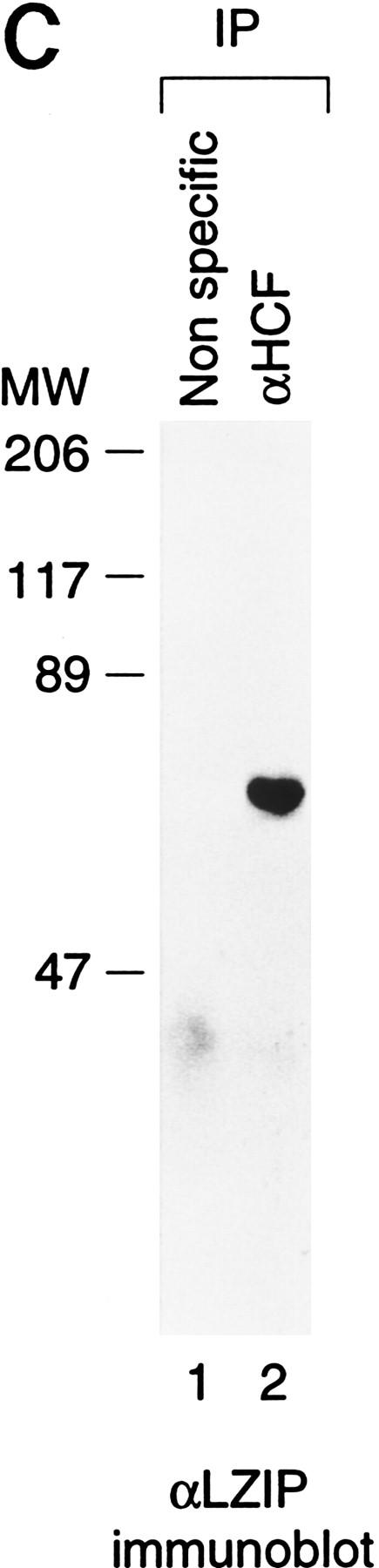
The bZIP protein LZIP interacts specifically with HCF. (A) Yeast two-hybrid assays showing specificity of the HCF–LZIP interaction. Growth of the yeast GAL1–HIS3 reporter strain transformed with both a Gal4 DNA-binding domain (DBD) fusion expression plasmid and a Gal4 activation domain (AD) fusion expression plasmid is shown. The identity of the fusion constructs is indicated in the diagram. Growth with histidine (+histidine) demonstrates that the expression plasmids are not deleterious to yeast growth, and growth without (−histidine) demonstrates activation of the GAL1–HIS3 reporter gene. The yeast were grown for 7 days at 30°C in both the presence and absence of histidine. (B) Amino acid sequence comparison of human LZIP (hLZIP) with mouse LZIP (mLZIP; Burbelo et al. 1994) and two regions of the LZIP-related Drosophila protein BBF-2/dCREB-A (Abel et al. 1992; Smolik et al. 1992, labeled dBBF-2). Sequence identities to hLZIP are indicated by dots in the comparison. The DNA-binding basic region and dimerization–interface leucine zipper are bracketed, with an asterisk above the seven hydrophobic residues that make up the extended leucine zipper of LZIP. The location of an additional 25 amino acids introduced by an alternate splice variant of mouse LZIP (Burbelo et al. 1994) is indicated (▾). The AD fusion points of the original two-hybrid screen isolates are indicated by the arrows above the top line. (HBM) HCF-binding motif. (C) Association of endogenous HCF and LZIP from human cells. Human 293 whole-cell nuclear extracts were precipitated with the nonspecific 12CA5 influenza hemagglutinin monoclonal antibody (lane 1) or the M2 anti-HCF monoclonal antibody (lane 2), and immunoblots of the precipitates were probed with the C7 LZIP antipeptide antibody.
Figure 1B compares the sequences of human and mouse LZIP. Basic DNA-binding and leucine-zipper dimer-formation regions typical of bZIP proteins are located in the center of LZIP. These regions are 93% identical between human and mouse LZIP and include unusually long leucine-zipper segments with seven imperfect leucine heptad repeats. Outside of the basic leucine-zipper region, the sequences are of similar size but display less sequence identity.
In a Northern hybridization analysis (data not shown; see also Lu et al. 1997), hLZIP mRNA was detected in all tissues tested, suggesting that, like HCF (Wilson et al. 1995a), hLZIP is broadly expressed. Figure 1C shows that the natural human HCF and LZIP proteins associate with one another: Immunoprecipitation from a human 293-cell extract with an anti-HCF antibody (lane 2), but not with a nonspecific antibody (lane 1), resulted in recovery of endogenous LZIP as evidenced in an immunoblot with anti-LZIP antisera.
The HCF tsBN67 cell proliferation mutant fails to interact with LZIP
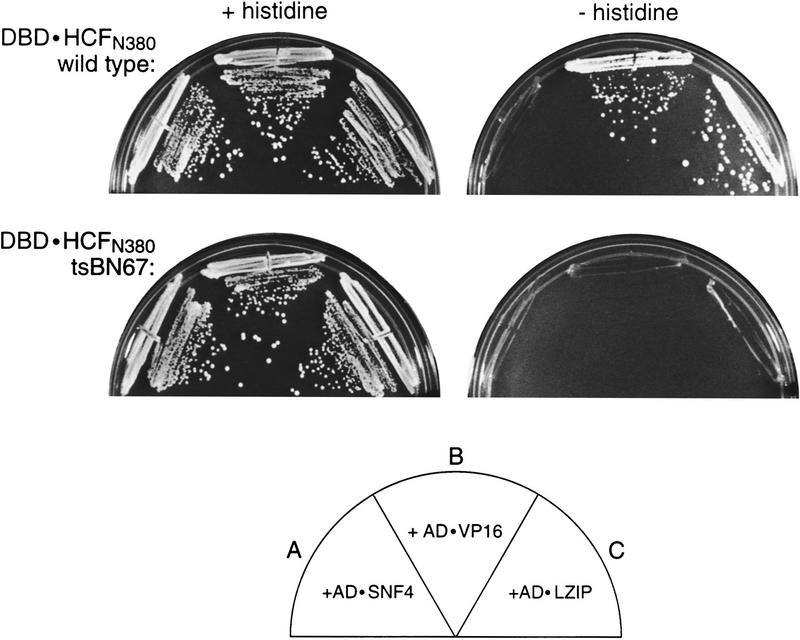
HCF is involved in promoting cell proliferation: A single proline-to-serine substitution at position 134 (P134S) of the HCFVIC domain causes a temperature-sensitive cell cycle arrest (Goto et al. 1997) and disrupts the ability of VP16 to bind HCF (Wilson et al. 1997). If VP16 mimics LZIP, LZIP should associate with the minimal 380-residue HCFVIC domain (HCFN380), and the tsBN67 mutation should prevent association. Figure 2 shows that these predictions are correct: In the two-hybrid assay, VP16 and LZIP both interact with wild-type HCFN380 (sectors B and C, top) but not tsBN67 HCFN380 (sectors B and C, bottom), even though both the wild-type and tsBN67 HCF fusion proteins were expressed at similar levels as determined by immunoblot analysis (data not shown). The tsBN67 effect on LZIP association makes LZIP a candidate target of HCF in controlling cell proliferation.
Figure 2.
The HCF tsBN67 cell-cycle arrest mutation disrupts interaction with LZIP. Yeast two-hybrid interaction assays with wild-type HCFN380 (DBD ⋅ HCFN380, top panels) and tsBN67 HCFN380 (DBD ⋅ HCFN380P134S, bottom panels) were performed as indicated in the diagram. The yeast were grown for 5 days at 30°C in both the presence (left) and absence (right) of histidine.
LZIP and VP16 associate similarly with the HCFVIC domain
To contrast VP16 and LZIP association with the HCFVIC domain more extensively, we assayed the effects of a systematic set of point mutations in the HCFVIC domain on HCF association with VP16 and LZIP. The HCFVIC domain consists of six sequence repeats related to a repeat motif found in the Drosophila protein Kelch, which are called HCFKEL1 to HCFKEL6 (Wilson et al. 1997). Sequence similarity to other proteins suggests that Kelch repeats form four-stranded β-sheets which come together to form a propeller-like structure, called a β-propeller, in which each repeat resembles a blade of the propeller (Bork and Doolittle 1994). The P134S tsBN67 mutation replaces a proline in HCFKEL3 that is universally conserved in all six repeats (Wilson et al. 1997) without affecting HCF stability in vivo (Goto et al. 1997) or protease sensitivity in vitro (R. Freiman and W. Herr, unpubl.). Therefore, we assayed the effects of substituting the corresponding proline in each HCFKEL repeat with alanine on association with LZIP and VP16, as shown in Figure 3.
Figure 3.
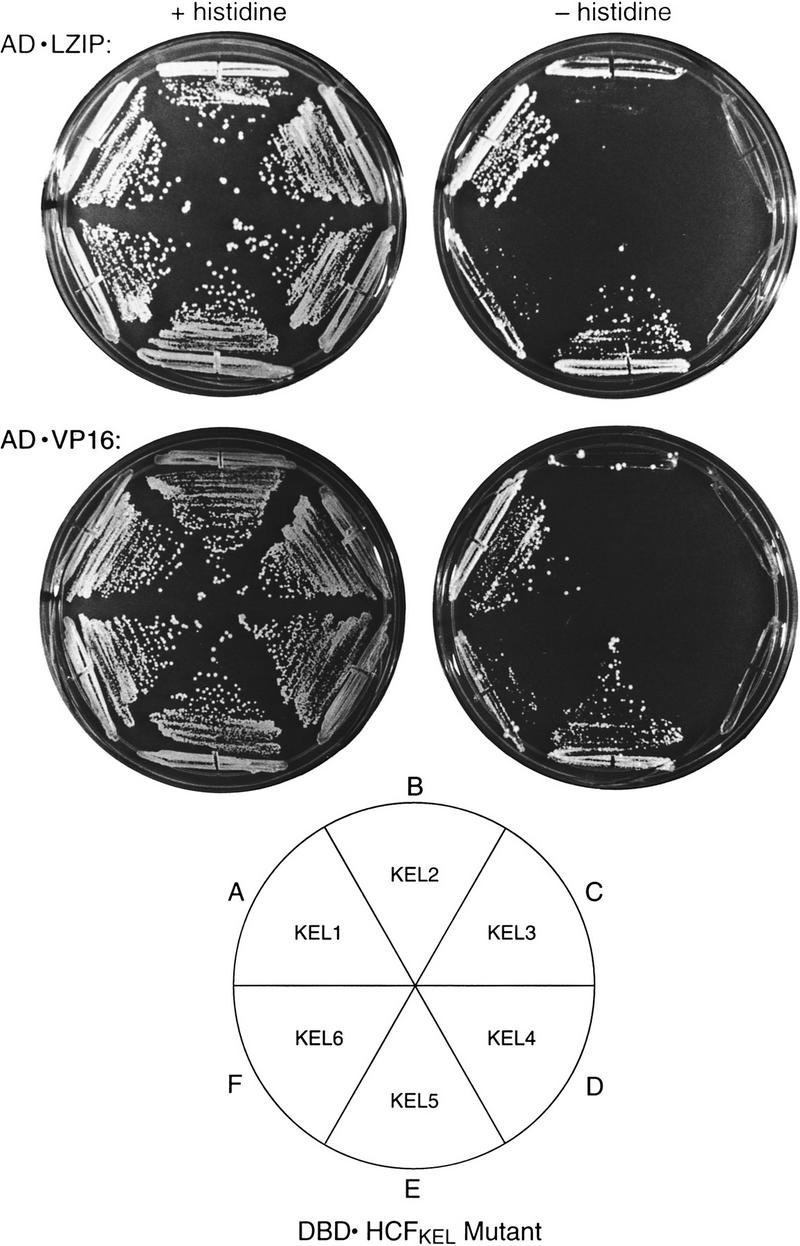
LZIP and VP16 recognize HCF similarly. Yeast two-hybrid interactions between mutated HCFN380 and LZIP (top) and VP16 (bottom). The DBD ⋅ HCFKEL 1–6 mutants (see diagram) contain individual proline-to-alanine substitutions as follows: KEL1, P30A; KEL2, P79A; KEL3, P134A; KEL4, P197A; KEL5, P252A; KEL6, P319A. The yeast were grown for 7 days at 30°C in both the presence (left) and absence (right) of histidine. A control with wild-type DBD ⋅ HCFN380 with AD ⋅ LZIP and AD ⋅ VP16 grown in parallel showed the same growth rate as the cells grown in sectors A and E.
Consistent with VP16 mimicry of the LZIP interaction with HCF, the HCFKEL repeat substitutions had parallel effects on VP16 and LZIP association: Substitution of the conserved proline in HCFKEL1 and HCFKEL5 had no evident effect on either LZIP or VP16 interaction with HCF (sectors A and E), demonstrating that substitution of this universally conserved proline by alanine does not necessarily lead to universal disruption of the HCFVIC domain structure. The HCFKEL2 and HCFKEL6 substitutions had an intermediate effect on LZIP and VP16 interaction (sectors B and F); only the HCFKEL3 and HCFKEL4 substitutions prevented effective LZIP or VP16 interaction with the HCFVIC domain (sectors C and D). The dominant effects of the mutations in HCFKEL3 and HCFKEL4 suggest that LZIP and VP16 bind the HCFVIC domain assymetrically.
LZIP and VP16 share a tetrapeptide motif important for association with HCF
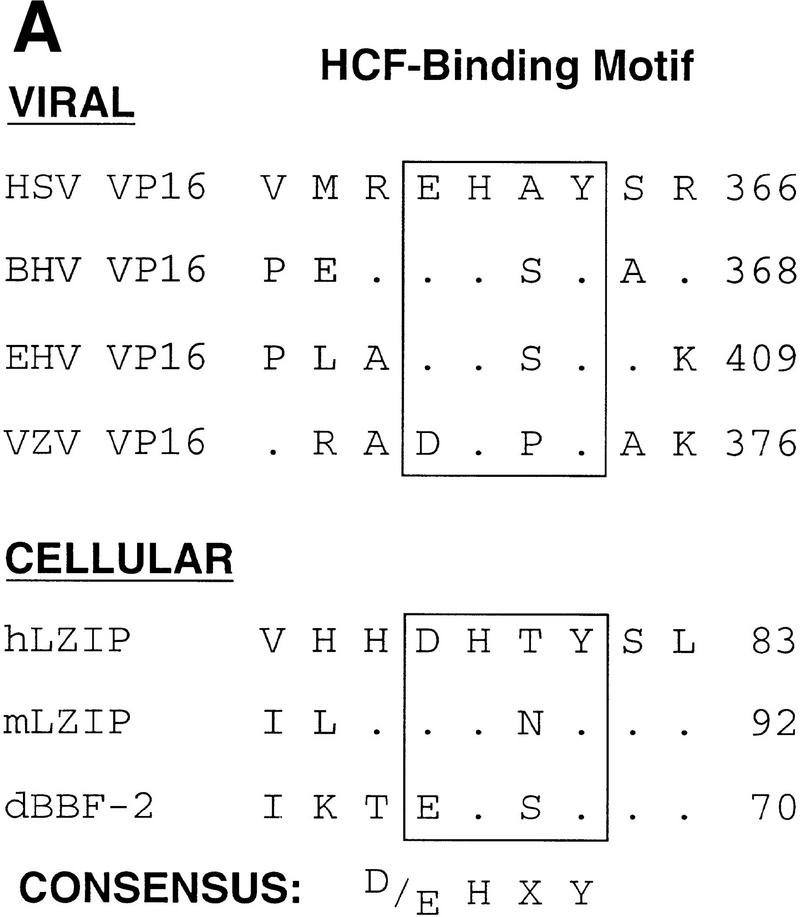
VP16 and LZIP display little sequence similarity (data not shown). There is, however, similarity to a short sequence in VP16—EHAY—that is involved in HCF association (Haigh et al. 1990; Hayes and O’Hare 1993; Wu et al. 1994; Simmen et al. 1997; Lai and Herr 1997). As shown in Figure 4A, this sequence is conserved in VP16 homologs from different herpesviruses, forming the consensus sequence D/EHXY, where the first position can be either aspartic or glutamic acid and the third position is variable. The human and mouse LZIP sequences share this tetrapeptide consensus [see Fig. 1B (labeled HBM) and Fig. 4A].
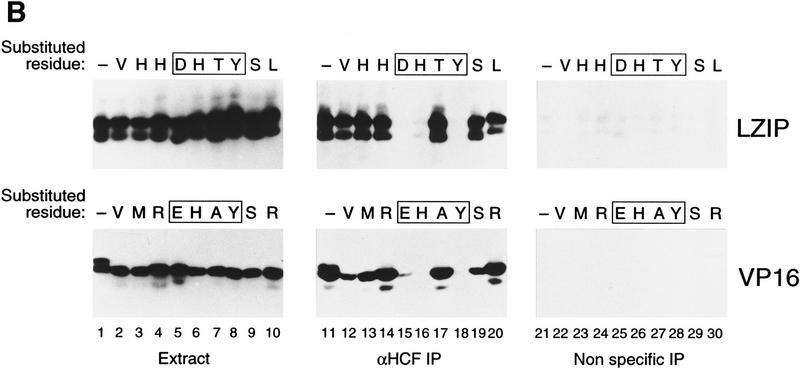
Figure 4.
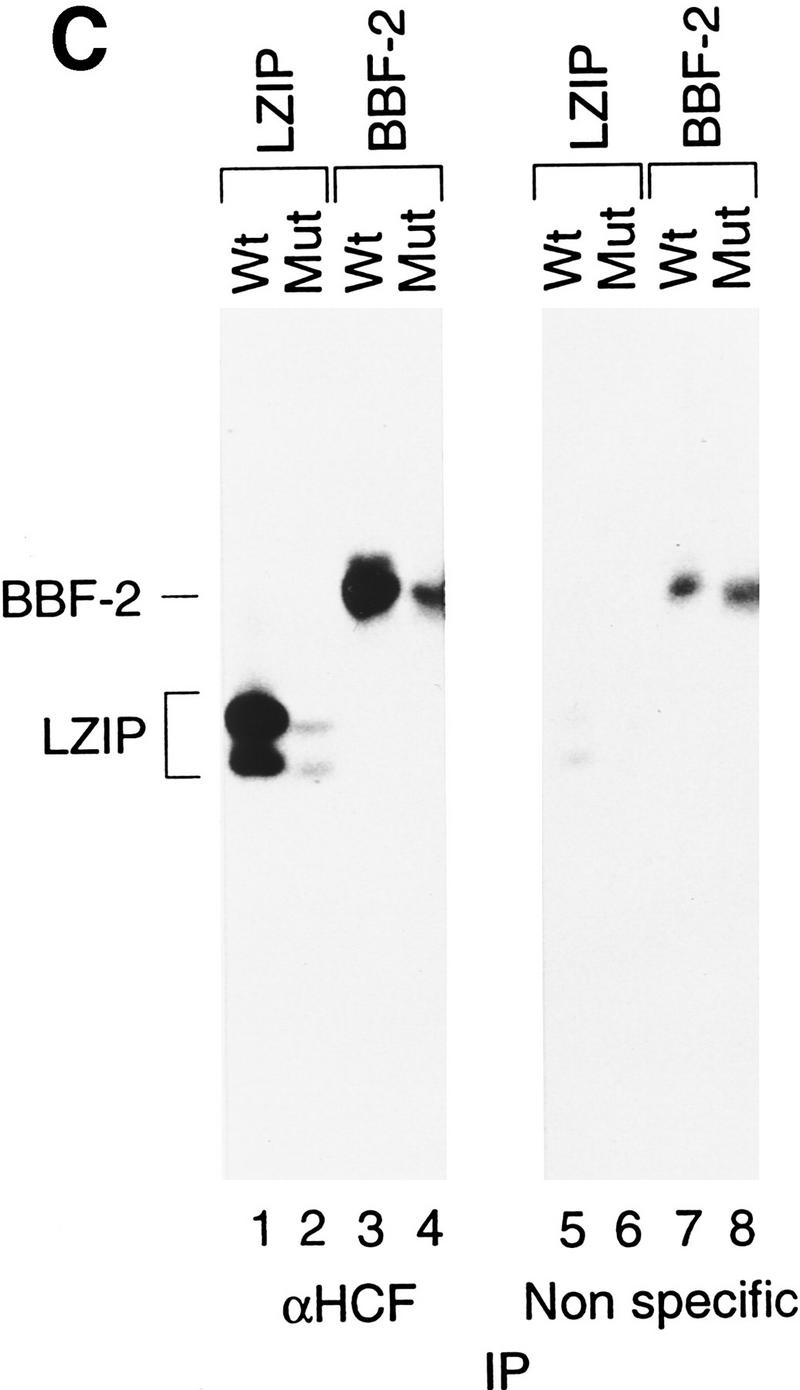
Mammalian LZIP, Drosophila BBF-2, and VP16 share a small HCF-binding motif. (A) HCF-binding motif sequence alignment. The VP16 sequences are derived from homologs of VP16 from bovine herpesvirus (GenBank accession no. Z54206; BTIF), equine herpesvirus (GenBank accession no. L16590), and varicella-zoster virus (GenBank accession no. X04370; ORF 10). Dots indicate sequence identities with the HSV VP16 (viral) and hLZIP (cellular) sequences shown. The position of the carboxy-terminal residue within the parental sequence is indicated to the right. The HCF-binding motif is boxed and a consensus is shown at the bottom. (D/E) Either aspartic or glutamic acid; (X) nonconserved variable residue. (B) HCF association with wild-type and mutant LZIP and VP16. Epitope-tagged wild-type or nine substitution mutants of LZIP (top panels) and VP16 (bottom panels) were transiently expressed in human 293 cells. The mutated residue in each mutant is indicated above each lane. In LZIP, all residues were changed to alanine, except for the threonine, which was changed to asparagine. In VP16, all nonalanine residues were changed to alanine, and the alanine was changed to serine. Expression of each mutant protein is shown by direct immunoblot with the anti-T7 epitope antibody of the 293 cell extract (lanes 1–10). Extracts were precipitated with the N18 HCF antipeptide antibody (lanes 11–20) or the nonspecific anti-N-Oct-3 antibody (lanes 21–30). In each panel, LZIP or VP16 was detected by immunoblot analysis with the anti T7 epitope tag antibody. (C) HCF recognizes the Drosophila BBF-2 HCF-binding motif. T7-epitope-tagged wild-type and mutant LZIP and BBF-2 were transiently expressed in human 293 cells. Position 1 of the HCF-binding motif in LZIP (D to A) and BBF-2 (E to A) were mutated (Mut). 293 cell extracts were immunoprecipitated with the N18 HCF antibody (lanes 1–4) or the nonspecific anti-N-Oct-3 (lanes 5–8) and the precipitates analyzed by immunoblot with the anti-T7 epitope tag antibody. Direct immunoblot with the anti T7 epitope tag antibody showed that for both LZIP and BBF-2 the wild-type and mutant proteins were expressed at the same level, but the BBF-2 proteins were expressed at higher levels than LZIP (data not shown).
To test the functional significance of this tetrapeptide motif in LZIP, we compared the effect of individual substitutions in a 9-amino-acid segment encompassing the human LZIP and HSV VP16 D/EHXY motifs on association with HCF. To measure association, we assayed coimmunoprecipitation of epitope-tagged wild-type and mutant LZIP and VP16 proteins with endogenous HCF after transient expression in human 293 cells, as shown in Figure 4B. Direct epitope tag immunoblot analysis of the extracts showed that the mutant LZIP and VP16 proteins were expressed at levels similar to their wild-type counterparts (lanes 1–10). Little protein, however, was recovered after immunoprecipitation with an irrelevant antibody (lanes 21–30). Strikingly, with both human LZIP and HSV VP16, mutation of the same three positions—the first, second, and fourth positions of the D/EHXY motif—interfered with HCF association, whereas mutation of the other residues, including the variable third position in the D/EHXY motif, did not prevent LZIP or VP16 association with HCF (lanes 11–20). These results suggest that VP16 mimics a small motif in LZIP—an HCF-binding motif (HBM)—to promote association with HCF. Consistent with this hypothesis, VP16 and LZIP compete with one another for association with HCF (Lu et al. 1997; C. Grozinger, R. Freiman, and W. Herr, unpubl.).
A D/EHXY motif in Drosophila BBF-2 promotes association with HCF
The ability to interact with VP16 is shared by metazoan HCFs (Kristie et al. 1989; Wilson et al. 1993b), suggesting that the HCF interaction with a cellular protein mimicked by VP16 is conserved in vertebrates and invertebrates. To test this hypothesis, we asked whether human HCF can associate with an LZIP-related protein from Drosophila. The known Drosophila protein most closely related to LZIP is the bZIP protein BBF-2 (Abel et al. 1992; also called dCREB-A, Smolik et al. 1992); this protein is 81% identical to human LZIP in the basic region and shares an extended imperfect leucine-zipper segment as shown in Figure 1B. Outside of the bZIP region, BBF-2 shares only limited similarity to the mammalian LZIP proteins, but it does contain a consensus D/EHXY motif near its amino terminus as shown in Figures 1B and 4A. Therefore, using the same assay shown in Figure 4B, we tested native human HCF association with both wild-type BBF-2 and a BBF-2 mutant containing a single alanine substitution of the first position of the D/EHXY motif, as shown in Figure 4C. In contrast to wild-type and mutant human LZIP, a low level of wild-type and mutant BBF-2 was recovered in the immunoprecipitation with the irrelevant antibody (lanes 5–8). Nevertheless, wild-type LZIP and BBF-2 proteins were recovered more effectively than the mutant forms after coprecipitation with HCF (cf. lanes 1 and 2, and 3 and 4), suggesting that HCF can associate with BBF-2 through a D/EHXY HCF-binding motif. Together, the sequence similarity between LZIP and BBF-2 and their common mode of interaction with HCF suggest that LZIP and BBF-2 are homologs serving similar functions in mammals and insects, such as promoting cell proliferation.
Discussion
We have identified related human and Drosophila bZIP proteins that associate with the cell proliferation factor HCF. The HSV regulatory protein VP16 mimics how these proteins—LZIP in humans and BBF-2 in Drosophila—associate with HCF: Like LZIP and BBF-2, VP16 contains a short D/EHXY HCF-binding motif, and LZIP and VP16 interactions with HCF are similarly affected by mutations in HCF, including the temperature-sensitive cell-cycle arrest mutation tsBN67. Because LZIP and BBF-2 interact with HCF similarly and VP16 copies this interaction, perhaps the conserved LZIP and BBF-2 interaction with HCF is the proposed conserved cellular interaction mimicked by VP16. The association with a bZIP protein implicates HCF in regulating cellular as well as viral patterns of gene transcription.
The common association with HCF suggests that LZIP and BBF-2 are human and Drosophila homologs. Consistent with this hypothesis the basic region DNA-binding domains of LZIP and BBF-2 are closely related, and both proteins contain unusually long and imperfect leucine-zipper segments (see Fig. 1B). The closest related proteins to LZIP and BBF-2 are cAMP response element-binding (CREB) and activating transcription factor 1 (ATF-1) in mammals and dCREB-B in Drosophila (for review, see Hurst 1995), suggesting that the HCF-interacting LZIP and BBF-2 proteins are members of a subfamily of bZIP proteins that includes the CREB/ATF-1-related proteins. Interestingly, both LZIP and CREB associate with large nuclear proteins: HCF in the case of LZIP, and the transcriptional coregulator CREB-binding protein (CBP) in the case of CREB (Chrivia et al. 1993).
Although LZIP can bind to CREB- and other bZIP-binding sites (Burbelo et al. 1994; Lu et al. 1997; C. Grozinger, R. Freiman, and W. Herr, unpubl.), the natural targets of LZIP regulation are unknown. In Drosophila, the natural targets of transcriptional regulation by BBF-2 are also not well understood, although disruption of the BBF-2/dCREB-A gene in Drosophila has specific effects on embryonic development (Andrew et al. 1997; Rose et al. 1997). Because LZIP fails to associate with the HCF cell-cycle arrest mutation tsBN67, LZIP may be involved in regulating cell-cycle progression together with HCF. tsBN67 cells arrest in a G0- or early G1-like state, which implicates HCF in preventing entry into G0 and/or promoting passage through early G1 (Goto et al. 1997). Thus, genes involved in progression through G0 and early G1 are attractive targets of LZIP regulation.
One of the characteristics of the HCF tsBN67 phenotype is a delay of one or a few cell divisions before stable growth arrest occurs (Goto et al. 1997). The implication of HCF association with a transcription factor to regulate cell-cycle progression offers an explanation for this phenotype: HCF and LZIP regulate—by activation or repression—expression of a gene whose product regulates cell-cycle progression; when the HCF or LZIP interaction is disrupted at the nonpermissive temperature by the tsBN67 mutation, the HCF–LZIP-regulated gene product has to be depleted or inactivated before cell-cycle arrest can occur.
Unlike obligate lytic viruses, HSV can undergo either a productive lytic infection or remain latent for long periods. VP16, which is important for initiating the cascade of HSV gene expression that results in productive infection, may serve as a sensor by which the virus is able to coordinate virus replication with the cell-cycle or differentiation state of the infected cell. The requirement for productive association with two cellular factors, Oct-1 and HCF, before VP16 can initiate HSV immediate-early gene transcription suggests that it may be these interactions with which VP16 senses the status of the cell. The studies described here suggest that in both cases VP16 is targeting the cellular transcriptional machinery: It associates with Oct-1, a POU domain transcription factor involved in histone and small nuclear RNA gene expression, and HCF, which is now implicated in regulating transcription through association with LZIP. VP16 may target the cellular transcriptional apparatus because this machinery changes rapidly as it responds to or directs changes in cell status.
Materials and methods
Yeast two-hybrid interaction assay
The GAL4 DNA-binding domain fusion expression plasmid pGBT9, containing the TRP1 gene, was used to construct GAL4 DBD fusions (residues 1–94) to HCF (the pGDBD–HCF series). pGDBD–HCFN450 (DBD ⋅ HCFN450) and pGDBD–HCFN380 (DBD ⋅ HCFN380) contain human HCF residues 2–450 and 2–380, respectively. pGDBD–HCFN450 was used to screen a human HeLa cDNA–GAL4 activation domain (AD; residues 768–881) fusion library constructed in pGADGH, which contains the LEU2 gene (a gift of G. Hannon, Cold Spring Harbor Laboratory). Both the DBD and AD plasmids were cotransformed in the reporter strain Hf7c containing both GAL4-responsive his3 and lacZ reporter genes. Yeast (1 × 106) transformants were screened for His+ growth. Of 41 His+ colonies recovered after 7–10 days growth at 30°C on synthetic media plates lacking trytophan, leucine, and histidine, only 5 displayed activation of the lacZ reporter. The AD plasmids were recovered from these five positives and retransformed into the reporter strain YGH1. Only three clones (3-1, 4-1, and 9-1) activated the Gal1–His3 reporter in the presence but not in the absence of the HCFN450 bait. Plasmids expressing the GAL4 DBD (DBD ⋅ empty) or AD alone (AD ⋅ empty), or fused to the yeast proteins SNF1 (DBD ⋅ SNF1) and SNF4 (AD ⋅ SNF4) were used as negative controls (Fields and Song 1989).
Cloning full-length human LZIP
Sequence analysis revealed that two of the three HCF-interacting fusion proteins were identical and that all three were AD fusions to either residue 43 (3-1 and 9-1) or 54 (4-1) of the same mouse LZIP-related gene (Burbelo et al. 1994). The full-length LZIP cDNA was cloned by PCR amplification of a human λgt10 phage cDNA library with two nested LZIP-specific primers and one λ-specific primer. The extended 5′ sequence revealed an in-frame initiator ATG codon; the sequence has been deposited in GenBank (accession no. AF029674}.
Plasmid constructions and mutagenesis
The GAL4 DBD–HCF fusion constructs—the pGDBD–HCF series—are all derived from pGBT9 but only encode GAL4 residues 1–94. This AD fusion vector, pGADGHX2, a pGADGH derivative for in-frame insertion of XbaI–BamHI fragments, was used to create pGADGHVP16ΔC (AD ⋅ VP16), which contains VP16 residues 5–412 lacking the carboxy-terminal VP16 activation domain, and pGADGHLZIPFL (AD ⋅ LZIP) which contains LZIP residues 2–371. In Figure 1A, the AD ⋅ LZIP construct contains residues 43–371 (clone 3-1). T7-epitope-tagged VP16, LZIP, and BBF-2 were expressed from pCGT plasmids. VP16 was expressed from pCGTVP16ΔC (Wilson et al. 1997). pCGTLZIPFL contains hLZIP residues 2–371, and pCGTBBF-2FL contains Drosophila BBF-2 residues 2–515. Amino acid substitutions were generated by oligonucleotide-directed mutagenesis as described previously (Kunkel et al. 1987).
Antisera and immunoprecipitation
The C7 anti-LZIP antiserum was raised in rabbits against the octapeptide CLQDRYSG containing the carboxy-terminal 7 amino acids of hLZIP (underlined) coupled to keyhole limpet hemocyanin and inoculated into rabbits as described (Atanasoski et al. 1997). The N18 anti-peptide HCF (Goto et al. 1997) and N-Oct 3 (Atanasoski et al. 1997) antisera and M2 anti-HCF monoclonal antibody (Wilson et al. 1995a) have been described previously. To assay endogenous HCF and LZIP interaction, human 293 extracts were prepared in extraction buffer (200 mm KCl, 100 mm Tris at pH 8.0, 10% glycerol, 0.1% NP-40, 20 mm EDTA, and 1 mm PMSF). A 1:50 dilution of the anti-HCF monoclonal antibody M2 (Wilson et al. 1995a) was incubated with the extract for 90 min at 4°C with rotation. Immune complexes were recovered with protein G–agarose beads, washed four times in the same buffer without glycerol and 0.05% NP-40, and resolved by 12% SDS-PAGE for immunoblot analysis. A 1:1000 dilution of the C7 LZIP antiserum was used to probe the immunoblot.
Transient expression, coprecipitation, and immunoblotting
To express T7 epitope-tagged LZIP, VP16, and BBF-2 proteins, pCGTLZIPFL (20 μg), pCGTVP16ΔC (0.2 μg), and pCGTBBF-2FL (2.0 μg) were transfected into human 293 cells by electroporation, and extracts prepared 36 hr post-transfection as described (Wilson et al. 1997) in 1 ml of extraction buffer containing 1.0% NP-40, 0.5% DOC, and 0.1% SDS. The anti-HCF N18 or anti-N-Oct 3 polyclonal sera were cross-linked to protein A–agarose as described (Harlow and Lane 1988). Ten microliters of antibody beads (50% slurry) was added to 200 μl of extract and incubated for 90 min at 4°C, recovered by centrifugation, and washed three times in extraction buffer. Immune complexes were resolved on a 10% SDS-PAGE and immunobots probed with the anti-T7 epitope tag monoclonal antibody (Novagen) at a 1:5000 dilution. The levels of T7 epitope-tagged LZIP and VP16 expression were assayed by direct immunoblotting of 1/20 and 1/100 of the extract, respectively.
Acknowledgments
We thank A. Wilson and T. Nishimoto for helpful discussions and for sharing results prior to publication; G. Hannon and L. Van Aelst for advice and reagents for the yeast two-hybrid assay; B. Ren and T. Maniatis for a BBF-2-encoding cDNA clone; D. Aufiero for technical assistance; N. Hernandez for critical reading of the manuscript; and J. Duffy, M. Ockler, and P. Renna for artwork. This study was supported by U.S. Public Health Service grant CA13106 from the National Cancer Institute.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
[Key Words: HCF protein; VP16; LZIP; protein–protein interactions; transcription]
E-MAIL herr@cshl.org; FAX (516) 367-8454.
References
- Abel T, Bhatt R, Maniatis T. A Drosophila CREB/ATF transcriptional activator binds to both fat body- and liver-specific regulatory elements. Genes & Dev. 1992;6:466–480. doi: 10.1101/gad.6.3.466. [DOI] [PubMed] [Google Scholar]
- Andrew DJ, Baig A, Bhanot P, Smolik SM, Henderson KD. The Drosophila dCREB-A gene is required for dorsal/ventral patterning of the larval cuticle. Development. 1997;124:181–183. doi: 10.1242/dev.124.1.181. [DOI] [PubMed] [Google Scholar]
- Atanasoski S, Schreiber E, Fontana A, Herr W. N-Oct 5 is generated by in vitro proteolysis of the neural POU-domain protein N-Oct 3. Oncogene. 1997;14:1287–1294. doi: 10.1038/sj.onc.1200953. [DOI] [PubMed] [Google Scholar]
- Bork P, Doolittle RF. Drosophila kelch motif is derived from a common enzyme fold. J Mol Biol. 1994;236:1277–1282. doi: 10.1016/0022-2836(94)90056-6. [DOI] [PubMed] [Google Scholar]
- Burbelo PD, Gabriel GC, Kibbey MC, Yamada Y, Kleinman HK, Weeks BS. LZIP-1 and LZIP-2:two novel members of the bZIP family. Gene. 1994;139:241–245. doi: 10.1016/0378-1119(94)90763-3. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Cleary MA, Stern S, Tanaka M, Herr W. Differential positive control by Oct-1 and Oct-2: activation of a transcriptionally silent motif through Oct-1 and VP16 corecruitment. Genes & Dev. 1993;7:72–83. doi: 10.1101/gad.7.1.72. [DOI] [PubMed] [Google Scholar]
- Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Goto H, Motomura S, Wilson AC, Freiman RN, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. A single-point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes & Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- Haigh A, Greaves R, O’Hare P. Interference with the assembly of a virus-host transcription complex by peptide competition. Nature. 1990;344:257–259. doi: 10.1038/344257a0. [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D. Antibodies: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- Hayes S, O’Hare P. Mapping of a major surface-exposed site in herpes simplex virus protein Vmw65 to a region of direct interaction in a transcription complex assembly. J Virol. 1993;67:852–862. doi: 10.1128/jvi.67.2.852-862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CC, Herr W. Differential control of transcription by homologous homeodomain coregulators. Mol Cell Biol. 1996;16:2967–2976. doi: 10.1128/mcb.16.6.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst HC. Transcription factors 1:bZIP proteins. Protein Profile. 1995;2:105–168. [PubMed] [Google Scholar]
- Kristie TM, Lebowitz JH, Sharp PA. The octamer-binding proteins form multi-protein-DNA complexes with the HSV a-TIF regulatory protein. EMBO J. 1989;8:4229–4238. doi: 10.1002/j.1460-2075.1989.tb08608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristie TM, Pomerantz JL, Twomey TC, Parent SA, Sharp PA. The cellular C1 factor of the herpes simplex virus enhancer complex is a family of polypeptides. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- Kunkel TA, Roberts JD, Zakour RA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Lai J-S, Herr W. Interdigitated residues within a small region of VP16 interact with Oct-1, HCF, and DNA. Mol Cell Biol. 1997;17:3937–3946. doi: 10.1128/mcb.17.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Yang P, O’Hare P, Misra V. Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra V, Walker S, Yang P, Hayes S, O’Hare P. Conformational alteration of Oct-1 upon DNA binding dictates selectivity in differential interactions with related transcriptional coactivator. Mol Cell Biol. 1996;16:4404–4413. doi: 10.1128/mcb.16.8.4404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hare P. The virion transactivator of herpes simplex virus. The Alpha-herpesviruses. Semin Virol. 1993;4:145–155. [Google Scholar]
- Rose RE, Gallaher NM, Andrew DJ, Goodman RH, Smolik SM. The CRE-binding protein dCREB-A is required for Drosophila embryonic development. Genetics. 1997;146:595–606. doi: 10.1093/genetics/146.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmen KA, Newell A, Robinson M, Mills JS, Canning G, Handa R, Parkes K, Borkakoti N, Jupp R. Protein interactions in the herpes simplex virus type 1 VP16-induced complex: VP16 peptide inhibition and mutational analysis of host cell factor requirements. J Virol. 1997;71:3886–3894. doi: 10.1128/jvi.71.5.3886-3894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolik SM, Rose RE, Goodman RH. A cyclic AMP-responsive element-binding transcriptional activator in Drosophila melanogaster, dCREB-A, is a member of the leucine zipper family. Mol Cell Biol. 1992;12:4123–4131. doi: 10.1128/mcb.12.9.4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki N, Peter W, Ciesiolka T, Gruss P, Schöler HR. Mouse Oct-1 contains a composite homeodomain of human Oct-1 and Oct-2. Nucleic Acids Res. 1993;21:245–252. doi: 10.1093/nar/21.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CC, McKnight SL. Anatomy of an enhancer. Trends Genet. 1992;8:232–237. [Google Scholar]
- Wilson AC, LaMarco K, Peterson MG, Herr W. The VP16 accessory protein HCF is a family of polypeptides processed from a large precursor protein. Cell. 1993a;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Cleary MA, Lai J-S, LaMarco K, Peterson MG, Herr W. Combinatorial control of transcription: The herpes simplex virus VP16-induced complex. Cold Spring Harbor Symp Quant Biol. 1993b;58:167–178. doi: 10.1101/sqb.1993.058.01.021. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Parrish JE, Massa HF, Nelson DL, Trask BJ, Herr W. The gene encoding the VP16-accessory protein HCF (HCFC1) resides in human Xq28 and is highly expressed in fetal tissues and the adult kidney. Genomics. 1995a;25:462–468. doi: 10.1016/0888-7543(95)80046-o. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Peterson MG, Herr W. The HCF repeat is an unusual proteolytic cleavage signal. Genes & Dev. 1995b;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Freiman RN, Goto H, Nishimoto T, Herr W. VP16 targets an amino-terminal domain of HCF involved in cell cycle progression. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TJ, Monokian G, Mark DF, Wobbe CR. Transcriptional activation by herpes simplex virus type 1 VP16 in vitro and its inhibition by oligopeptides. Mol Cell Biol. 1994;14:3484–3493. doi: 10.1128/mcb.14.5.3484. [DOI] [PMC free article] [PubMed] [Google Scholar]